Summary
Some types of glia play an active role in neuronal signaling by modifying their activity although little is known about their role in sensory information signaling at the receptor level. In this research, we report a functional role for the glia that surround the soma of the olfactory receptor neurons (OSNs) in adult Drosophila. Specific genetic modifications have been targeted to this cell type to obtain live individuals who are tested for olfactory preference and display changes both increasing and reducing sensitivity. A closer look at the antenna by Ca2+ imaging shows that odor activates the OSNs, which subsequently produce an opposite and smaller effect in the glia that partially counterbalances neuronal activation. Therefore, these glia may play a dual role in preventing excessive activation of the OSNs at high odorant concentrations and tuning the chemosensory window for the individual according to the network structure in the receptor organ.
Subject areas: Behavioral neuroscience, Cellular neuroscience, Sensory neuroscience
Graphical abstract
Highlights
-
•
Perineural glia interact with olfactory receptor neurons modifying perception
-
•
Changes in perception can occur either to increase or decrease sensitivity to odorants
-
•
Glia affect olfaction especially at high odorant concentrations
-
•
Glial response to odors is opposite and depends on the receptor neuron activation
Behavioral neuroscience; Cellular neuroscience; Sensory neuroscience
Introduction
In both vertebrates and invertebrates, glia constitute a large part of the nervous system and perform many essential roles. Traditionally, the role of glia has been associated with the development and support of neuronal cells, but more recently, a more active role has been shown, especially for astrocytes of the central nervous system (CNS).1,2,3,4 In the peripheral nervous system, special attention has been given in vertebrates to the role of satellite glial cells (SGCs), which interact with neurons in the sensory ganglia that mediate pain sensation.5,6 Recently, a comparative review analyzed the similarities and differences of both types of glial cells: astrocytes and SGCs.7
In Drosophila flies, the importance of glia in processes from development8,9,10 to pathological neuronal degeneration in the adult brain has been clearly demonstrated.11 A series of reports show that neuron-glia interactions are important for behavior, affecting circadian rhythms,12,13 sleep,14,15 and memory formation.16,17
Many classes of glial cells in Drosophila adults have morphological and molecular similarities with their corresponding glial types in mammals.9,18,19,20,21,22,23 As in mammals, individual astrocyte-like cells with branches cover a relatively large territory in Drosophila, with the potential to regulate many different neuronal synapses in the brain. Other types of glia in Drosophila are also a part of the so-called tripartite synapses that regulate neurotransmission.24,25
At the sensory level, the effects of glia on Drosophila vision-processing circuits26,27,28 and smell10,29,30,31,32 have been described. However, less information is known about the neuron-glia interaction in the peripheral nervous system and more specifically in sensory signaling at the receptor level.
In Caenorhabditis elegans, some peripheral sensory glia have been shown to drive olfactory adaptation33 in a multimodal nociceptive sensory system, but the correspondence with other species in which individual sensory modalities are clearly differentiated cannot be directly inferred.
In this report, we address the neuron-glia interaction at the receptor level and its role in olfactory signaling in adult Drosophila.
Olfactory reception in Drosophila takes place mainly in the third antennal segment, where the olfactory receptor cells are housed inside special structures called sensilla.34,35 In the antenna, two types of glia have been described.36 First, a central type originates in the brain and migrates toward the antenna during development, surrounding the axons of the olfactory receptor neurons (OSNs) and resembling the Schwann cells of vertebrates. The second type has an antennal origin and accounts for approximately 70% of antennal glial cells. It corresponds to the Mz317-Gal4 driver; is found closely surrounding the OSN soma, similar to vertebrate SGCs; and extends to surround the axonal fascicles with its central glial envelope.
Taking advantage of the vast array of genetic tools available in Drosophila, we approach the analysis of neuron-glia interactions in olfactory reception by targeting genetic expression changes in the Mz317-type glia using the Gal4/UAS method36 and investigating changes in olfactory performance in whole, live adults without surgical manipulations. Functional measurements include behavioral preference tests and antennal Ca2+ imaging.
Results
Mz317-type glia in the antennae
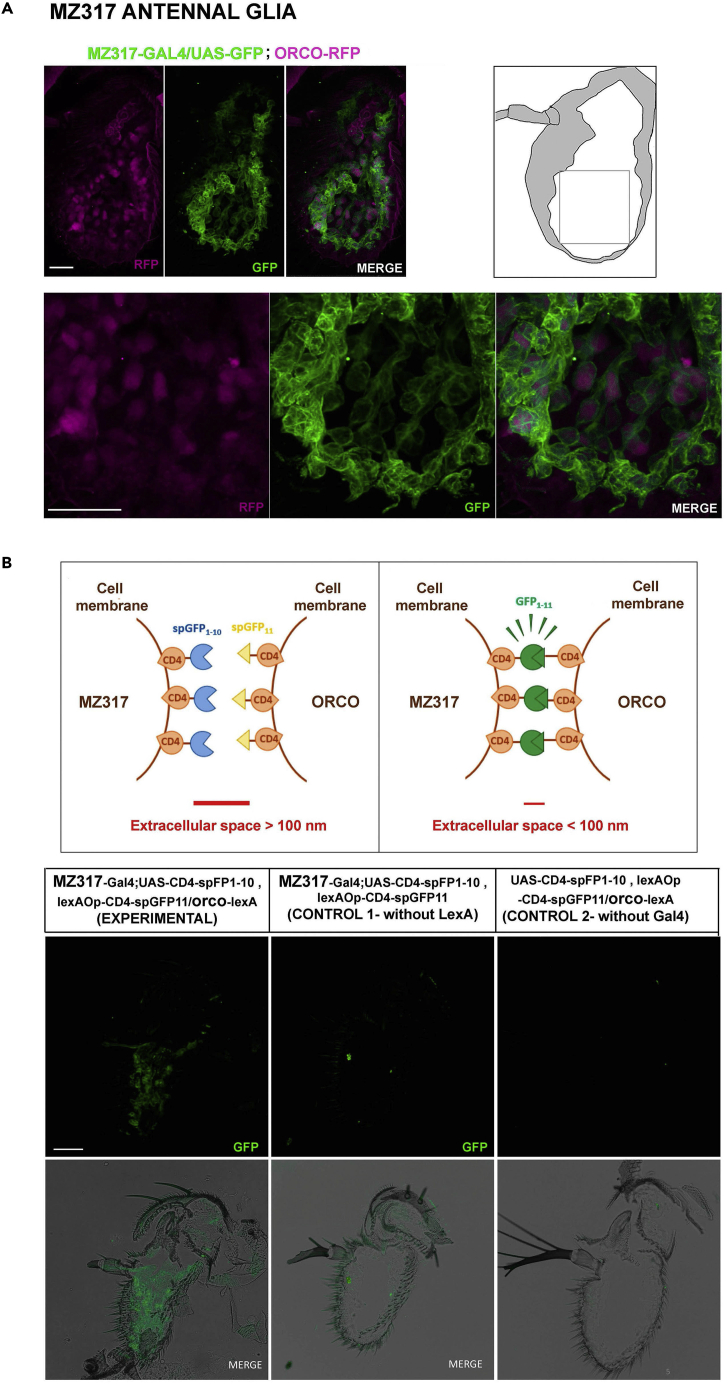
Thick cryosections of the third antennal segment of Mz317-Gal4/UAS-GFP; Orco-RFP flies confirm that Mz317 cells present a thin membranous structure that closely surrounds the cell bodies of the OSNs forming a network structure along the third antennal segment (Figure 1A). Moreover, a new experiment using the GRASP (GFP reconstitution across synaptic partners) technique,37 with expression of one part of the GFP marker in the ORNs (Orco expressing OSNs) and the other part of the GFP marker in the Mz317 cells, generated fluorescence through the assembly of the complete GFP molecule around the OSN soma (Figure 1B). This indicates that both structures, neurons and Mz317 cells, are in close contact (<100 nm) and that there is no intermediate structure between them.
Figure 1.
Characterization of the Mz317 glia in the third antennal segment
(A) Double labeling of OR neurons expressing the ORNs (in magenta) and Mz317 glia (in green) in a 40-μm-thick cryosection observed by confocal microscopy. Above the image of ORNs and Mz317 in the third antennal segment. The drawing on the right represents the third antennal segment, and a magnification of the region in the square is presented below. The Mz317 glia closely surround the soma of the ORNs, forming a network structure.
(B) Above, schematic design of the GRASP experiment expressing the spGFP1-10 semimarker in Mz317 glia and the spGFP11 semimarker in ORNs. GFP labeling is only observed when both semimarkers bind. Bottom, 14-μm antennal cryosections with GFP labeling (in green) in the third antennal segment of experimental individuals but not in controls expressing only one of the semimarkers. In all photos, the scale bar indicates 40 μm.
The morphological characteristics of MZ317 glia in the Drosophila main olfactory receptor organ resemble those of the subperineural glia of a polymodal class IV dendritic arborization neuron of the peripheral nervous system that responds to harmful stimuli in Drosophila larvae.38 In the vertebrate peripheral nervous system, the SGCs of the spinal root ganglion surround neuronal cell bodies. Although OSNs are bipolar neurons in Drosophila while the neurons surrounded by SGCs are pseudomonopolar in vertebrates,39 in both cases, these glia are located far from the synaptic junction between first-order and second-order sensory neurons.
Furthermore, the distance between SGCs and the neurons they surround has been established at 20 nm, allowing mutual neuron-glia interactions.5
Additional confirmation of the glial characteristics of Mz317 cells in the antenna is presented in Figure S1, where the GFP marker is present in the third antennal segment of Mz317-Gal4/UAS-GFP and Mz317-Gal4/elav-Gal80; UAS-GFP flies, where the GFP marker signals to nonneuronal cells. However, GFP marker expression disappears completely in Mz317-Gal4/repo-Gal80; UAS-GFP flies, where the expression is shown only in nonglial cells.40
Behavioral effects of Mz317 cell ablation
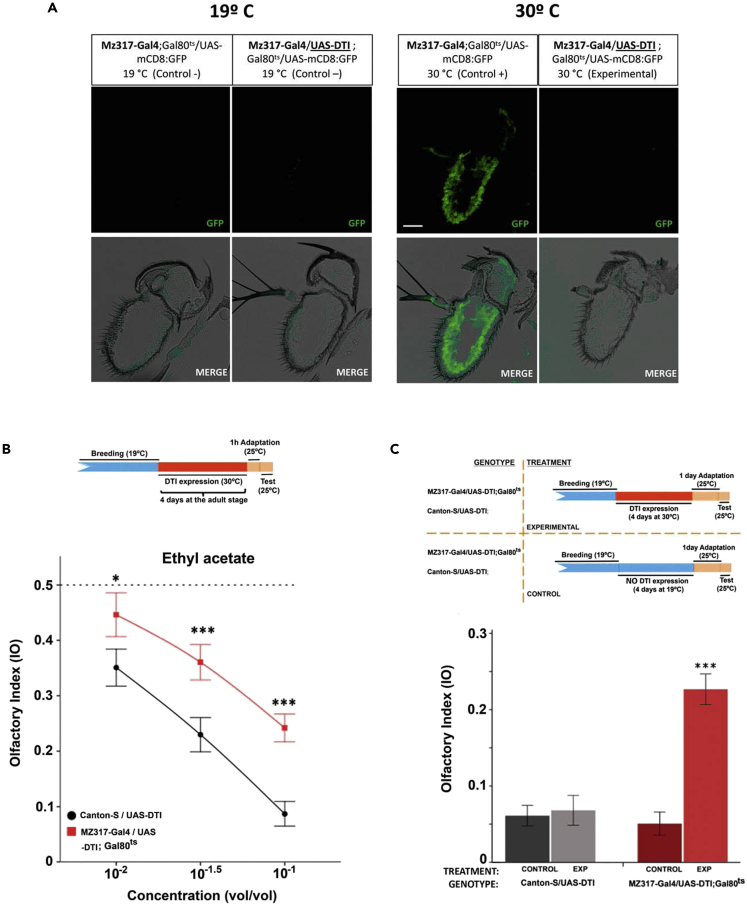
We induced cell ablation by using the Mz317-Gal4 line to direct the expression of DTl (the diphtherian toxin light chain) in Mz317 cells. Direct crossing of Mz317-Gal4 and UAS-DTl lines did not produce any hybrid offspring because in these individuals, expression of the toxin following the space-temporal pattern of the Mz317 driver becomes lethal at some point in development. Restricted expression of the diphtheria toxin, using a thermosensitive Gal80 inhibitor, following the Mz317-driver pattern for 4 days in the adult stage, was enough to produce cell ablation in the antenna without individual lethality (see the disappearance of the GFP marker in the antennae of Mz317-Gal4/UAS-DTl; UAS-GFP/Gal80ts adult flies at the restrictive temperature, Figure 2A).
Figure 2.
Behavioral response in the T-maze after targeted cell ablation of Mz317 cells
(A) Temporal control of expression directed by the Mz317-Gal4 driver using the Gal80ts element for producing viable fly adults with cell ablation. The Gal80ts element blocks the action of the Gal4 element at low temperatures (19°C), but at high temperatures (30°C), the Gal80ts element is inactivated, allowing the action of Gal4 to direct the expression of the UAS-gene insert. In these panels, we tested the presence of Mz317 cells in the third antennal segment after cell ablation for 4 days in the adult stage with diphtheria toxin (DTl). At 19°C, the GFP reporter present in the UAS-GFP insert is not expressed. At 30°C, we can see GFP expression in cells that do not express the diphtheria toxin, but Mz317 cells do not appear in the Mz317-Gal4/UAS-DTl; Gal80ts/UAS-mcD8:GFP antenna because the expression of DTl kills these cells. Scale bar, 40 μm
(B) The breeding protocol indicates the applied temperatures during different time periods. Cell ablation is controlled by temperature blocking of the restrictive action of Gal80ts at 30°C. Below, the dose-response curve of control flies compared with the experimental flies that underwent Mz317 cell ablation for 4 days in the adult stage (for Canton-S/UAS-DTl, N10−2 = 22, N10−1.5 = 22, N10−1 = 27; for Mz317-Gal4/UAS-DTI; Gal80ts, N10−2 = 20, N10−1.5 = 20, N10−1 = 25). Each data point represents the mean +/- SEM.
(C) Above, experimental procedure indicating the breeding treatment applied to both genotypes. Below, olfactory preference response to the odorant ethyl acetate at the 10−1 concentration in the T-maze. Only the flies with MZ317 cell ablation differed significantly from the other 3 groups (for Canton-S/UAS-DTl, NCONTROL = 15, NEXP = 14; for Mz317-Gal4/UAS-DTl; Gal80ts, NCONTROL = 15, NEXP = 13). In all cases, each data point represents the mean ± SEM of 20 replicate tests with 20 flies each.
∗∗∗ = p < 0.001, ∗ = p < 0.05.
Figure 2B shows the dose-response curve obtained in the T-maze in response to three concentrations of the odorant ethyl acetate (EA) in Mz317-Gal4/UAS-DTl; Gal80ts flies subjected to targeted cell ablation to Mz317 cells for 4 days in the adult stage by inactivating the Gal80ts element with heat shock treatment. We observed that less sensitive odor perception (a higher concentration than normal is required to obtain the same olfactory index [IO] value observed in the control flies) occurs in the experimental flies at all concentrations tested. Statistical significance was studied for each concentration between the experimental and control IO values; for EA 10−2, t40 = −2.638∗, p < 0.05; for EA 10−1.5, t40 = −4.150∗∗∗, p < 0.001; for EA 10−1, t50 = −5.676∗∗∗, p < 0.001).
Temperature treatments have been previously reported to be responsible for odorant sensitivity changes even in wild-type flies.41 To avoid incorrect conclusions, an additional experiment in control and experimental flies subjected or not to heat shocks but allowed prolonged recovery times after treatment was performed in response to a single concentration of odorant, EA (10−1, v/v) (Figure 2C). Again, we observed a significant decrease in sensitivity related to Mz317 glial ablation (F3,53 = 25.639∗∗∗, p < 0.001), which confirms Mz317 cell ablation as the cause of diminishing olfactory sensitivity.
Opposite behavioral effects of Mz317 optogenetic activation and inhibition
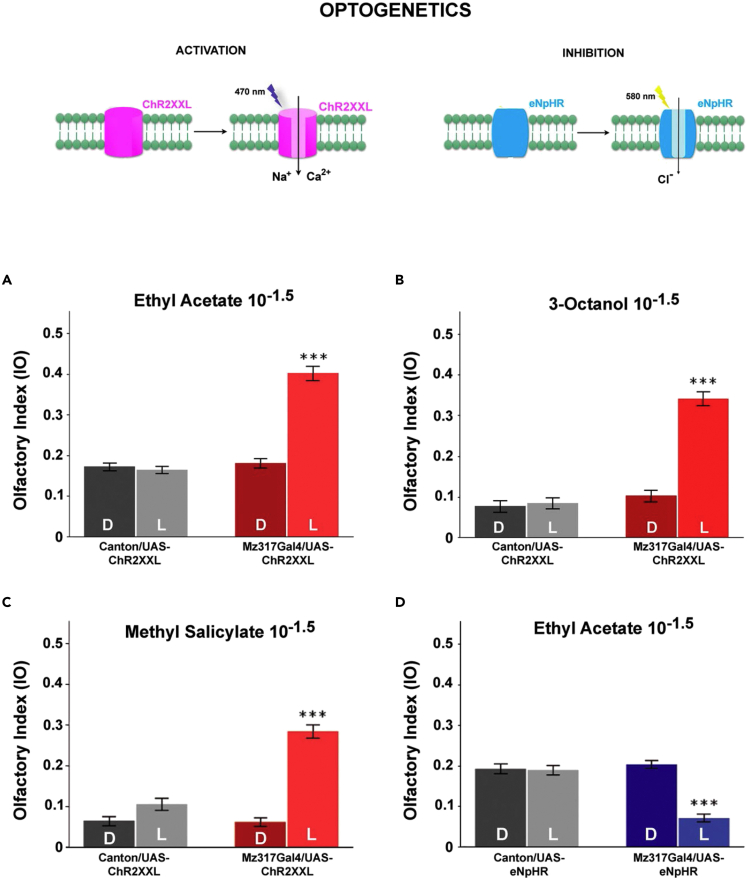
The Mz317 cells were subjected to optogenetic treatment. In this setup, we can alter the transmembrane potential of cells via targeted expression of light-sensitive channels in Mz317 cells, which open when we expose the flies to intense light. Optogenetic experiments have the advantage that modifications in membrane potential occur instantaneously when light is applied and that the same individuals tested in the dark can be used as controls because they possess the same genotype as the experimental individuals (exposed to light).
Depending on the channels expressed, we can achieve cell activation if we express cation channels (channelrhodopsin, ChR2XXL) or inhibition if we express Cl− channels (halorhodopsin, eNpHR) (Figure 3A). In our setup, intense light fully illuminates the T-maze when flies make their choice.42
Figure 3.
Effects of optogenetic Mz317 cell activation or inhibition on olfactory behavior in the T-maze
(A) Optogenetic activation is achieved in flies expressing channel rhodopsin in Mz317 cells by light. Only these flies (Mz317-Gal4/UAS-ChR2XXL) in the presence of light (L), N = 25, respond significantly differently from flies that either choose the odorant in the dark (D), N = 24, or cannot express the channel (Canton-S/UAS-ChR2XXL) tested both with light (L), N = 25, or in the dark (D), N = 24, in response to ethyl acetate (10−1.5).
(B) In response to 3-octanol (10−1.5), N = 25 for each class and condition
(C) In response to methyl salicylate (10−1.5), N = 25 for each class and condition.
(D) Optogenetic inhibition of flies expressing halorhodopsin in Mz317 cells (Mz317-Gal4/UAS-eNpHR) by light (L) in response to ethyl acetate (10−1.5) significantly increased sensitivity compared with the same flies in the dark (D) and that of the control flies in both conditions (D) and (L), in the opposite direction of optogenetic activation. N = 29 for each class except for Canton-S/UAS-eNpHR in the dark, where N = 27.
All values represent the mean ± SEM (∗∗∗ = p < 0.001).
Figures 3B–3D show that when light activation of the Mz317 cells occurs, perception of the odor decreases compared with the perception of the same odor in the dark. However, olfactory perception did not change from dark to light in control flies (for each experiment, analysis of variance was performed followed by a post hoc comparison of the means). This was true for all the odorants we tested, EA 10−1.5 (F3,93 = 86.415∗∗∗, p < 0.001), octanol 10−1.5 (F3,93 = 73.327∗∗∗, p < 0.001), and methylsalicylate 10−1.5 (F3,93 = 60.965∗∗∗, p < 0.001).
When we inhibited Mz317 cells with light (Figure 3E), the perception of odorant concentration changed compared with the perception of the same flies in the dark and to that of the control flies in both light and dark (F3,110 = 33.158∗∗∗, p < 0.001). It should be noted that the change occurred in the opposite direction of the previous case, i.e., increasing sensitivity to odor.
Although the results of these experiments, both cell ablation and optogenetics, seem clear, we must not forget that we are measuring olfactory behavior and that this involves both the receptor organ and the brain. To reliably connect changes in the glia of the receptor organs and olfactory perception, we must confirm the cell type targeted by the Mz317 Gal4 driver and determine whether this driver is specific to the antenna or exists in the brain.
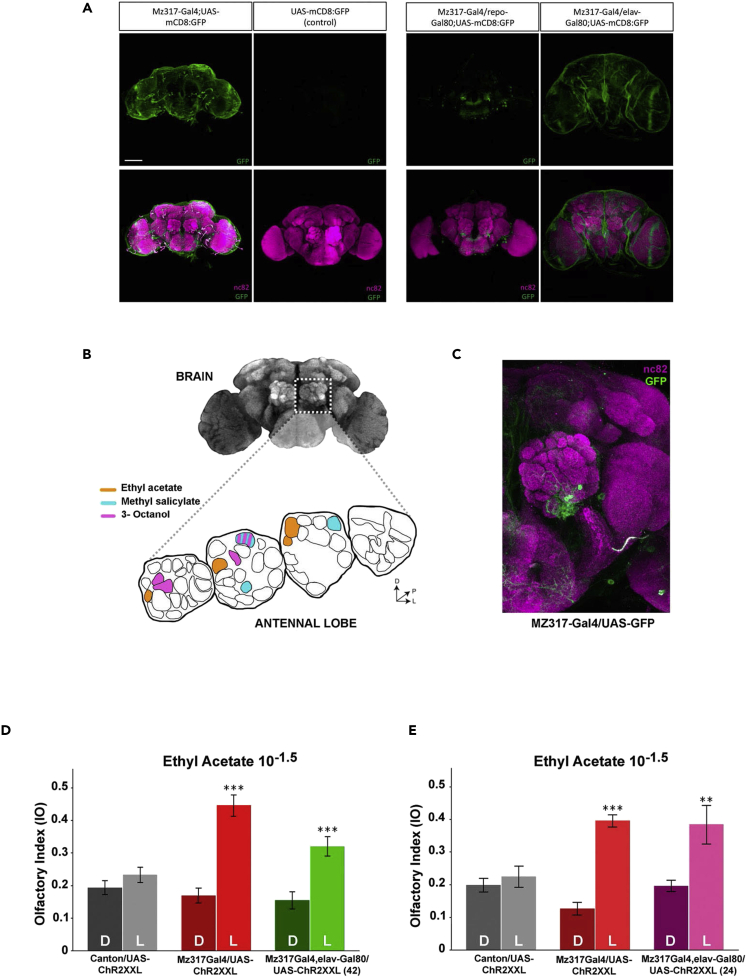
In the antenna, we have already shown that the labeling of Mz317 affects only glial cells (Figure S1). However, in the brain of Mz317-Gal4/UAS-GFP flies, we have a marker that affects glia, but it also affects a small number of neurons, some of them in a restricted region close to the antennal lobe (Figures 4A and 4C). The fact that the behavioral effects of optogenetic activation appear in response to various odors (EA, octanol, and methylsalicylate), which preferentially excite different combinations of glomeruli in the antennal lobe, all more or less distant from the region of neuronal labeling (Figure 4B), eliminates the influence of brain neurons on the behavioral response of experimental Mz317 flies.
Figure 4.
Ruling out the contribution of brain MZ317 neurons to olfactory perception changes
(A) Cellular characterization of the Mz317-Gal4 driver in the adult brain. Mz317-Gal4 driver expression in the brain corresponds to nonneuronal (glial) cells, as seen in the Mz317-Gal4/elav-Gal80; UAS-mcD8:GFP flies, but it also affects a limited number of nonglial (neuron) cells, as seen in the Mz317-Gal4/repo-Gal80; UAS-mcD8:GFP flies. Three neurons and their axons affect a small region close to the antennal glomeruli. Scale bar, 100 μm.
(B) The main glomeruli that responded to the 3 odorants used in optogenetic experiments are shown in color according to DOOR 2.0.43 Scheme modified from.44
(C) Detail of GFP marker expression close to the antennal lobe.
(D and E) Optogenetic activation of MZ317 glial cells, excluding neurons (in recombinant lines MZ317-Gal4, elav-Gal80; 42 in green and 24 in magenta), by channel rhodopsin recapitulates the response observed with the original Mz317-Gal4 driver to ethyl acetate (10−1.5), diminishing olfactory sensitivity (∗∗∗ = p < 0.001; ∗∗ = p < 0.01). All values represent the mean ± SEM of N = 20 for the experiment in (D), and in (E), N correspond to 10, 9, 6, 7, 8, and 7 replicate tests, respectively, for each group from left to right.
However, to rule out the effect of neurons on the described behavioral phenotype, optogenetic activation was repeated in two different fly groups (derived from lines 42 and 24). Lines 42 and 24, generated independently, contain a recombinant chromosome including the Mz317-Gal4 and elav-Gal80 inserts. Therefore, in hybrids with UAS-ChR2XXL, channel rhodopsin expression is restricted to nonneuronal cells within Mz317 cells.
It was observed (Figures 4D and 4E) that the response of the Mz317-Gal4, elav-Gal80/UAS-ChR2XXL flies replicates that of the original Mz317-Gal/UAS-ChR2XXL flies (for hybrids of the 42 line, F5,114 = 18.044∗∗∗, p < 0.001; for hybrids of the 24 line, where fewer replicate tests were performed, F5,41 = 11.329∗∗∗, p < 0.001), which confirms that neurons are not involved in the observed response changes.
Identification of genes involved in the Mz317-glia and OSN interaction by gene silencing with RNAi
Thus far, we have made modifications in Mz317 glial cells that affect the response of OSNs. These changes can be used to infer the existence of a relationship between OSNs and Mz317 glia and could also explain the appearance of these phenotypes in a disease scenario, but we cannot be sure that this interaction occurs during the normal functioning of olfactory reception. That is, we cannot be sure that Mz317 glia are involved in the olfactory signaling generated at the receptor level and transmitted to the brain under natural conditions.
To explore this possibility, we expressed interfering RNAs targeting several genes that have been reported to be related to different mechanisms for the interaction between SGCs and sensory neurons in the dorsal root ganglion of mammals6 using the Gal4/UAS system. In our case, the shakB, VGlut, Irk1, Irk2, and Eaat2 genes that affect gap junctions (shakB); the functioning of glutamatergic signaling (VGlut); K+ channels (Irk1, Irk2); and a taurine/aspartate transporter (Eaat2) that is orthologous to glutamate transporters in vertebrates45 were chosen as candidate genes.
These RNAis block the expression of the mRNAs of their own genes, so they will only have an effect on behavior if the RNA of the corresponding gene is already present in the cell. In addition, the investigation of specific genes can point to the pathways by which neuron-glia interactions occur.
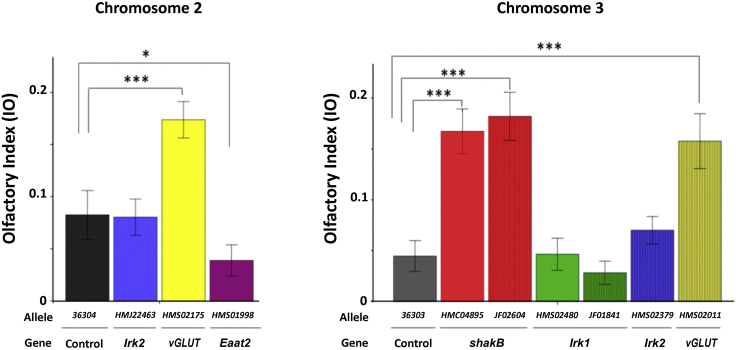
For every gene, we tested 2 different UAS-RNAi lines; some lines have the insertion containing the corresponding UAS-RNAi-gene construct in chromosome 2 and others in chromosome 3 of Drosophila. In only 1 case, the Eaat2 gene, a single line, was tested because it was the only line available. The olfactory behavior of the experimental hybrid Mz317-Gal4/UAS-RNAi-Gene in the T-maze was compared with that of the corresponding controls for chromosome 2 or chromosome 3 (Figure 5) generated with the same insertion program, TRiP,46,47 and enhanced with the expression of Dicer (see the STAR Methods section).
Figure 5.
Effects of RNAi silencing of several candidate genes in MZ317 cells on olfactory behavior in response to ethyl acetate (10−1.5)
RNAi lines were generated by the TRiP program, which also produced control lines for chromosome 2 and chromosome 3. We used 2 UAS-RNAi stocks for each gene except for Eaat2. In all cases, we compared the experimental flies, Mz317-Gal4/UAS-RNAi-GeneX, against the corresponding controls, Mz317-Gal4/UAS-36304 and MZ317-Gal4/UAS-36303, for chromosomes 2 and 3, respectively. The mean ± SEM values are presented. N = 20 for each fly type. T test values are from left to right; for chromosome 2, t-Irk238 = 0.107, n.s.; t-Eaat238 = 2.226∗; t-vGLUT38 = −4.417∗∗∗; for chromosome 3, t-shakB-HM38 = −6.491∗∗∗; t-shakB-JF38 = −6.912∗∗∗; t-vGLUT38 = −5.159∗∗∗; t-Irk1-HM38 = −0.194, n.s.; t-Irk1-JF38 = −1.122, n.s.; t-Irk2 38 = −1.172, n.s. Significant differences of each fly type compared with the control flies are indicated in the figure (∗∗∗ = p < 0.001; ∗∗ = p < 0.01; ∗ = p < 0.05).
For 3 of the 5 genes tested, there were significant differences in the olfactory perception of EA due to decreased gene expression induced by RNAis. In the cases in which two RNAi lines of a gene were tested, both lines had the same result. Thus, diminishing the gene expression of Irk1 and Irk2, which encode K+ channels, does not seem to affect the olfactory behavior under these conditions, but gene silencing of VGlut and ShakB decreases sensitivity, and Eaat2 increases it.
Therefore, neuron-glia interactions also seem to occur under natural conditions in the olfactory reception. Moreover, as we observed in the above optogenetic activation and inhibition experiments, modulation can occur in both directions, decreasing and increasing olfactory sensitivity.
Mz317 antennal glia response to odor is opposite and contingent to the response of the ORNs
To determine the possible origin of the neuron-glia interaction at the level of olfactory reception during olfactory perception, the functional changes produced in the antenna in response to odorant stimuli were observed by Ca2+ imaging. Selective expression of the Ca2+ sensor GCaMPf in OSNs or in Mz317 glia using a binary expression system, either the Gal4/UAS or the lexA/lexA-operator system, allowed us to identify the effects produced in each cell type in response to odor.
Measurements were performed on whole antennae of live, immobilized individuals subjected to gas-phase 1.5-s odor pulses, as in natural conditions.
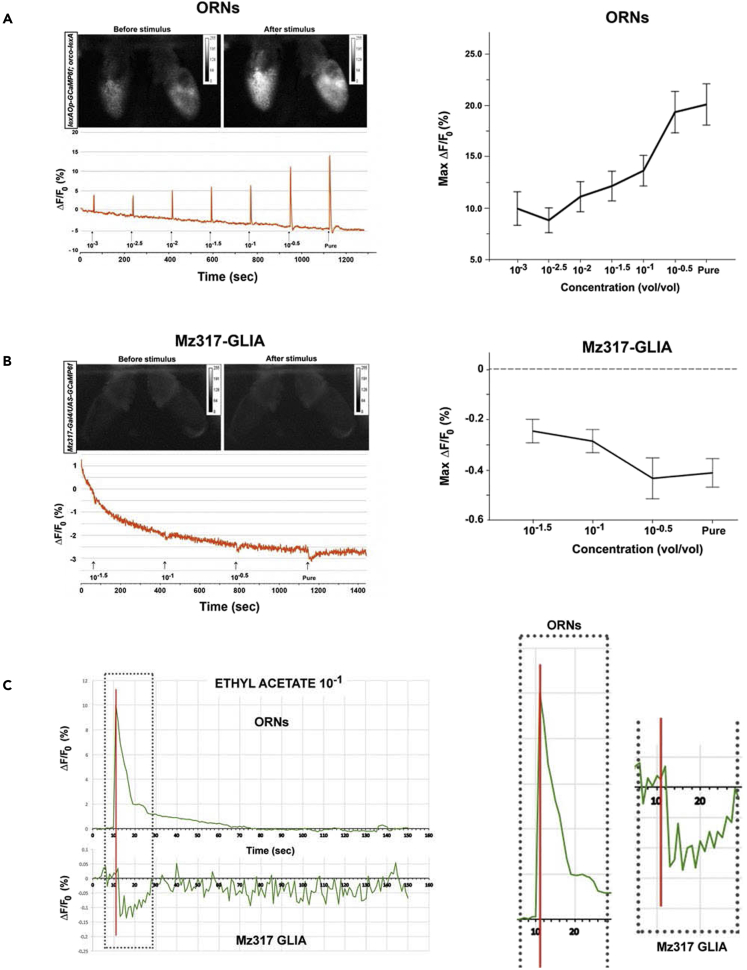
Figure 6A shows the response of ORNs (the OSNs that express the Orco coreceptor) to 1.5-s odor pulses of increasing concentrations of EA by directed expression of the GCaMPf Ca2+ sensor (lexAOp-GCaMPf; orco-lexA) in these cells, as indicated by changes in fluorescence. First, we observed a representative original trace from 1 antenna. On the right, the average dose-response curve obtained from a total of 20 antennae is displayed. A strong and increasing Ca2+ response to odor that was dependent on the odorant concentration was observed. A statistical analysis of the response showed significant differences between the responses to different odorant concentrations (F6,114 = 21.47∗∗∗, p < 0.001).
Figure 6.
Ca2+ imaging of the third antennal segment in response to the odorant ethyl acetate
(A) Left, representative fluorescence response of the ORNs of a fly subjected to increasing odorant concentrations. Right panel, dose–response curve of the maximal response plotted as the mean ± SEM of 20 antennae.
(B) Left, representative fluorescence response of the Mz317 glial cells of a fly subjected to increasing odorant concentrations. Right panel, dose-response curve of the maximal response (mean ± SEM of 20 antennae).
(C) Left, averaged rectified traces of 20 antennae each for ORNs and Mz317 glial cells in response to 1.5 s pulses of ethyl acetate (10−1). Note that the same scale is used for the x-axis (time) but not for the y-axis. Right, temporal details of the response kinetics for both cell types. The red line indicates the time when the response achieves the maximal value for the ORNs.
We also measured the changes in fluorescence produced in the Mz317 glial cells in response to increasing concentrations of odor, Mz317-Gal4/UAS-GCaMPf (Figure 6B). In this case, the odorant pulses induced a decrease in Ca2+ in the opposite direction of the neuronal response, but this effect also increased in response to increasing concentrations of the odorant EA (F3,57 = 7.86∗∗∗, p < 0.001). The response was much smaller than that observed in the ORNs, but the way the measurements were made may be, at least, partly responsible for this difference. We should not forget that the measurement was made over the entire area of the third antennal segment in each photo, and the structure of the Mz317 antennal glia is much thinner and represents a much smaller percentage of the antenna than the ORN somas. However, the response to odor pulses, although small, can be observed in each single trace (Figure 6B, left), and it is not due to the air pulse or any other artifact (see Figure S2 showing the responses to pulses of air, paraffin oil, and 2 different concentrations of the odorant EA).
Other measurement systems that directly analyze the response of ORN somas in sliced antennae48,49,50 cannot be used to analyze Mz317 glia, as the area encompassing the glia cannot be clearly defined. Furthermore, given the network structure of Mz317 antennal glia, the slicing of the antenna itself may affect the glial function.
The kinetics of the response in both cell types were analyzed on the basis of the mean rectified traces of the response of 20 antennae to odor. Figure 6C displays the responses of both cell types to EA (10−1), but the same conclusions can be drawn for the other concentrations. The details on the right show that the response of Mz317 glia occurs later since it has not yet started when the response of ORNs has reached its maximum. This may be because the two cell types have different response dynamics, but it may also indicate that the response of Mz317 glia is not autonomous but is produced by neuronal activation.
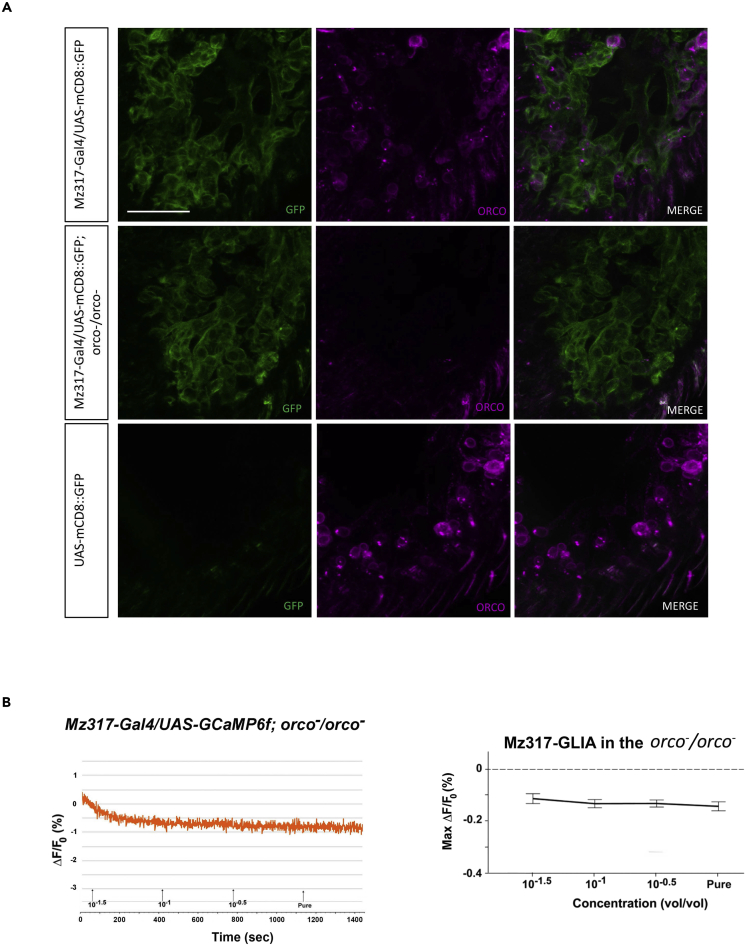
To test this hypothesis, we repeated the experiment by measuring fluorescence in Mz317 glial cells in flies deficient for the Orco coreceptor (Mz317-Gal4/UAS-GCaMPf; orco−/orco−) whose ORNs do not respond to odors. It has been shown that the absence of Orco does not affect the structure of Mz317 glia (Figure 7A). Figure 7B shows that there is no response to odors in the Mz317 glia. In fact, there were no significant differences among the responses to several concentrations of EA (F3,57 = 1.23, n.s.), which confirms that the glial response is not autonomous but is contingent on that of the ORNs. All this evidence suggests a regulation system in which Mz317 glia try to control the activation of ORNs by avoiding an excessive response that might become lethal.
Figure 7.
The absence of Orco in orco−/orco− mutants does not affect the structure of antennal Mz317 glia but eliminates the response of Mz317 antennal glial cells
(A) Detail of the immunostaining of 14-μm antennal cryosections of several fly genotypes. Green (GFP) immunostaining labeling of Mz317 glia and magenta labeling of the Orco coreceptor in Mz317-Gal4/UAS-mCD8:GFP flies were used as positive controls, Mz317-Gal4/UAS-mCD8:GFP; orco-/orco-experimental flies and UAS-mcD8:GFP as negative control flies. The scale bar indicates 40 μm. The same Mz317 glial structure appears in the positive control and the orco−/orco− mutants.
(B) Left, representative fluorescence response of the Mz317 glial cells of an orco−/orco− mutant fly submitted to increasing odorant concentrations. Right, dose-response curve of the maximal response (mean ± SEM of 20 antennae) at the times when the response of Mz317 glial cells was observed. No significant response was observed among different odorant concentrations, and the difference from the 0 value corresponds to signal noise.
Some of the possible mechanisms of this interaction will be related to those signaled by the genes in the RNAi experiment.
Discussion
Although an increasing number of articles have reported a functional role for glia in neuronal signaling in the CNS, less is known about neuron-glia interactions in the peripheral nervous system and, more specifically, concerning sensory systems, whether they affect sensory signaling at the receptor level and whether these changes are projected to the perception level.
In this report, we address this subject by questioning whether glia surrounding the OSN soma may play a role in the detection and/or signaling of odor information to the brain in a model species with a well-differentiated olfactory system, Drosophila melanogaster. Moreover, to elucidate the ultimate effect of peripheral neuron-glia interactions on olfactory perception, we generated individuals with targeted genetic modifications in Mz317-type glial cells and used whole, live flies as experimental subjects, taking advantage of the available genetic tools for this species (see the articles by Martín and del Valle Rodríguez et al.51,52).
Mz317 glia and olfactory perception
In this work, we used the Gal4/UAS binary system to control gene expression by using the Mz317-Gal4 driver. It directs the expression of markers, genes, and other molecules to a type of antennal perineural glia (aPNG) that surrounds the somas of olfactory receptor neurons (OSNs) and the axon bundles surrounded by another type of glia, the subperineural glia that can be detected in the antenna with the GH146-Gal4 driver.36
Regarding the types of manipulations performed on Mz317 cells, we started with those of greatest impact, eliminating the cells by targeted expression of the diphtheria toxin gene.
At the level of signal reception, other studies have reported that in vertebrates, reactive gliosis occurs, i.e., sudden death of glial cells can lead to neuronal malfunction. Thus, when damage occurs in the retina leading to gliosis, the membrane potentials of neurons are altered, leading to strong and long-lasting depolarization of neurons, accompanied by excessive Ca2+ influx and finally excitotoxic neuronal death.53
In our study, we observed that cell death of Mz317 cells starting from the embryonic stage leads to nonviability of individuals. However, death restricted to the adult stage is not lethal and allows us to test the olfactory behavioral response of genetically modified individuals. Thus, adult individuals lacking Mz317 cells, although still able to respond to odors in a double-choice T-maze, do so less efficiently, as higher concentrations of odorant are required to elicit the same response observed in control individuals. This alteration affects not only the most intense ones but also a wide range of odorant concentrations.
Thus, our data support the idea that Mz317 glial cells are involved in the generation of the olfactory signal that is reflected in the results of olfactory perception, which is the relevant biological response for the individual.
In an attempt to more precisely define the role of glia in olfactory perception, as detected by behavioral tests, we then used methods that affect Mz317 cells in a more timely manner and that can produce a reversible effect, optogenetic methods.54,55,56 In this case, we could test whether modifying the transmembrane potential of Mz317 cells via the targeted expression of ion channels that open under the effect of intense light, and whose duration we can control, affects the signaling of OSNs and, ultimately, the olfactory perception of individuals. Although glial cells do not produce action potentials,57 it was considered that this method could be valid in this case because it would affect glia directly through ionic exchange with the extracellular space, which Mz317 glia cells share with ORNs, or indirectly if the change in membrane potential triggers some of the mechanisms of glia-neuron interaction that also work through the common intercellular space. In fact, experiments studying the close contact between these two cell types using the GRASP technique37,58 indicate that Mz317 glia and ORNs are less than 100 nm apart and that there is no other cell or structure between them, which would confirm that the two cell types share intercellular space. We must recall that in the root ganglion of vertebrates, only 20 nm separate the SGCs from the neurons, and changes in the ionic environment as well as glutamate, aminopeptides, and ATP signaling mediate the glia-neuron somata interaction through this intercellular space.6
Activation of Mz317 cells by light was produced through the opening of rhodopsin channels, which leads to the entry of Na+ and Ca2+ cations, producing a significant decrease in the sensitivity of individuals to the odors studied, EA, 3-octanol, and methylsalicylate, especially at high odorant concentrations. It should be noted that each of these odors primarily activates distinct glomeruli in the antennal lobe.59,60 This finding suggests that this is a general neuron-glia interaction effect at the receptor level that modulates odor signaling.
However, inhibition by Cl− anions induced by light activation of halorhodopsin channels ectopically expressed in this type of glia produces the opposite effect, increasing the sensitivity of individuals and causing them to respond to an odorant concentration as if it were more intense than it truly is. It seems, therefore, that our data would support work advocating the importance of maintenance of the ionic environment for proper functioning and signaling of neurons in the brains of Drosophila.61 In C. elegans, such a relationship has been established between glial cells and mechanosensors.62 Moreover, in C. elegans, the peripheral glia AMsh surrounding the multimodal ASH neuron, a type of polymodal nociceptive neuron involved in the response to harmful stimulus, mediates olfactory adaptation through Ca2+ channels and GABA neurotransmitter signaling.33 Previous work in Drosophila adults using mutants also identified the role of the trp Ca2+ channel in olfactory adaptation at the receptor level.63
However, the expression level of the potassium channels encoded by Irk1 and Irk2 genes in the MZ317 glia does not seem to affect the olfactory response.
Previous reports in vertebrates have proposed changes in the ionic environment as a method, although not the only one by which SGCs interact with the neuron somata they envelope.6
Thus, Mz317 glia could modulate the activity of OSNs in both directions, increasing or decreasing sensitivity by altering the intracellular ionic content of the glial cells and the subsequent communication to the OSN somata, which would modify their responsiveness to stimuli.
Genes involved in the interaction of Mz317glia and OSN
In our case, five genes that have been shown to be expressed and involved in neuron-glia interactions in SGCs enveloping neuronal somata in the sensory ganglia of the spinal cord of humans related to pain,6 the shakB, VGlut, Irk1, Irk2, and Eaat2 genes were chosen as candidate genes.
In this study, a decrease in olfactory perception of EA was observed when the VGlut and shakB genes were silenced in the two RNAi studies for each gene. The shakB protein related to gap junctions has already been described as important in Drosophila olfactory perception at the level of the antennal lobe, specifically in the interaction between projection neurons and local excitatory neurons.30 In mammals, it has been proposed to be important in communication between glial cells that wrap the same neuron but not between glia and the neuronal soma they surround.6 Thus, under electronic microscopy, gap junctions have been found between SGCs but not between SGCs and neurons. However, in neuropathic pain conditions, both types of gap junctions have been found64,65 although the strongest unions were from the SGC-SGC type.66
In our case, further experiments should be performed to answer this question, but it will be difficult to explain changes in the Ca2+ concentration in opposite directions as the ones observed by Ca2+ imaging between cells united by gap junctions.
The importance of the VGlut gene for the functioning of glutamatergic neurons has also been noted, with one-third of the neurons present in the antennal lobe of Drosophila adults being glutamatergic.67,68 Our results suggest that a spectrum of activities occur through these genes at the level of reception in the antenna.
Furthermore, in this work, we describe an increase in olfactory sensitivity to EA when the Eaat2 gene is silenced, in the opposite direction to that observed for the previous two genes. This gene encodes a taurine/aspartate transporter and is an ortholog of glutamate transporters in vertebrates.45 Its expression has been described in glial cells of the CNS of Drosophila embryos,69 and it is an important gene in olfactory perception, as a mutant for the gene showed decreased the aversive behavior to propionic acid.70 Although we targeted gene silencing toward Mz317 cells in our study, other authors70 used a mutant for the gene in the whole organism. Similarly, the expression of the gene in Drosophila brain-wrapping glia has been reported to modulate sleep and the metabolic rate.71 Our study also links this gene to olfactory reception.
This result not only allows us to confirm that the neuron-glia interaction occurs under natural conditions in the olfactory system of adult D. melanogaster but also indicates the involvement of certain components and pathways in this interaction, the electrical synapses (ShakB) as well as other factors related to cell excitation (VGlut and Eaat2), and the possibility that the glia modulate neuronal activity in both directions, either increasing or decreasing their sensitivity.
A more extensive study of the genes expressed by Mz317 glial cells should be performed to expand our understanding of the mechanisms of the OSN-Mz317 glia interaction.
Constraints of behavioral studies with Mz317 cells and their relationship to changes associated with aPNG
However, although behavioral tests may be the most appropriate tests to determine the importance of the observed modifications at the cellular or molecular level on the functional capacity of the individual, they have limitations in terms of the conclusions that can be drawn. The behavior involves the entire sensory circuit, and to designate a particular part of this circuit as responsible for the observed behavioral modifications, it must be ensured that other parts of the circuit are not modified or, even if they are, that they do not alter the function.
In our case, we found that the Mz317-Gal4 insert we used as an expression driver not only labels the antennal glia but also shows labeling in some areas of the brain, in the glia surrounding some structures that do not seem to affect elements of the olfactory pathways and in a small number of neurons (Figure 4A), some of them related to a small portion of the antennal lobe that could affect the olfactory circuit.
Because labeling is an indicator of those cells in which there are Mz317-Gal4 target gene modifications, cell death, optogenetic variations, or RNAi expression also occurred in these cells in the brains of the individuals who showed behavioral differences. The possible role of neurons in the observed olfactory perception changes was ruled out with 2 different sets of data. First, we showed the same behavioral effect in response to three different odors, EA, 3 octanol, and metyl salycilate, after optogenetic activation. Because the main glomeruli that respond to each of the 3 odors are different and located in different regions of the antennal lobes and at different distances from the region where the branches of the labeled neurons reach, it is unlikely that optogenetic activation of the neurons is the cause of the observed behavioral differences.
Second, and more definitively, the behavior observed in experimental flies where the Mz317-Gal4 driver has been restricted to target only glial cells (Mz317-Gal4, elav-Gal80; UAS-ChR2XXL) replicates that of the original flies without such a restriction. Therefore, we showed that glial cells are responsible for the behavioral phenotype.
Analysis of cellular activity in the third antennal segment in response to odor pulses
Finally, if the Mz317 neuron-glia interaction at the level of olfactory receptor organs is the origin of the observed differences in olfactory perception, we must be able to relate the functional response at the antennal level of both cell types to the olfactory stimulus.
For this purpose, Ca2+ imaging measurements were performed in the third antennal segments of live and immobilized individuals in response to gaseous odor pulses as close as possible to natural conditions. In Drosophila, this technique has previously been used to view the activity of OSNs of the third antennal segment72 although it is mainly used to observe activity at the antennal lobe level in the brain73,74,75 and in the olfactory bulb of vertebrates.76,77 We performed the measurements on intact antennae from immobilized individuals, unlike other types of preparations, such as those performed by others48,49,50 on ex vivo antennal sections. This may affect the results obtained because, on the one hand, it is not as easy to define the cell boundaries in MZ317 glia as in ORNs to determine the specific region to explore fluorescence changes, and on the other hand, by sectioning the antenna, we would disrupt the tightly interrelated network structure of aPNG glia cells in the third antennal segment. In this sense, measuring fluorescence changes over the entire third antennal segment will give us smaller absolute values than those specifically targeting individual cells. However, the results we obtain are robust because they can be observed at the single trace level.
We showed that both ORNs and Mz317 glia respond to odor pulses and that the response is concentration-dependent. ORNs increase while Mz317 glia reduce the Ca2+ concentration. However, some important differences appear compared with the report on the response of receptor neurons and glia to aversive odorants in a model species, C. elegans.33 In Caenorhabditis, both ASH neurons (a type of polymodal nociceptive neuron) and AMsh glia sense aversive odorants but always in the direction of increasing Ca2+ concentration. In this case, each cell type presents its own odorant receptors and responds in a cell autonomous manner. This is not the case for Drosophila, where suppression of ORN activation in orco−/orco− mutants eliminates the response of Mz317 glia.
To understand these differences, we must consider the biology of both species. C. elegans is a simpler animal in which the sensory modalities are not as differentiated as in Drosophila. Thus, for example, ASH neurons are a type of multimodal nociceptive neuron that respond to various types of aversive, chemical, high osmotic, and mechanical stimuli.78,79,80,81 In addition, the structure formed by ASH neurons and AMsh glia is not reminiscent of Drosophila olfactory sensilla. Thus, in Caenorhabditis AMsh, glia surround the soma and dendrites of ASH neurons almost to the outside (see the articles by Bacaj et al. and Zhang and Yan82,83) while in Drosophila, the olfactory sensilla contain the OSNs and the accessory and supporting cells surrounding the neuron and part of the dendrites,34,35,84,85 isolating them from the intercellular space at the soma level. It is at this level that Mz317 glia act, closely surrounding the neuronal body, according to the GRASP experiment, and form a network structure with other glial cells surrounding the soma of other OSNs along the third antennal segment. Therefore, if Mz317 glia do not reach the outside of the sensilla and are isolated from the external lymph where olfactory stimuli arrive, it is logical that they do not have an autonomous but an indirect response to neuronal activation that also happens to be opposite. This kind of opposite response between glia and neurons has been recently reported for the microglia of rodents that show a response to chemotactic cues in the CNS. In that case, and opposite to neurons, the driving force for the response is hyperpolarization, which involves Ca2+ entry from the intracellular space, possibly mediated by ATP.86
It was reported long ago in Drosophila87 that OSNs show a greater resistance to excessive excitation than photoreceptors. Thus, mutants of the rdgB gene cause photoreceptor death and degeneration after the first response to light, while the OSNs showed only delayed recovery after intense odorant stimulation. Our results allow us to propose that the action of the MZ317 may be at least in part responsible for the adaptation phenomenon at the receptor level that prevents this overstimulation.
In C. elegans, the AMsh glial response can inhibit the activation of ASH neurons mediating adaptation33; in our case, moreover, the global network structure of the Mz317 glia throughout the third antennal segment further suggests a broader role in tuning olfactory activation in highly odorous environments.
Limitations of the study
The topic addresses the role of glial cells as active agents in neuronal signaling in a well-organized sensory system. The approach involves the use of living, whole individuals in which genetic modifications targeting glia have been made so that the function of glia can be studied without additional manipulations in experimental animals. This allows us to answer the question whether alterations at the receptor level are relevant for the final sensory coding of an olfactory stimulus and are reflected in the sensory perception of individuals. This is deduced by behavioral tests in which the animals move freely. Therefore, the conclusions correspond to what occurs under natural conditions.
However, this approach also has its limitations. On the one hand, the use of the Gal4/UAS binary system of directed gene expression targets the expression to all cells indicated by the Gal4 insert, which, in our case, include both cells of the third antennal segment and others in the brain, such as glial cells and some neurons. In this report, we have paid special attention to rule out the effect of cells other than those belonging to the olfactory receptor organ on the observed behavioral responses.
On the other hand, the network nature of MZ317 glia in the third antennal segment limits the type of Ca2+ imaging studies that can be performed. Thus, imaging studies of olfactory function in sliced antenna performed by other authors48,49,50 that allow a more detailed study in single cells and the use of a pharmacological approach with inhibitors cannot be used in the present work without compromising the pooled glial function.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Invitrogen | Cat#A6455; RRID: AB_221570 |
| Alexa 488 polyclonal anti-rabbit | Invitrogen | Cat# A11008; RRID: AB_143165 |
| Mouse monoclonal anti-nc82 | Developmental Studies Hybridoma | Cat# Supernatant 1 ml; RRID: AB_2314866 |
| Cy3 polyclonal anti-mouse | Jackson Immunoresearch Laboratories | Cat# 715-165-151; RRID: AB_2315777 |
| Mouse monoclonal anti-GFP | Invitrogen | Cat#A11120; RRID: AB_221568 |
| Rabbit polyclonal anti-ORCO | Larsson et al.88 | N/A |
| Alexa 488 polyclonal anti-mouse | Invitrogen | Cat#A21131; RRID: AB_2535771 |
| Cy3 polyclonal anti-rabbit | Jackson Immunoresearch Laboratories | Cat#111-165-003; RRID: AB_2338000 |
| Chemicals, peptides, and recombinant proteins | ||
| Ethyl acetate | Sigma-Aldrich | Cat# 1.00863.0500; CAS: 141-78-6 |
| 3-octanol | Sigma-Aldrich | Cat# 218405-50G; CAS: 589-98-0 |
| Methylsalicylate | Fluka | Cat# 76631; CAS: 119-36-8 |
| Paraffin oil | Merck | Cat# 1.07174.2500; CAS: 8042-47-5 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: Canton-S | Bloomington Drosophila Stock Center | BDSC:64349; FlyBase: FBst0064349 |
| D. melanogaster: Mz317-Gal4: w; Mz317-Gal4/CyO | Ito et al.89 | N/A |
| D. melanogaster: orco-lexA: w; Bl/CyO; orco-lexA/TM6B | Lai and Lee90 | N/A |
| D. melanogaster: repo-Gal80: w; repo-Gal80 (N18)/CyO | Awasaki et al.91 | N/A |
| D. melanogaster: elav-Gal80: w; elav-Gal80/CyO; rotund-Gal4, UAS-GFPnls/TM6 | Casas-Tintó et al.92 | N/A |
| D. melanogaster: Gal80ts: w[∗]; P{w[+mC] = tubP-GAL80[ts]}2/TM2 | Bloomington Drosophila Stock Center | BDSC:7017; FlyBase: FBst0007017 |
| D. melanogaster: UAS-DTI: w[∗]; P{w[+mC] = UAS-Cbeta∖DT.I}18/CyO | Bloomington Drosophila Stock Center | BDSC:25039; FlyBase: FBst0025039 |
| D. melanogaster: UAS-ChR2XXL: y[1]w[1118]; PBac{y[+mDint2] w[+mC] = UAS-ChR2.XXL}VK00018 | Bloomington Drosophila Stock Center | BDSC:58374; FlyBase: FBst0058374 |
| D. melanogaster: UAS-eNpHR: y[1] w[∗]; wg[Sp-1]/CyO, P{Wee-P.ph0}Bacc[Wee-P20]; P{y[+t7.7] w[+mC] = 20XUAS-eNpHR3.0.YFP}attP2/TM6C, Sb[1] Tb[1] | Bloomington Drosophila Stock Center | BDSC:36368; FlyBase: FBst0036368 |
| D. melanogaster: RNAi of dEaat2: y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HMS01998}attP40/CyO | Bloomington Drosophila Stock Center | BDSC:40832; FlyBase: FBst0040832 |
| D. melanogaster: RNAi of shakB V20: y[1] sc[∗] v[1] sev[21]; P{y[+t7.7]v[+t1.8] = TRiP.HMC04895}attP2 | Bloomington Drosophila Stock Center | BDSC:57706; FlyBase: FBst0057706 |
| D. melanogaster: RNAi of shakB V10: y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.JF02604}attP2 | Bloomington Drosophila Stock Center | BDSC: 27292; FlyBase: FBst0027292 |
| D. melanogaster: RNAi of VGlut chr 2: y[1] sc[∗] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.HMS02175}attP40 | Bloomington Drosophila Stock Center | BDSC: 40927; FlyBase: FBst0040927 |
| D. melanogaster: RNAi of VGlut chr 3: y[1] sc[∗] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.HMS02011}attP2 | Bloomington Drosophila Stock Center | BDSC: 40845; FlyBase: FBst0040845 |
| D. melanogaster: RNAi of Irk1 v20: y[1] sc[∗] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.HMS02480}attP2 | Bloomington Drosophila Stock Center | BDSC: 42644; FlyBase: FBst0042644 |
| D. melanogaster: RNAi of UAS-Irk1 v10: y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.JF01841}attP2 | Bloomington Drosophila Stock Center | BDSC: 25823; FlyBase: FBst0025823 |
| D. melanogaster: RNAi of Irk2 chr 2: y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HMJ22463}attP40 | Bloomington Drosophila Stock Center | BDSC: 58333; FlyBase: FBst0058333 |
| D. melanogaster: RNAi of Irk2 chr 3: y[1] sc[∗] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.HMS02379}attP2 | Bloomington Drosophila Stock Center | BDSC: 41981; FlyBase: FBst0041981 |
| D. melanogaster: Control TRiP chr 2: y[1] v[1]; P{y[+t7.7] = CaryP}attP40 | Bloomington Drosophila Stock Center | BDSC: 36304; FlyBase: FBst0036304 |
| D. melanogaster: Control TRiP chr 3: y[1] v[1]; P{y[+t7.7] = CaryP}attP2 | Bloomington Drosophila Stock Center | BDSC: 36303; FlyBase: FBst0036303 |
| D. melanogaster: UAS-Dicer: P{w[+mC] = UAS-Dcr-2.D}1, w[1118]; betaTub60D[Pin-1]/CyO | Bloomington Drosophila Stock Center | BDSC: 24644; FlyBase: FBst0024644 |
| D. melanogaster: UAS-GCaMP6f: w[1118]; P{y[+t7.7] w[+mC] = 20XUAS-IVS-GCaMP6f}attP40 | Bloomington Drosophila Stock Center | BDSC: 42747; FlyBase: FBst0042747 |
| D. melanogaster: lexAOp-GCaMP6f: w[1118]; P{y[+t7.7] w[+mC] = 13XLexAop2-IVS-GCaMP6f-p10}su(Hw)attP5 | Bloomington Drosophila Stock Center | BDSC: 44277; FlyBase: FBst0044277 |
| D. melanogaster: UAS-mCD8::GFP chr2: y[1] w[∗]; P{w[+mC] = UAS-mCD8::GFP.L}LL5, P{UAS-mCD8::GFP.L}2 | Bloomington Drosophila Stock Center | BDSC: 5137; FlyBase: FBst0005137 |
| D. melanogaster: UAS-mCD8::GFP chr3: y[1] w[∗]; betaTub60D[Pin-Yt]/CyO; P{w[+mC] = UAS-mCD8::GFP.L}LL6 | Bloomington Drosophila Stock Center | BDSC: 5130; FlyBase: FBst0005130 |
| D. melanogaster: orco-RFP: w[∗]; P{w[+mC] = UAS-mCD8::GFP.L}LL5/CyO; P{w[+mW.hs] = Orco-RFP.K}10D | Bloomington Drosophila Stock Center | BDSC: 63045; FlyBase: FBst0063045 |
| D. melanogaster: UAS-spGFP1-10, lexAOp-spGFP11: w[∗]; P{y[+t7.7] w[+mC] = CoinFLP-LexA::GAD.GAL4}attP40 P{w[+mC] = lexAop-rCD2.RFP}2; P{w[+mC] = UAS-CD4-spGFP1-10}3, P{w[+mC] = lexAop-CD4-spGFP11}3/TM6C, Sb[1] | Bloomington Drosophila Stock Center | BDSC: 58755; FlyBase: FBst0058755 |
| Software and algorithms | ||
| ImageJ | Schneider et al.93 | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information should be directed to Dr. Esther Alcorta (ealcorta@uniovi.es).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
The experimental model used was Drosophila melanogaster. All genotypes used have been listed in the Reagents and Resources section. Female flies were used in functional studies as they present stable and robust olfactory responses.42
Two-to four-day-old females fasted for the previous 24 h were tested for olfactory preference in the T-maze.94,95 For the Ca2+ imaging experiments two-to ten-day-old females were used.
Flies and crosses were maintained in 220 c.c. bottles with standard yeast-sucrose medium in a thermoregulated chamber at 25 ± 1°C following a 12:12 h light-dark cycle. For the optogenetics experiments the medium was supplemented with all-trans-retinal 300 μM (Sigma-Aldrich, Germany) and the flies were kept in the dark as previously described.42
The Spanish Ministry for Ecological Transition and Demographic Challenge granted all permits needed for the use of genetically modified flies (A/ES/21/I-30).
Method details
Odor presentation
For the behavioral experiments, odorant stimuli and controls were provided in filter paper present in the corresponding S and C tubes.
For the Ca2+ imaging experiments, we used air currents instead. Individuals were maintained in a constant air flow of approximately 300 mL/min. Odor pulses were generated by partially diverting that flow into a tube with 25 μL of the appropriate dilution of the odorant on filter paper with an electrically activated valve (150 mL/min, 50% of the total flow). This system prevents artifacts associated with the cessation of air at any point.96 Odor pulses occurred for 1.5 s. The time between pulses varied between 120 and 360 s depending on the genotype used because of the different kinetics of the response and recovery after the stimulus.
Behavioral assay
Flies introduced to the initial tube (I) were forced to a chamber that acted as an elevator and placed the flies between a stimulus tube (S) containing 0.5 mL of a certain concentration (v/v) of odorant diluted in paraffin oil and a control tube (C) containing only 0.5 mL of the solvent. Flies were allowed to choose one of the tubes for 1 min. The olfactory index (IO) is defined as the ratio of the flies in S compared to the total number of flies that chose one of the two tubes, S or C. IO values between 0 and 0.5 indicate repulsion, and IO values between 0.5 and 1 indicate attraction by the odor. The 0.5 value means indifference. Most experiments were carried out at odorant concentrations that correspond to the repellent region of the dose–response curve because in the attractive zone, this curve has a bell shape, and 2 different odorant concentrations may evoke the same IO value and induce confusion, which is not the case in the repellent region, where each IO value corresponds to a single odorant concentration.
All replicate tests were carried out at constant temperature and relative humidity (24 ± 1°C and 40-60% RH) between 4 and 7 p.m. To avoid cross effects on the response due to lateral preference, the stimulus was placed alternately on the right and left side of the maze. In addition, the control and experimental groups were alternated to avoid a possible "time effect" throughout the testing period.
For optogenetic experiments, a previously described setup was used.42 In short, an intense white LED light of 200 W, 4000 K and 18,000 lumens (Alverlamp LSPR0200W40, Valencia, Spain), which serves to stimulate both channel rhodopsin and halorhodopsin, was placed 22 cm perpendicular to and illuminated both election tubes. Flies subjected to the light stimulus were subjected to 5 min of light before the test because it yielded more consistent results.
Ca2+ imaging
Drosophila individuals were immobilized in a yellow pipette tip and placed on a slide. The third antennal segments were also immobilized with the help of a stretched glass electrode.
For imaging, we used a Nikon Eclipse FN-1 microscope with a 10× Nikon Plan Fluor objective equipped with a white light source (Nikon Intensilight C-HGFI). An excitation filter of 460–500 nm and an emission filter of 510–550 nm were applied for the GCaMP6f protein used as a Ca2+ sensor and selectively expressed in different cellular subgroups. Monochrome images of 680 × 512 pixels were collected at a 1 Hz frequency with a ProgRes CF cool CCD camera (JENOPTIK, Germany). Odor pulse emission and image capture were synchronized by a computer.
Immunohistochemistry
For antennal preparations, adult flies were anesthetized and placed in a collar,97 in which they were submerged in OCT and frozen for subsequent cryocutting. Two different protocols were used to obtain antennal cryosections, and thick (40 μm) and thin (14 μm) cryosections were made.
The thick sections were used to observe native fluorescence generated by GFP and RFP gene expression under a confocal microscope (Figure 1A). For the thin sections, immunohistochemistry was applied to detect the GFP marker according to a previous protocol.98 Brain preparation squashes were performed following a previous protocol.60
The thick antennal cryosections were used to observe native fluorescence generated by GFP and RFP gene expression under a confocal microscope (Figure 1A). For the thin sections, immunohistochemistry was applied to detect the GFP marker according to a previous protocol.98 The primary antibody used was rabbit anti-GFP (1:5000; Invitrogen), and the secondary antibody was Alexa 488 anti-rabbit (1:1000; Invitrogen).
Rabbit anti-GFP (1:2500; Invitrogen) and mouse anti-nc82 (1:10; Developmental Studies Hybridoma Immunohistochemical assays for GFP expression in brain preparations were performed following.60 Rabbit anti-GFP and mouse anti-nc82 were used as primary antibodies, and Alexa 488 anti-rabbit (1:100; Invitrogen) and Cy3 anti-mouse (1:100; Jackson ImmunoResearch Laboratories) were used as secondary antibodies. Images of antennal cryosections and brain squashes were collected with a Leica TCS-SP8X Confocal Laser Microscope (Leica Microsystems) and analyzed with ImageJ software (NIH).
To analyze the structure of MZ317 antennal glial cells in the ortho background, we used mouse anti-GFP (1:5000; Invitrogen) and rabbit anti-ORCO (1:10,000; kindly provided by Dr. L. Vosshall, The Rockefeller University, New York) diluted in PTS as primary antibodies. The secondary antibodies Alexa 488 anti-mouse (1:1000; Invitrogen) and Cy3 anti-rabbit (1:1000; Jackson ImmunoResearch Laboratories) were used to show green and magenta immunostaining, respectively.
Quantification and statistical analysis
Behavioral tests
As a rule, 20 replication tests were performed for each concentration, condition and genotype. ANOVA was performed for statistical analysis, followed by post hoc comparison of means a posteriori. When only 2 lines were compared, Student’s t test was used. In the case of gene silencing with interfering RNA, direct comparisons were made between the hybrids of each gene and the corresponding control hybrids, either of chromosome 2 or chromosome 3, using Student’s t test. Calculations were computed using SPSS software (IBM, USA).
Ca2+ imaging
The images were analyzed for the third antennal segment area using ImageJ (NIH)) to obtain the mean fluorescence intensity values on a scale between 0 and 255. Functional changes were measured by the change in fluorescence as follows48,49,72,99:
where F0 represents the average fluorescence for the 10 frames prior to the odorant pulse and F the fluorescence at a given time. Each antenna was recorded for several odorant concentrations in an increasing direction.
For the dose–response curve, the maximal deflection in response to odor pulses was measured and averaged for a total of 20 antennae. To obtain the averaged traces, signals were rectified according to the line generated by the mean of the 10 frames prior to the stimulus and the last 10 frames of the recording for each odorant concentration and antenna.
For statistical analysis, we performed an ANOVA of repeated measures because the same antenna was tested for increasing concentrations of odorant stimuli. This was followed by a post hoc analysis of the means to establish significant differences between each pair of concentrations.
Acknowledgments
We thank W. Leiserson (Yale University, CT, USA), R. Benton (University of Lausanne, Switzerland), T. Lee (Janelia Research Campus, VA, USA), L. Vosshall (The Rockefeller University, New York, USA), and S. Casas-Tintó (IIER-AGH, Instituto de Salud Carlos III, Madrid, Spain) for kindly providing fly stocks and reagents. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This work was supported by the Spanish Ministry of Science and Innovation (SAF2013-48759-P and BFU2017-85882-P) to E.A. and C.G.-D.; the University of Oviedo, Spain (PAPI-17-PEMERG-2 and PAPI-GR-2016-0012) to C.G.-D., F.M., and E.A.; and the Principality of Asturias Government (IDI-2018-000182) to E.A., (PA-20-PF-BF19-040) to A.C.-S., and (PA-20-PF-BF19-042) to L.C.-C.
Author contributions
F.M., C.G.-D., and E.A. designed the research and contributed new setup and analytical tools; L.C.-C., L.R.M., R.C., and A.C.-S. performed the research; L.C.-C. and E.A. analyzed the data; and E.A. wrote the article.
Declaration of interests
The authors declare no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105837.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code. All codes are available via open access tools and resources listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Perea G., Araque A. Glial calcium signaling and neuron-glia communication. Cell Calcium. 2005;38:375–382. doi: 10.1016/j.ceca.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 2.Perea G., Araque A. Synaptic information processing by astrocytes. J. Physiol. Paris. 2006;99:92–97. doi: 10.1016/j.jphysparis.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Kettenmann H., Verkhratsky A. [Neuroglia--living nerve glue] Fortschr. Neurol. Psychiatr. 2011;79:588–597. doi: 10.1055/s-0031-1281704. [DOI] [PubMed] [Google Scholar]
- 4.Martin-Fernandez M., Jamison S., Robin L.M., Zhao Z., Martin E.D., Aguilar J., Benneyworth M.A., Marsicano G., Araque A. Synapse-specific astrocyte gating of amygdala-related behavior. Nat. Neurosci. 2017;20:1540–1548. doi: 10.1038/nn.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanani M., Spray D.C. Emerging importance of satellite glia in nervous system function and dysfunction. Nat. Rev. Neurosci. 2020;21:485–498. doi: 10.1038/s41583-020-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L.-Y.M., Gu Y., Chen Y. Communication between neuronal somata and satellite glial cells in sensory ganglia. Glia. 2013;61:1571–1581. doi: 10.1002/glia.22541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanani M., Verkhratsky A. Satellite glial cells and astrocytes, a comparative review. Neurochem. Res. 2021;46:2525–2537. doi: 10.1007/s11064-021-03255-8. [DOI] [PubMed] [Google Scholar]
- 8.Silies M., Klämbt C. Adhesion and signaling between neurons and glial cells in Drosophila. Curr. Opin. Neurobiol. 2011;21:11–16. doi: 10.1016/j.conb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Stork T., Bernardos R., Freeman M.R. Analysis of glial cell development and function in Drosophila. Cold Spring Harb. Protoc. 2012;2012:1–17. doi: 10.1101/pdb.top067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu B., Li J., Chou Y.-H., Luginbuhl D., Luo L. Fibroblast growth factor signaling instructs ensheathing glia wrapping of Drosophila olfactory glomeruli. Proc. Natl. Acad. Sci. USA. 2017;114:7505–7512. doi: 10.1073/pnas.1706533114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., Sanz E., Li Z., Hui J., Graham B.H., Quintana A., et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng F.S., Tangredi M.M., Jackson F.R. Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr. Biol. 2011;21:625–634. doi: 10.1016/j.cub.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson F.R., Ng F.S., Sengupta S., You S., Huang Y. Glial cell regulation of rhythmic behavior. Methods Enzymol. 2015;552:45–73. doi: 10.1016/bs.mie.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seugnet L., Suzuki Y., Merlin G., Gottschalk L., Duntley S.P., Shaw P.J. Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr. Biol. 2011;21:835–840. doi: 10.1016/j.cub.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W.-F., Maguire S., Sowcik M., Luo W., Koh K., Sehgal A. A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol. Psychiatry. 2015;20:240–251. doi: 10.1038/mp.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki D., Horiuchi J., Ueno K., Ueno T., Saeki S., Matsuno M., Naganos S., Miyashita T., Hirano Y., Nishikawa H., et al. Glial dysfunction causes age-related memory impairment in Drosophila. Neuron. 2014;84:753–763. doi: 10.1016/j.neuron.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 17.Matsuno M., Horiuchi J., Yuasa Y., Ofusa K., Miyashita T., Masuda T., Saitoe M. Long-term memory formation in Drosophila requires training-dependent glial transcription. J. Neurosci. 2015;35:5557–5565. doi: 10.1523/JNEUROSCI.3865-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awasaki T., Lai S.-L., Ito K., Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J. Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doherty J., Logan M.A., Taşdemir O.E., Freeman M.R. Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman M.R. Drosophila central nervous system glia. Cold Spring Harb. Perspect. Biol. 2015;7:a020552. doi: 10.1101/cshperspect.a020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omoto J.J., Yogi P., Hartenstein V. Origin and development of neuropil glia of the Drosophila larval and adult brain: two distinct glial populations derived from separate progenitors. Dev. Biol. 2015;404:2–20. doi: 10.1016/j.ydbio.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremer M.C., Jung C., Batelli S., Rubin G.M., Gaul U. The glia of the adult Drosophila nervous system. Glia. 2017;65:606–638. doi: 10.1002/glia.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildirim K., Petri J., Kottmeier R., Klämbt C. Drosophila glia: few cell types and many conserved functions. Glia. 2019;67:5–26. doi: 10.1002/glia.23459. [DOI] [PubMed] [Google Scholar]
- 24.Danjo R., Kawasaki F., Ordway R.W. A tripartite synapse model in Drosophila. PLoS One. 2011;6:e17131. doi: 10.1371/journal.pone.0017131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss A.L., Kawasaki F., Ordway R.W. A distinct perisynaptic glial cell type forms tripartite neuromuscular synapses in the Drosophila adult. PLoS One. 2015;10:e0129957. doi: 10.1371/journal.pone.0129957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borycz J., Borycz J.A., Edwards T.N., Boulianne G.L., Meinertzhagen I.A. The metabolism of histamine in the Drosophila optic lobe involves an ommatidial pathway: β-alanine recycles through the retina. J. Exp. Biol. 2012;215:1399–1411. doi: 10.1242/jeb.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahman M., Ham H., Liu X., Sugiura Y., Orth K., Krämer H. Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat. Neurosci. 2012;15:871–875. doi: 10.1038/nn.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y., An F., Borycz J.A., Borycz J., Meinertzhagen I.A., Wang T. Histamine recycling is mediated by CarT, a carcinine transporter in Drosophila photoreceptors. PLoS Genet. 2015;11:e1005764. doi: 10.1371/journal.pgen.1005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazama H., Yaksi E., Wilson R.I. Cell death triggers olfactory circuit plasticity via glial signaling in Drosophila. J. Neurosci. 2011;31:7619–7630. doi: 10.1523/JNEUROSCI.5984-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaksi E., Wilson R.I. Electrical coupling between olfactory glomeruli. Neuron. 2010;67:1034–1047. doi: 10.1016/j.neuron.2010.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H., Zhou B., Yan W., Lei Z., Zhao X., Zhang K., Guo A. Astrocyte-like glial cells physiologically regulate olfactory processing through the modification of ORN-PN synaptic strength in Drosophila. Eur. J. Neurosci. 2014;40:2744–2754. doi: 10.1111/ejn.12646. [DOI] [PubMed] [Google Scholar]
- 32.Ma Z., Stork T., Bergles D.E., Freeman M.R. Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature. 2016;539:428–432. doi: 10.1038/nature20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan D., Zhang H., Yue X., Fan Y., Xue Y., Shao J., Ding G., Chen D., Li S., Cheng H., et al. Sensory glia detect repulsive odorants and drive olfactory adaptation. Neuron. 2020;108:707–721.e8. doi: 10.1016/j.neuron.2020.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Shanbhag S.R., Müller B., Steinbrecht R.A. Atlas of olfactory organs of Drosophila melanogaster: 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 1999;28:377–397. doi: 10.1016/S0020-7322(99)00039-2. [DOI] [Google Scholar]
- 35.Shanbhag S.R., Müller B., Steinbrecht R.A. Atlas of olfactory organs of Drosophila melanogaster 2. Internal organization and cellular architecture of olfactory sensilla. Arthropod Struct. Dev. 2000;29:211–229. doi: 10.1016/s1467-8039(00)00028-1. [DOI] [PubMed] [Google Scholar]
- 36.Sen A., Shetty C., Jhaveri D., Rodrigues V. Distinct types of glial cells populate the Drosophila antenna. BMC Dev. Biol. 2005;5:25. doi: 10.1186/1471-213X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feinberg E.H., Vanhoven M.K., Bendesky A., Wang G., Fetter R.D., Shen K., Bargmann C.I. GFP Reconstitution across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 38.Yadav S., Younger S.H., Zhang L., Thompson-Peer K.L., Li T., Jan L.Y., Jan Y.N. Glial ensheathment of the somatodendritic compartment regulates sensory neuron structure and activity. Proc. Natl. Acad. Sci. USA. 2019;116:5126–5134. doi: 10.1073/pnas.1814456116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanani M. Satellite glial cells in sensory ganglia: from form to function. Brain Res. Brain Res. Rev. 2005;48:457–476. doi: 10.1016/j.brainresrev.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Castañeda-Sampedro A., Calvin-Cejudo L., Martin F., Gomez-Diaz C., Alcorta E. The Ntan1 gene is expressed in perineural glia and neurons of adult Drosophila. Sci. Rep. 2022;12:14749. doi: 10.1038/s41598-022-18999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riveron J., Boto T., Alcorta E. The effect of environmental temperature on olfactory perception in Drosophila melanogaster. J. Insect Physiol. 2009;55:943–951. doi: 10.1016/j.jinsphys.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Coya R., Martin F., Calvin-Cejudo L., Gomez-Diaz C., Alcorta E. Validation of an optogenetic approach to the study of olfactory behavior in the T-maze of Drosophila melanogaster adults. Insects. 2022;13:662. doi: 10.3390/insects13080662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Münch D., Galizia C.G. DoOR 2.0--comprehensive mapping of Drosophila melanogaster odorant responses. Sci. Rep. 2016;6:21841. doi: 10.1038/srep21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Task D., Lin C.-C., Vulpe A., Afify A., Ballou S., Brbic M., Schlegel P., Raji J., Jefferis G., Li H., et al. Chemoreceptor co-expression in Drosophila melanogaster olfactory neurons. Elife. 2022;11:e72599. doi: 10.7554/eLife.72599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besson M.T., Soustelle L., Birman S. Identification and structural characterization of two genes encoding glutamate transporter homologues differently expressed in the nervous system of Drosophila melanogaster. FEBS Lett. 1999;443:97–104. doi: 10.1016/s0014-5793(98)01695-0. [DOI] [PubMed] [Google Scholar]
- 46.Ni J.-Q., Markstein M., Binari R., Pfeiffer B., Liu L.-P., Villalta C., Booker M., Perkins L., Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.-S., et al. The transgenic RNAi project at Harvard medical school: resources and validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukunda L., Miazzi F., Kaltofen S., Hansson B.S., Wicher D. Calmodulin modulates insect odorant receptor function. Cell Calcium. 2014;55:191–199. doi: 10.1016/j.ceca.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Mukunda L., Miazzi F., Sargsyan V., Hansson B.S., Wicher D. Calmodulin affects sensitization of Drosophila melanogaster odorant receptors. Front. Cell. Neurosci. 2016;10:28. doi: 10.3389/fncel.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miazzi F., Jain K., Kaltofen S., Bello J.E., Hansson B.S., Wicher D. Targeting insect olfaction in vivo and in vitro using functional imaging. Front. Cell. Neurosci. 2022;16:839811. doi: 10.3389/fncel.2022.839811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martín F., Alcorta E. Novel genetic approaches to behavior in Drosophila. J. Neurogenet. 2017;31:288–299. doi: 10.1080/01677063.2017.1395875. [DOI] [PubMed] [Google Scholar]
- 52.del Valle Rodríguez A., Didiano D., Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods. 2011;9:47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giaume C., Kirchhoff F., Matute C., Reichenbach A., Verkhratsky A. Glia: the fulcrum of brain diseases. Cell Death Differ. 2007;14:1324–1335. doi: 10.1038/sj.cdd.4402144. [DOI] [PubMed] [Google Scholar]
- 54.Boyden E.S., Zhang F., Bamberg E., Nagel G., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 55.Lima S.Q., Miesenböck G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 56.Fiala A., Suska A., Schlüter O.M. Optogenetic approaches in neuroscience. Curr. Biol. 2010;20:R897–R903. doi: 10.1016/j.cub.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 57.Sancho L., Contreras M., Allen N.J. Glia as sculptors of synaptic plasticity. Neurosci. Res. 2021;167:17–29. doi: 10.1016/j.neures.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shearin H.K., Quinn C.D., Mackin R.D., Macdonald I.S., Stowers R.S. t-GRASP, a targeted GRASP for assessing neuronal connectivity. J. Neurosci. Methods. 2018;306:94–102. doi: 10.1016/j.jneumeth.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fishilevich E., Vosshall L.B. Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 60.Silbering A.F., Rytz R., Grosjean Y., Abuin L., Ramdya P., Jefferis G.S.X.E., Benton R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiss S., Melom J.E., Ormerod K.G., Zhang Y.V., Littleton J.T. Glial Ca2+signaling links endocytosis to K+ buffering around neuronal somas to regulate excitability. Elife. 2019;8:e44186. doi: 10.7554/eLife.44186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson C.K., Fernandez-Abascal J., Wang Y., Wang L., Bianchi L. The Na+-K+-ATPase is needed in glia of touch receptors for responses to touch in C. elegans. J. Neurophysiol. 2020;123:2064–2074. doi: 10.1152/jn.00636.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Störtkuhl K.F., Hovemann B.T., Carlson J.R. Olfactory adaptation depends on the Trp Ca2+ channel in Drosophila. J. Neurosci. 1999;19:4839–4846. doi: 10.1523/JNEUROSCI.19-12-04839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang T.Y., Hanani M., Ledda M., De Palo S., Pannese E. Aging is associated with an increase in dye coupling and in gap junction number in satellite glial cells of murine dorsal root ganglia. Neuroscience. 2006;137:1185–1192. doi: 10.1016/j.neuroscience.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 65.Pannese E. The structure of the perineuronal sheath of satellite glial cells (SGCs) in sensory ganglia. Neuron Glia Biol. 2010;6:3–10. doi: 10.1017/S1740925X10000037. [DOI] [PubMed] [Google Scholar]
- 66.Spray D.C., Iglesias R., Shraer N., Suadicani S.O., Belzer V., Hanstein R., Hanani M. Gap junction mediated signaling between satellite glia and neurons in trigeminal ganglia. Glia. 2019;67:791–801. doi: 10.1002/glia.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahr A., Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr. Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 68.Liu W.W., Wilson R.I. Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc. Natl. Acad. Sci. USA. 2013;110:10294–10299. doi: 10.1073/pnas.1220560110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soustelle L., Besson M.-T., Rival T., Birman S. Terminal glial differentiation involves regulated expression of the excitatory amino acid transporters in the Drosophila embryonic CNS. Dev. Biol. 2002;248:294–306. doi: 10.1006/dbio.2002.0742. [DOI] [PubMed] [Google Scholar]
- 70.Besson M.T., Sinakevitch I., Melon C., Iché-Torres M., Birman S. Involvement of the Drosophila taurine/aspartate transporter dEAAT2 in selective olfactory and gustatory perceptions. J. Comp. Neurol. 2011;519:2734–2757. doi: 10.1002/cne.22649. [DOI] [PubMed] [Google Scholar]
- 71.Stahl B.A., Peco E., Davla S., Murakami K., Caicedo Moreno N.A., van Meyel D.J., Keene A.C. The taurine transporter Eaat2 functions in ensheathing glia to modulate sleep and metabolic rate. Curr. Biol. 2018;28:3700–3708.e4. doi: 10.1016/j.cub.2018.10.039. [DOI] [PubMed] [Google Scholar]
- 72.Martelli C., Fiala A. Slow presynaptic mechanisms that mediate adaptation in the olfactory pathway of Drosophila. Elife. 2019;8:e43735. doi: 10.7554/eLife.43735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim S.M., Wang J.W. Calcium imaging of pheromone responses in the insect antennal lobe. Methods Mol. Biol. 2013;1068:179–187. doi: 10.1007/978-1-62703-619-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silbering A.F., Bell R., Galizia C.G., Benton R. Calcium imaging of odor-evoked responses in the Drosophila antennal lobe. J. Vis. Exp. 2012:2976. doi: 10.3791/2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strauch M., Galizia C.G. Fast PCA for processing calcium-imaging data from the brain of Drosophila melanogaster. BMC Med. Inform. Decis. Mak. 2012;12:S2. doi: 10.1186/1472-6947-12-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ung K., Tepe B., Pekarek B., Arenkiel B.R., Deneen B. Parallel astrocyte calcium signaling modulates olfactory bulb responses. J. Neurosci. Res. 2020;98:1605–1618. doi: 10.1002/jnr.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ung K., Huang T.-W., Lozzi B., Woo J., Hanson E., Pekarek B., Tepe B., Sardar D., Cheng Y.-T., Liu G., et al. Olfactory bulb astrocytes mediate sensory circuit processing through Sox9 in the mouse brain. Nat. Commun. 2021;12:5230. doi: 10.1038/s41467-021-25444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bargmann C.I., Thomas J.H., Horvitz H.R. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 1990;55:529–538. doi: 10.1101/sqb.1990.055.01.051. [DOI] [PubMed] [Google Scholar]
- 79.Kaplan J.M., Horvitz H.R. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1993;90:2227–2231. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Troemel E.R., Kimmel B.E., Bargmann C.I. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 81.Hilliard M.A., Apicella A.J., Kerr R., Suzuki H., Bazzicalupo P., Schafer W.R. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. EMBO J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bacaj T., Tevlin M., Lu Y., Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang A., Yan D. C. elegans as a model to study glial development. FEBS J. 2022;289:1476–1485. doi: 10.1111/febs.15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nava Gonzales C., McKaughan Q., Bushong E.A., Cauwenberghs K., Ng R., Madany M., Ellisman M.H., Su C.-Y. Systematic morphological and morphometric analysis of identified olfactory receptor neurons in Drosophila melanogaster. Elife. 2021;10:e69896. doi: 10.7554/eLife.69896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prelic S., Pal Mahadevan V., Venkateswaran V., Lavista-Llanos S., Hansson B.S., Wicher D. Functional interaction between Drosophila olfactory sensory neurons and their support cells. Front. Cell. Neurosci. 2021;15:789086. doi: 10.3389/fncel.2021.789086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laprell L., Schulze C., Brehme M.-L., Oertner T.G. The role of microglia membrane potential in chemotaxis. J. Neuroinflammation. 2021;18:21. doi: 10.1186/s12974-020-02048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Woodard C., Alcorta E., Carlson J. The rdgB gene of Drosophila: a link between vision and olfaction. J. Neurogenet. 1992;8:17–31. doi: 10.3109/01677069209167269. [DOI] [PubMed] [Google Scholar]
- 88.Larsson M.C., Domingos A.I., Jones W.D., Chiappe M.E., Amrein H., Vosshall L.B. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 89.Ito K., Urban J., Technau G.M. Distribution, classification, and development ofDrosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch. Dev. Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- 90.Lai S.-L., Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 91.Awasaki T., Huang Y., O’Connor M.B., Lee T. Glia instruct developmental neuronal remodeling through TGF-β signaling. Nat. Neurosci. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Casas-Tintó S., Arnés M., Ferrús A. Drosophila enhancer-Gal4 lines show ectopic expression during development. R. Soc. Open Sci. 2017;4:170039. doi: 10.1098/rsos.170039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tully T., Quinn W.G. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J. Comp. Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- 95.Devaud J.M. Experimental studies of adult Drosophila chemosensory behaviour. Behav. Processes. 2003;64:177–196. doi: 10.1016/s0376-6357(03)00134-7. [DOI] [PubMed] [Google Scholar]
- 96.Martin F., Alcorta E. Measuring activity in olfactory receptor neurons in Drosophila: focus on spike amplitude. J. Insect Physiol. 2016;95:23–41. doi: 10.1016/j.jinsphys.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 97.Saina M., Benton R. Visualizing olfactory receptor expression and localization in Drosophila. Methods Mol. Biol. 2013;1003:211–228. doi: 10.1007/978-1-62703-377-0_16. [DOI] [PubMed] [Google Scholar]
- 98.Benton R., Sachse S., Michnick S.W., Vosshall L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code. All codes are available via open access tools and resources listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.