Abstract
1. Providing an alternative to pyrethroids, organophosphates, and carbamates, the neonicotinoids are now the most widely used insecticides in the world. They are water soluble and relatively stable in soil and water which allows for run-off through surface waters and thus potentially impacting aquatic species and environments.
2. While the mammalian metabolism of neonicotinoids has been studied extensively, there is a lack of understanding of their metabolism in fish species. The current study constitutes the first report of the metabolism of imidacloprid (IMI) and acetamiprid (AC) in rainbow trout.
3. Formation of respective metabolites 5-hydroxy-imidacloprid and N-desmethyl-acetamiprid was conserved across orders of biological organization in both microsomal and liver slice assays.
4. Michaelis–Menten kinetics were determined for the microsomal conversion of IMI to 5-hydroxy-imidacloprid in rainbow trout (Km = 79.2 μM; Vmax = 0.75 pmole/min/mg) and rat (Km = 158.7 μM; Vmax = 38.4 pmole/min/mg). Kinetics for the microsomal demethylation of AC to N-desmethyl-acetamiprid were determined in the rat (Km = 70.9 μM; Vmax = 10 pmoles/min/mg). N-desmethyl-acetamiprid was found in detectable but below quantifiable levels across the range of test concentrations which precluded a calculation of kinetic rate constants in rainbow trout (RBT).
5. Ultimately, the formation of the metabolites 5-hydroxy-imidacloprid and N-desmethyl-acetamiprid was conserved across RBT and rat species.
Keywords: Metabolism, acetamiprid, imidacloprid, liver slice, microsomes, rainbow trout, rat, neonicotinoid
Introduction
Imidacloprid (IMI, a N-nitroguanidine) and acetamiprid (AC, a N-cyanoamidine) are insecticides belonging to the chemical class, neonicotinoids. These chemicals were first introduced in the early 1990s as alternatives to organophosphates, pyrethroids and carbamates which were losing effectiveness due to insect resistance. Originally developed as systemic insecticides, they feature higher selectivity factors such as toxicity for insects as compared to mammals based upon a stronger affinity for the insect versus the mammal nicotinic acetylcholine receptor (nAChR) (Liu & Casida, 1993; Tomizawa & Casida, 2003). Since their introduction, neonicotinoids have become the most heavily used insecticides in the world (Simon-Delso et al., 2015). Specifically, these chemicals are agonists of the nicotinic acetylcholine receptors of target insects that function as ligand-gated ion channels responsible for rapid neurotransmission (Tomizawa & Yamamoto, 1993).
Neonicotinoids as a group are relatively high in water solubility (2.95 and 0.61 g/L for AC and IMI), generally have long half-lives in soil and water, are stable to hydrolysis at pH below nine (UOH, 2019) and thus have the potential for transport to surface and ground water. Persistence varies among the classes of neonicotinoids, the nitroguanidine-substituted (e.g. IMI) with aerobic and aquatic degradation half-lives >6 months versus the the substantially less <30 days presented by cyano-substituted (e.g. AC) members. Median neonicotinoid surface water concentrations are on the order of ng/L with peak concentrations in the low ug/L levels (Sanchez-Bayo et al., 2016; Struger et al., 2017). Specifically, the Aquatic Risk Assessment released in December 2016 listed detection frequency (5–67%) and maximum residue (7.9 μg/L) for IMI as part of the United States Geological Survey (USGS) non-target monitoring data. Target monitoring data for IMI resulted in a detection frequency of 0–89% and maximum residue of 12.7 μg/L (USEPA, 2016). As a result, nontarget aquatic organisms are exposed to fluxuating concentrations of run-off pesticide resulting in a potential for adverse effects.
Considered as highly toxic, a wide range of acute toxicity values (LC50’s) for neonicotinoids to aquatic insects and invertebrates covering six orders of magnitude (<1 ug/L to >100,000 ug/L) has been reported (Morrissey et al., 2015; Raby et al., 2018). In contrast, fish neonicotinoid LC50’s for rainbow trout and zebrafish are in the range of 83–281 mg/L (Ding et al., 2004; Tomlin, 2009). While considered slightly or not acutely toxic to fish, chronic effects such as genotoxicity, oxidative stress and early life stage developmental toxicity caused by neonicotinoids have been observed for fish, specifically for IMI in the range of 0.1 to 15 mg/L (Crosby et al., 2015; Ge et al., 2015; Iturburu et al., 2017).
It is the negatively charged nitro or cyano group on the neonicotinoid insecticides that interact with a unique, positively charged amino acid residue present on insect, but not mammalian nAChRs (Tomizawa & Casida, 2003). Presumably, this is also the case for fish as well as vertebrates in general which may explain the reduced acute toxicity of neonicotinoids. However, environmental breakdown and metabolism products in the form of positively charged nitrogens show a high affinity to mammalian nAChR (Tomizawa, 2004). From studies conducted with other species, metabolism of IMI is generally thought to go through two major routes; oxidation to form 5-hydroxy and the olefin or nitroreduction to form the nitroso, guanidine, and urea of IMI. The olefin formed from the hydroxylation product has been shown to be 10 times more toxic to whitefly and aphids than the parent IMI (Dai et al., 2006; Nauen et al., 2001). In contrast, the guanidine and urea products formed via nitroreduction do not impart any insecticidal properties. On the other hand, the guanidine metabolite has been reported to exhibit higher levels of mammalian toxicity than parent IMI (Tomizawa & Casida, 2003; 2005). The desnitro-imidacloprid has been shown to be higher intoxicity to mice with a reported LD50 of 16–24 mg/kg as compared to parent IMI (LD50 = 35–49 mg/kg) (Lee Chao & Casida, 1997). Relatively little neonicotinoid metabolism information for fish is available. A greater understanding of metabolism in fish is needed to determine if bioactivation to more toxic metabolites acting through the nAChR occurs.
The current study was undertaken to identify and compare neonicotinoid metabolic pathways of in vitro microsomal systems in the rainbow trout and rat. Concentrations of IMI and AC necessary to achieve enzyme saturation were used to characterize formation rate and biotransformation capacity. Additional experiments with the trout liver slice model (Tapper et al., 2018) were utilized to compare in vitro to ex-vivo results. This research sought to characterize the metabolites formed to better understand where chemical biotransformation may lead to enhanced toxicity or potential detoxification. The effort included comparison of metabolism pathways across species to better understand where similarities and differences exist in biotransformation reactions.
Materials and methods
Chemicals
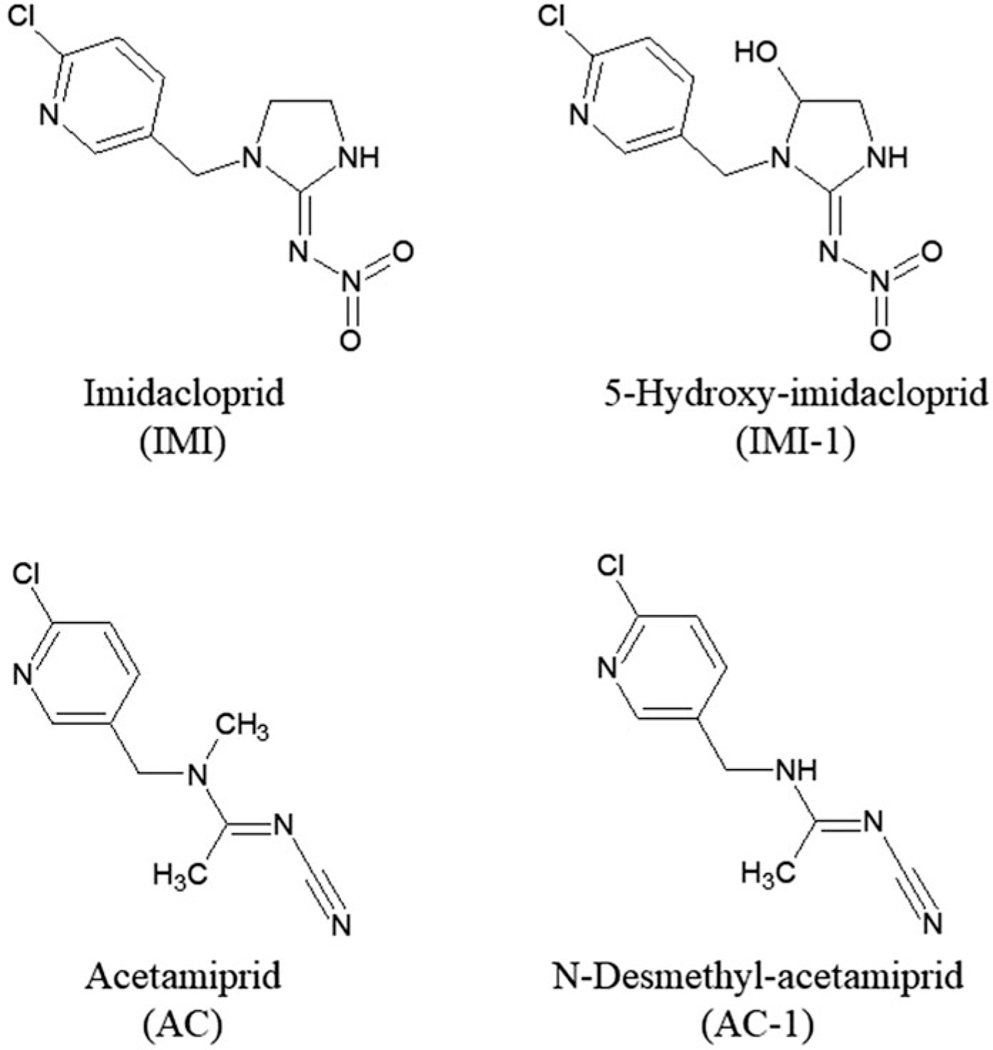
Parent chemicals and metabolites (Figure 1), along with the acronyms used to identify the chemicals, Chemical Abstract Services Registry Number (CASRN), source, and purity, are as follows: Acetamiprid, AC, 135410–20-7, Nippon Soda Company (Tokyo, Japan), 100%; Imidacloprid, IMI, 138261–41-3, Bayer Crop Science Division (Kansas City, MO, USA), 98.8%; Acetamiprid-N-desmethyl, AC-1, 190604–92-3, Nippon Soda Company, 99.7%; 5-Hydroxy-imidacloprid, IMI-1, 155802–61-2, AccuStandard (New Haven, CT, USA), 100%. All other chemicals used for biological and chemical analysis were purchased from Sigma Aldrich (Sigma Aldrich, St Louis, MO, USA) unless otherwise specified.
Figure 1.
Parent chemicals imidacloprid (IMI), acetamiprid (AC) and metabolite structures.
Fish
Immature rainbow trout (Oncorhynchus mykiss, RBT), Erwin/Arlee strain, used for production of liver slices and subcellular microsome fractions, were obtained from US Geological Survey, Upper Midwest Environmental Science Center (La Crosse, WI, USA). Fish were acclimated for at least 2 weeks before use in Lake Superior water (2-μm filtered, ultraviolet light treated, 11 °C, pH = 7.7, hardness = 45 mg/L) and kept on a constant photoperiod of 16-h light: 8-h dark. Fish body weights ranged from 196 to 976 g and liver weights from 1.9 to 9.1 g. Hepatic-somatic index ranged from 0.8% to 1.2%. Male and female trout were used for this study. Sex was determined by visual inspection of gonads. Previous investigations in this laboratory have observed no apparent differences in Phase I biotransformation for sexually immature rainbow trout regardless of size and gender (unpublished observations). Data for individual fish used in liver slice and microsome preparations are specified in Supplemental Table S1.
RBT liver slice exposures
Slice preparation
Precision-cut trout liver slices were prepared and exposed to test chemical as previously described for various lengths of time at 11 °C in 12-well tissue culture plate (Schmieder et al., 2000, 2004). Liver cores (8 mm) were prepared using a modified tissue coring press (Model MD2000; Alabama Research and Development, Munford, AL, USA). Cores were cut into 200 μm thick slices using a Krumdieck precision tissue slicer (Alabama Research and Development, Munford, AL, USA). Slices from a single fish were used in each test.
Test solution preparation
AC 3.67 M and IMI) 0.367 M stocks were prepared in dimethyl sulfoxide (DMSO). Serial dilutions in DMSO of these stocks were prepared for the slice viability tests. Stocks were diluted in exposure media consisting of phenol red-free L-15 media (Gibco, Life Technologies, Grand Island, NY, USA), 10% fetal bovine serum (FBS), and penicillin (100 U/mL)/streptomycin (100 μg/mL) to produce the test solutions.
Liver slice viability
Toxicity of AC and IMI was measured in tests separate from the metabolism assays. Samples for toxicity measurements were taken following 48 hours incubation at 11 °C in the same type of 12 well plates used for the metabolism tests. Each well contained one slice and 1.7 mL of exposure media/test chemical. The mitochondrial dehydrogenase assay (MTT) using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide as substrate was used to assess cytotoxicity in the liver slices (Tapper et al., 2018; Vistica et al., 1991). As an additional toxicity measurement, the amount of lactate dehydrogenase enzyme (LDH) leakage from liver slices into media was measured following the 48-h incubation and compared with total slice LDH activity in control slices at 48 h as previously described (Bergmeyer & Bernt, 1974; Schmieder et al., 2000, 2004). Toxicity in liver slices was assessed at 10, 100, 200, 500, 1000, and 2000 μM AC; and 1, 10, 20, 50, 100, 200 μM IMI.
Liver slice metabolism exposures
Media concentrations of parent chemical and metabolites were measured in three RBT liver slice tests (see Chemical and metabolite analyses section for details). In all tests, slices were incubated at 11 °C in covered 12 well plates containing 1.7 mL of exposure media on an orbital shaker (Tapper et al., 2018). In Test-1, a chemical concentration rangefinder, one or two slices per well were exposed to 1000 and 2000 μM AC or 100 and 200 μM IMI for 96 hours. Test-2 expanded upon the use of multiple slices per well to maximize metabolite yield and therefore two, three, four, or five slices per well, were exposed to 2000 μM AC or 200 μM IMI for 120 hours. Liver slices, four per well, were exposed to 2000 μM AC or 200 μM IMI for 96, or 120 hours during Test-3 to maximize yield of metabolites for the purpose of identification. DMSO solvent controls, 0.054% final concentration, were incubated under the same conditions and sampled at the same time points. There were at least two treatment replicates at each time point in each of the three tests.
Chemical test solutions were prepared and then dispensed into the appropriate wells. For example, 200 μM IMI was prepared by the addition of 5.4 μL of 367 mM IMI to 10 mL of exposure media. Then 1.7 mL of this test solution was added to the appropriate well. Test solutions in 12-well plates were equilibrated to incubation temperature before the addition of liver slices to wells. Plates were incubated at 11 °C on an orbital shaker. Incubation was terminated by removing 500 μL of exposure media from well, then adding it to a microfuge tube containing 200 μL ACN. These samples were processed for chemical evaluation (see Chemical and metabolite analyses section).
Subcelluar fractions preparation
RBT microsome preparation
Microsome fraction preparations were produced from pooled livers from five to nine immature female and male rainbow trout (Dady et al., 1991; Kolanczyk et al., 1999). Four different microsome preparations were used for this study. Fish were euthanized before excision of livers. Livers were minced in three volumes of 4 °C KCl 1.15% (3 mL/g liver tissue). Minced tissue was homogenized on ice in a chilled glass/teflon pestle. Homogenate was centrifuged at 10000×g for 20 min at 4 °C. The supernatant was removed and centrifuged at 105000g for 60 min at 4 °C. The resulting pellet was resuspended, at the original KCl volume, with 4 °C, pH 7.4, 0.1 M K2HPO4, 1 mM EDTA buffer then homogenized as previously described. Homogenate was centrifuged at 105000×g for 60 min at 4 °C. The pellet was resuspended in half the original KCl volume (0.5 mL/g liver tissue) with 4 °C, Tris-HCl, EDTA 1 mM, pH 7.4 buffer. Microsome preparations were quick froze and stored at −80 °C until used for chemical exposure. Protein concentration of microsome preparations were determined by the Bradford (1976) method.
Rat microsome source
Rat liver microsomes were purchased from Xenotech (Lenexa, KS, USA). Microsomes were prepared from livers of untreated male Sprague Dawley rats. Each lot or preparation was produced from a pool of 300 to 495 rat livers. Microsome preparations were stored at −80 °C until used for chemical exposure.
Microsome exposures
RBT kinetic exposures
Kinetic metabolism exposures of RBT microsome to IMI were performed utilizing the following procedure. Four different RBT microsomes preparations were exposed to 3.125, 6.25, 12.5, 25, 50, 100, 150, 200 μM IMI. Reaction mixtures composed of 100 mM K2HPO4 buffer pH 7.4, RBT microsomes 4 mg/mL, and NADPH 5 mM (Oriental Yeast Co., 3–6-10 Azusawa, Itabashi-ku, Tokyo 174–8505, Japan) were prepared in 2-mL capped microfuge tubes and acclimated to incubation temperature. To these tubes, the appropriated volume of IMI or carrier control DMSO were added to achieve desired IMI concentration. Two replicates were prepared for each treatment. Final reaction volume was 500 μL for all treatments. Capped tubes were placed on an orbital shaker and incubated for 120 min at 11 °C. Incubation was terminated with addition of ACN (200 μL), then samples were processed for chemical evaluation (see Chemical and metabolite analyses section).
Rat kinetic exposures
Rat microsome kinetic metabolism exposures were performed as done for the RBT microsome exposure with the following exceptions: the protein concentration was 2 mg/mL and incubation temperature was 37 °C. Three different rat microsome preparations were exposed to 3.125, 6.25, 12.5, 50, 100, 150, 200, 300, 350 μM IMI. These same rat microsome preparation were also exposed to 3.125, 6.25, 12.5, 50, 100, 150, 200, 300, 350 μM AC.
RBT metabolism exposures
Metabolism exposures of RBT microsomes to AC and IMI was performed as described for RBT microsome kinetic exposures with the following exceptions: microsomes were exposed to only 100 μM of AC or IMI, and samples were taken at multiple incubation lengths including 30, 60, 120, 180, 240 and 300 min.
Rat metabolism exposures
Metabolism exposures of rat microsomes to AC and IMI was performed as described for rat microsome kinetic exposures with the following exceptions: microsomes were exposed to only 100 μM of AC or IMI, and samples were taken at multiple incubation lengths including 30, 60, 120, 180 and 240 min.
Chemical and metabolite analyses
The ACN extraction efficiencies of the parent chemicals IMI and AC from exposure media were measured at a concentration range of 1–500 μM resulting in recoveries between 92–96% for both compounds. Media from exposure and control wells (500 μL) was analyzed for IMI, AC and for the emergence of distinct or reported metabolites. After exposures, chemical reactions were stopped with 200 μL of ACN, and mixed with 25 μL of saturated barium hydroxide and zinc sulfate (25% w/v), followed by mixing and centrifugation to precipitate proteins and debris as previously described by Tapper et al. (2018). Supernatants were diluted to 2.5% ACN with water and transferred to amber vials for qualitative and quantitative LC analyses with UV detection. Detected metabolites were recovered and concentrated for targeted liquid chromatography mass spectrometry (LC-MS) identification experiments by pooling unused supernatants and performing sample fractionation, gentle drying with N2 and reconstitution in appropriate LC solvent (see Supplemental Figure S1).
Chemical analyses and metabolite characterization were performed on an Ultimate 3000 Thermo ultra-high pressure liquid chromatography system (μHPLC; Thermo-Electron Scientific, Ft. Lauderdale, FL, USA). Further metabolite identity validation was done on an Agilent 1200 series μHPLC coupled to a 6410 series Triple Quadrupole LC-MS (Agilent Technologies, Wood Dale, IL, USA). Systems components and experimental conditions used are specified in Supplemental Tables S2 and S3. Qualitative analysis of IMI, AC and main metabolites was done with LC software tools by comparison of retention times, absorbance properties and Log Kow to available standards. Quantitative analyses of parent chemicals and AC-1 were performed with standards and calibration curves. However, because of the initial difficulty of obtaining a suitable standard for IMI-1 identity confirmation (see Metabolite identification section), the quantification of this metabolite was performed semi-quantitatively using area response correlations as previously described by Serrano et al. (2019).
Data analysis
Data were plotted or reported as mean ± standard deviation (SD). GraphPad Prism for Windows (V5.02; GraphPad software, San Diego, CA, USA) was used for curve fitting and statistical analysis. Significant toxicity responses in the slice assay (p < 0.05) were determined by ANOVA analysis with a Dunnett’s multiple comparison test and two-tailed t-tests. Microsomal kinetic data were determined through Michaelis-Menten curve-fitting within GraphPad Prism.
Chromeleon Thermo-Dionex LC software (v2.0; Thermo Electron-Dionex, Ft Lauderdale, FL, USA) was used for calibration curve generation, chemical and Std concentration calculation and for assessment of limits of detection for LC analyses (LOD; analyte concentration corresponding to signal/noise (S/N) >3 measured as peak height). Accuracy was determined by assessing the % difference between nominal and measured chemical concentrations. Lower limits of quantification (LLQ) were determined at a S/N of at least 12:1 for each chemical. Intra-day variability and inter-day reproducibility were determined as previously reported (Serrano et al., 2019). For all chemicals, a linear correlation (coefficient >0.999) was obtained between response and concentration over the selected calibration range 0.5 to 250 μM for IMI and 1 to 500 μM for AC. Intra- and inter-day variability were <2% for all tests. LLQ and LOD for all analytes were 1.15 and 0.70 nmole/mL, respectively.
Results
AC and IMI toxicity
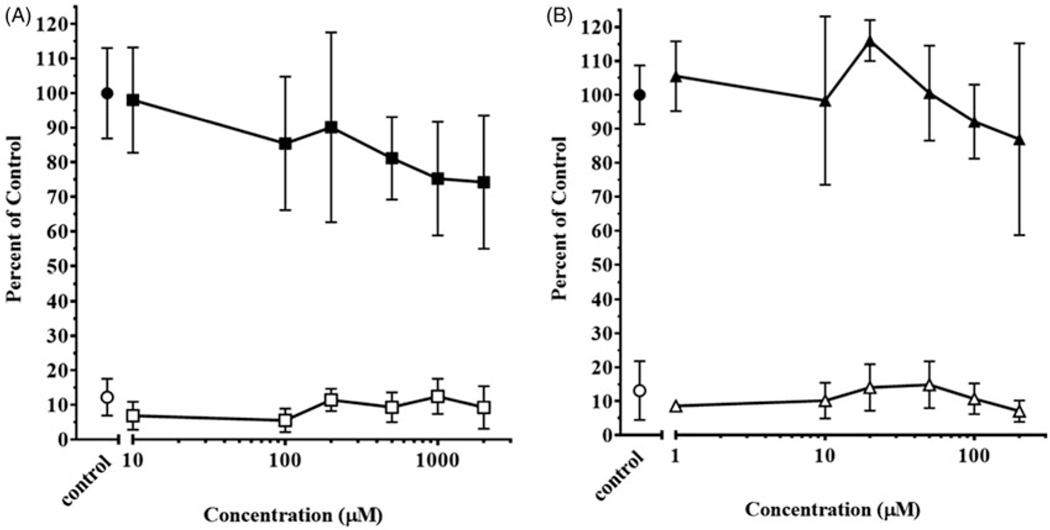
The viability of liver slices was assessed over the course of 48 hours using both MTT and LDH assays. No significant toxicity was measured in either assay for AC concentrations up to 2000 mM (Figure 2(A)). Likewise, there was no evidence of toxicity to slices exposed to concentrations of IMI up to 200 μM (Figure 2(B)). Therefore, further liver slice metabolism experiments were conducted at 1000, 2000 μM AC and 100, 200 μM IMI.
Figure 2.
Toxicity of (A) acetamiprid (AC) and (B) imidacloprid (IMI) in rainbow trout liver slices. Determination of MTT activity in slices exposed to carrier control ethanol (●) and AC (■) or IMI (▲). Data are expressed as percentage of absorbance at 595 nm of control slices. Determination of LDH leakage into the exposure media from slices exposed to carrier control ethanol (○) and AC (⎕) or IMI (Δ). Data are expressed as percentage of total LDH activity in control slices. Symbols represent mean ± SD of six replicate slices from six separate wells.
RBT liver slice metabolism
The media concentration of parent chemical and metabolites were measured during RBT liver slice assays incubated at 11 °C in covered 12-well plates containing 1.7 mL of exposure media on an orbital shaker. An initial test with 1000 and 2000 μM AC was performed with either one or two slices per well for 96 hours (Table 1). AC measured concentrations were close to the nominal values of 1000 and 2000 μM. The HPLC analysis of samples resulted in detection of a single metabolite, AC-1, in addition to parent chemical. In a single slice exposure 0.084 ± 0.021 and 0.131 ± 0.029 μM AC-1 was formed for the respective 1000 and 2000 μM AC exposures (n = 3). The doubling of slices per well resulted in roughly a doubling of media metabolite production of AC-1 to 0.200 and 0.383 μM in the 1000 and 2000 μM AC exposures (n = 1).
Table 1.
Metabolism of 1000 and 2000 μM nominal concentrations of acetamiprid (AC) or 100 and 200 μM nominal concentrations of imidacloprid (IMI) with rainbow trout liver slices in exposure media following a 96-h incubation at 11 °C. Single metabolites (AC-1) and (IMI-1) were detected in exposure media. Values listed as μM mean ± SD, one slice/well experiment (n = 3), two slice/well experiment (n = 1).
| One Slice |
Two Slices |
|||
|---|---|---|---|---|
| 1000 μM | 2000 μM | 1000 μM | 2000 μM | |
|
| ||||
| AC (parent) | 1078.3 ± 41.6 | 2119.7 ± 121.2 | 1184.2 | 2142.3 |
| AC-1 (metabolite) | 0.084 ± 0.021 | 0.131 ± 0.029 | 0.200 | 0.383 |
| 100 μM | 200 μM | 100 μM | 200 μM | |
|
| ||||
| IMI (parent) | 107.3 ± 3.4 | 203.4 ± 2.2 | 113.1 | 227.3 |
| IMI-1 (metabolite) | 1.056 ± 0.186 | 2.279 ± 0.188 | 1.578 | 4.003 |
One or two slices per well were exposed to 100 and 200 μM IMI for 96 hours (Table 1). Measured IMI concentrations in the exposure media were close to the nominal values of 100 and 200 μM. Analysis of media resulted in a single metabolite, IMI-1, in addition to parent chemical IMI. In the single slice exposures, 1.056 ± 0.186 and 2.279 ± 0.188 μM IMI-1 was detected respectively in the 100 and 200 μM IMI exposure media samples (n = 3). Upon doubling the slices per well, IMI-1 was detected at 1.578 and 4.003 μM in the 100 and 200 μM IMI exposure media (n = 1).
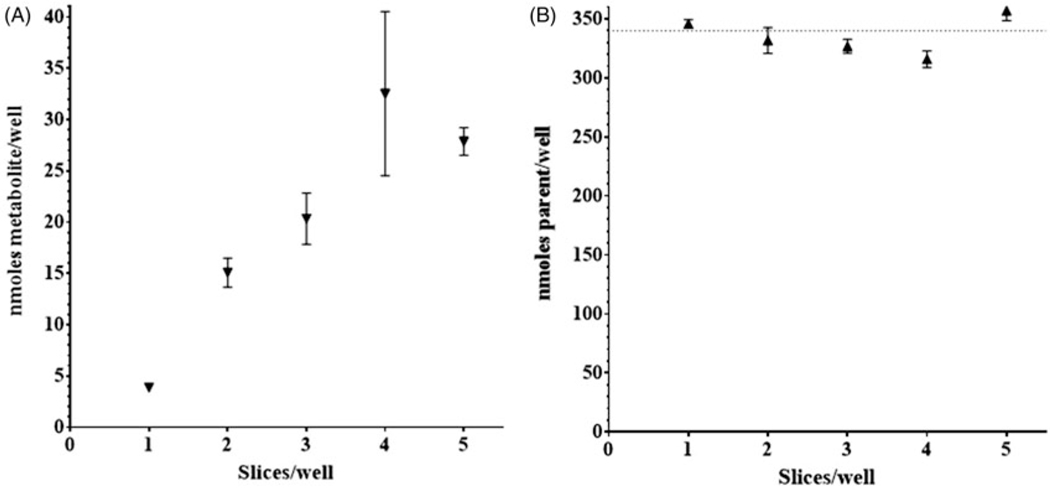
There was an apparent doubling of metabolite production in both the AC and IMI initial experiments with the addition of a second slice per well. This prompted an experiment with 200 μM IMI to measure the production of metabolites with increasing number of slices per well over 96 hours (n = 2). A proportional increase of metabolite, IMI-1, was observed up to four slices per well (Figure 3(A)). Measurement of parent chemical, IMI, in media samples was consistent across the number of slices per well with a downward trend through four slices per well (Figure 3(B)).
Figure 3.
RBT liver slice metabolism. Concentration of parent chemical imidacloprid (IMI,▲) and metabolite IMI-1 (▼) in exposure media incubated in the presence of rainbow trout liver slices. IMI-1 (A) and IMI (B) concentration in exposure media after 96-h incubation at 11 °C. Nominal IMI concentration was 340 nmoles/well (200 μM). Symbols represent mean ± SD of two replicate samples.
Having determined that four slices per well is optimum for the production of metabolite, an experiment was run with IMI over 96 and 120 hours. Measured concentrations of parent chemical in the exposure media were 193.4 ± 2.6 and 197.2 ± 2.4 μM (n = 3) across the two respective time points and matched closely the nominal concentration of 200 μM (Table 2). The 96-h IMI-1 metabolite found in the media was measured at 6.908 ± 0.536 μM and continued to increase with time, 10.821 ± 0.185 μM at 120 hours.
Table 2.
Metabolism of 200 μM nominal concentration of imidacloprid (IMI) or a 2000 μM nominal concentration of Acetamiprid (AC) with rainbow trout liver slices (4 slices/well) in exposure media following 96 and 120 h incubations at 11 °C.
| 96 h | 120 h | |
|---|---|---|
|
| ||
| IMI (parent) | 193.4 ± 2.6 | 197.2 ± 2.4 |
| IMI-1 (metabolite) | 6.908 ± 0.536 | 10.821 ± 0.185 |
| AC (parent) | 2231.3 ± 282.2 | 2307.5 ± 283.6 |
| AC-1 (metabolite) | 0.802 ± 0.016 | 0.857 ± 0.068 |
Single metabolites (IMI-1) and (AC-1) were detected in exposure media. Values listed as μM mean ± SD (n = 3).
An experiment with 2000 μM AC in exposure media with four slices per well produced 0.802 ± 0.016 μM AC-1 at 96 hours (Table 2). Production of AC-1 metabolite at 120 hours did not increase, 0.857 ± 0.068 μM. The measured AC parent concentrations in media were 2231.3 ± 282.2 and 2307.5 ± 283.6 μM at 96 and 120 hours respectively (n = 3).
Microsomal metabolism studies
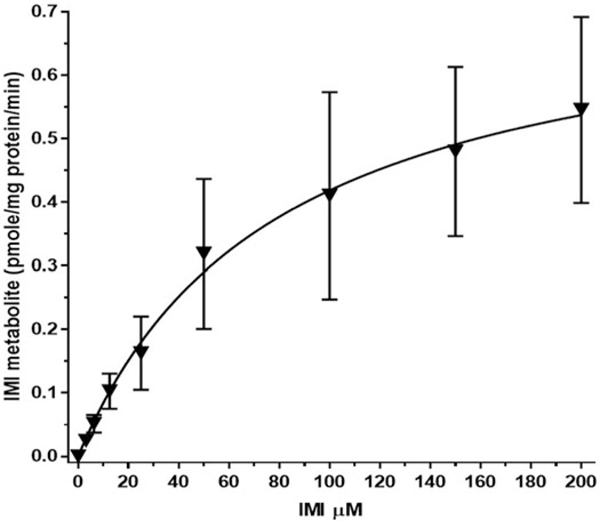
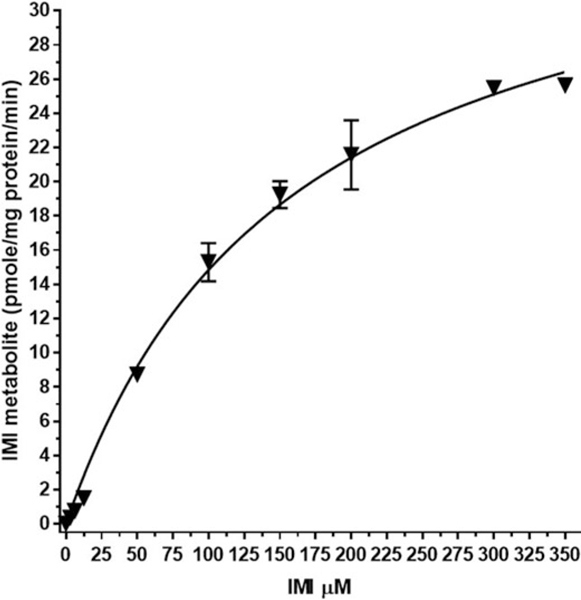
Incubations of RBT liver microsomes with IMI (3.125 – 200 μM) were conducted at the physiologically relevant temperature of 11 °C over 120 minutes (Figure 4). The figure shows the mean of values ± standard error (SE) for IMI-1 metabolite formation. Fitting of this curve to Michaelis–Menten kinetic rates across preparations resulted in a Km of 79.2 μM and Vmax 0.75 pmol/min/mg for the reaction to produce IMI-1 metabolite. Table 3 shows the rate data for each of four individual preparations of RBT microsomes. The Km ranged from 57.7 to 96.6 μM, while Vmax ranged from 0.21 to 1.30 pmol/min/mg. A mean of the individual preparation rates was 75.9 μM for Km and 0.75 pmol/min/mg for Vmax.
Figure 4.
RBT microsome imidacloprid (IMI) enzyme kinetic graph. Graph represents the formation of IMI-1 during the 120 min, 11 °C incubation relative to the initial exposure of IMI. Mean of separate tests with four different microsome preparations ± SE.
Table 3.
Michaelis–Menten kinetics for formation of IMI-1 metabolite in RBT microsomes over a range of IMI concentrations (3.125–200 μM) at 11 °C, 120 min 4 mg/mL protein, 5 mM NADPH, 100 mM K2HPO4 pH 7.4.
| PREPARATION | Km (μM)±SE | Vmax (pmol/min/mg)±SE |
|---|---|---|
|
| ||
| #1 – April 9 2018 | 77.1 ± 9.1 | 0.21 ± 0.01 |
| #2 – August 27 2018 | 57.7 ± 7.4 | 0.75 ± 0.04 |
| #3 – September 4 2018 | 96.6 ± 12.4 | 1.30 ± 0.08 |
| #4 – July 16 2018 | 72.1 ± 8.2 | 0.73 ± 0.03 |
| Mean of Individual Rates | 75.9 ± 16.1 | 0.75 ± 0.44 |
| Mean Rate Across Preparations | 79.2 ± 52.0 | 0.75 ± 0.20 |
For the purpose of species comparison, incubations of rat liver microsomes with IMI (3.125 – 350 μM) were run at the physiologically relevant temperature of 37 C over 120 min (Figure 5). The figure shows the mean of values ± SE for the production of IMI-1 metabolite. Fitting of this curve to Michaelis–Menten kinetic rates across preparations resulted in a Km of 158.7 μM and Vmax 38.4 pmol/min/mg for the reaction to produce IMI-1 metabolite. Table 4 shows the rate data for each of three individual preparations of rat microsomes. The Km ranged from 151.7 to 165.8 μM, while Vmax ranged from 37.5 to 39.9 pmol/min/mg. A mean of the individual preparation rates was 158.9 μM for Km and 38.4 pmol/min/mg for Vmax.
Figure 5.
Rat microsome imidacloprid (IMI) enzyme kinetic graph. Graph represents the formation of IMI-1 during the 120 min, 37 °C incubation relative to the initial exposure of IMI. Mean of separate tests with three different microsome preparations ± SE.
Table 4.
Michaelis–Menten kinetics for formation of IMI-1 metabolite in rat microsomes over a range of IMI concentrations (3.125–350 μM) at 37 °C, 120 min, 2 mg/mL protein, 5 mM NADPH, 100 mM K2HPO4 pH 7.4.
| PREPARATION | Km (μM)±SE | Vmax (pmol/min/mg)±SE |
|---|---|---|
|
| ||
| #1 – Lot# 1610290 | 165.8 ± 21.0 | 39.9 ± 2.2 |
| #2 – Lot# 1410271 | 151.7 ± 21.5 | 37.9 ± 2.3 |
| #3 – Lot# 1310214 | 159.3 ± 33.0 | 37.5 ± 3.4 |
| Mean of Individual Rates | 158.9 ± 7.1 | 38.4 ± 1.3 |
| Mean Rate Across Preparations | 158.7 ± 20.3 | 38.4 ± 2.1 |
Incubations of RBT microsomes with AC at 11 °C over 120 minutes resulted in the formation of metabolite AC-1 at detectable but below quantifiable levels at the high concentrations. Therefore, it was not possible to determine the kinetics for formation of AC-1.
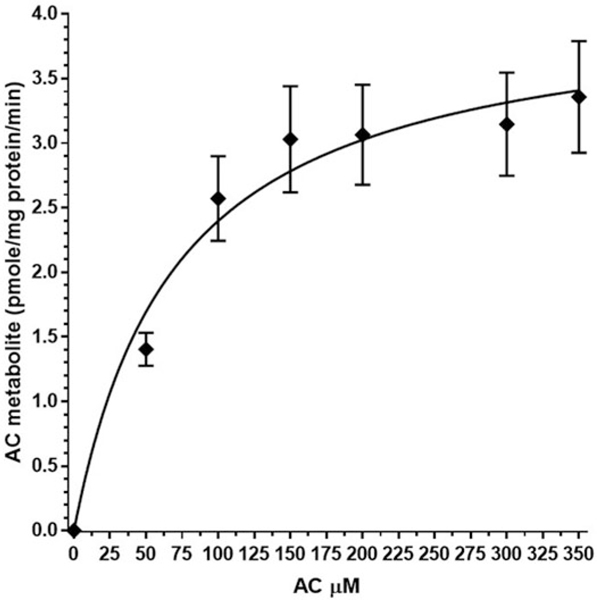
Rat liver microsomes were incubated with AC (3.125–350 μM) at the physiologically relevant temperature of 37 °C over 120 minutes (Figure 6). The figure shows the mean of values ± SE for the production of AC-1 metabolite. Fitting of this curve to Michaelis–Menten kinetic rates across preparations resulted in a Km of 71.0 μM and Vmax 4.1 pmol/min/mg for the reaction to produce AC-1 metabolite. Table 5 shows the rate data for each of three individual preparations of rat microsomes. The Km ranged from 64.0 to 76.0 μM while Vmax ranged from 3.0 to 4.7 pmol/min/mg. A mean of the individual preparation rates was 70.2 μM for Km and 4.1 pmol/min/mg for Vmax.
Figure 6.
Rat microsome acetamiprid (AC) enzyme kinetic graph. Graph represents the formation of AC-1 during the 120 min, 37 °C incubation relative to the initial exposure of AC. Mean of separate tests with three different microsome preparations ± SE.
Table 5.
Michaelis–Menten kinetics for formation of AC-1 metabolite in rat microsomes over a range of AC concentrations (3.125–350 μM) at 37 °C, 120 min, 2 mg/mL protein, 5 mM NADPH, 100 mM K2HPO4 pH 7.4.
| PREPARATION | Km (μM)±SE | Vmax (pmol/min/mg)±SE |
|---|---|---|
|
| ||
| #1 – Lot# 1410271 | 70.6 ± 12.6 | 4.62 ± 0.24 |
| #2 – Lot# 1610290 | 76.0 ± 14.6 | 4.70 ± 0.27 |
| #3 – Lot# 1310214 | 64.0 ± 8.3 | 2.98 ± 0.11 |
| Mean of Individual Rates | 70.2 ± 6.0 | 4.10 ± 0.97 |
| Mean Rate Across Preparations | 71.0 ± 27.7 | 4.10 ± 0.47 |
Metabolite identification
HPLC data demonstrated that both IMI and AC produced only one distinct, but species-conserved metabolite after media incubations with either rat and RBT microsomes, or RBT slices (LC-DAD chromatograms provided in Supplemental Figures S4 and S5). Potential standards for the identification of AC-1 were available at the early stages of the study that matched the main hypothesized AC metabolites. However, the identity of the metabolite formed from IMI (IMI-1) was difficult to infer from various potential conformations, requiring more experimental information to hypothesize the structure and investigate Std availability. Earlier elution times (Rt) observed for the distinct metabolites in comparison with the parent compounds by reversed-phase LC (4.36, 4.68, 4.70 and 5.01 min for IM-I, AC-1, IMI and AC, respectively), indicated an increase in polarity for both the IMI-1 and AC-1 as compared to the starting chemicals. Three standards were initially available for AC-1 identification with lower calculated log Kow than AC and were tested in our LC system (Supplemental Table S4). Comparison of AC-1 absorbance max (245 nm) and Rt data to the standards suggested that Acetamiprid-N-desmethyl (MW = 208.6) was the distinct metabolite. Similarly, two suitable IMI metabolite standards were obtained with suspected higher polarity (<log Kow) than IMI (5-hydroxy-IMI and IMI-olefin; Supplemental Table S3). In this case, comparison of IMI-1 experimental absorbance max (266 nm), absorbance profile and Rt data to the standards suggested that the conserved IMI metabolite was 5-hydroxy-IMI (MW = 271.6). The identity of IMI-1 and AC-1 was validated by LC-MS data comparison of the metabolites and the selected standards (Supplemental Figures S2 and S3).
Discussion
Neonicotinoids known mode of action is through irreversible binding of the chemical to the nicotinic acetylcholine receptor (nAChR) located in cells of the central nervous system in target insects (Ware & Whitacre, 2004). At high chemical concentration levels, overstimulation and blocking of the receptors causes paralysis and eventual death. Neonicotinoids are highly water soluble and stable in soil and water which allows for movement to surface waters, thus potentially impacting aquatic species and their environment. While highly toxic to aquatic invertebrates and insects, neonicotinoids are considered slightly or not acutely toxic to fish. This selective toxicity results from the different composition of the nAChR in insects and vertebrates. The insect form of nAChR contains a unique positively charged amino acid residue that favors the binding of neonicotinoids containing a negatively charged nitro or cyano group (Tomizawa & Casida, 2003). Modification of neonicotinoid nitro or cyano groups through biotransformation has resulted in metabolites with a positively charged nitrogen and therefore a greater affinity for the nAChR and higher toxicity in mammals (Lee Chao & Casida, 1997; Tomizawa, 2004). While the mammalian metabolism of neonicotinoids has been studied, there is a lack of understanding of their metabolism in fish including potential bioactivation to more toxic metabolites acting through the nAChR.
A previous in vivo toxicokinetic study using IMI and RBT, showed no biotransformation of this chemical in liver S9 fraction (Frew et al., 2018). We report for the first time in fish, the production of a single metabolite, IMI-1, from in vitro microsomal and ex vivo liver slice incubations with IMI. In the Frew study, the in vitro metabolism was evaluated at 11 °C with 1 μM IMI using a substrate-depletion approach. Focus of the current study was on the formation of metabolite IMI-1 at expected low concentrations after pilot studies. Specifically, production of metabolite IMI-1 from IMI was only 0.1% in the microsomal assay and 1% in a single liver slice experiment (~5% with multiple slices). When considering these low conversion rates, it is understandable that a depletion of substrate was not detected by Frew et al. In the current study, a distinguishable reduction of parent IMI concentration was only seen in the liver slice experiments with optimized four slices per well.
The single metabolite found in the IMI microsomal studies was conserved across RBT and rat species as well as the levels of biological organization (microsomes and liver slices). Suspected transformation pathways were proposed but not limited to (1) hydroxylation of the imidazolidine to 5-hydroxy-imidacloprid followed by dehydrogenation to the imidacloprid-olefin, (2) reduction and cleavage of the nitroimine substituent to form nitrosimine, guanidine, and urea or (3) oxidative cleavage of the methylene bridge to form 6-chloronicotinic acid. Standards of 6-chloronicotinic acid, 5-hydroxy-imidacloprid and imidacloprid-olefin were purchased. Identification of the IMI metabolite was initially achieved through a retention time match to an authentic standard of 5-hydroxy-imidacloprid. The identification of this metabolite was further verified by fraction collection and concentration of the IMI-1 peak in samples from the four slice/well experiments and comparison of the LC-MS product ion mass spectrum of m/z 272 ([M + H]+) with authentic 5-hydroxy-imidacloprid (Supplemental Figure S2) as an exact match.
Two major routes of IMI in vivo metabolism in the rat have been previously reported (Klein, 1987; Klein & Karl, 1990). In the first pathway, IMI may be broken by oxidative cleavage to 6–chloronicotinic acid and imidazolidine. Imidazolidine is excreted in the urine, and 6-chloronicotinic acid undergoes further metabolism via glutathione conjugation to form mercaptonicotinic acid and a hippuric acid. IMI may also be metabolized by hydroxylation of the imidazolidine ring in the second major pathway. Metabolic products from the second pathway include 5-hydroxy- and olefin derivatives. It is the latter of the two reactions that we observed in our in vitro experiments with the RBT and rat while producing the 5-hydroxy-imidacloprid.
Kinetic data was determined for the microsomal conversion of IMI to IMI-1 in both the RBT and rat. Comparison of IMI-1 kinetics showed a lower Km for the fish (79.2 μM) versus rat (158.7 μM) and hence a greater enzymatic affinity for IMI substrate for this reaction. A much higher rate of formation (Vmax) for this reaction was observed in the rat (38.4 pmole/min/mg) versus the fish (0.75 pmole/min/mg). A calculation ignoring species differences for Q10 resulted in a value of 4.5 which is outside of the typical range of Q10 =2 to 3. There is an inherent problem with comparing rates across species with vastly different physiological temperatures at different levels of evolution as well as comparison of an ectotherm with an endotherm.
A single metabolite AC-1 was found in both the RBT and rat microsomal assay with AC. Several AC metabolite standards were available and AC-1 was identified through retention time match to N-desmethyl-acetamiprid. The identification of the metabolite was further verified by fraction collection and concentration of the AC-1 peak in liver slice samples (four slices/well) and comparison of the LC-MS product ion mass spectrum of m/z 209 ([M + H]+) with authentic N-desmethyl-acetamiprid (Supplemental Figure S3) as an exact match. AC-1 was found in detectable but not quantifiable amounts in the RBT microsomal assay with AC. Kinetic constants 70.9 mM (Km) and 4.10 pmole/min/mg (Vmax) were determined for the conversion of AC to AC-1 in the rat microsomal model. Appearance of the metabolite AC-1 in the rainbow trout study was consistent with findings of Tanoue & Mori (1997) and Premkumar et al. (1995) during rat in vivo studies. The discrepancy between the studies was that additional metabolites were found in the excretia of the rat including 5–10% parent AC, 15–25% AC-1 demethylation product, 25–30% 6-chloronicotinic acid cleavage product, and several other components (<2%). The in vitro rainbow trout study with both liver microsomes and slices produced a single metabolite, AC-1, and at a much lower conversion rate. In addition to the inherant species difference, the physiological temperature difference of 11 °C versus 37 °C may have been a factor. The methods of detection were also different with the trout study LC-UV and rat studies using 14 C radioisotope analysis which is better for mass balance and potentially greater sensitivity. Lastly, the level of biological organization may have been a factor with the in vivo studies including a potential for extrahepatic metabolism. This study introduced the option of using multiple slices per well in the liver slice assay for the purpose of enhancing product yields for metabolite identification. Microsomal studies with AC resulted in detectable but below quantifiable levels of AC-1 production. By utilizing multiple slices per well in the liver slice assay, enough metabolite could be obtained for positive identification through LC-MS. This study further exemplifies the utility of the liver slice assay for enhancing the study of metabolism in rainbow trout as introduced by Tapper et al. (2018).
Conclusions
Imidacloprid (IMI) and acetamiprid (AC) are metabolized in fish.
A single metabolite identified as N-desmethyl-acetamiprid (AC-1) was characterized for AC.
A single metabolite identified as 5-hydroxy-imidacloprid (IMI-1) was characterized for IMI.
Formation of respective metabolites 5-hydroxy-imidacloprid and N-desmethyl-acetamiprid was conserved across RBT and rat species.
Formation of respective metabolites 5-hydroxy-imidacloprid and N-desmethyl-acetamiprid was conserved across levels of biological organization (microsomes/liver slices) in fish.
Kinetic rates were reported for the hydroxylation of IMI in RBT (Km = 79.2 μM; Vmax = 0.75 pmoles/min/mg) and rat (Km = 158.7 μM; Vmax=38.4 pmoles/min/mg) liver microsomes.
Kinetic rates for the demethylation of AC were reported for rat liver microsomes (Km = 70.9 μM; Vmax = 4.10 pmoles/min/mg).
Supplementary Material
Acknowledgements
The authors thank Keith Sappington and Ideliz Negrón-Encarnación (USEPA/OCSPP/OPP) for inciteful review of the manuscript, as well as Dr. Brett Blackwell (USEPA/CCTE/GLTED) for technical collaboration.
Footnotes
Disclosure statement
All research reported in this manuscript was funded by the US Environmental Protection Agency (USEPA) and conducted by or under the supervision of USEPA employees. Manuscript review was performed following provisions of the USEPA Office of Research and Development. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. Views presented in the text are those of the authors and not necessarily the opinion of the USEPA. No potential conflict of interest was reported by the authors.
This work was authored as part of the Contributor’s official duties as an Employee of the United States Government and is therefore a work of the United States Government. In accordance with 17 USC. 105, no copyright protection is available for such works under US Law.
References
- Bergmeyer HU, Bernt E. (1974). Lactate dehydrogenase. In: Bergmeyer HU, ed. Methods of enzymatic analysis. Vol. 2. New York: Academic Press, 574–579. [Google Scholar]
- Bradford MM. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–54. [DOI] [PubMed] [Google Scholar]
- Crosby EB, Bailey JM, Oliveri AN, Levin ED. (2015). Neurobehavioral impairments caused by developmental imidacloprid exposure in zebrafish. Neurotox. Teratol 49:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dady JM, Bradbury SP, Hoffman AD, et al. (1991). Hepatic microsomal N-hydroxylation of aniline and 4-chloroaniline by rainbow trout (Oncorhynchus mykiss). Xenobiotica 21:1605–20. [DOI] [PubMed] [Google Scholar]
- Dai YJ, Yuan S, Ge F, et al. (2006). Microbial hydroxylation of imidacloprid for the synthesis of highly insecticidal olefin imidacloprid. Appl Microbiol Biotechnol 71:927–34. [DOI] [PubMed] [Google Scholar]
- Ding Z, Yang Y, Jin H, et al. (2004). Acute toxicity and bioconcentration factor of three pesticides on Brachydanio rerio. Ying Yong Sheng Tai Xue Bao 15:888–90. [PubMed] [Google Scholar]
- Frew JA, Brown JT, Fitzsimmons PN, et al. (2018). Toxicokinetics of the neonicotinoid insecticide imidacloprid in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol C 205:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Yan S, Wang J, et al. (2015). Oxidative stress and DNA damage induced by imidacloprid in zebrafish (Danio rerio). J Agric Food Chem 63:1856–62. [DOI] [PubMed] [Google Scholar]
- Iturburu FG, Zomisch M, Panzeri AM, et al. (2017). Uptake, distribution in different tissues, and genotoxicity of imidacloprid in the freshwater fish. Australoheros Facetus Environ Toxicol Chem 36:699–708. [DOI] [PubMed] [Google Scholar]
- Klein O.(1987). [14C]-NTN 33893: Biokinetic part of the ‘general metabolism study’ in the rat. Unpublished Report no. PF2889, 1987, submitted to WHO by Bayer AG, Mannheim, Germany. INCHEM Toxicological Evaluations: Imidacloprid; International Programme on Chemical Safety, World Health Organization: Geneva, Switzerland. [Google Scholar]
- Klein O, Karl W.(1990). Methylene [14C] imidacloprid: metabolism part of the general metabolism study in the rat. Unpublished Report no. PF 3316, 1990, submitted to WHO by Bayer AG, Mannheim, Germany. INCHEM Toxicological Evaluations: Imidacloprid; World Health Organization, International Programme on Chemical Safety. [Google Scholar]
- Kolanczyk R, Schmieder P, Bradbury S, Spizzo T. (1999). Biotransformation of 4-methoxyphenol in rainbow trout (Oncorhynchus mykiss) hepatic microsomes. Aq Toxicol 45:47–61. [Google Scholar]
- Lee Chao S, Casida JE. (1997). Interaction of imidacloprid metabolites and analogs with the nicotinic acetylcholine receptor of mouse brain in relation to toxicity. Pest Biochem Physiol 58:77–88. [Google Scholar]
- Liu MY, Casida JE. (1993). High affinity binding of [3H]-imidacloprid in the insect acetylcholine receptor. Pestic Biochem Physiol 46:40–6. [Google Scholar]
- Morrissey CA, Mineau P, Devries JH, et al. (2015). Neonicotinoid contamination of global surface waters and associated risk to aquatic invertebrates: a review. Environ Internat 74:291–303. [DOI] [PubMed] [Google Scholar]
- Nauen R, Ebbinghaus-Kintscher U, Schmuck R. (2001). Toxicity and nicotinic acetylcholine receptor interaction of imidacloprid and its metabolites in Apis mellifera (Hymenoptera: Apidae). Pest Manage Sci 57: 577–86. [DOI] [PubMed] [Google Scholar]
- Premkumar ND, Guo CY, Vegurlekar SS. (1995). Absorption, distribution, metabolism, elimination and pharmacokinetics after chronic dosing of [14C]acetamiprid in rat. Unpublished ABC Laboratory Study No. 42207. Submitted to WHO by Nippon Soda Co., Ltd, Tokyo, Japan. [Google Scholar]
- Raby M, Nowierski M, Perlov D, et al. (2018). Acute toxicity of 6 neonicotinoid insecticides to freshwater invertebrates. Environ Toxicol. Chem 37: 1430–45. [DOI] [PubMed] [Google Scholar]
- Sanchez-Bayo F, Goka K, Hayasaka D. (2016). Contamination of the aquatic environment with neonicotinoids and its implication for eco-systems. Frontiers Env Sci 4:1–14. [Google Scholar]
- Schmieder PK, Tapper MA, Denny JS, et al. (2004). Use of trout liver slices to enhance mechanistic interpretation of estrogen receptor binding for cost-effective prioritization of chemicals within large inventories. Environ Sci Technol 38:6333–42. [DOI] [PubMed] [Google Scholar]
- Schmieder P, Tapper M, Linnum A, et al. (2000). Optimization of a precision-cut trout liver tissue slice assay as a screen for vitellogenin induction: comparison of slice incubation techniques. Aq Toxicol 49:251–68. [DOI] [PubMed] [Google Scholar]
- Serrano J, Tapper MA, Kolanczyk RC, et al. (2019). Metabolism of cyclic phenones in rainbow trout in vitro assays. Xenobiotica. doi: 10.1080/00498254.2019.1596331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Delso N, Amaral-Rogerrs V, Belzunces LP, et al. (2015). Systemic insecticides (neonicotinoids and fipronil): trends, uses, mode of action and metabolites. Environ Sci Pollut Res 22:5–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struger J, Grabuski J, Cagampan S, et al. (2017). Factors influencing the occurance and distribution of neonicotinoid insecticides in surface waters of southern Ontario, Canada. Chemosphere 169:516–23. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Mori H. (1997). 14C-NI-25—Metabolism study in rats. Unpublished report No. EC-724 from Nisso Chemical Analysis Service Co., Ltd, Odawara, Kanagawa, Japan. Submitted to WHO by Nippon Soda Co., Ltd, Tokyo, Japan. [Google Scholar]
- Tapper MA, Serrano JA, Schmieder PK, et al. (2018). Metabolism of diazinon in rainbow trout liver slices. Appl In Vitro Toxicol 4:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa M.(2004). Neonicotinoids and derivatives: effects in mammalian cells and mice. J Pestic Sci 29:177–83. [Google Scholar]
- Tomizawa M, Casida JE. (2003). Selective toxicity of neonicotinoids attributable to specificity of insect and mammalian nicotinic receptors. Annu Rev Entomol 48:339–64. [DOI] [PubMed] [Google Scholar]
- Tomizawa M, Casida JE. (2005). Neonicotinoid insecticide toxicology: mechanisms of selective action. Annu Rev Pharmacol Toxicol 45: 247–68. [DOI] [PubMed] [Google Scholar]
- Tomizawa M, Yamamoto I. (1993). Structure-activity relationships of nicotinoids and imidacloprid analogs. J Pestic Sci 18:91–8. [Google Scholar]
- Tomlin CDS, ed. (2009). The e-pesticide manual. 12th edition. Surrey: British Crop Protection Council. [Google Scholar]
- UOH. (2019). Pesticide Properties Database, October 2019 version. Agriculture and Environment Research Unit (AERU), Science and Technology Research Institute, University of Hertfordshire, U.K. Available from: https://sitem.herts.ac.uk/aeru/ppdb/en/atoz.htm#I [last accessed 28 Oct 2019]. [Google Scholar]
- USEPA. (2016). Preliminary Aquatic Risk Assessment to Support the Registration Review of Imidacloprid. Office of Chemical Safety and Pollution Prevention. Office of Pesticides Program. Washington D.C. Available at: https://www.regulations.gov/document?D=EPA-HQ-OPP-2008-0844-1086 [last accessed 22 Aug 2019]. [Google Scholar]
- Vistica DT, Skehan P, Scudiero D, et al. (1991). Tetrazolium-based assays for cellular viability: a critical examination of selected parameters affecting formazan production. Cancer Res 51:2515–20. [PubMed] [Google Scholar]
- Ware GW, Whitacre DM. (2004). The pesticide book. Willoughby, OH: MeisterPro Information Resources. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.