SUMMARY
In albinism, aberrations in the ipsi-/contralateral retinal ganglion cell (RGC) ratio compromise the functional integrity of the binocular circuit. Here we focus on the mouse ciliary margin zone (CMZ), a neurogenic niche at the embryonic peripheral retina, to investigate developmental processes regulating RGC neurogenesis and identity acquisition. We found that the mouse ventral CMZ generates predominantly ipsilaterally-projecting RGCs, but this output is altered in the albino visual system due to CyclinD2 downregulation and disturbed timing of the cell cycle. Consequently, albino as well as CyclinD2-deficient pigmented mice exhibit a diminished ipsilateral retinogeniculate projection and poor depth perception. In albino mice, pharmacological stimulation of calcium channels, known to upregulate CyclinD2 in other cell types, augmented CyclinD2-dependent neurogenesis of ipsilateral RGCs and improved stereopsis. Together, these results implicate CMZ neurogenesis and its regulators as critical for the formation and function of the mammalian binocular circuit.
Graphical Abstract
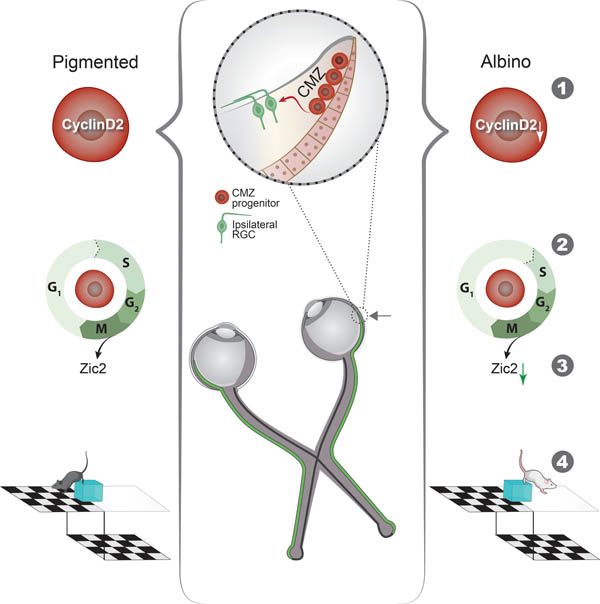
eTOC
This study is an in-depth investigation of molecular alterations underlying chiasmatic misrouting in the albino binocular circuit. During development, in a specialized retinal niche, the ciliary margin, fewer cells express the cell cycle regulator CyclinD2. Consequently, the cell cycle is elongated resulting in fewer ipsilaterally-projecting RGCs and perturbed binocular vision.
INTRODUCTION
A key evolutionary feature of the mammalian visual system is a higher degree of binocular overlap, attributed to a change in the eye position from the sides to the front of the head. To properly form the binocular circuit, retinal ganglion cells (RGCs) from each retina project axons in specific proportions to either the ipsilateral or the contralateral brain hemisphere via a partial decussation at the optic chiasm (Petros et al., 2008, Murcia-Belmonte and Erskine, 2019, Herrera et al., 2019). The resultant convergence and precise integration of visual information from both eyes to binocularly-driven cortical neurons constitute the foundation of stereopsis (Cumming and DeAngelis, 2001, Scholl et al., 2013, Howarth et al., 2014). While genes specific to ipsi- and contralateral RGCs are being uncovered (Wang et al., 2016, Lo Giudice et al., 2019, Kuwajima et al., 2017, Morenilla-Palao et al., 2020, Su et al., 2021, Shekhar et al., 2021, Fries et al., 2022, Fernandez-Nogales et al., 2022), a mechanism that links early regulatory processes driving the production of ipsi- and contralateral RGCs in appropriate proportions to functional aspects of binocular vision is not fully understood.
Classic birthdating experiments show that RGC neurogenesis begins at the center of the retina at the junction of the optic cup and optic stalk, and spreads as a wave toward the periphery (Young, 1985). In fish, amphibian tadpoles, and birds, retinal neurons can also be generated at the peripheral-most region of the embryonic retina, the ciliary margin zone (CMZ), facilitating eye growth (Kubota et al., 2002, Fischer and Reh, 2000, Wan et al., 2016, Fernandez-Nogales et al., 2019). Recent studies describe a combinatorial code of growth factors driving CMZ vs neural retina specification in the vertebrate eye cup (Balasubramanian et al., 2021), and report that the mouse CMZ contains a progenitor population with distinct molecular identity that gives rise to various retinal cell types, including RGCs (Marcucci et al., 2016, Belanger et al., 2017). However, crucial questions regarding the neurogenic properties of the mammalian CMZ remain unresolved. First, are there CMZ-specific mechanisms that regulate RGC fate acquisition and projection path to the brain? Second, is RGC neurogenesis from the CMZ critical to the formation of the binocular circuit, thereby supporting depth perception?
In humans with albinism, a genetic disorder affecting melanin biosynthesis, stereoscopic vision is impaired. Albinism is characterized by delayed center-to-periphery retinal neurogenesis and diminished expression of Zic2, a key transcription factor conveying ipsilateral RGC fate, resulting in an abnormally small ipsilateral eye-to-brain projection and aberrant targeting of recipient regions in the dorsal lateral geniculate nucleus (dLGN) (Herrera et al., 2003, Rebsam et al., 2012, Bhansali et al., 2014). Ipsilateral RGCs are positioned adjacent to the ventral CMZ. Consequently, we asked whether and how the capacity of the CMZ to generate RGCs, especially the ipsilateral population, is affected in the albino mouse, and interrogate its impact on the functional integrity of the binocular visual system.
Here we demonstrate that the mouse ventral CMZ is a rich source of ipsilateral RGCs. We then focus on the temporal control of the cell cycle in this region. Unlike the neural retina, the CMZ is enriched in CyclinD2, a cell cycle protein that regulates the proliferation and fate of radial glial cells and intermediate progenitors in cortical and subcortical neurogenic zones (Glickstein et al., 2009, Tsunekawa et al., 2012, Tsunekawa et al., 2014, Pilaz et al., 2016a, Marcucci et al., 2016). Our results reveal that expression of CyclinD2 in CMZ progenitors is required for appropriate G1/S phase transition. In the albino eye, downregulation of CyclinD2 leads to an asynchronous cell cycle and reduced RGC neurogenesis from the CMZ during the time window critical for Zic2 expression. Consequently, both albino and conditional CyclinD2-deficient pigmented mice exhibit depth perception deficits due to disproportionate binocular inputs to the dLGN. Lastly, we show that artificial activation of calcium channels modulates CyclinD2 expression in the CMZ, increases ipsilateral RGCs, prevents axon misrouting in the albino binocular circuit, and restores depth perception.
RESULTS
Ipsilateral RGC neurogenesis is prominent in the ventral CMZ in pigmented mice but reduced in the albino
Cells of the CMZ expressing the transcription factor Msx1 have been described as a source of retinal neurons during mouse development. To characterize the output of CMZ neurogenesis, we used Msx1 CreERT2; tdTomato pigmented (TyrWT) and albino (Tyrc2j) mice as a tool to fate map progenitors of the CMZ, and examined RGC production in the time window critical for ipsilateral RGC fate acquisition (embryonic days E13.5-E15.5) (Herrera et al., 2003, Marcucci et al., 2018) (Fig. 1A-A’). At each of the three time-points (E13.5, 14.5, 15.5) of Msx1 CreERT2 activation, the number of Msx1+ cells was similar between pigmented and albino ventral CMZ (Fig. 1B-B’).
Figure 1: Ipsilateral RGC neurogenesis is prominent in the ventral CMZ in pigmented mice but reduced in the albino.
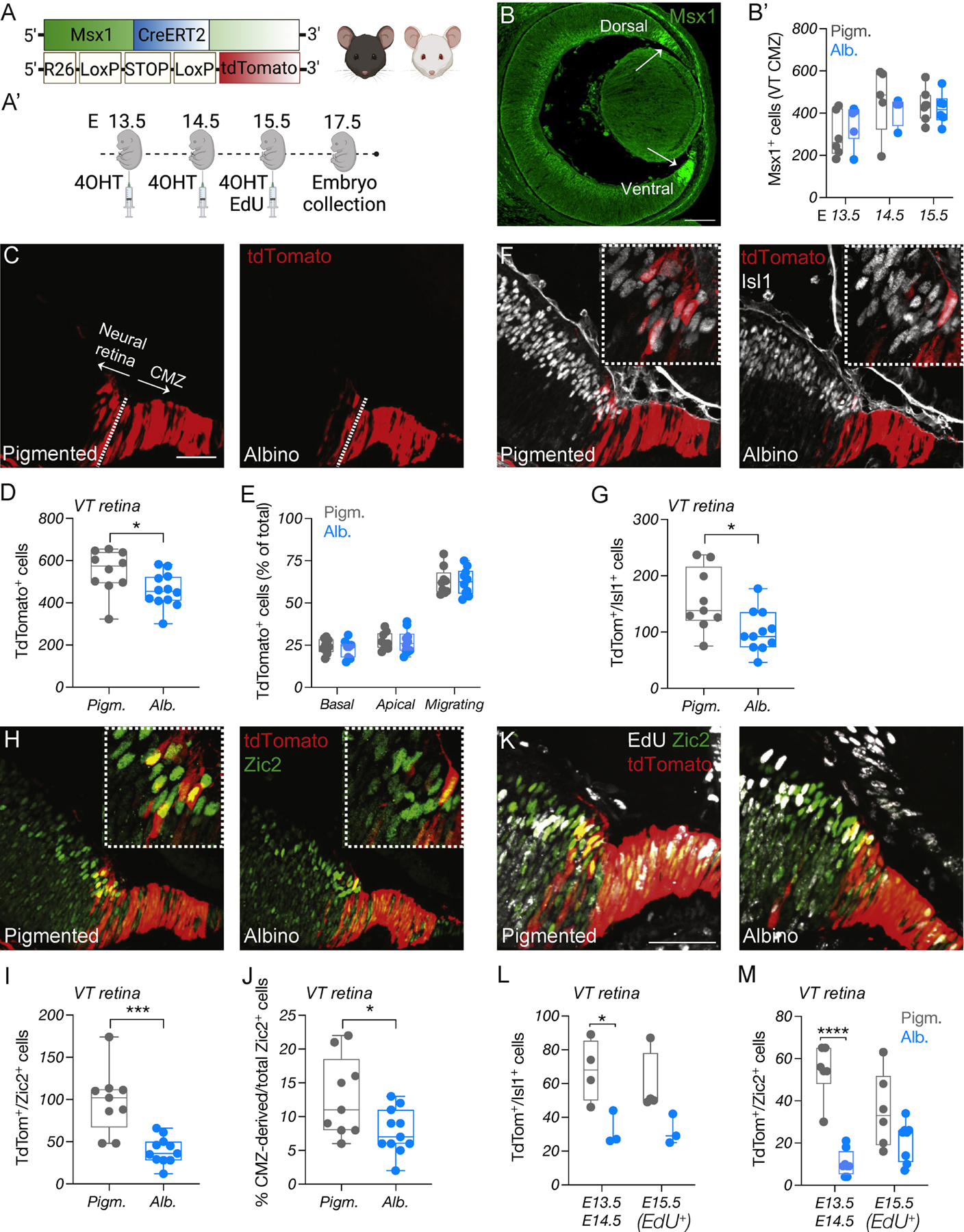
A) Schema of elements used to generate Msx1 CreERT2; tdTomato; Tyr mice.
A’) Timeline of 4-OHT and EdU injections and sample collection.
B) Immunostaining of Msx1 at E14.5. Scale bar: 100μm.
B’) Quantification of Msx1+ cells in the pigmented and albino ventral CMZ at E13.5, E14.5 and E15.5.
C, F, H) Immunostaining of tdTomato (C), Islet1 (F), and Zic2 (H) in the pigmented and albino ventrotemporal CMZ and retinal crescent at E17.5. Scale bar: 50μm.
D, G, I) Quantification of tdTomato+ (D), tdTomato+/Islet1+ (G), tdTomato+/Zic2+ (I) cells in the pigmented and albino ventrotemporal retina at E17.5.
E) Quantification of tdTomato+ cells (% of total) residing in the apical, basal or intermediate surface of the pigmented and albino retina at E17.5.
J) Quantification of the proportion (% of total) of Zic2+ cells deriving from the CMZ.
K) Immunostaining of tdTomato, EdU, and Zic2 in the pigmented and albino ventrotemporal retina and retinal crescent E17.5. Scale bar: 50μm.
L-M) Quantification of tdTomato+/Islet1+ (L) and tdTomato+/Zic2+ (M) cells born at E13.5–14.5 (EdU-) or E15.5 (EdU+) in the pigmented and albino ventrotemporal retina at E17.5.
Upon tamoxifen administration, by E17.5, the Msx1+ domain of the CMZ was well-labeled with tdTomato, and some tdTomato+ cells were localized in the adjacent neural retina, in agreement with previous studies (Marcucci et al., 2016, Belanger et al., 2017) (Fig. 1C). Neurogenesis from the ventral CMZ increased gradually along the nasal-temporal axis of the eye, with more tdTomato+ cells found in the temporal segment of the retina (Fig. 1 C and Fig. S1A). Although this pattern was detected in both pigmented and albino mice, a ~16% reduction in the total number of CMZ-derived cells was found in the albino compared to pigmented neural retina (Fig. 1D). Nevertheless, the tdTomato+ cell distribution in the apical/ basal retinal surfaces and cells in the process of migration were similar between the two groups (Fig. 1E).
To detect and quantify the retinal cell types that derive from the ventral CMZ, we examined the co-expression of tdTomato with Islet1, AP-2α, Prox1, and Otx2, marking postmitotic RGCs, amacrine cells, horizontal cells, and bipolar cells/cones, respectively. As noted above, RGC neurogenesis from the CMZ was elevated in the temporal compared with the nasal retina of pigmented and albino mice (Fig. S1B). The albino neural retina, however, contained ~35% fewer tdTomato+/Islet1+ RGCs (Fig. 1F-G), suggesting a compromised capacity for RGC neurogenesis in the albino ventral CMZ. In contrast to RGCs, there was minimal production of other retinal cell types from the CMZ in pigmented mice at this particular time window (Fig. 1A’) and these numbers were similar in albino mice (Fig. S1C-D).
Next, we asked whether the curtailed RGC neurogenesis in the albino CMZ impacts the ipsilateral (Zic2+), the contralateral (Brn3a+) RGC population, or both. We focused on the ventrotemporal quadrant of the retina, the locus where ipsilateral RGCs are generated from E13.5 to E16.5, followed by late-born contralateral RGCs from E15.5 until birth (Marcucci et al., 2019). The pigmented and albino CMZ indeed produced ipsilateral RGCs. However, the number of Zic2+ RGCs from the albino CMZ was reduced compared to the pigmented CMZ (Fig. 1H-J and Fig. S1E). In contrast, extremely few Brn3a+/tdTomato+ cells were detected in both the pigmented and albino retina (Fig. S1F-H), suggesting that the majority of RGCs derived from the ventrotemporal CMZ are ipsilateral (Fig. S1I), and indicating that between E13.5 and E17.5 contralateral RGCs originate mainly from the neural retina.
To investigate whether the ventral CMZ has unique neurogenic properties that are not shared with the remainder of the CMZ, or whether ventral vs dorsal CMZ origin determines the ipsilateral vs contralateral RGC fate, respectively, we extended our analyses to the dorsal CMZ segment. Unlike the ventral segment, very few CMZ-derived tdTomato+ cells were observed in both the pigmented and albino dorsal neural retina (Fig. S1J), and even fewer of them were Islet1+ and/or Brn3a+ (Fig. S1K-L), with no differences being noted between the two groups. These results suggest that the dorsal CMZ has limited neurogenic capacity and does not share the neurogenic properties of the ventral CMZ at the developmental time points examined (Fig. 1A’).
To probe for differences in the pace of neurogenesis between the pigmented and albino ventral CMZ, that might be responsible for the reduction in albino CMZ-derived Zic2+ RGCs, we combined fate mapping with birthdating by injecting EdU at E15.5 (Fig. 1A’, 1K). This allowed us to distinguish tdTomato+ cells born at E13.5 and E14.5 (EdU-) from those born at or after E15.5 (EdU+). We found that more than half of the pigmented CMZ-derived Zic2+ RGCs are born at E13.5-E14.5, whereas at the same time window the albino CMZ produced significantly fewer Islet+ and Zic2+ RGCs (Fig. 1L-M). The apoptotic rate of CMZ progenitors was indistinguishable between the two groups, arguing that cell death is not responsible for this difference in neurogenic output (Fig. S1M).
Collectively, our findings show that the ventral CMZ mainly produces ipsilateral RGCs between E13.5 and E15.5, and that reduced neurogenesis in the CMZ partially accounts for the diminished ipsilateral RGCs population that marks albinism.
scRNA-Seq of the pigmented and albino CMZ
To uncover transcriptomic alterations in the albino CMZ that could account for its compromised neurogenic properties, we conducted scRNA-Seq on FACS-isolated cells from E13.5 pigmented (TyrWT) and albino (Tyrc2j) mouse peripheral retinae (Fig. 2A-B) utilizing αCre; tdTomato mice that allowed for enrichment of CMZ cells (Marquardt et al., 2001, Fan et al., 2022). Unsupervised clustering and UMAP representation of the developing peripheral retina (Fig. 2C-D and Fig. S2A) revealed eleven cell clusters (Lo Giudice et al., 2019, Clark et al., 2019, Balasubramanian et al., 2021). First, a group of progenitor cells (positive for Sfrp2, Hes1, Fgf15) expressed markers specific to the CMZ (cluster 1) (Ccnd2, Wfdc1, Msx1, Gja1). Adjacent to the CMZ cluster, three groups of progenitor cells (RPC1–3; clusters 0, 2, 9) (positive for Sfrp2, Ccnd1, Hes1, Fgf15) could be distinguished from each other based on distinct patterns of cell cycle gene expression (Fig. S2B-C). The RPC clusters were connected to a young neurogenic cell population (cluster 3) expressing Atoh7, Hes6, and Neurog2, as well as cell cycle genes (H1 histone family genes, Top2a), followed by a mid- (Atoh7, Hes6) and a more mature neurogenic cluster (clusters 10, 5), enriched in Atoh7 and Hes6, as well as in RGC, horizontal and amacrine cell genes. Emanating from the neurogenic cells, and opposite to the CMZ cluster, we observed two separate branches, one with young and old RGCs (clusters 4, 7) (Isl1, Pou4f2-enriched), and the other that included a horizontal/amacrine cell cluster (cluster 8) (Ptf1a, Lhx1, Onecut1/2-enriched) as well as a photoreceptor precursors/cone cluster (cluster 11) (Otx1, Neurod1/4, Crx-enriched).
Figure 2: scRNA-Seq in the pigmented and albino CMZ.
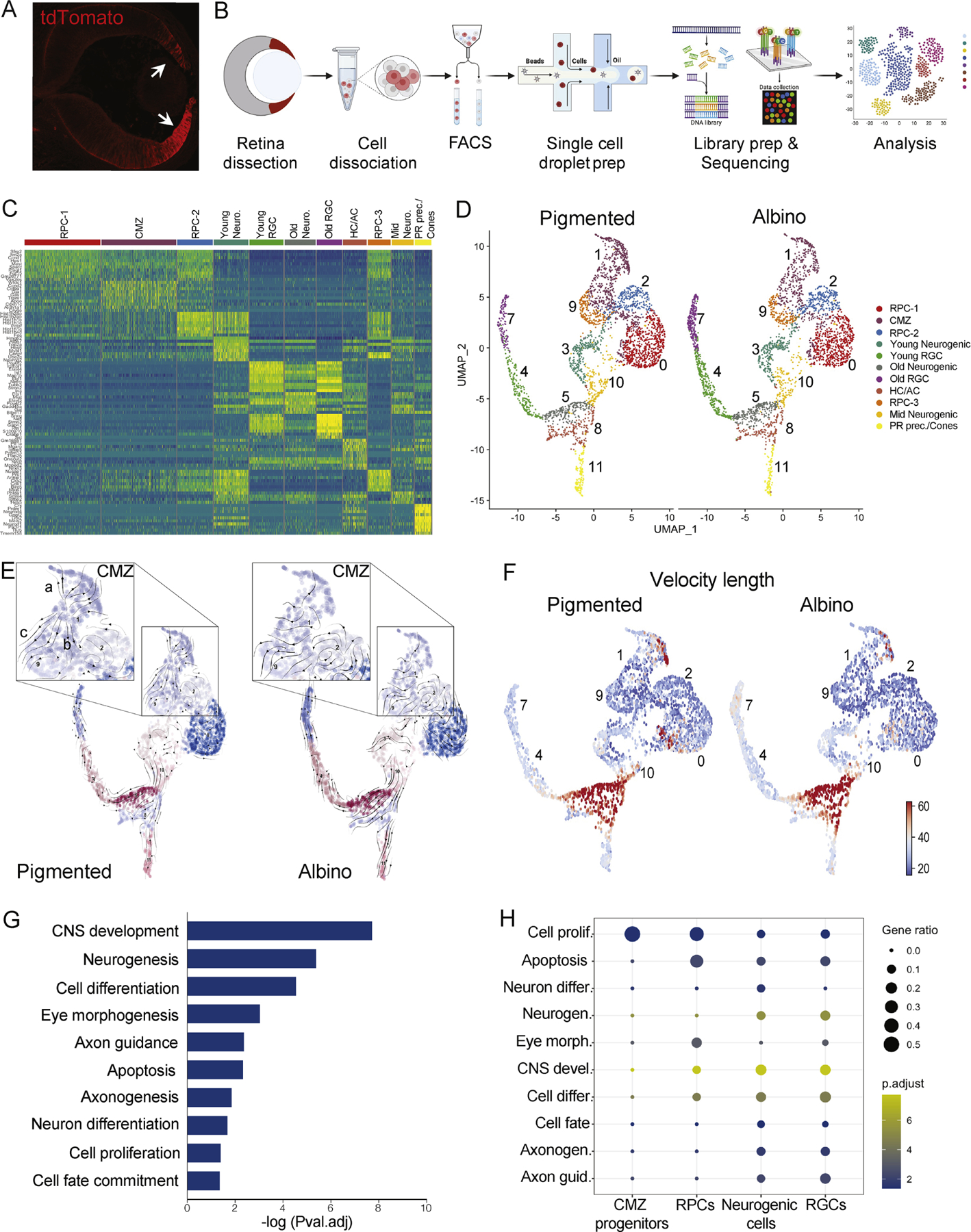
A) tdTomato expression in the CMZ and peripheral retina of αCre; tdTomato mice at E13.5.
B) scRNA-Seq work flow.
C) Differential gene expression in single-cell clusters.
D) UMAP representation of single-cell clusters in the pigmented and albino datasets.
E) RNA Velocity vector stream projected on UMAP of single-cell clusters in the pigmented and albino datasets.
F) Length of velocity vector quantifying arrow length, embedded on UMAP of single-cell clusters in the pigmented and albino datasets.
G) GO terms corresponding to all DEG between pigmented and albino datasets.
H) GO term (from G) enrichment across cell states.
RPC: retinal progenitor cell, CMZ: ciliary margin zone cell, Neuro: neurogenic cell, RGC: retinal ganglion cell, HC: horizontal cell, AC: amacrine cell, PR: photoreceptor
We next investigated changes to the rate and direction of cell state transitions using RNA velocity analysis (Fig. 2E-F and Fig. S3A). Cells within the CMZ cluster of the pigmented eye (cluster 1) had three distinct predicted trajectories (Fig. 2E), which reflected transition towards: a) terminally differentiated CMZ cells indicated by arrows moving upwards towards the tip of cluster 1 (CMZ), self-renewing RPCs indicated by arrows moving towards clusters 2 and 9 (RPC-2, RPC-3), or neurogenic states as indicated by arrows exiting towards cluster 7 (RGCs). In contrast, velocity across all three predicted trajectories was altered in cells of the albino CMZ (Fig. 2E), potentially reflecting a disruption in the progression of differentiation. Specifically, the velocity of cells transitioning into mature CMZ cells (e.g., expressing Mitf, Wls) was reduced as seen by the quantification of vector length (Fig. 2F and Fig. S3A). The albino RPC-1 population (cluster 0) also displayed slower velocities of cell state progression towards neurogenic fate (cluster 10) compared to the pigmented RPC-1 population (Fig. 2F and Fig. S3A). Together, these data suggest that neurogenesis from the albino CMZ does not follow the directionality and tempo of the pigmented CMZ.
Lastly, we performed a gene ontology (GO) analysis of differentially expressed genes (DEG) in all clusters between pigmented and albino cells (Fig. 2G) and further examined how the identified GO terms are represented across the CMZ, RPC, neurogenic, and RGC states (Fig. 2H). The GO term “regulation of cell proliferation” was the major difference between the pigmented and albino CMZ and retinal progenitors (Fig. 2H and Fig. S3B), supporting the alterations in RNA velocity length. The GO terms of “differentiation”, “neurogenesis”, and cell “fate commitment” were enriched in neurogenic cells, whereas DEG characterizing RGC neurogenesis, axonogenesis, and axon development were enriched in RGCs (Fig. 2H), in line with the transcriptional changes reflecting cell state transitions during retinal development. Genes associated with ipsilateral RGC fate were downregulated in the albino neurogenic and RGC clusters, reflecting a reduced population of RGCs with ipsilateral fate, whereas genes linked to contralateral fate were upregulated (Fig. S3C-F) (Fries et al., 2022, Fernandez-Nogales et al., 2022, Herrera et al., 2003, Lo Giudice et al., 2019, Wang et al., 2016).
Cell cycle is perturbed in the albino CMZ
Studies on the albino retina have associated lack of pigment with spatiotemporal defects in retinal neurogenesis, such as mitotic spindle orientation, and delayed pace of center-to-periphery cell production and maturation (Cayouette et al., 2001, Rachel et al., 2002, Tibber et al., 2006, Bhansali et al., 2014). It is not clear, however, whether regulation of specific cell cycle phases is linked to RGC fate specification in the CMZ.
The finding that the expression of genes regulating cell proliferation was altered in the albino CMZ prompted us to compare the pigmented and albino scRNA-Seq datasets based on S and G2/M phase scores (Fig. 3A). Cell cycle scoring revealed a reduction in the number of cells in both S and G2/M phases in the albino compared to pigmented peripheral retina (Fig. 3B). To validate this observation and to further probe for differences in the progression of pigmented and albino CMZ cells through the cell cycle, we performed dual-pulse birthdating at E13.5 and E14.5 using BrdU and EdU. To identify cells within the cell cycle we additionally stained for Ki67 (Fig. 3C). We observed no significant differences in the total number of Ki67+ CMZ cells between the two groups (Fig. 3D). However, the albino CMZ contained a ~32–38% greater number of cells in G1 phase compared to the pigmented CMZ, and ~19–34% fewer cells in the S phase (Fig. 3E). Next, we quantified the number of CMZ cells in M phase using the mitotic marker PH3 (Fig. 3F) and found ~32–39% fewer mitotic cells in the albino compared to pigmented CMZ at E13.5 and E14.5 (Fig. 3G). Therefore, progenitors in the albino CMZ progress through the cell cycle and reach mitotic exit at a slower pace (Fig. 3H) due to a delay in their transition from G1 to S phase.
Figure 3: Cell cycle is perturbed in the albino CMZ.
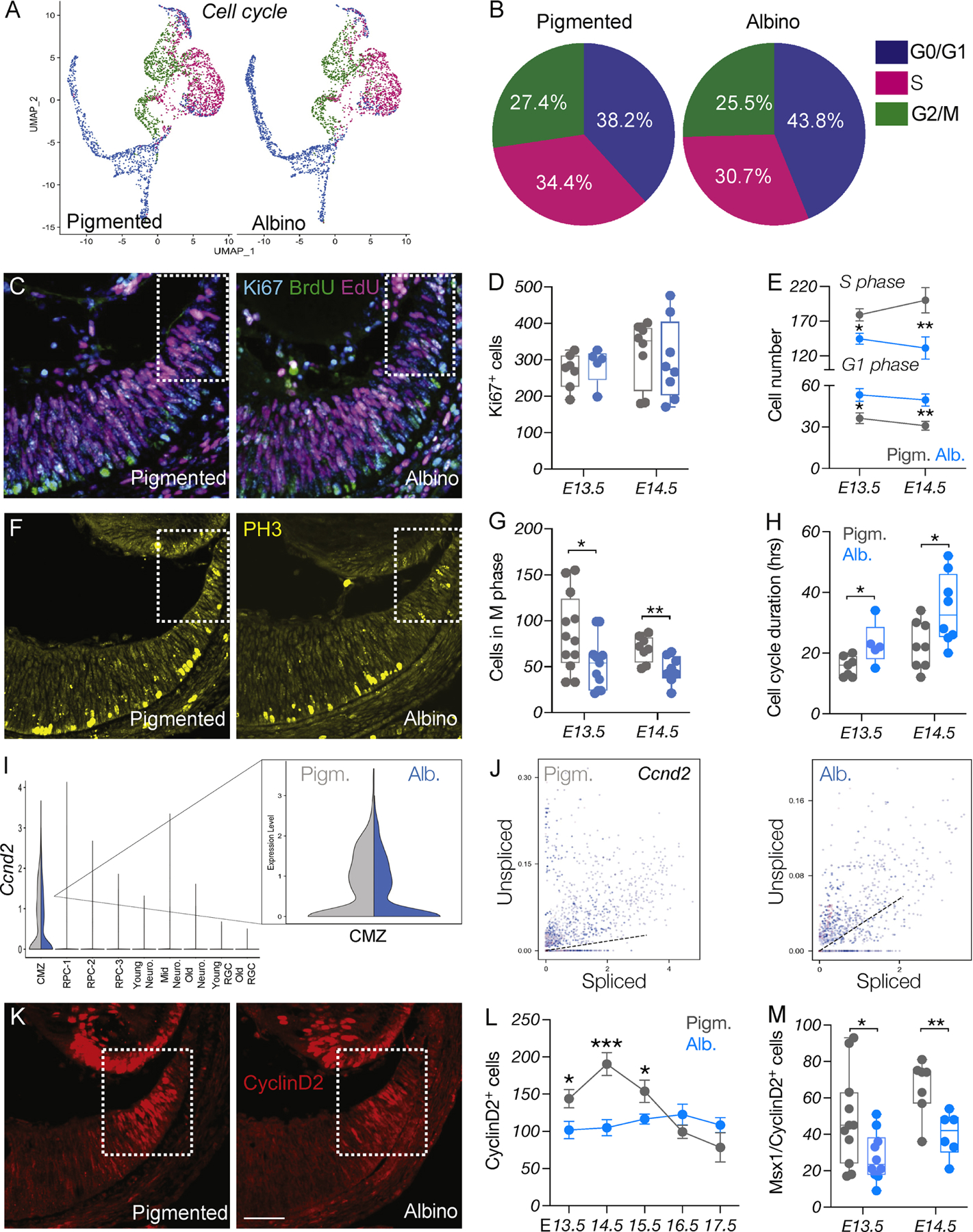
A) UMAP representation of single cells based on cell cycle phase.
B) Quantification of cells in the G0-G1, S or G2/M phase of the cell cycle, in pigmented and albino datasets.
C) Dual-pulse birthdating: Immunostaining of Ki67, BrdU, and EdU in the pigmented and albino ventrotemporal CMZ at E14.5.
D) Quantification of Ki67+ (cycling) cells at E13.5 and E14.5.
E) Quantification of cells in the G1 and S phases of the cell cycle at E13.5 and E14.5.
F) Immunostaining of the mitotic marker PH3 in the pigmented and albino ventrotemporal CMZ at E14.5.
G) Quantification of cells in the M phase of the cell cycle (PH3+) at E13.5 and E14.5.
H) Quantification of cell cycle duration (hours) at E13.5 and E14.5.
I) Violin plot showing downregulation of Ccnd2 in the albino CMZ cell cluster.
J) Transcriptional dynamics of Ccnd2 as a ratio of unspliced to spliced transcripts in the pigmented and albino peripheral retina.
K) Immunostaining of CyclinD2 in the pigmented and albino ventrotemporal CMZ at E14.5. Scale bar: 100μm.
L-M) Quantification of CyclinD2+ (L) from E13.5 to E17.5 and of CyclinD2+/Msx1+ (M) cells at E13.5 and E14.5.
To seek the cause of this delay, we further analyzed the scRNA-Seq data and identified Ccnd2 as the predominant DEG in the CMZ cluster (Fig. S3B and Fig. 3I). Ccnd2 encodes for CyclinD2, a cell cycle-regulatory protein that has been associated with neurogenesis in the CMZ (Marcucci et al., 2018, Wang et al., 2016). Cells of the pigmented peripheral retina had a positive Ccnd2 velocity profile that indicates transcriptional activation, whereas in the albino, a greater number of cells had negative Ccnd2 velocity profile, indicating its transcriptional downregulation (Fig. 3J). Consistent with our scRNA-Seq findings, the albino CMZ contained fewer CyclinD2+ cells compared to pigmented at E13.5, and this reduction was maintained throughout the time window of CyclinD2 expression, until E17.5 (Fig. 3K-L). In addition, CyclinD2 expression was reduced by ~39–42% in the albino Msx1+ CMZ progenitor pool (Fig. 3M).
These results demonstrate that diminished CyclinD2 expression in retinal progenitors within the albino CMZ is correlated with disrupted transitioning from the G1 to S phase of the cell cycle, subsequently impacting the transition to M phase.
CyclinD2-dependent G1/S transition regulates ipsilateral RGC neurogenesis
We hypothesized that delayed cell cycle G1/S progression at E13.5 and E14.5 due to insufficient CyclinD2 expression leads to delayed or reduced neurogenesis during the critical and narrow time window for the expression of Zic2 and production of ipsilateral RGCs. To determine a causal role of CyclinD2 in the timing of cell cycle phase transitions in the CMZ and in ipsilateral RGC neurogenesis, we compared pigmented CyclinD2flox (CyclinD2WT) mice with pigmented αCre; CyclinD2flox mice lacking CyclinD2 specifically in the CMZ (CyclinD2cKO) (Fig. S4A). First, we repeated dual pulse labeling (Fig. 4A and Fig. S4B). Similar to the albino CMZ, the CyclinD2cKO CMZ had ~35% more cells in G1 and ~20% fewer in S phase compared to CyclinD2WT (Fig. 4B and Fig. S4C-D). Furthermore, in the CyclinD2cKO CMZ we detected a ~30% reduction of PH3+ cells (Fig. S4E-F) whereas the apoptotic rate of CMZ progenitors was unaffected (Fig. S4G), suggesting that decrease in CyclinD2 expression in the CMZ is associated with a prolonged G1/S transition, preventing cells from exiting the cell cycle, as in the albino retina (Fig. 3C-E).
Figure 4: CyclinD2-dependent G1/S transition regulates ipsilateral RGC neurogenesis.
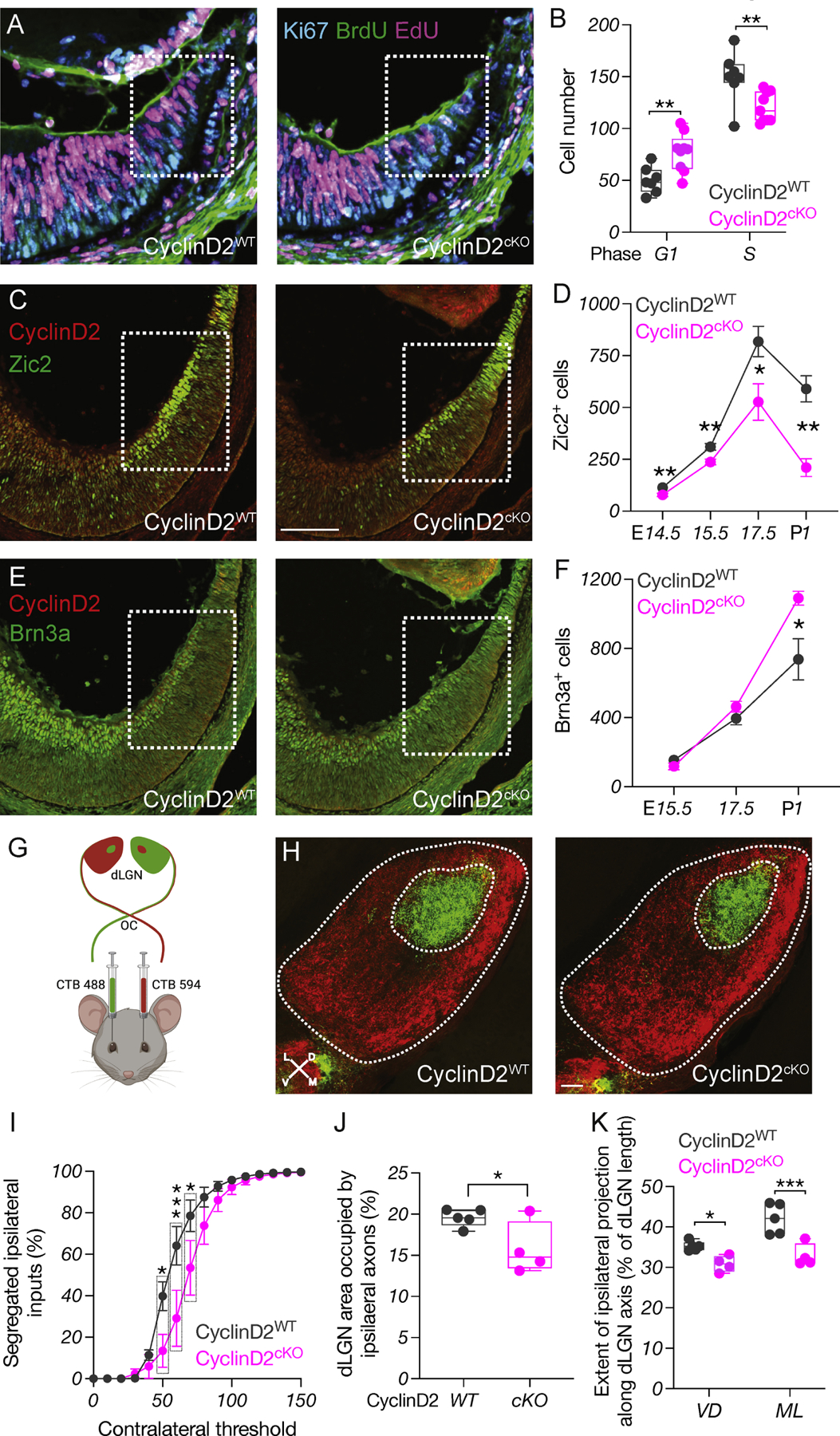
A) Dual-pulse birthdating: Immunostaining of Ki67, BrdU, and EdU in the CyclinD2WT and CyclinD2cKO ventrotemporal CMZ at E14.5. Scale bar: 100μm.
B) Quantification of cells in the G1 and S phases of the cell cycle at E14.5.
C) Immunostaining of CyclinD2 and Zic2 in the CyclinD2WT and CyclinD2cKO ventrotemporal retina and CMZ at E15.5. Scale bar: 100μm.
D) Quantification of Zic2+ cells at E14.5, E15.5, E17.5 and P1.
E) Immunostaining of CyclinD2 and Brn3a in the CyclinD2WT and CyclinD2cKO ventrotemporal retina and CMZ at E15.5.
F) Quantification of Brn3a+ cells at E15.5, E17.5 and P1.
G) Strategy used for RGC axon tracing to the dLGN.
H) Immunostaining of dLGN sections from CyclinD2WT and CyclinD2cKO mice labeled with 488- and 594-CTB at P30. The entire dLGN, as well as the dLGN core receiving ipsilateral input, are outlined. Scale bar: 200μm. D: dorsal, V: ventral, M: medial, L: lateral. Axes indicate the orientation used for quantification along the DV and ML planes.
I) Segregation plot: Percent of segregated inputs as a function of contralateral threshold.
J) Area of the ipsilateral projection as percentage of total dLGN area.
K) Extent of the ipsilateral projection as percentage of length along the DV and ML dLGN axes.
Because temporal control of cell birth has been linked to the regulation of neurogenesis and cell fate specification (Cepko et al., 1996, Tripodi et al., 2011, Osterhout et al., 2014, Pilaz et al., 2016b, Rossi et al., 2017, Xu et al., 2020, Cavalieri et al., 2021), we tested whether CyclinD2 deficiency impedes cells from acquiring ipsilateral RGC identity through Zic2 expression. We noted a significant reduction in the number of Zic2+ RGCs in the CyclinD2cKO ventrotemporal neural retina from E14.5 until P1, resembling the albino phenotype (Fig. 4C-D and Fig. S4H). We also probed for differences in contralateral RGCs between CyclinD2WT and CyclinD2cKO mice (Fig. 4E-F and S4H). We performed our measurements from E15.5 - a time when Brn3a+ cells appear in the ventrotemporal neural retina among the Zic2+ population - until P1, to capture the late-born contralateral RGCs (Marcucci et al., 2019). During embryogenesis (E15.5-E17.5), CyclinD2 deletion from the CMZ did not lead to changes in the number of Brn3a+ cells (Fig. 4F and Fig. S4H), consistent with our finding that the ventral CMZ generates primarily ipsilateral RGCs (Fig. 1). Although this result is at odds with what was previously reported (Marcucci et al., 2016), the discrepancy can be explained by the different transgenic mouse line used. However, we see that by P1 the number of late-born Brn3a+ RGCs residing within the peripheral ventrotemporal retina was ~32% higher in the CyclinD2cKO (Fig. 4F and Fig. S4H), reflecting an ultimate compensation for the overall reduction in the ipsilateral RGC population by contralateral RGCs.
To determine whether CyclinD2-deficiency from the CMZ influences the neurogenesis of retinal cell types beyond ipsilateral RGCs, we counted cells positive for amacrine, horizontal, and cone/bipolar cell markers in the E17.5 peripheral ventrotemporal retina. Within the retinal segment extending 150μm from the end of the CMZ we did not find significant differences between CyclinD2WT and CyclinD2cKO mice (Fig. S4I-J), in agreement with our fate mapping results showing that the ventrotemporal CMZ contributes minimally to the neurogenesis of these retinal neurons at the developmental time point we examined (Fig. S1C-D).
We next determined whether the decrease in Zic2+ RGCs in the CyclinD2cKO retina is reflected in defects in afferent targeting of the dLGN, as described previously in albino mice (Rebsam et al., 2012). We used whole-eye CTB labeling (Fig. 4G) and traced RGC axons in target regions of the CyclinD2WT and CyclinD2cKO dLGN at P30. We found that both the ipsilateral and the contralateral retinogeniculate projections of CyclinD2cKO mice appropriately innervate their respective target areas (Fig. 4H). However, the area receiving ipsilateral retinal input was markedly smaller (~20%) in the CyclinD2cKO dLGN compared with CyclinD2WT (Fig. 4I-J). This effect extended along both the mediolateral and dorsoventral dLGN axes (Fig. 4K).
Thus, our results indicate that CyclinD2 is a crucial component of a CMZ-specific molecular machinery that links cell cycle progression with ipsilateral RGC neurogenesis, and is necessary for proper retinogeniculate targeting.
Depth perception requires CyclinD2 expression in the CMZ
We hypothesized that alterations in the proportions of ipsi- and contralaterally-projecting RGCs in albino and CyclinD2cKO mice could lead to deficits in binocular vision. To test this, we used the binocularly-driven “visual cliff” behavioral task, a well-established paradigm that measures depth perception in both humans and rodents (Lim et al., 2016) (Fig. 5A). We first compared the performance of pigmented (TyrWT) and albino (Tyrc2j) mice. Pigmented mice almost always stepped onto the shallow side, whereas albino mice failed to distinguish between the shallow and deep side of the platform (Fig. 5B, and Fig. S5A). Both pigmented and albino mice spent the same time making a decision to step on the platform (Fig. 5C). We next compared CyclinD2WT and CyclinD2cKO mice. CyclinD2cKO mice exhibited poor depth perception, indicated by the lower number of trials in which they stepped onto the shallow side and longer time to reach a decision, compared to CyclinD2WT littermates which performed similarly to pigmented mice (Fig. 5D-E and Fig. S5B).
Figure 5: Depth perception requires CyclinD2 expression in the CMZ.
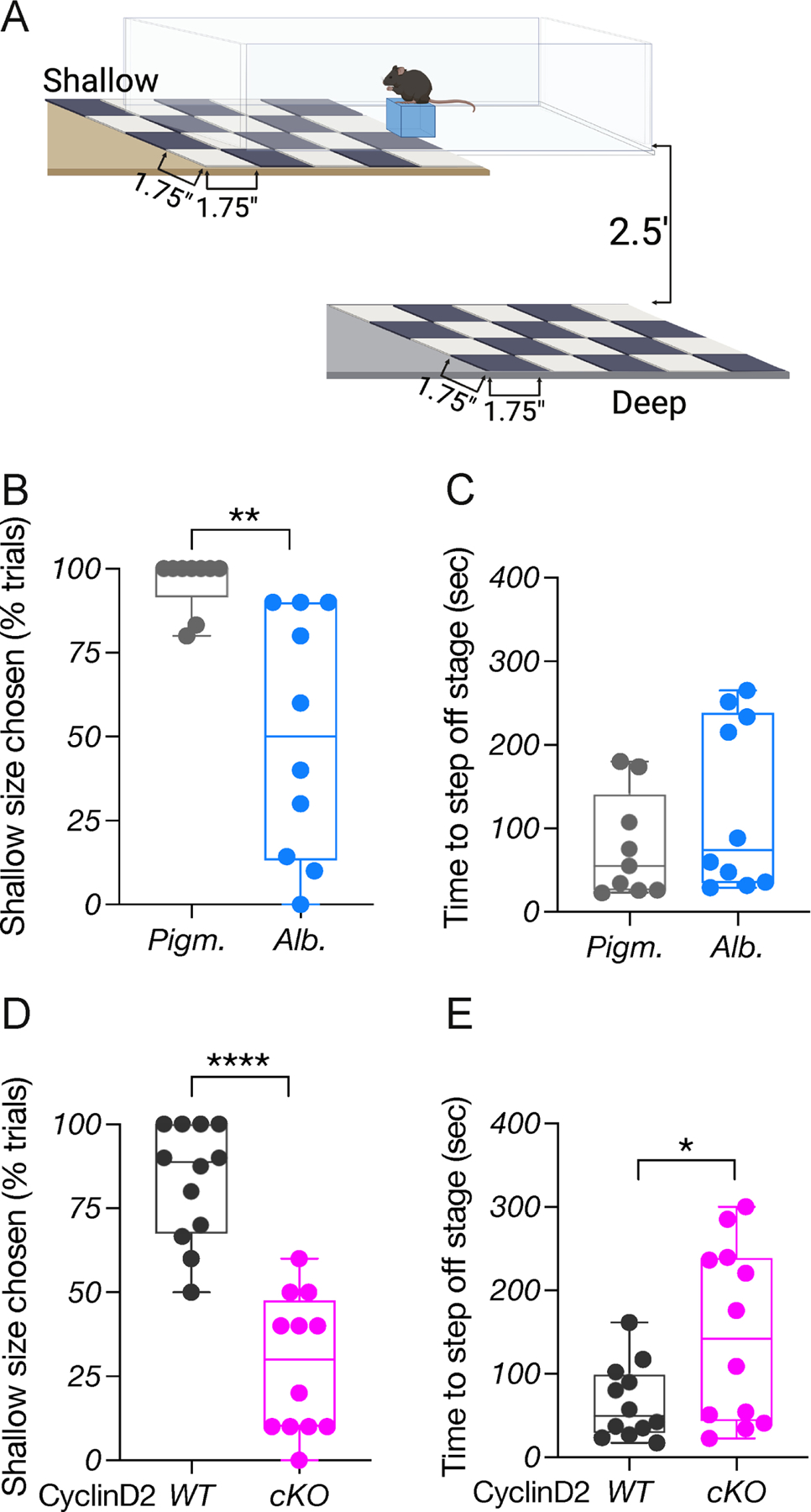
A) Behavioral platform used for the binocularly-driven visual cliff assay.
B) Quantification of depth perception comparing pigmented and albino mice.
C) Quantification of time (sec) needed to step off the stage to either side, comparing pigmented and albino mice.
D) Quantification of depth perception comparing CyclinD2WT and CyclinD2cKO mice.
E) Quantification of time (sec) needed to step off the stage to either side, comparing CyclinD2WT and CyclinD2cKO mice.
To ensure that the behavior of CyclinD2cKO mice was indicative of reduced depth perception, and not due to other visual deficits that could prevent them from discriminating the black-and-white grids clearly, we tested CyclinD2WT and CyclinD2cKO mice for contrast sensitivity and visual acuity using the Optodrum (Fig. S5C). Both groups were able to track the stimulus until its contrast dropped to 4.92% (Fig. S5D), and had average visual acuity 0.45 cycles/degree (Fig. S5E).
Together, our results show that perturbations in the ratio of ipsi- to contralateral RGC projection lead to poor depth perception both in albino and CyclinD2cKO mice, indicated by their inferior performance in the visual cliff task. Thus, CyclinD2 expression within the CMZ, and its involvement in regulating ipsi-/contralateral RGC neurogenesis, is critical for establishing the circuit for stereopsis.
CyclinD2 upregulation restores binocular circuit formation and function in albino mice
Given the instrumental role of CyclinD2 in the proper formation of the binocular circuit, we asked whether we could rescue the reduced ipsilateral RGC population and impaired visually-guided behavior of albino mice by normalizing CyclinD2 expression in the CMZ.
To design a strategy to modulate CyclinD2 expression, we took advantage of our scRNA-Seq datasets, focusing on the CMZ cluster. DEG analysis between pigmented and albino CMZ cells revealed a significant downregulation of Fos and Jun (Fig. S6A-B), two transcriptional regulators of CyclinD proteins (Brown et al., 1998, Turchi et al., 2000, Bakiri et al., 2000). In turn, Fos and Jun can be regulated by second messengers such as intracellular calcium (Leclerc et al., 1999, Cruzalegui et al., 1999). Therefore, we hypothesized that calcium channel activation could lead to Fos and/or Jun elevation, resulting directly or indirectly in an induction of CyclinD2 expression in the albino CMZ. Our scRNA-Seq revealed an enrichment of Cacna1d in both the pigmented and albino CMZ and RPC clusters but not in more mature states. Validation experiments showed that CaV1.3, the L-type voltage gated calcium channel subunit encoded by Cacna1d, is expressed within the neurogenic (Msx1+) domain of both the pigmented and albino CMZ (Fig. S6C-D), at early stages of retinogenesis. L-type calcium channel activation by BayK-8644 has been reported to increase CyclinD2 protein levels in the pancreas (Salpeter et al., 2011). Therefore, to manipulate CyclinD2 levels, we injected pregnant dams with BayK-8644, and used sham-injected mice as controls.
We first tested the effect of BayK on Fos and Jun in the pigmented (TyrWT) and albino (Tyrc2j) CMZ. Administration of BayK at E13.5 and E14 led to an upregulation of both Fos and Jun in the albino CMZ (Fig. S6E-H), consistent with the role of CaV1 channels in immediate early gene (IEG) activation (Samak et al., 2011, Yang et al., 2022, Servili et al., 2018, Wheeler et al., 2012). BayK injection at E13.5 and E14 also elevated the expression of CyclinD2 in the albino ventral CMZ by ~50% at E14.5 and ~45% at E15.5, bringing it to pigmented-like levels (Fig. 6A-C). Following the normalization in the levels of CyclinD2, the number of albino CMZ cells in G1, S, and M phases of the cell cycle approached those of the pigmented CMZ (Fig. S7A-E).
Figure 6: CyclinD2 upregulation restores binocular circuit formation and function in albino mice.
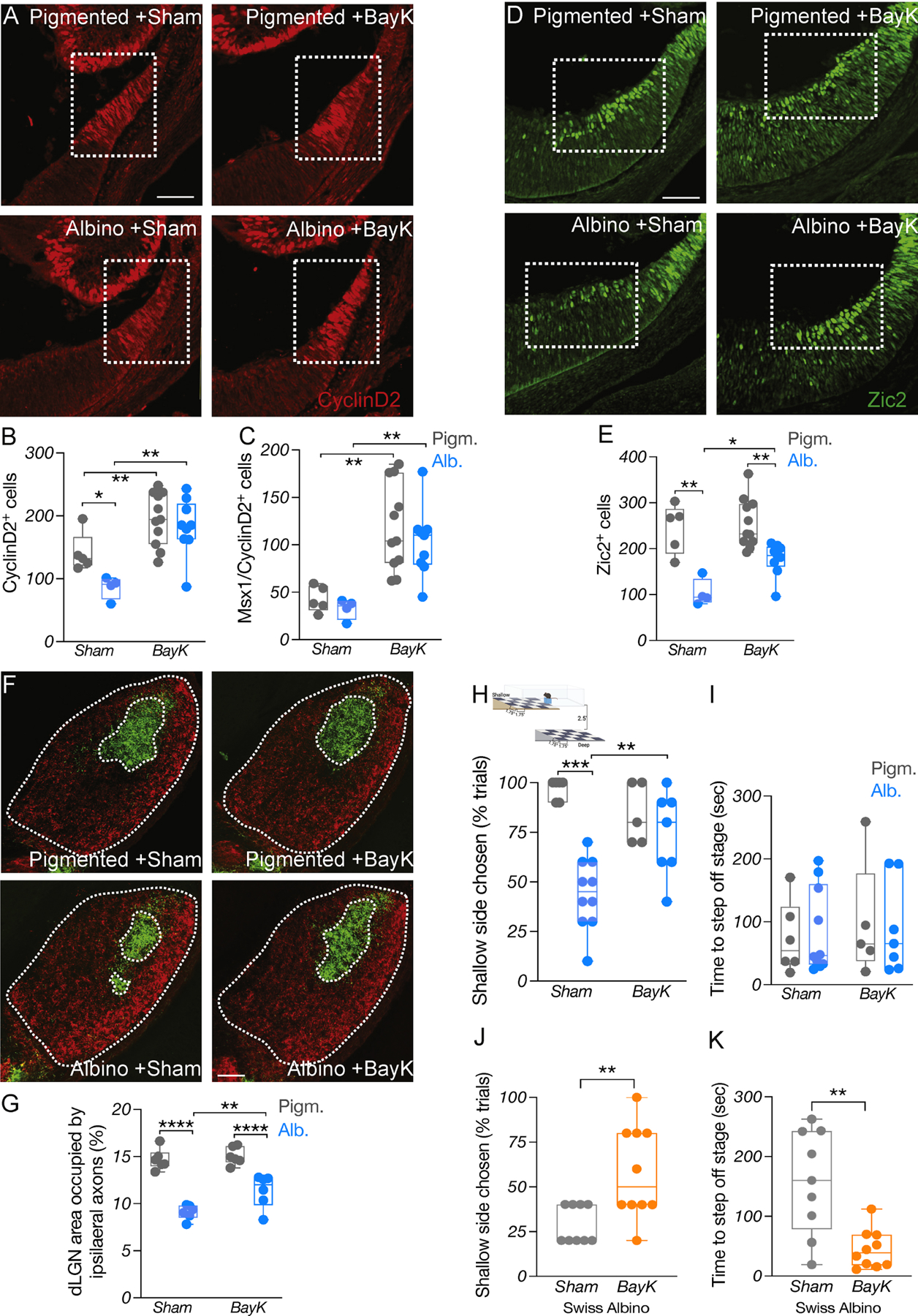
A) Immunostaining of CyclinD2 in the ventrotemporal CMZ of pigmented and albino Sham vs BayK treated mice at E15.5. Scale bar: 100μm.
B-C) Quantification of CyclinD2+ (B) and CyclinD2+/Msx1+ (C) cells at E15.5.
D) Immunostaining of Zic2 in the ventrotemporal retina of pigmented and albino Sham vs BayK treated mice at E15.5. Scale bar: 100μm.
E) Quantification of Zic2+ cells at E15.5.
F) Immunostaining of coronal dLGN sections from pigmented and albino Sham vs BayK treated mice labeled with 488- and 647-CTB at P60. The entire dLGN, as well as the dLGN core receiving ipsilateral input, are outlined. Scale bar: 100μm.
G) Area of the ipsilateral projection as percentage of total dLGN area.
H) Depth perception comparing pigmented and albino Sham vs BayK treated mice.
I) Quantification of time (sec) needed to step off the stage to either side during the visual cliff task.
J) Quantification of depth perception comparing Swiss albino Sham vs BayK treated mice.
K) Quantification of time (sec) needed to step off the stage to either side in the visual cliff task, comparing Swiss albino Sham vs BayK treated mice.
In addition to an elevation in CyclinD2, BayK led to a significant (~50%) increase of Zic2+ RGCs in the ventrotemporal retina, compared to sham-treated albino mice (Fig. 6D-E), without changing the apoptotic rate within the Zic2+ RGC domain in either the pigmented or the albino group (Fig. S7F). However, BayK had no effect on the Zic2+ RGC population of CyclinD2cKO mice, which remained diminished compared to CyclinD2WT (Fig. S7G-H), suggesting that the BayK-induced Zic2+ RGC augmentation in the albino retina requires CyclinD2.
We then used whole-eye CTB labeling to assess whether the ipsilateral innervation of the albino dLGN was also ameliorated upon BayK treatment. As expected, the dLGN core area receiving ipsilateral retinal input was smaller in sham-treated albino mice compared to pigmented mice. This defect was partially reversed in BayK-treated albino mice, in which the ipsilateral retinorecipient region of the dLGN was significantly (~40%) increased compared to the sham albino group (Fig. 6F-G).
To test whether the restoration of the ipsilateral retinogeniculate projection ultimately leads to improved depth perception, we subjected sham- and BayK-treated pigmented and albino mice to the visual cliff task. Albino mice that received sham treatment during development exhibited poor depth perception compared to pigmented littermates, as expected. Strikingly, albino mice that received BayK performed similarly to pigmented mice (Fig. 6H-I). In contrast to albino mice, CyclinD2cKO mice did not improve their behavior at the visual cliff task, in agreement with our finding that the Zic2+ RGC population remained reduced in these mice after BayK treatment (Fig. S7I). Lastly, we tested whether the BayK-induced improvement in depth perception could be observed in albino mice of a different genetic background. Similar to albino mice, Swiss albinos that received BayK during development exhibited considerably improved behavior in the visual cliff test compared to those that received sham (Fig. 6J-K).
In summary, our results revealed a CMZ-specific mechanism that controls neurogenesis of the ipsilateral population of RGCs in the proper proportion, and showed that pharmacological stimulation of this mechanism during development can restore the binocular circuit and depth perception in albinism (Fig. 7).
Figure 7: Summary.
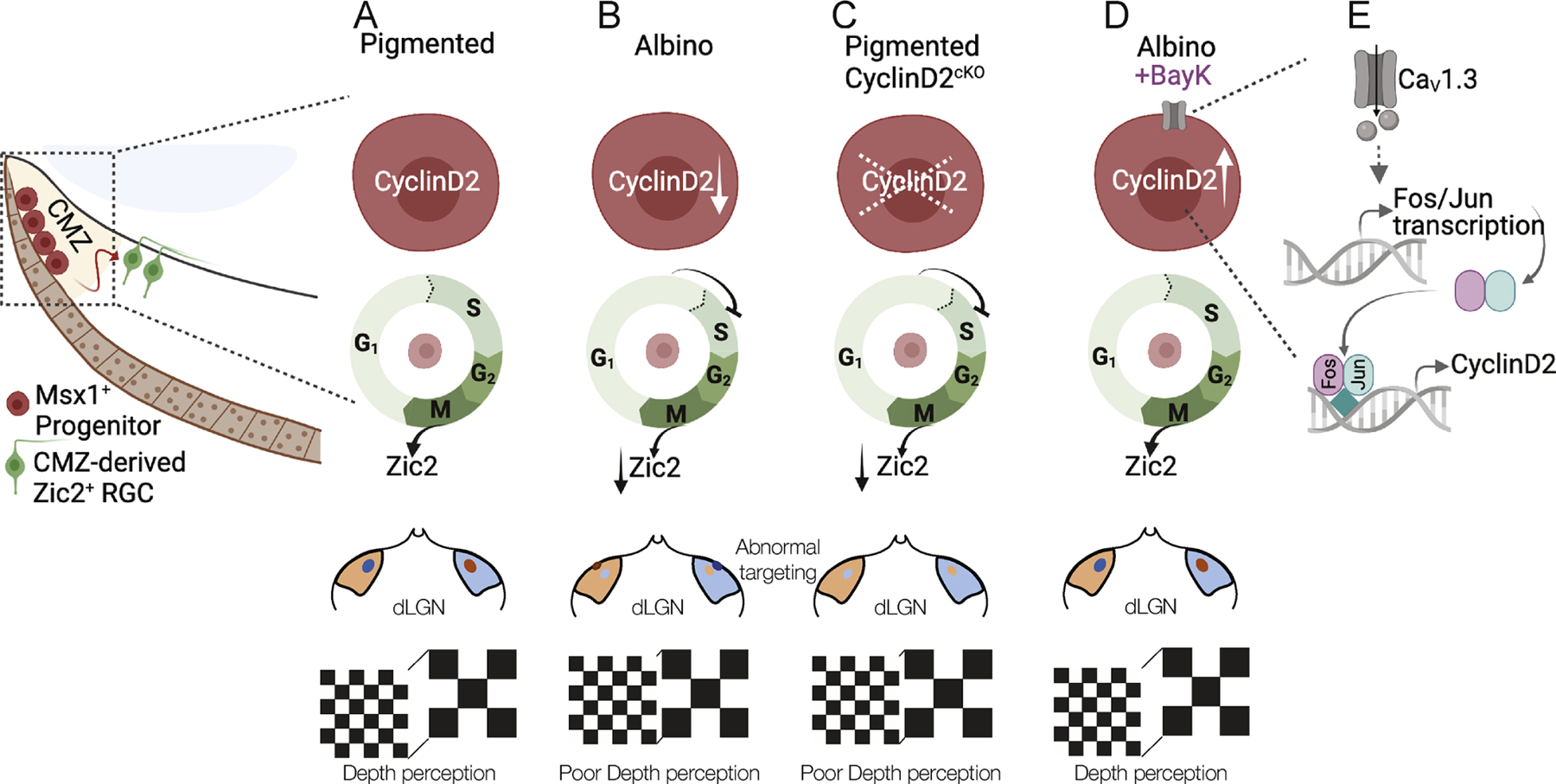
A) The link between CyclinD2 and cell cycle, the ipsilateral RGC hallmark gene (Zic2), retinogeniculate projection, and binocular vision.
B) In the albino, CyclinD2 reduction leads to delay in G1/S phase progression from, reduced numbers of Zic2+ RGCs, reduced ipsilateral retinogeniculate projection, and poor depth perception.
C) This phenotype is recapitulated when CyclinD2 is deleted from the pigmented peripheral retina.
D-E) The binocular circuit-related deficits in the albino retina can be rescued by the delivery of the CaV1 channel agonist BayK-8644, possibly via Fos/Jun transcriptional activation.
DISCUSSION
In this study, we provide evidence for CMZ neurogenesis as a source of ipsilateral RGCs. Our work highlights CyclinD2 as a key regulator of the neurogenic output of the CMZ by controlling aspects of the cell cycle during a developmental time window critical for ipsilateral RGC neurogenesis and assumption of Zic2 expression. In addition, we establish a critical link between calcium channel function and RGC fate acquisition, providing an inroad to unravel the relationship between perturbed melanogenesis in the RPE and aberrations of the eye to brain circuitry seen in albinism.
The ciliary margin zone as a source of retinal neurons
During retinal development, most neurons are generated following a center-to-periphery gradient. Two previous studies (Marcucci et al., 2016, Belanger et al., 2017) showed that the embryonic mammalian CMZ contains a progenitor population with distinct molecular identity, and can generate a variety of retinal cell types, including RGCs. However, because of the small number of CMZ-derived retinal neurons traced in these studies, the identity of the CMZ-derived RGCs and the overall functional importance of the CMZ in mammalian visual circuit development could not be explored. The present research builds upon these findings and expands our view of the neurogenic role of CMZ, by revealing the identity of CMZ-derived RGCs and their role in binocular vision. Our findings strongly suggest that the anomalies of the binocular circuit in albinism are rooted in the CMZ niche, and provide evidence that RGC neurogenesis from the CMZ is indispensable for the proper formation and function of the binocular circuit.
Although few contralateral RGCs originated from CMZ in our experiments, it remains to be determined whether CMZ progenitors produce contralateral RGCs at later time-points not tested here (i.e., E17.5 until birth), or instead, whether they are intrinsically distinct from progenitors in the neural retina, in that they have lost their multipotency and are committed to an ipsilateral fate while still in the cell cycle. Indications that the ventral CMZ has neurogenic properties that are not shared with the rest of the CMZ stem from our results showing that the dorsal CMZ has low potential to generate retinal neurons, at least during the developmental time window of ipsilateral RGC neurogenesis.
Another outstanding question is whether the CMZ-derived ipsilateral RGCs have special light response properties or topographic representation within the dLGN. A recent study (Johnson et al., 2021) revealed that ipsilateral RGCs are represented by only 9 out of the 40 functional RGC subtypes to support binocular vision in mice. Determining whether ipsilateral RGCs born in the CMZ belong to conventional contrast-encoding, melanopsin-expressing luminance-encoding, and/or tSbC types, and whether their projections are confined to a specific dLGN region will further illuminate the functional importance of the CMZ in key aspects of vision such as binocularity.
Regulation of ipsilateral RGC neurogenesis
Previously, we reported that CyclinD2 plays a role in neurogenesis from the CMZ and the production of ipsilateral RGCs from this niche (Marcucci et al., 2019, Marcucci et al., 2016). Our present scRNA-Seq and velocity analysis, together with our birthdating experiments and cell cycle analysis in albino and CyclinD2cKO, go further than these previous studies and demonstrate that in controlling cell cycle phase transitions, CyclinD2 is a set point for the functional integrity of the binocular circuit.
Conditional CyclinD2-deficient mice largely recapitulate the albino phenotype in several aspects of cell cycle and mitotic exit, Zic2+ RGC neurogenesis, and binocular circuit architecture and function. However, the reduction in the Zic2+ RGC population is less severe in the CyclinD2cKO compared to the albino, and the thalamic area receiving ipsilateral retinal input is less impacted. These results agree with the view that the mouse retina is governed by two distinct routes of ipsilateral RGC neurogenesis: one that originates in the CMZ and is controlled by CyclinD2, and the other that originates from RPCs in the neural retina.
Depth perception in albino mice
Binocular disparity-tuned neurons have been characterized in the mouse visual cortex, enabling the discrimination of stereoscopic depth (Samonds et al., 2019, Tan et al., 2022), which in turn guides behavior both in natural scenes, i.e., during prey approach and contact (Johnson et al., 2021), and in non-natural settings, i.e., during a pole descent cliff assay (Boone et al., 2021). The cliff response is one of the few innate visual behaviors in mice (Fox, 1965). Similar to observations in albino rats (Walk and Gibson, 1961), we find that both albino and CyclinD2cKO mice perform poorly in the visual cliff paradigm. This finding highlights the importance of ipsilateral RGC neurogenesis from the CMZ in the formation of the binocular circuit, and the functional impact of alterations in this process on albinism.
One question is whether the performance at the visual cliff varies along with the size of the Zic2+ RGC population. Our experiments suggest that the relationship between ipsilateral RGC numbers and visual cliff performance is not linear; instead, a certain “threshold” of ipsilateral RGCs is required for adequate depth discrimination. Lastly, to fully address the issue of depth perception in albinism and its impact on the interactions with the surrounding environment, it would be of interest to characterize the behavior of these mice during other visually-guided tasks, such as prey hunting (Johnson et al., 2021).
The role of the retinal pigment epithelium in retinal development
A lingering question is how the alterations in melanin biogenesis in albinism lead to disruptions in CyclinD2-mediated control of the cell cycle within the CMZ, and ultimately to diminished ipsilateral RGCs. One possibility suggested by the present data is that the RPE provides signals to the adjacent CMZ that regulate the tempo of the cell cycle during neurogenesis of RGCs. In albinism, RPE-originating signals may be either insufficient or excessive, due to metabolic and cellular aberrations caused by the lack of pigment (Iwai-Takekoshi et al., 2016, Iwai-Takekoshi et al., 2018).
Our current work points to calcium as a potential signal that influences the output of CMZ neurogenesis. Melanin in the RPE has calcium buffering capacity and has been associated with the tight regulation of calcium levels available to the neural retina (Drager, 1985, Bellono and Oancea, 2014, Ambrosio et al., 2016, Salceda and Sanchez-Chavez, 2000, Bush and Simon, 2007, Pearson et al., 2004). In turn, calcium transients have been shown to regulate retinal development by inducing progenitor proliferation (Pearson et al., 2002). The mechanism by which calcium triggers CyclinD2 expression, potentially involving Fos and Jun, calls for further investigation.
Limitations of study
Our findings illuminate mechanisms leading to the albino RGC phenotype; nonetheless, limitations exist: We examined a limited time window of neurogenesis, such that certain retinal cell types and subtypes from the CMZ may have been missed. Second, our study does not directly show that BayK leads to calcium influx in albino CMZ cells to induce Fos/Jun and CyclinD2 expression. Moreover, two constraints in the field are the lack of histological markers for ipsilateral RGC subtypes, and a limited understanding of the transcriptional program driving precursors to ipsi- or contralateral fate.
STAR METHODS
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Carol Ann Mason cam4@columbia.edu
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT/RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Organisms | ||
| Mouse: C57Bl/6J | Jackson Labs | RRID:IMSR_JAX:000664 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Labs | RRID:IMSR_JAX:007914 |
| Mouse: B6(Cg)-Tyrc−2J/J | Jackson Labs | RRID:IMSR_JAX:000058 |
| Mouse: J:ARC(S) | Jackson Labs | RRID:IMSR_JAX:034608 |
| Mouse: Msx1-CreERT2 | gift from Dr. Xin Zhang | Columbia University |
| Mouse: αCre | gift from Dr. Xin Zhang | Columbia University |
| Mouse: CyclinD2flox | gift from Dr. Elizabeth Ross | Weill Cornell |
| Primary Antibodies | ||
| mouse anti-Brn3a | EMD Millipore | RRID:AB_94166 |
| rat anti-CyclinD2 | Santa Cruz | RRID:AB_627350 |
| rat anti-Ki67 | Thermo Fisher | RRID:AB_2865120 |
| mouse anti-BrdU | Thermo Fisher | RRID:AB_2536432 |
| rabbit anti-phosphoHistone3 | Thermo Fisher | RRID:AB_10984484 |
| goat anti-Msx1 | R&D systems | RRID:AB_2148804 |
| mouse anti-Cav1.3 | GeneTex | RRID:AB_10731346 |
| mouse anti-Cre recombinase | Millipore-Sigma | RRID:AB_2085748 |
| mouse-anti AP2α | DSHB | RRID:AB_528084 |
| goat-anti Prox1 | R&D systems | RRID:AB_2170716 |
| rabbit anti-Otx2 | Thermo Fisher | RRID:AB_2792910 |
| rabbit anti-Zic2 | gift from Dr. Steve Brown | University of Vermont |
| mouse anti-Islet1/2 | gift from Susan Morton | Columbia University |
| guinea pig anti-RFP | gift from Susan Morton | Columbia University |
| Secondary Antibodies | ||
| donkey anti-rat AlexaFluor594 | Jackson Immunoresearch | RRID:AB_2340689 |
| donkey anti-mouse AlexaFluor647 | Jackson Immunoresearch | Cat. No.: 703–605-155 |
| donkey anti-mouse AlexaFluor405 | Jackson Immunoresearch | RRID:AB_2340840 |
| donkey anti-mouse AlexaFluor488 | Jackson Immunoresearch | RRID:AB_2340846 |
| donkey anti-rabbit AlexaFluor488 | Jackson Immunoresearch | RRID:AB_2313584 |
| donkey anti-rabbit AlexaFluor647 | Jackson Immunoresearch | RRID:AB_2492288 |
| donkey anti-guinea pig AlexaFluor594 | Jackson Immunoresearch | RRID:AB_2340474 |
| donkey anti-goat AlexaFluor647 | Jackson Immunoresearch | RRID:AB_2340437 |
| donkey anti-goat AlexaFluor488 | Jackson Immunoresearch | RRID:AB_2336933 |
| Chemicals | ||
| Cholera Toxin Subunit B, Alexa Fluor 488 Conjugate | Thermo Fisher | Cat. No.: C22841 |
| Cholera Toxin Subunit B, Alexa Fluor 594 Conjugate | Thermo Fisher | Cat. No.: C22842 |
| Cholera Toxin Subunit B, Alexa Fluor 647 Conjugate | Thermo Fisher | Cat. No.: C34778 |
| (±)-Bay K8644 | Sigma-Aldrich | Cat. No.: B112 |
| BrdU (5-Bromo-2´-Deoxyuridine) | Fisher Scientific | Cat. No.: B23151 |
| EdU (5-ethynyl-2’-deoxyuridine) | Fisher Scientific | Cat. No.: A10044 |
| (Z)-4-Hydroxytamoxifen | Tocris Bioscience | Cat. No.: 34–125-0 |
| Commercial assays/ Kits | ||
| Click-iT Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor647 dye | Thermo Fisher | Cat. No.: C10640 |
| RNAscope Fluorescent Multiplex Reagent Kit | ACD | Cat. No.: 320850 |
| Papain Dissociation System | Worthington | Cat. No.: LK003150 |
| DeadEnd Fluorometric TUNEL System | Promega | Cat. No.: G3250 |
| RNA scope probes | ||
| RNAscope Probe- Mm-Fos-C3 | ACD | Cat. No.: 316921-C3 |
| RNAscope Probe- Mm-Jun-C2 | ACD | Cat. No.: 453561-C2 |
| RNAscope Probe- Mm-Igfbp5-C1 | ACD | Cat No.: 425731 |
| Software and Algorithms | ||
| ImageJ (Fiji) | N/A | https://imagej.net/Fiji |
| Prism 9.0 | Graphpad software | https://www.graphpad.com/ |
| R for statistical computing | N/A | https://cran.r-project.org/ |
| MetaMorph | N/A | |
| Deposited data | ||
| GEO | GSE209764 | Reviewer token: yzgjqcuslhejdyv |
Method details
Animals
Mice were maintained under standard conditions approved by the Institutional Animal Care and Use Committee (IACUC) of Columbia University. Animals were housed in a specific pathogen–free animal house under a 12/12-hour light/dark cycle and were provided food and water. Msx1-CreERT2 mice in C57Bl/6J background and αCre mice on a mixed genetic background were a gift from Dr. Xin Zhang, Columbia University. αCre mice were backcrossed for several generations with wild type C57Bl/6J mice. CyclinD2flox mice on a C57Bl/6J background were a gift from Dr. Elizabeth Ross, Weill Cornell. tdTomato Ai14, Tyrc2j, and Swiss/ICR albino mice were purchased from Jackson Laboratories. Tyrc2j mice were bred with wild type C57Bl/6J mice to obtain pigmented TyrWT/c2j controls. Animals of both sexes were used in our experiments.
Anesthesia
Ketamine and Xylazine were diluted in saline, and administered at 100mg/kg and 10mg/kg, respectively. Toe pinch was used to confirm full anesthesia.
Single cell dissociation and sorting
We could not isolate CMZ cells from the inducible Msx1 CreERT2; tdTomato line because Msx1 expression begins at E13.5 and thus very few cells are labeled at this early time point. Instead, we utilized αCre; tdTomato mice that allowed for enrichment of CMZ cells, as upon activation of the retina-specific regulatory element of Pax6 α-enhancer, the retinal periphery including the CMZ is fluorescently labeled (Marquardt et al., 2001, Fan et al., 2022). E13.5 retinae with attached RPE from 4 pigmented and 5 albino αCre; tdTomato+ littermate embryos each were harvested in ice cold Dulbecco’s modified Eagle’s medium (DMEM) after removal of the lens. Dissociation was performed in activated papain solution including Deoxyribonuclease I, using a papain dissociation system (Worthington Biochemical) for 10 min at 37°C and stopped using DMEM + 10% fetal bovine serum (FBS). Cells were gently triturated using a 22G needle, centrifuged at 300g for 5min, and washed with DMEM + 10% FBS. After passing through a 70μm filter and washing again in DMEM + 10% FBS, the cell suspension was stained with DAPI (0.2μg/ml) to label dead cells. Flow cytometry was performed at the core facility of Zuckerman Institute, Columbia University. Single cells were collected with an Astrios cell sorter and gated for live cells and TdTomato+ cells using FlowJo software. Approximately 25,000 pigmented and 15,000 albino cells were collected into 1.5-ml tubes precoated with FBS and DMEM + 10% FBS for submission for single-cell sequencing.
scRNA-Sequencing
Single-cell sequencing was performed at the single-cell sequencing core in Columbia Genome Center. Flow-sorted single cells were loaded into chromium microfluidic chips with v3 chemistry and barcoded with a 10× chromium controller (10× Genomics). RNA from the barcoded cells was reverse-transcribed, and sequencing libraries were constructed with a Chromium Single Cell v3 reagent kit (10× Genomics). Sequencing was performed on NovaSeq 6000 (Illumina). We sequenced 6,089 pigmented and 3,676 albino cells at a mean depth of 3,200 genes per cell.
Raw reads mapped to the mm10 reference genome by 10× Genomics Cell Ranger pipeline (v2.1.1) using default parameters were used for all downstream analyses using Seurat v3 (Stuart et al., 2019), Velocyto and scVelo. Briefly, the dataset was filtered to contain cells with at least 200 expressed genes and genes with expression in more than three cells. Cells were also filtered for mitochondrial gene expression (<30%). The dataset was log-normalized and scaled. Unsupervised clustering was performed initially, followed by manual annotation of Seurat clusters. An integration analysis was performed to compare and analyze the control and mutant gene expression matrices, which included normalization for cell numbers. The unsupervised clustering with a resolution parameter of 0.5 for both control and mutant cells was represented on a UMAP space, and cluster identity was assigned. Expression of various known genes was used to determine cluster identities. Doublets identified based on biological incompatibility were manually excluded from the dataset. Extra-retinal contaminant cells were manually removed from the dataset. The cell cycle was assessed using the vignette from Seurat. For RNA velocity analysis, loom files were constructed by extracting the reads from the 10× single-cell dataset. After gene filtering, spliced and unspliced counts were normalized based on total counts per cell. Velocity plots are represented as the transcriptomic integration of control and mutant datasets to represent similarities and differences in their velocities, respectively. Transcriptional dynamics were determined by ratio of spliced to unspliced counts fitted to a generalized linear regression model (Bergen et al., 2020, La Manno et al., 2018). Single cell data will be made available in GEO database and the interactive single cell portal upon acceptance of manuscript.
Intraperitoneal injections into pregnant dams
4-Hydroxytamoxifen (4-OHT) was dissolved in corn oil (Millipore-Sigma) at final concentration 5mg/ml and was administered intraperitoneally at 25μg/g body weight. BrdU was dissolved in 10mM Tris-HCl pH7.6 (Millipore-Sigma) at stock concentration 40mg/ml. Prior to injection, stock was further diluted in PBS and administered at 100μg/g body weight. EdU was dissolved in PBS at stock concentration 2.5mg/ml. Prior to injection, stock was further diluted in PBS and administered at 10μg/g body weight. BayK-8644 was dissolved in 1ml DMSO (Millipore-Sigma) at stock for concentration 5mg/ml. Prior to injection, stock was further diluted 1:10 in saline and administered at 2 mg/kg body weight. DMSO diluted 1:10 in saline was used as sham, at the same injection volume as BayK-8644.
Fate mapping
Animal breedings were Msx1Cre/+ERT2; tdTomatoflox/flox x Msx1Cre/+ERT2; tdTomatoflox/flox. Three doses of 4-OHT were administered intraperitoneally to pregnant dams at E13.5, E14.5, and E15.5. EdU was also administered at E15.5. Msx1Cre/CreERT2; tdTomatoflox/flox embryos were collected at E17.5 and processed for immunohistochemistry. Beyond this time point, these embryos are lethal.
Dual pulse birthdating
At the day of the experiment (E13.5 or E14.5), BrdU was injected intraperitoneally into the pregnant dam. 2 hours later EdU was administered in the same route. Embryos were collected 30min later and processed for immunohistochemistry. Cells in G1 phase were counted based on the expression of Ki67 and absence of BrdU and EdU labeling (Ki67+ only). Cells in S phase were counted based on Ki67 and EdU labeling with or without BrdU (Ki67+/EdU+/BrdU±). Cell cycle duration (hours) was calculated using the formula: Tc = 2*(Ki67+cells / BrdU only+ cells)
Anterograde labeling of retinogeniculate projections
Cholera toxin subunit B conjugated to Alexa Fluor 488, 594 or 647 was dissolved in PBS with 0.1% DMSO at stock concentration 1mg/ml. Mice were fully anesthetized and 1–2μl of CTB were injected intravitreally with a glass micropipette, until the whole eye was filled. Mouse brains were processed 48 hours later to obtain floating sections.
Tissue processing for cryosections
Pregnant dams were anesthetized and embryos were collected in ice cold PBS. E13.5-E15.5 embryo heads were fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) in PBS for 2 hours at 4°C. E17.5 embryos were transcardially perfused with cold 4% PFA and heads were post-fixed in 4% PFA overnight at 4°C. PFA was rinsed with PBS, and heads were incubated in 10% sucrose in PBS overnight at 4°C, followed by 20% sucrose in PBS overnight at 4°C, and 30% sucrose in PBS overnight at 4°C. Heads were cryomolded in O.C.T. Compound (Thermo Fisher Scientific) in dry ice and stored at −80°C. Coronal sections were obtained at 12μm thickness on Superfrost microscope slides (Thermo Fisher Scientific).
Tissue processing for brain floating sections
Adult mice were transcardially perfused with 10ml cold PBS followed by 30ml cold 4% PFA, and brains were dissected and post-fixed in 4% PFA overnight at 4°C. PFA was rinsed with PBS, and brains were molded in 3% agarose in PBS. Vibratome coronal sections were obtained at 150μm thickness in ice cold PBS and mounted on Superfrost microscope slides.
Immunohistochemistry and Imaging
Frozen sections were rinsed with PBS. Antigen retrieval was performed in an 85°C water bath for 15 minutes using pre-heated sodium citrate+0.05% Tween20, pH6. Slides remained incubated in the sodium citrate solution at room temperature for 20min, then were washed three times with PBS and incubated with blocking solution of 10% normal donkey serum (Jackson immunoResearch) in PBS with 0.2% Tween20, for 1 hour at room temperature inside a humid chamber protected from light. After blocking, primary antibody solution (1% NDS in PBS-T) was applied overnight at 4°C. Primary antibodies used were: rabbit anti-Zic2 (1:10,000), mouse anti-Islet1/2 (1:100), guinea pig anti-RFP (1:30,000), mouse anti-Brn3a (1:50), rat anti-CyclinD2 (1:50), rat anti-Ki67 (1:100), mouse anti-BrdU (1:200), rabbit anti-phosphoHistone3 (1:300), goat anti-Msx1 (1:100), mouse anti-Cav1.3 (1:100), mouse anti-Cre recombinase (1:100), mouse anti-AP2α (1:100), goat anti-Prox1 (1:200), rabbit anti-Otx2 (1:100). Primary antibody solution was washed three times with PBS-T and secondary antibody solution (1:500 in 1% NDS in PBS-T) was applied for 2 hours at room temperature, protected from light. Secondary antibodies (1:500) used were: donkey anti-rat AlexaFluor594, donkey anti-mouse AlexaFluor647, donkey anti-mouse AlexaFluor405, donkey anti-mouse AlexaFluor488, donkey anti-rabbit AlexaFluor488, donkey anti-rabbit AlexaFluor647, donkey anti-guinea pig AlexaFluor594, donkey anti-goat AlexaFluor647, donkey anti-goat AlexaFluor488. Secondary antibodies were washed three times with PBS. For EdU visualization, Click-iT reaction was performed after secondary antibody incubation, for 30min at room temperature according to kit instructions. Slides were incubated with DAPI (Fisher Scientific) 1:1,000 in PBS for 10min at room temperature, and mounted using Fluoro-Gel Medium (Electron Microscopy Sciences). Fluorescent imaging was performed on a W1-Yokogawa spinning disk confocal microscope. Images for Z-stacks were obtained at 0.5μm step size.
RNA scope
Sections were processed strictly according the instructions of the RNAscope Fluorescent Multiplex kit (ACDbio). No modifications were made. The RNAscope probes (ACD bio) used were: Mm-Fos-C3, Mm-Jun-C2, Mm-Igfbp5-C1. Fluorescent imaging was performed on a W1-Yokogawa spinning disk confocal microscope, at a 100x magnification using silicone oil. Images for Z-stacks were obtained at 0.15μm step size. Analysis was performed by averaging the number of RNA puncta per nucleus.
TUNEL assay
Sections were washed with PBS and permeabilized with 0.1% Triton for 2 hours at room temperature, then washed three times with PBS. Before permeabilization, positive control slides were treated with DNase I buffer for 5 minutes at room temperature. Equilibration buffer was applied for 10 minutes at room temperature. The reaction mixture was prepared according the instructions of the DeadEnd Fluorometric TUNEL System (Promega), then applied to the sections for 1 hour at 37°C protected from light. To terminate the reaction, 2X SCC solution was prepared according to instructions of the DeadEnd Fluorometric TUNEL System and applied to the sections for 15 minutes at room temperature. Sections were washed three times with PBS and mounted.
Behavioral paradigms
For the visual cliff task, the apparatus consisted of a 60 × 30 × 30cm open-top platform with a clear bottom. The “shallow side” laid on a flat surface and displayed directly a 4.45 × 4.45 cm black and white grid. The “deep side” of the platform extended outside the flat surface and displayed the same grid at 2.5ft depth. Each mouse was placed on a 5 × 5 × 5 cm stage in the center of the platform and, behaving freely, could step onto either side. A camera was placed above the apparatus. Mice performed 10 trials within 3 days. Every trial lasted until the mouse stepped down from the stage and onto the platform, or for 5min in trials when the mice did not step down. Trials were scored as “depth perceived” when mice stepped on the “shallow side” of the platform, or as “depth not perceived” when the mice either stepped on the “deep side” or did not step down after 5min (Lim et al., 2016). Three albino mice did not step down in all 10 trials and were excluded from our analysis. The orientation of the platform was changed randomly between trials, to exclude behavioral influences by environmental cues. Mice used were littermates and 7–8 weeks of age. For measurements of contrast sensitivity and visual acuity, we used the OptoDrum (Stria.Tech). Awake mice were placed on the platform inside the arena, and the OptoDrum will presented rotating stripe patterns to the animal. Stimulus rotation was set at 12°/sec. For visual acuity measurements, stimulus contrast was set at 99.72% and its spatial frequency drifted across trials from 0.061 cycles/° to 0.500 cycles/°. For contrast sensitivity measurements, stimulus spatial frequency was set at 0.061cycles/° and its contrast across trials dropped from 99.72% to 0.84%. Mouse behavior was monitored by a camera.
Quantification and Statistical analysis
Cell number quantification
For ages E13.5-E15.5 the CMZ in each section was defined as the region extending 150μm from the distal-most tip of the retina. From E17.5 until P1 the CMZ was clearly recognizable based on its morphology. This area was chosen based on the average extend of CyclinD2 expression in the pigmented wild-type eye. For quantifications in the ventrotemporal retina, we focused on the segment where Zic2+ RGCs reside, extending 150μm from the end of the CMZ, as this is the maximum distance in which we detected CMZ-derived (tdTomato+) cells with our fate mapping experiment.
Cells were counted manually using the cell counter plugin in Fiji software. Individual cells were distinguished by DAPI staining. In the entire series of sections, the first one that included the optic nerve head opening was assigned as “0μm distance from the optic nerve head”. Because sections were 12μm thick, every section along the rostro-caudal axis was assigned as “n+12μm distance from the optic nerve head”, with n being the distance of the previous section. The counts obtained from all sections of the temporal retina were summed. For E13.5 and E14.5 eyes, 6 serial sections were analyzed, starting from the one at 0μm distance from the optic nerve head. For E15.5 eyes, 9 serial sections were analyzed, starting from the one at 0μm distance from the optic nerve head. For E17.5 eyes, 32 serial sections were analyzed, starting from the one at 0μm distance from the optic nerve head. Left and right eyes were aligned based on distance from the optic nerve head and their counts were averaged.
Analysis of ipsilateral input to the dLGN
Quantification of eye-specific axon terminals was performed on 10x fluorescent images from three consecutive coronal 100μm sections through the region of the dLGN containing the greatest extent of the ipsilateral projection, as described previously (Rebsam et al., 2012). The extent of eye-specific projections within the dLGN along the dorsoventral (DV) and mediolateral (ML) axes of the dLGN was measured by tracing a line along each axis of the dLGN. The extent of the ipsilateral projection was measured by tracing a line between the two points delineating the maximal extent of the ipsilateral signal along the chosen axis. The length of the ipsilateral projection was divided by the length of the dLGN for each axis. To calculate the area of the dLGN receiving ipsilateral input we traced the boundary of the dLGN was outlined, excluding the intrageniculate leaflet, the ventral lateral geniculate nucleus, and the optic tract, as well as the boundary of the central ipsilateral patch, and identified the pixels included in each traced region. The pixels included in the traced ipsilateral region were divided by the pixels included in the traced dLGN region. We conducted an additional eye-specific segregation analysis, using MetaMorph software (Molecular Devices). We used variable thresholds for the contralateral projection and a fixed threshold for the ipsilateral projection. The boundary of the dLGN was outlined, as described above. The intensity threshold for the ipsilateral projection was chosen when the signal-to-background ratio was at least 1.2. The overlap between ipsilateral and contralateral projections (pixel overlap) was measured at every 10th threshold value for the contralateral image to obtain the function of overlap between ipsilateral and contralateral projections. The proportion of dLGN occupied exclusively by ipsilateral axons was measured as a ratio of ipsilateral pixels to the total number of pixels in the dLGN region.
Statistical analysis
Statistical analyses were performed using the Graph Pad Prism9 software. Results are presented as mean ± SEM. In all graphs, points correspond to individual mice. Two-sided unpaired t test, and two-way ANOVA (with Sidak test applied for multiple comparisons) were used with statistical significance set at p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Supplementary Material
HIGHLIGHTS.
The mouse CMZ is functionally important for the development of binocular vision.
In albinism, CyclinD2 downregulation in the CMZ prolongs G1/S phase transition.
CyclinD2 deficiency compromises ipsilateral RGC neurogenesis and depth perception.
Modulation of calcium channels in the albino CMZ restores binocular vision.
ACKNOWLEDGEMENTS
We thank Ira Schieren in the Flow Cytometry Platform, Susan Morton in the Antibody Platform at the Zuckerman Institute, and the Sulzberger Genome Center at Columbia University. Imaging was performed with support from the Zuckerman Institute’s Cellular Imaging Platform, and genomics with the Genomics and High Throughput Screening Shared Resource. We thank Xin Zhang (Dept. of Ophthalmology, Columbia University) for providing the Msx1 Cre and αCre mice, Stephen Brown (University of Vermont) for providing the Zic2 antibody, Sania Khalid for technical help, and Jane Dodd and Xin Zhang for their input. This work was funded by NIH R01 EY015290, R01 EY12736, António Champalimaud Vision Award, Simons Foundation Senior Fellow Award, and Vision of Children (CAM), Fight for Sight and a Georgakopoulos Family Fellowship (NS), BrightFocus Foundation Grant G2021007S (RB), NIH R01 EY032062, R01 EY032507, and Precision Medicine Initiative at Columbia University (SWMJ), R01 NS105477 (MER), Vision Core Grant P30 EY019007 (M. Goldberg), NIH/NCI Cancer Center Support Grant P30CA013696 and National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1TR001873 (Sulzberger Genome Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
Authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse and equitable conduct of research.
REFERENCES
- AMBROSIO AL, BOYLE JA, ARADI AE, CHRISTIAN KA & DI PIETRO SM 2016. TPC2 controls pigmentation by regulating melanosome pH and size. Proc Natl Acad Sci U S A, 113, 5622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAKIRI L, LALLEMAND D, BOSSY-WETZEL E & YANIV M 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J, 19, 2056–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALASUBRAMANIAN R, MIN X, QUINN PMJ, LO GIUDICE Q, TAO C, POLANCO K, MAKRIDES N, PEREGRIN J, BOUAZIZ M, MAO Y, WANG Q, DA COSTA BL, BUENAVENTURA D, WANG F, MA L, TSANG SH, FABRE PJ & ZHANG X 2021. Phase transition specified by a binary code patterns the vertebrate eye cup. Science Advances, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELANGER MC, ROBERT B & CAYOUETTE M 2017. Msx1-Positive Progenitors in the Retinal Ciliary Margin Give Rise to Both Neural and Non-neural Progenies in Mammals. Dev Cell, 40, 137–150. [DOI] [PubMed] [Google Scholar]
- BELLONO NW & OANCEA EV 2014. Ion transport in pigmentation. Arch Biochem Biophys, 563, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERGEN V, LANGE M, PEIDLI S, WOLF FA & THEIS FJ 2020. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol, 38, 1408–1414. [DOI] [PubMed] [Google Scholar]
- BHANSALI P, RAYPORT I, REBSAM A & MASON C 2014. Delayed neurogenesis leads to altered specification of ventrotemporal retinal ganglion cells in albino mice. Neural Dev, 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOONE HC, SAMONDS JM, CROUSE EC, BARR C, PRIEBE NJ & MCGEE AW 2021. Natural binocular depth discrimination behavior in mice explained by visual cortical activity. Curr Biol, 31, 2191–2198 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN JR, NIGH E, LEE RJ, YE H, THOMPSON MA, SAUDOU F, PESTELL RG & GREENBERG ME 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol, 18, 5609–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSH WD & SIMON JD 2007. Quantification of Ca(2+) binding to melanin supports the hypothesis that melanosomes serve a functional role in regulating calcium homeostasis. Pigment Cell Res, 20, 134–9. [DOI] [PubMed] [Google Scholar]
- CAVALIERI D, ANGELOVA A, ISLAH A, LOPEZ C, BOCCHIO M, BOLLMANN Y, BAUDE A & COSSART R 2021. CA1 pyramidal cell diversity is rooted in the time of neurogenesis. Elife, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAYOUETTE M, WHITMORE AV, JEFFERY G & RAFF M 2001. Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J Neurosci, 21, 5643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CEPKO CL, AUSTIN CP, YANG X, ALEXIADES M & EZZEDDINE D 1996. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A, 93, 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK BS, STEIN-O’BRIEN GL, SHIAU F, CANNON GH, DAVIS-MARCISAK E, SHERMAN T, SANTIAGO CP, HOANG TV, RAJAII F, JAMES-ESPOSITO RE, GRONOSTAJSKI RM, FERTIG EJ, GOFF LA & BLACKSHAW S 2019. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron, 102, 1111–1126 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRUZALEGUI FH, HARDINGHAM GE & BADING H 1999. c-Jun functions as a calcium-regulated transcriptional activator in the absence of JNK/SAPK1 activation. EMBO J, 18, 1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUMMING BG & DEANGELIS GC 2001. The physiology of stereopsis. Annu Rev Neurosci, 24, 203–38. [DOI] [PubMed] [Google Scholar]
- DRAGER UC 1985. Calcium binding in pigmented and albino eyes. Proc Natl Acad Sci U S A, 82, 6716–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN Y, CHEN W, WEI R, QIANG W, PEARSON JD, YU T, BREMNER R & CHEN D 2022. Mapping transgene insertion sites reveals the alpha-Cre transgene expression in both developing retina and olfactory neurons. Commun Biol, 5, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERNANDEZ-NOGALES M, LOPEZ-CASCALES MT, MURCIA-BELMONTE V, ESCALANTE A, FERNANDEZ-ALBERT J, MUNOZ-VIANA R, BARCO A & HERRERA E 2022. Multiomic Analysis of Neurons with Divergent Projection Patterns Identifies Novel Regulators of Axon Pathfinding. Adv Sci (Weinh), e2200615. [DOI] [PMC free article] [PubMed]
- FERNANDEZ-NOGALES M, MURCIA-BELMONTE V, CHEN HY & HERRERA E 2019. The peripheral eye: A neurogenic area with potential to treat retinal pathologies? Prog Retin Eye Res, 68, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISCHER AJ & REH TA 2000. Identification of a proliferating marginal zone of retinal progenitors in postnatal chickens. Dev Biol, 220, 197–210. [DOI] [PubMed] [Google Scholar]
- FOX MW 1965. The visual cliff test for the study of visual depth perception in the mouse. Anim Behav, 13, 232–3. [DOI] [PubMed] [Google Scholar]
- FRIES M, BROWN TW, JOLICOEUR C, BOUDREAU-PINSONNEAULT C, JAVED A, ABRAM P & CAYOUETTE M 2022. Pou3f1 orchestrates a gene regulatory network controlling contralateral retinogeniculate projections. bioRxiv, 2022.06.28.497982. [DOI] [PubMed]
- GLICKSTEIN SB, MONAGHAN JA, KOELLER HB, JONES TK & ROSS ME 2009. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci, 29, 9614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERRERA E, BROWN L, ARUGA J, RACHEL RA, DOLEN G, MIKOSHIBA K, BROWN S & MASON CA 2003. Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell, 114, 545–57. [DOI] [PubMed] [Google Scholar]
- HERRERA E, ERSKINE L & MORENILLA-PALAO C 2019. Guidance of retinal axons in mammals. Semin Cell Dev Biol, 85, 48–59. [DOI] [PubMed] [Google Scholar]
- HOWARTH M, WALMSLEY L & BROWN TM 2014. Binocular integration in the mouse lateral geniculate nuclei. Curr Biol, 24, 1241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWAI-TAKEKOSHI L, BALASUBRAMANIAN R, SITKO A, KHAN R, WEINREB S, ROBINSON K & MASON C 2018. Activation of Wnt signaling reduces ipsilaterally projecting retinal ganglion cells in pigmented retina. Development, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWAI-TAKEKOSHI L, RAMOS A, SCHALER A, WEINREB S, BLAZESKI R & MASON C 2016. Retinal pigment epithelial integrity is compromised in the developing albino mouse retina. J Comp Neurol, 524, 3696–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON KP, FITZPATRICK MJ, ZHAO L, WANG B, MCCRACKEN S, WILLIAMS PR & KERSCHENSTEINER D 2021. Cell-type-specific binocular vision guides predation in mice. Neuron, 109, 1527–1539 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUBOTA R, HOKOC JN, MOSHIRI A, MCGUIRE C & REH TA 2002. A comparative study of neurogenesis in the retinal ciliary marginal zone of homeothermic vertebrates. Brain Res Dev Brain Res, 134, 31–41. [DOI] [PubMed] [Google Scholar]
- KUWAJIMA T, SOARES CA, SITKO AA, LEFEBVRE V & MASON C 2017. SoxC Transcription Factors Promote Contralateral Retinal Ganglion Cell Differentiation and Axon Guidance in the Mouse Visual System. Neuron, 93, 1110–1125 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LA MANNO G, SOLDATOV R, ZEISEL A, BRAUN E, HOCHGERNER H, PETUKHOV V, LIDSCHREIBER K, KASTRITI ME, LONNERBERG P, FURLAN A, FAN J, BORM LE, LIU Z, VAN BRUGGEN D, GUO J, HE X, BARKER R, SUNDSTROM E, CASTELO-BRANCO G, CRAMER P, ADAMEYKO I, LINNARSSON S & KHARCHENKO PV 2018. RNA velocity of single cells. Nature, 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LECLERC C, DUPRAT AM & MOREAU M 1999. Noggin upregulates Fos expression by a calcium-mediated pathway in amphibian embryos. Dev Growth Differ, 41, 227–38. [DOI] [PubMed] [Google Scholar]
- LIM JH, STAFFORD BK, NGUYEN PL, LIEN BV, WANG C, ZUKOR K, HE Z & HUBERMAN AD 2016. Neural activity promotes long-distance, target-specific regeneration of adult retinal axons. Nat Neurosci, 19, 1073–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LO GIUDICE Q, LELEU M, LA MANNO G & FABRE PJ 2019. Single-cell transcriptional logic of cell-fate specification and axon guidance in early born retinal neurons. Development [DOI] [PubMed]
- MARCUCCI F, MURCIA-BELMONTE V, WANG Q, COCA Y, FERREIRO-GALVE S, KUWAJIMA T, KHALID S, ROSS ME, MASON C & HERRERA E 2016. The Ciliary Margin Zone of the Mammalian Retina Generates Retinal Ganglion Cells. Cell Rep, 17, 3153–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCUCCI F, SOARES CA & MASON C 2018. Distinct timing of neurogenesis of ipsilateral and contralateral retinal ganglion cells. J Comp Neurol [DOI] [PMC free article] [PubMed]
- MARCUCCI F, SOARES CA & MASON C 2019. Distinct timing of neurogenesis of ipsilateral and contralateral retinal ganglion cells. J Comp Neurol, 527, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARQUARDT T, ASHERY-PADAN R, ANDREJEWSKI N, SCARDIGLI R, GUILLEMOT F & GRUSS P 2001. Pax6 is required for the multipotent state of retinal progenitor cells. Cell, 105, 43–55. [DOI] [PubMed] [Google Scholar]
- MORENILLA-PALAO C, LOPEZ-CASCALES MT, LOPEZ-ATALAYA JP, BAEZA D, CALVO-DIAZ L, BARCO A & HERRERA E 2020. A Zic2-regulated switch in a noncanonical Wnt/betacatenin pathway is essential for the formation of bilateral circuits. Sci Adv, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURCIA-BELMONTE V & ERSKINE L 2019. Wiring the Binocular Visual Pathways. Int J Mol Sci, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSTERHOUT JA, EL-DANAF RN, NGUYEN PL & HUBERMAN AD 2014. Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Rep, 8, 1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEARSON R, CATSICAS M, BECKER D & MOBBS P 2002. Purinergic and muscarinic modulation of the cell cycle and calcium signaling in the chick retinal ventricular zone. J Neurosci, 22, 7569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEARSON RA, CATSICAS M, BECKER DL, BAYLEY P, LUNEBORG NL & MOBBS P 2004. Ca(2+) signalling and gap junction coupling within and between pigment epithelium and neural retina in the developing chick. Eur J Neurosci, 19, 2435–45. [DOI] [PubMed] [Google Scholar]
- PETROS TJ, REBSAM A & MASON CA 2008. Retinal axon growth at the optic chiasm: to cross or not to cross. Annu Rev Neurosci, 31, 295–315. [DOI] [PubMed] [Google Scholar]
- PILAZ LJ, LENNOX AL, ROUANET JP & SILVER DL 2016a. Dynamic mRNA Transport and Local Translation in Radial Glial Progenitors of the Developing Brain. Curr Biol, 26, 3383–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PILAZ LJ, MCMAHON JJ, MILLER EE, LENNOX AL, SUZUKI A, SALMON E & SILVER DL 2016b. Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron, 89, 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RACHEL RA, DOLEN G, HAYES NL, LU A, ERSKINE L, NOWAKOWSKI RS & MASON CA 2002. Spatiotemporal features of early neuronogenesis differ in wild-type and albino mouse retina. J Neurosci, 22, 4249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REBSAM A, BHANSALI P & MASON CA 2012. Eye-specific projections of retinogeniculate axons are altered in albino mice. J Neurosci, 32, 4821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSSI AM, FERNANDES VM & DESPLAN C 2017. Timing temporal transitions during brain development. Curr Opin Neurobiol, 42, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALCEDA R & SANCHEZ-CHAVEZ G 2000. Calcium uptake, release and ryanodine binding in melanosomes from retinal pigment epithelium. Cell Calcium, 27, 223–9. [DOI] [PubMed] [Google Scholar]
- SALPETER SJ, KLOCHENDLER A, WEINBERG-COREM N, PORAT S, GRANOT Z, SHAPIRO AM, MAGNUSON MA, EDEN A, GRIMSBY J, GLASER B & DOR Y 2011. Glucose regulates cyclin D2 expression in quiescent and replicating pancreatic beta-cells through glycolysis and calcium channels. Endocrinology, 152, 2589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMAK G, NARAYANAN D, JAGGAR JH & RAO R 2011. CaV1.3 channels and intracellular calcium mediate osmotic stress-induced N-terminal c-Jun kinase activation and disruption of tight junctions in Caco-2 CELL MONOLAYERS. J Biol Chem, 286, 30232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMONDS JM, CHOI V & PRIEBE NJ 2019. Mice Discriminate Stereoscopic Surfaces Without Fixating in Depth. J Neurosci, 39, 8024–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHOLL B, BURGE J & PRIEBE NJ 2013. Binocular integration and disparity selectivity in mouse primary visual cortex. J Neurophysiol, 109, 3013–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERVILI E, TRUS M, MAAYAN D & ATLAS D 2018. beta-Subunit of the voltage-gated Ca(2+) channel Cav1.2 drives signaling to the nucleus via H-Ras. Proc Natl Acad Sci U S A, 115, E8624–E8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEKHAR K, WHITNEY IE, BUTRUS S, PENG Y-R & SANES JR 2021. Diversification of multipotential postmitotic mouse retinal ganglion cell precursors into discrete types. bioRxiv, 2021.10.21.465277. [DOI] [PMC free article] [PubMed]
- STUART T, BUTLER A, HOFFMAN P, HAFEMEISTER C, PAPALEXI E, MAUCK WM 3RD, HAO Y, STOECKIUS M, SMIBERT P & SATIJA R 2019. Comprehensive Integration of Single-Cell Data. Cell, 177, 1888–1902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SU J, SABBAGH U, LIANG Y, OLEJNIKOVA L, DIXON KG, RUSSELL AL, CHEN J, PAN YA, TRIPLETT JW & FOX MA 2021. A cell-ECM mechanism for connecting the ipsilateral eye to the brain. Proc Natl Acad Sci U S A, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN L, RINGACH DL & TRACHTENBERG JT 2022. The Development of Receptive Field Tuning Properties in Mouse Binocular Primary Visual Cortex. J Neurosci, 42, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIBBER MS, WHITMORE AV & JEFFERY G 2006. Cell division and cleavage orientation in the developing retina are regulated by L-DOPA. J Comp Neurol, 496, 369–81. [DOI] [PubMed] [Google Scholar]
- TRIPODI M, STEPIEN AE & ARBER S 2011. Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature, 479, 61–6. [DOI] [PubMed] [Google Scholar]
- TSUNEKAWA Y, BRITTO JM, TAKAHASHI M, POLLEUX F, TAN SS & OSUMI N 2012. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J, 31, 1879–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSUNEKAWA Y, KIKKAWA T & OSUMI N 2014. Asymmetric inheritance of Cyclin D2 maintains proliferative neural stem/progenitor cells: a critical event in brain development and evolution. Dev Growth Differ, 56, 349–57. [DOI] [PubMed] [Google Scholar]
- TURCHI L, LOUBAT A, ROCHET N, ROSSI B & PONZIO G 2000. Evidence for a direct correlation between c-Jun NH2 terminal kinase 1 activation, cyclin D2 expression, and G(1)/S phase transition in the murine hybridoma 7TD1 cells. Exp Cell Res, 261, 220–8. [DOI] [PubMed] [Google Scholar]
- WALK RD & GIBSON EJ 1961. A comparative and analytical study of visual depth discrimination. Psychol. Monogram
- WAN Y, ALMEIDA AD, RULANDS S, CHALOUR N, MURESAN L, WU Y, SIMONS BD, HE J & HARRIS WA 2016. The ciliary marginal zone of the zebrafish retina: clonal and time-lapse analysis of a continuously growing tissue. Development, 143, 1099–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Q, MARCUCCI F, CERULLO I & MASON C 2016. Ipsilateral and Contralateral Retinal Ganglion Cells Express Distinct Genes during Decussation at the Optic Chiasm. eNeuro, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHEELER DG, GROTH RD, MA H, BARRETT CF, OWEN SF, SAFA P & TSIEN RW 2012. Ca(V)1 and Ca(V)2 channels engage distinct modes of Ca(2+) signaling to control CREB-dependent gene expression. Cell, 149, 1112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU B, TANG X, JIN M, ZHANG H, DU L, YU S & HE J 2020. Unifying developmental programs for embryonic and postembryonic neurogenesis in the zebrafish retina. Development, 147. [DOI] [PubMed] [Google Scholar]
- YANG Y, YU Z, GENG J, LIU M, LIU N, LI P, HONG W, YUE S, JIANG H, GE H, QIAN F, XIONG W, WANG P, SONG S, LI X, FAN Y & LIU X 2022. Cytosolic peptides encoding CaV1 C-termini downregulate the calcium channel activity-neuritogenesis coupling. Commun Biol, 5, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOUNG RW 1985. Cell differentiation in the retina of the mouse. Anat Rec, 212, 199–205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT/RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Organisms | ||
| Mouse: C57Bl/6J | Jackson Labs | RRID:IMSR_JAX:000664 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Labs | RRID:IMSR_JAX:007914 |
| Mouse: B6(Cg)-Tyrc−2J/J | Jackson Labs | RRID:IMSR_JAX:000058 |
| Mouse: J:ARC(S) | Jackson Labs | RRID:IMSR_JAX:034608 |
| Mouse: Msx1-CreERT2 | gift from Dr. Xin Zhang | Columbia University |
| Mouse: αCre | gift from Dr. Xin Zhang | Columbia University |
| Mouse: CyclinD2flox | gift from Dr. Elizabeth Ross | Weill Cornell |
| Primary Antibodies | ||
| mouse anti-Brn3a | EMD Millipore | RRID:AB_94166 |
| rat anti-CyclinD2 | Santa Cruz | RRID:AB_627350 |
| rat anti-Ki67 | Thermo Fisher | RRID:AB_2865120 |
| mouse anti-BrdU | Thermo Fisher | RRID:AB_2536432 |
| rabbit anti-phosphoHistone3 | Thermo Fisher | RRID:AB_10984484 |
| goat anti-Msx1 | R&D systems | RRID:AB_2148804 |
| mouse anti-Cav1.3 | GeneTex | RRID:AB_10731346 |
| mouse anti-Cre recombinase | Millipore-Sigma | RRID:AB_2085748 |
| mouse-anti AP2α | DSHB | RRID:AB_528084 |
| goat-anti Prox1 | R&D systems | RRID:AB_2170716 |
| rabbit anti-Otx2 | Thermo Fisher | RRID:AB_2792910 |
| rabbit anti-Zic2 | gift from Dr. Steve Brown | University of Vermont |
| mouse anti-Islet1/2 | gift from Susan Morton | Columbia University |
| guinea pig anti-RFP | gift from Susan Morton | Columbia University |
| Secondary Antibodies | ||
| donkey anti-rat AlexaFluor594 | Jackson Immunoresearch | RRID:AB_2340689 |
| donkey anti-mouse AlexaFluor647 | Jackson Immunoresearch | Cat. No.: 703–605-155 |
| donkey anti-mouse AlexaFluor405 | Jackson Immunoresearch | RRID:AB_2340840 |
| donkey anti-mouse AlexaFluor488 | Jackson Immunoresearch | RRID:AB_2340846 |
| donkey anti-rabbit AlexaFluor488 | Jackson Immunoresearch | RRID:AB_2313584 |
| donkey anti-rabbit AlexaFluor647 | Jackson Immunoresearch | RRID:AB_2492288 |
| donkey anti-guinea pig AlexaFluor594 | Jackson Immunoresearch | RRID:AB_2340474 |
| donkey anti-goat AlexaFluor647 | Jackson Immunoresearch | RRID:AB_2340437 |
| donkey anti-goat AlexaFluor488 | Jackson Immunoresearch | RRID:AB_2336933 |
| Chemicals | ||
| Cholera Toxin Subunit B, Alexa Fluor 488 Conjugate | Thermo Fisher | Cat. No.: C22841 |
| Cholera Toxin Subunit B, Alexa Fluor 594 Conjugate | Thermo Fisher | Cat. No.: C22842 |
| Cholera Toxin Subunit B, Alexa Fluor 647 Conjugate | Thermo Fisher | Cat. No.: C34778 |
| (±)-Bay K8644 | Sigma-Aldrich | Cat. No.: B112 |
| BrdU (5-Bromo-2´-Deoxyuridine) | Fisher Scientific | Cat. No.: B23151 |
| EdU (5-ethynyl-2’-deoxyuridine) | Fisher Scientific | Cat. No.: A10044 |
| (Z)-4-Hydroxytamoxifen | Tocris Bioscience | Cat. No.: 34–125-0 |
| Commercial assays/ Kits | ||
| Click-iT Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor647 dye | Thermo Fisher | Cat. No.: C10640 |
| RNAscope Fluorescent Multiplex Reagent Kit | ACD | Cat. No.: 320850 |
| Papain Dissociation System | Worthington | Cat. No.: LK003150 |
| DeadEnd Fluorometric TUNEL System | Promega | Cat. No.: G3250 |
| RNA scope probes | ||
| RNAscope Probe- Mm-Fos-C3 | ACD | Cat. No.: 316921-C3 |
| RNAscope Probe- Mm-Jun-C2 | ACD | Cat. No.: 453561-C2 |
| RNAscope Probe- Mm-Igfbp5-C1 | ACD | Cat No.: 425731 |
| Software and Algorithms | ||
| ImageJ (Fiji) | N/A | https://imagej.net/Fiji |
| Prism 9.0 | Graphpad software | https://www.graphpad.com/ |
| R for statistical computing | N/A | https://cran.r-project.org/ |
| MetaMorph | N/A | |
| Deposited data | ||
| GEO | GSE209764 | Reviewer token: yzgjqcuslhejdyv |


