Abstract
Coccidioides immitis antigens which stimulate a T helper cell 1 (Th1) pathway of host immune response are considered to be essential components of a vaccine against coccidioidomycosis. Recombinant urease (rURE) and recombinant heat shock protein 60 (rHSP60) of C. immitis were expressed in Escherichia coli and tested as vaccine candidates in BALB/c mice. A synthetic oligodeoxynucleotide which contained unmethylated CpG dinucleotides and was previously shown to enhance a murine Th1 response was used as an immunoadjuvant. T cells isolated from the spleens and lymph nodes of the rURE- and rHSP60-immune mice showed in vitro proliferative responses to the respective recombinant protein, but only those T lymphocytes from rURE-immunized mice revealed markedly elevated levels of expression of selected Th1-type cytokine genes. BALB/c mice immunized subcutaneously with rURE and subsequently challenged by the intraperitoneal (i.p.) route with a lethal inoculum of C. immitis arthroconidia demonstrated a significant reduction in the level of C. immitis infection compared to control animals. rHSP60 was much less effective as a protective antigen. Evaluation of cytokine gene expression in lung tissue and levels of recombinant urease-specific immunoglobulins (immunoglobulin G1 [IgG1] versus IgG2a) in murine sera at 12 days after challenge provided additional evidence that immunization with rURE stimulated a Th1 response to the pathogen. Urease was further evaluated by expression of the URE gene in a mammalian plasmid vector (pSecTag2A.URE) which was used to immunize mice by the intradermal route. In this case, 82% of the vector construct-immunized animals survived more than 40 days after i.p. infection, compared to only 10% of the mice immunized with the vector alone. In addition, 87% of the pSecTag2A.URE-immunized survivors had sterile lungs and spleens. These data support the need for further evaluation of the C. immitis urease as a candidate vaccine against coccidioidomycosis.
Coccidioidomycosis (San Joaquin Valley fever) is a fungal respiratory disease of humans which is endemic to southwestern United States, northern Mexico, and numerous semiarid areas of Central and South America (34). Infection occurs by inhalation of airborne spores (arthroconidia) produced by the saprobic phase of Coccidioides immitis which grows in alkaline desert soil. It is estimated that 100,000 new cases of this disease occur annually within the rapidly growing population of people who live in regions of the United States between southwest Texas and southern California, where the disease is endemic (15). Although the majority of immunocompetent individuals are able to resolve their C. immitis infection spontaneously, the level of morbidity associated even with the primary form of this respiratory mycosis warrants consideration of a vaccine against the disease. Immunocompromised patients, including those infected with human immunodeficiency virus, are at high risk to contract disseminated coccidioidomycosis (3). It is also apparent from results of several clinical studies that African-Americans and Asians are genetically predisposed to development of the potentially fatal, disseminated form of the respiratory disease (14). A history of recurrent epidemics of this mycosis in recreational and urban areas of the San Joaquin Valley and parts of Arizona has helped to stimulate new research on improved therapy and vaccine development (15). The rationale for commitment of research efforts to develop a C. immitis vaccine is based on clinical evidence that individuals who recover from the respiratory disease retain long-term cellular immunity against future infections by the pathogen (37).
T lymphocytes are known to play a key role in acquired immunity against C. immitis infection (5, 6, 37). Recent investigations of potential C. immitis vaccine candidates have focused on purified T-cell-reactive antigens expressed in vitro by the parasitic phase of the fungus (27). Two such antigens have been cloned, and the recombinant proteins have been tested for their ability to protect mice against a lethal challenge of C. immitis (1, 22, 25, 26). In this report, we compare the T-cell-mediated immune responses of BALB/c mice to two additional C. immitis antigens, urease and a 60-kDa heat shock protein (HSP60). The genes which encode these two antigens were previously cloned and expressed in Escherichia coli (40, 43), and the recombinant proteins (rURE and rHSP60) have been shown to stimulate proliferative response of murine immune T cells in cellular immunoassays (K. Li, J.-J. Yu, and G. T. Cole, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemothr. 1999, abstr. 453, p. 552, 1999) (40).
MATERIALS AND METHODS
Purification of recombinant proteins.
The protocols for expression and purification of rURE and rHSP60 of C. immitis have been reported elsewhere (40, 43). Endotoxin contamination of each stock solution of recombinant protein (1 mg/ml) solubilized in phosphate-buffered saline (PBS; 0.1 M, pH 7.4) was assayed using a Limulus ameboyte lysate kit (QCL-1000; BioWhittaker, Walkersville, Md.). All preparations had fewer than 30 endotoxin units (150 ng of endotoxin) per μg of protein.
FKES.
Endosporulating spherules were obtained from parasitic phase cultures of C. immitis (strain C735) grown in modified Converse medium (28) for 132 h as previously described (19). The cells were chemically fixed in 0.5% formalin (Sigma, St. Louis, Mo.) for 3 days (4°C) and washed three times with PBS, and the formalin-killed endosporulating spherules (FKES) were either used directly to immunize mice by the subcutaneous (s.c.) route or stored at −70°C until used for T-cell proliferation assays as described below. Aliquots of FKES were plated on GYE agar (19) to confirm the absence of viable cells.
CpG DNA.
Unmethylated CpG dinucleotides present in a synthetic oligodeoxynucleotide (ODN) preparation (CpG ODN; Integrated DNA Technologies, Inc., Coralville, Iowa) were used as an immunoadjuvant in this study as previously described (32). The CpG ODN sequence used to immunize mice (TCCATGACGTTCCTGACGTT [CpG motifs are underlined]) was the same as that reported by Chu et al. (7) (ODN 1826). The ODN was phosphorothioate modified to increase resistance to nuclease degradation (7). The CpG ODN preparation was dissolved in PBS (1 mg/ml) and used as a stock solution for subsequent immunizations.
Animals.
All T-cell proliferation assays and protection experiments were conducted with 10-week-old, female BALB/c mice supplied by the National Cancer Institute (Bethesda, Md.). BALB/c mice have been shown to be highly susceptible to C. immitis infection (24).
Protein immunization and T-cell proliferation assay.
Four groups of four female mice each were immunized s.c. with either one of the two recombinant proteins (15 μg of rURE or 15 μg of rHSP60 plus 15 μg of CpG ODN), FKES (106 cells plus 15 μg of CpG ODN), or bovine serum albumin (BSA; fraction V; Sigma) (15 μg of BSA plus 15 μg of CpG ODN)(each being prepared in 100 μl of incomplete Freund's adjuvant (IFA; Sigma) (7). The mice were boosted by the s.c. route 14 days later with the same amount of each antigen plus immunoadjuvant. The mice were sacrificed 14 days after the last immunization, and the spleen as well as the inguinal and axillary lymph nodes were removed. The spleens and lymph nodes of each group of mice were pooled and macerated; separation of lymphocytes from the cell suspensions was conducted by isopycnic centrifugation over Ficoll-Paque (Pharmacia, Piscataway, N.J.) as previously described (10). The lymphocyte fraction was then passed through a nylon wool column (Polysciences, Inc., Warrington, Pa.) to enrich for T cells. The relative number of T cells and B cells in this preparation was determined by flow cytometry as described below. The lymphocytes (approximately 2 × 105 cells) were cultured with 5 × 105 irradiated, syngeneic spleen cells for 3 days (37°C, 5% CO2), with or without antigen, in 96-well flat-bottom plates (catalog no. 3596; Costar, Cambridge, Mass.). The wells contained 0.2 ml of RPMI 1640 tissue culture medium (Sigma) plus 10% (vol/vol) fetal calf serum (FCS; Hyclone Laboratories, Logan, Utah), 50 μM 2-mercaptoethanol, and 50 μg of gentamicin (Sigma) per ml. At the end of 3 days, 1.0 μCi of [3H]thymidine (specific activity, 6.7 Ci/mmol; NEN, Boston, Mass.) was added to each well, and the plates were incubated under the same conditions for an additional 18 h. The cells were then harvested onto glass fiber sheets (BioWhittaker) as previously described (9). The filters were placed in scintillation fluid, and incorporation of the radioisotope into T cells was determined in a liquid scintillation counter (model LS 3801; Beckman Coulter, Inc., Fullerton, Calif.). Three replicate wells were used for each concentration of antigen tested. The data for each sample are expressed as the mean counts per minute ± standard error of the means.
Flow cytometry.
Immune lymphocytes isolated as described above were adjusted to 106 cells/well in a 96-well U-bottom plate (Falcon no. 3022; Becton Dickinson, Franklin Lakes, N.J.). All cell staining reactions and washing steps were performed at 4°C in Hanks balanced salt solution containing 1% BSA and 0.1% (wt/vol) NaN3. The lymphocytes were subjected to two color staining for flow cytometry analysis as described by Coligan et al. (10). The cells were first incubated with 10 μg of rat anti-mouse CD16/CD32 (anti-FcγII/III receptor; PharMingen, San Diego, Calif.) per ml for 15 min to block any subsequent nonspecific binding of conjugated antibody. After three washes, the cells were reacted with 5 μg each of fluorescein isothiocyanate-labeled hamster anti-mouse CD3ɛ and phycoerythrin-labeled rat anti-mouse CD45R/B220 monoclonal antibodies (PharMingen) per ml for 30 min. Isotype-matched immunoglobulin (Ig) preparations (hamster IgG and rat IgG2aκ) were used as negative staining controls. After three washes, the cells were suspended in 200 μl of fixative (1% paraformaldehyde in Hanks balanced salt solution) and then pipetted through a 100-μm-pore-size nylon mesh and analyzed in an Epics Elite flow cytometer (Beckman Coulter, Miami, Fla.).
Cytokine expression assays using RT-PCR.
Antigen-exposed T cells cultured in six-well plates (Costar) as described above for proliferation assays were isolated by isopycnic centrifugation over Ficoll-Paque as previously described to separate live cells from cell debris. The T cells exposed to different antigens served as a source of total RNA for reverse transcription (RT)-PCR assays. Lung tissues obtained from BALB/c mice which were immunized as described above and sacrificed 12 days after C. immitis challenge as described below were also used as a source of total RNA for separate RT-PCR assays. Total RNA was extracted in each case using the Trizol total RNA isolation reagent and procedure recommended by the manufacturer (Gibco BRL Life Technologies, Grand Island, N.Y.). The crude extract was digested with RQ1 RNase-free DNase (Promega, Madison, Wis.) and the RNA was purified using an RNA cleanup protocol (Plant RNeasy mini kit; Qiagen, Chatsworth, Calif.). The purity and quantity of the RNA preparations were monitored by UV absorbance. A ratio of optical density at 280 nm (OD280) to OD260 that was >1.9 was obtained for each preparation.
Expression of cytokine genes which are representative of the T helper cell 1 (Th1) or Th2 pathway of immune response were examined by comparison to the level of mRNA that encodes the constitutively expressed murine hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene (39). The RT-PCR protocol used was essentially the same as previously reported (18). The amount of total RNA was equalized for each RT reaction. In brief, the cDNA was synthesized in a 50-μl solution containing 50 mM Tris-HCl (pH 8.3) plus 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol (Sigma), 0.5 μM (each) deoxynucleoside triphosphates, 200 μM PCR oligo(dT)17 adapter primer (13), 1 μg of T-cell-derived total RNA, 0.5 μl of 200 RNase inhibitor (200 U/ml; Gibco BRL), and 400 U of SuperScript II RNase H− reverse transcriptase (Gibco BRL). The reaction mixture was incubated at 42°C for 50 min and then shifted to 70°C for 10 min to denature the reverse transcriptase. The PCR mixture contained 1 μl of the cDNA plus 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphates, 5 μM (each) primers for amplification of the selected cDNAs, and 1 U of Taq DNA polymerase (Sigma) in a total volume of 25 μl.
The PCR primer pairs (sense and antisense) used to amplify the HPRT and selected cytokine cDNAs were as follows: HPRT, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ and 5′-GATTCAACTTGCGCTCATCTTAGGC-3′; gamma interferon (IFN-γ), 5′-TGAACGCTACACACTGCATCTTGG-3′ and 5′-CGACTCCTTTTCCGCTTCCTGAG-3′; interleukin-2 (IL-2), 5′-GTCAACAGCGCACCCACTTCAA-3′ and 5′-TTGAGATGATGCTTTGACAGAA-3′; IL-4, 5′-ACGGAGATGGATGTGCCAAACGT-3′ and 5′-CTCAGTACTACGAGTAATCCA-3′; IL-5, 5′-AGAAGGATGCTTCTGCACTTGAGT-3′ and 5′-CAAGGAACTCTTGCAGGTAATCCA-3′; and IL-10, 5′-ATGCAGGACTTTAAGGGTTACTTG-3′ and 5′-TTGGTCTTGGAGCTTATTAAAATC-3′. Thirty-five cycles were conducted for amplification of the template cDNA with each of the primer pairs. Initial denaturation was performed at 94°C for 3 min. Each subsequent cycle consisted of a melting step (94°C, 1 min), an annealing step (60°C, 1 min), and an extension step (72°C, 1 min). The final extension step of the cycle was conducted at 72°C for 10 min. The predicted sizes of the amplified products are as follows: HPRT, 165 bp; IFN-γ, 460 bp; IL-2, 445 bp; IL-4, 289 bp; IL-5, 364 bp; and IL-10, 244 bp. The amplicons were distinguished as distinct bands after separation of the PCR products by agarose electrophoresis (2% [wt/vol] gel) and staining with ethidium bromide (36).
ELISA.
The indirect enzyme-linked immunosorbent assay (ELISA) was performed for examination of Ig titers to selected antigens in immunized and C. immitis-infected mice as previously described (1). Heat-inactivated sera from mice immunized with rURE, rHSP60, or BSA plus CpG ODN as described above were tested for reactivity with the respective, purified antigens. Sera were collected separately from three mice in each group by heart puncture exsanguination at 12 days after intraperitoneal (i.p.) challenge with C. immitis as described below. Control sera were obtained from three nonimmunized, infected mice. The final concentration of each purified recombinant antigen and BSA applied to the wells of the microtiter plates (Falcon no. 3077; Becton Dickinson) was 10 ng in 100 μl of PBS. The plates were washed with PBS which contained 0.33% (vol/vol) Brij 35 detergent (Sigma) and blocked with the same buffer plus 5% (vol/vol) FCS (Sigma). A fourfold serial dilution (1:10 to 1:10,240) of each serum sample in PBS was used to test binding of Igs (total Ig, IgG2a, and IgG1) to the purified antigens as previously described (1). After incubation of the sera with the antigens in the microtiter plates (24°C, 2 h), the wells were washed with PBS that contained Brij 35 as above. Alkaline phosphatase conjugated with goat anti-mouse Ig (IgM, IgG, and IgA; heavy and light chains), goat anti-mouse IgG2a, or goat anti-mouse IgG1 (Southern Biotechnology Associates, Inc., Birmingham, Ala.) was added to the wells at a dilution of 1:1,000 in PBS to detect bound, primary antibody. The plates were incubated at room temperature for 15 min, and the reaction was stopped by addition of 1.0 N H2SO4. Antibody adsorption was determined by OD at 405 nm. Nonspecific antibody adsorption to each antigen was determined by OD after incubation of the nonimmune, control serum samples diluted 1/2,000 in antigen-coated wells of the microtiter plates as above. The antibody titer of each serum sample from the immunized, infected mice is defined as the dilution that yielded an OD in the ELISA that was 50% of the maximum OD for each antigen after subtraction of the value for nonspecific antibody adsorption. The data are presented as mean values for the three test sera in each group ± standard error of the mean.
Protection assays.
Results of protection assays presented in this report are based on infection with a single strain (C735) of C. immitis, which we have previously shown is highly virulent in BALB/c mice (8). C. immitis strain C735 was grown on GYE plates at 30°C for 6 to 8 weeks, and arthroconidia were then harvested by passing a suspension of the cells in sterile saline through an autoclaved glass wool column to remove hyphal fragments. The concentration of the stock suspension of arthroconidia was adjusted to 103 viable cells/ml of saline. BALB/c mice were immunized with selected antigens using the protocol described above or with DNA as described below. The animals were then challenged by the i.p. route at 14 days after the last protein immunization or 30 days after the last DNA immunization. The inoculum contained 100 viable arthroconidia in 100 μl of PBS. As previously argued (26), the i.p. route of inoculation permitted more precise control of the size of the inoculum which was actually delivered to host tissues and better reproducibility of levels of coccidioidal infection in the lungs than the intranasal route of challenge. The i.p. route of challenge has been used to evaluate other cloned antigens of C. immitis as potential vaccine candidates (1, 22, 25, 26). Protection was evaluated both by residual C. immitis burden at 12 days postinfection and by the ability of immunized mice to survive to at least 40 days after challenge. For fungal burden evaluations, the lung and spleen homogenates of surviving mice were subjected to quantitative analyses of CFU in dilution plate cultures as previously described (26). For both fungal burden and survival studies, each group of test and control animals consisted of at least 12 mice.
Construction and purification of the pSecTag2A.URE plasmid.
A 1.6-kb fragment of the URE gene (nucleotides 1917 to 3561) (43), which was essentially the same as that used for expression of rURE, was subcloned into a mammalian plasmid expression vector (pSecTag2A; Invitrogen, Carlsbad, Calif.). The vector contains a cytomegalovirus promoter and both ampicillin and Zeocin resistance genes for selection in E. coli and mammalian cells. The 1,645-bp URE gene fragment was amplified by PCR using sense and antisense primers (5′-CCAAGCTTGGACTTCGACCGT-3′ and 5′-CTCAGCGCAGATGGCTCGAGC-3′, respectively) that contained engineered HindIII and XhoI restriction sites (underlined). The amplified fragment was subcloned into the HindIII-XhoI-digested pSecTag2A vector to create the pSecTag2A.URE construct. Both this construct and pSecTag2A vector alone were used separately to transform E. coli strain Top 10 (4). Transformants were selected from Luria broth agar plates supplemented with ampicillin (4). Plasmid DNA was purified from bacterial lysates using a Qiagen plasmid purification system. The plasmid DNA preparations used in this study had 0.2 to 2.0 endotoxin units (1 to 10 ng of endotoxin) per μg of DNA.
In vitro transfection and immunoblot analysis.
In vitro transfection of a murine fibroblast cell line (NIH 3T3; provided by Manohar Ratnam, Medical College of Ohio) with pSecTag2A.URE was conducted to confirm expression of the rURE by the mammalian cells prior to DNA immunization of mice. The cells were grown in Dulbecco's modified Eagle's medium (Sigma) containing 10% (vol/vol) FCS (Sigma). The fibroblast cells were incubated in six-well tissue culture plates (Costar) at 37°C (5% CO2) for 24 h, after which time the cell density reached approximately 3 × 105 cells/well. Immediately prior to transfection, the medium was aspirated from wells, and plasmid DNA (construct or vector alone) was delivered to the cells by bombardment using a Helios gene gun system (Bio-Rad, Hercules, Calif.) as described by the manufacturer. The conditions of bombardment were essentially the same as previously described (42) except that the plasmid DNA was coated onto 1.6-μm-diameter gold beads and the gene gun was pressurized with helium at 200 lb/in2. After DNA delivery, the wells of the culture plates were quickly flooded with Dulbecco's modified Eagle's medium plus FCS, and the plates were incubated for 72 h under the conditions described above. Fresh growth medium which contained Zeocin (600 μg/ml; Invitrogen) was then added to the wells, the plates were incubated for an additional 72 h, and the viable cells in each well were subjected to limiting dilution in 48-well plates (Costar) for isolation of transfected clones. Control fibroblasts were transfected with pSecTag2A vector alone, and Zeocin-resistant clones were selected as described above.
Two selected clones transfected with the vector construct or vector alone were examined for rURE expression by RT-PCR and immunoblot analysis. Total RNA was isolated from the transfected murine fibroblast clones as described above, and RT-PCR was performed using the same set of primers as used for amplification of the 1.6-kb URE gene fragment. The PCR conditions were the same as described above except that the annealing temperature was 52°C and the extension step was conducted for 2 min. The PCR products were examined by agarose gel electrophoresis as above. For examination of protein production, the total lysate of NIH 3T3 cells transfected with either pSecTag2A.URE or vector alone were first separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% gel), and then subjected to immunoblot analysis using murine antibody raised against rURE as previously described (43).
DNA immunization and T-cell proliferation–cytokine expression assays.
Two groups of BALB/c mice (15 animals each) were immunized intradermally (i.d.) with plasmid DNA-coated 1.6-μm-diameter gold particles (plasmid construct or vector alone), using the gene gun pressurized at 400 lb/in2. Nair lotion (Carter-Wallace, New York, N.Y.) was first applied to the abdomen of each mouse to remove its hair, and then a total of 2 μg of plasmid DNA was delivered i.d. to two sites on the abdominal region. Booster immunizations of 1 μg of DNA each were performed at 30 days and 60 days after the first immunization. Thirty days after the last immunization, three mice from each group were sacrificed and T cells were isolated for examination in T-cell proliferation and cytokine expression assays as described above. The remaining immunized mice (12 per group) were challenged with C. immitis by the i.p. route as previously described and scored for survival over the subsequent 40 days. The duration between DNA immunizations and challenge in this study was based on a previously reported gene gun immunization experiment using BALB/c mice (41). All mice which survived to 42 days postchallenge were sacrificed, and their lungs and spleens were homogenized and subjected to dilution plate analysis to determine residual C. immitis burden as described above.
Statistical analyses.
The numbers of CFU per organ were expressed on a log scale. Because these values did not fall into a normal distribution, the Mann-Whitney U test was used to compare medians in all cases. Survival differences between groups of mice were calculated for statistical significance by the Kaplan-Meier method. All statistical analyses were performed using the SPSS version 9.0 statistical package for Windows (SPSS Inc., Chicago, Ill.).
RESULTS
T-cell proliferation assays of recombinant protein-immunized mice.
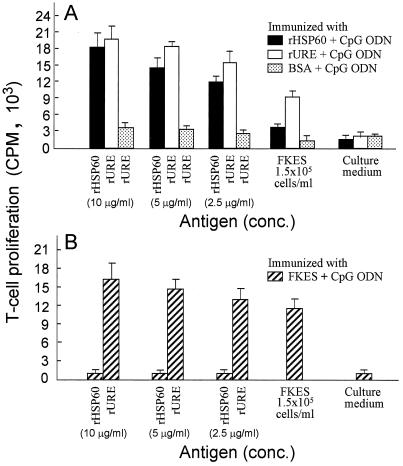
T lymphocytes were obtained from spleen and lymph node tissues of separate groups of mice 14 days after the final immunization with rHSP60, rURE, BSA, or FKES plus the CpG ODN immunoadjuvant. The T cells were tested in vitro for proliferative responses to homologous and heterologous antigens (Fig. 1). T cells isolated from rHSP60- or rURE-immune mice were incubated with either the corresponding homologous antigen (10, 5, or 2.5 μg/ml) or FKES (105 cells/ml). T cells from rURE-immunized mice showed slightly higher response to the homologous antigen and markedly greater proliferative response to FKES than T cells from the rHSP60-immune mice (Fig. 1A). Control T cells from BSA-immunized mice stimulated in vitro with rURE, FKES, or culture medium consistently showed little response, which indicates that the endotoxin present in the recombinant protein preparation did not significantly influence the results of this assay. Mice were also immunized with FKES, and T cells isolated from these animals were tested for their in vitro response to different concentrations of rHSP60 and rURE (Fig. 1B). In this case, the difference in proliferative response to the two recombinant proteins was clearly distinguished. At each concentration tested, rURE elicited a much greater in vitro response than rHSP60. rURE at a concentration of 10 μg/ml stimulated a slightly greater proliferative response of the FKES-immune T cells than the homologous antigen (105 FKES/ml).
FIG. 1.
Proliferative response of T cells pooled from lymph nodes and spleens of mice immunized as indicated. The maximum response to each antigen is shown. Mean values and standard deviations of three determinations are reported.
Expression of cytokine genes by in vitro-stimulated immune T cells.
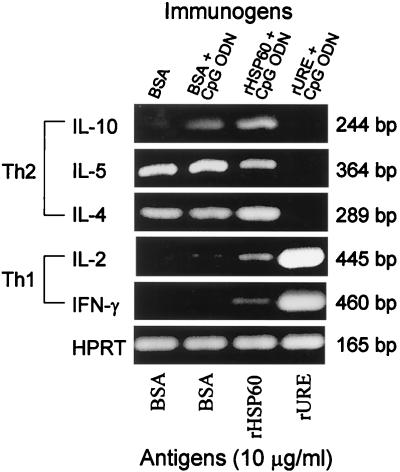
Immune T cells were obtained from mice immunized with BSA alone, BSA plus CpG ODN, rHSP60 plus CpG ODN, or rURE plus CpG ODN. Total RNA was separately isolated from the four preparations of T lymphocytes which had been incubated for 4 days with the corresponding homologous antigen as described above. The RNAs were subjected to RT-PCR analysis using cytokine gene-specific primers (Fig. 2). Gene expression levels were compared to that of the constitutive HPRT gene. The results showed that T cells isolated from rHSP60-immune mice and then stimulated in vitro with the homologous antigen elicited expression of IL-10, IL-5, and IL-4 cytokine mRNAs, which indicates the activation of a Th2 pathway of immune response. The relatively low expression of IL-2 and IFN-γ mRNAs suggests that the HSP60-immune T cells showed a weak activation of a Th1 pathway of immune response. In contrast, T cells obtained from rURE-immune mice which were stimulated in vitro with the rURE antigen at the same concentration as rHSP60 revealed relatively high levels of expression of IL-2 and IFN-γ genes but no detectable expression of the IL-10, IL-5, or IL-4 gene. The stimulated rURE-immune cells demonstrated a dominant Th1 response.
FIG. 2.
In vitro expression of selected cytokines and HPRT mRNA in T cells isolated from lymph nodes and spleens of immunized mice. Total RNA preparations were obtained from T cells isolated from four mice in each immunized group. Mice were immunized with BSA or recombinant protein as indicated, and isolated T cells were stimulated in vitro under the conditions described in Materials and Methods for the proliferation assay reported in Fig. 1. RT-PCR was performed using specific primer pairs for Th1 (IFN-γ and IL-2) and Th2 cytokines (IL-4, IL-5, and IL-10). The HPRT amplicon was included as an internal control. The PCR products were electrophoresed in 2% agarose gels, stained with ethidium bromide, and observed with a UV transilluminator. The results are representative of three separate experiments.
In vivo immune responses.
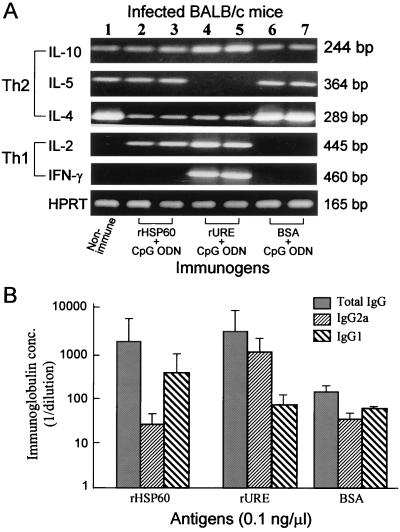
Total RNA from lung tissue of uninfected, naive mice (Fig. 3A, lane 1) and lung tissue of mice immunized and infected with C. immitis by the i.p. route and sacrificed 12 days postchallenge (lanes 2 to 7) were subjected to RT-PCR analysis of expression of selected cytokine genes. The levels of expression, based on intensity of the ethidium bromide-stained bands in agarose gels, were compared to that of the constitutively expressed HPRT gene. Lung tissues from two mice of each group immunized with rHSP60 (lanes 2 and 3), rURE (lanes 4 and 5), or BSA (lanes 6 and 7) plus CpG ODN were examined. The results showed that only the rURE-immune, infected mice were characterized by expression of IFN-γ mRNA and the absence of IL-5 gene expression. These same mice also showed slightly elevated levels of IL-2 and IL-10 gene expression compared to the naive and other immunized mice. In contrast, the rHSP60-immune mice showed expression of IL-5 mRNA but no detectable IFN-γ gene expression. Unlike the naive and BSA-immunized mice, the rHSP60-immune mice also revealed expression of the IL-2 gene. The more dominant Th1 pattern of expression of cytokine genes in lung tissue of the rURE-immune, infected mice distinguished this group of animals from others tested in this assay.
FIG. 3.
(A) Expression of Th1 and Th2 cytokines and HPRT mRNA in lung tissue of infected mice which either had not been immunized (lane 1) or had been immunized as described in Materials and Methods with one of the selected antigens plus CpG ODN adjuvant (i.e., rHSP60 [lanes 2 and 3], rURE [lanes 4 and 5], BSA [lane 6 and 7]). The RT-PCR and electrophoresis conditions were the same as those used for analysis of cytokine expression in Fig. 2. HPRT was used as an internal control. The results are representative of three separate experiments. (B) Total Ig and IgG subclass responses of immunized and infected BALB/c mice to the indicated antigens. Mice were sacrificed and terminally bled as described in Materials and Methods; plasma samples were tested for antigen-specific total Ig, IgG2a, and IgG1 by ELISA as indicated. The results are expressed as reciprocal titers (1/dilution) and presented as means plus standard deviations for three test sera in each group of immunized, infected mice.
Sera obtained from mice immunized and infected as above and sacrificed at 12 days postchallenge were compared by indirect ELISA determinations of titers of total Ig and antibody isotypes to rHSP60, rURE, and BSA (Fig. 3B). The mean titers of serum samples from three groups of mice (three animals each) which were immunized with either rHSP60, rURE, or BSA plus CpG ODN are presented for each homologous test antigen in the ELISA. Mice immunized with rHSP60 showed an IgG1 titer to the homologous antigen which was approximately twice that of the IgG2a titer. In contrast, the IgG2a titer to rURE determined for sera from rURE-immune mice was markedly higher than the IgG1 titer. Sera from control animals immunized with BSA plus immunoadjuvant showed essentially no difference in the titers of IgG2a and IgG1. These data provide additional evidence for a difference in the nature of the host immune response to rHSP60 and rURE.
Fungal burden in recombinant protein-immunized mice.
Four groups of mice (12 per group) were immunized with BSA or recombinant protein, challenged with 100 arthroconidia of C. immitis by the i.p. route, and sacrificed 12 days later to compare the number of residual organisms present in lungs and spleens. The results of a vaccination experiment are shown in Fig. 4. The rURE-immune mice showed significantly lower counts of C. immitis in both lungs and spleens compared to control mice immunized with BSA plus CpG ODN. The Mann-Whitney U test for statistical significance of the difference between CFU in organs of these two groups of mice yielded P values of 0.0001 and 0.002. The numbers of organisms present in lungs and spleens of rHSP60-immune mice did not show a statistically significant difference compared to CFU in lungs and spleens of control animals immunized with BSA plus CpG ODN (P = 0.268 and 0.690, respectively.) The median number of organisms in lung homogenates of rURE-immune mice was 1.3 (log10) CFU (range, 0.1 to 5.3) compared to rHSP60-immune mice, with a median of 5.5 (log10) CFU (range, 0.9 to 6.2). This difference in CFU between rURE- and rHSP60-immune mice was statistically significant (P = 0.023). A similar difference was seen in the spleen. The median CFU in rURE-immune mice was 1.5 (log10) (range, 1.4 to 5.8), compared to the median CFU in rHSP60-immune mice of 5.2 (log10) (range, 4.9 to 6.2). The difference between CFU in the spleens of rURE- and rHSP60-immune mice was also statistically significant (P = 0.013). The difference between the median CFU and range of counts of organisms in lungs and spleens of control mice immunized with BSA alone compared to BSA plus CpG ODN was not statistically significant. Similar data were obtained in a second vaccination experiment with rURE compared to BSA plus CpG ODN immunization of BALB/c mice. The median CFU in the control mice was 5.0 (range, 3.0 to 6.2) in the lungs and 5.5 (range, 4.5 to 6.3) in the spleen. The medians (and ranges) of CFU in the rURE-immune mice were 0.0 (0.0 to 5.5) and 0.0 (0.0 to 6.0), respectively. The P values based on the difference between these counts in immunized versus control mice were 0.008 and 0.007, respectively, which are statistically significant.
FIG. 4.
Box plot representation of the numbers of CFU detected in lungs and spleen of mice immunized with selected reagents as indicated and described in Materials and Methods, infected with C. immitis by the i.p. route, and sacrificed 12 days after infection. Boxes indicate the 25th and 75th percentiles; bars show the 5th and 95th percentiles. The line within each box indicates the median.
Survival of mice immunized with recombinant protein.
Mice immunized with BSA or recombinant protein plus CpG ODN and challenged by the i.p route as above (12 mice per group) were scored for survival over a 40-day period postchallenge. The results of a representative survival experiment are shown in Fig. 5. Control mice immunized with BSA plus immunoadjuvant typically began to die as a result of C. immitis infection at about 12 days postchallenge, and the number of survivors sharply decreased over the following 10 days. Both rHSP60- and rURE-immune mice showed prolonged survival after i.p. infection. However, the rate of mortality among the rHSP60-immune mice during the 8-day period after the first fatality (i.e., days 16 to 23 postchallenge) was more comparable to that of the control mice than that of the rURE-immune mice. At 40 days postchallenge, only 15% of the mice immunized with rHSP60 survived, compared to 42% of the rURE-immune mice. The difference between the percentage of rURE-immune mice which survived over the 40-day period compared to control mice immunized with BSA plus CpG ODN was highly significant as determined by the Kaplan-Meier test (P = 0.0002). The difference in survival between rURE- and rHSP60-immunized mice was also statistically significant (P = 0.013). The results of a repeated survival experiment in which the same immunization and challenge protocols were used revealed that the percentage of rURE-immune survivors compared to that of control mice was highly significant (P < 0.0001). We tested a range of doses of rURE and rHSP60 (5, 15, 30, and 60 μg [data not shown]) in these survival experiments. Immunization with 5 μg of rURE using the protocol described in Materials and Methods resulted in survival of only 25% of the mice at 40 days after i.p. challenge. Immunization with 30 or 60 μg of rURE resulted in 44 or 47% survival, respectively. Immunization of BALBc mice with 30 or 60 μg of rHSP60 resulted in survival of 12 or 16%, respectively, of the animals at 40 days postchallenge. These additional survival studies using doses of 30 and 60 μg of immunogen supported our conclusion that rURE was a better vaccine candidate than rHSP60. Recombinant protein-immunized mice which survived to 42 days postchallenge were sacrificed, and their lungs and spleens were excised, homogenized, and dilution plated to determine whether C. immitis was present. Both lungs and spleens of rURE- and rHSP60-immune mice were positive in the survival experiments. Despite this lack of clearance of the pathogen, recombinant urease was identified as a potential vaccine candidate based on results of its evaluation in T-cell proliferation assays, cytokine expression studies, and survival experiments. The urease antigen was further tested for its ability to protect mice against C. immitis infection by immunization with a mammalian plasmid expression vector which contained the same URE gene fragment as used for transformation of E. coli and expression of the rURE.
FIG. 5.
Comparison of protective efficacy of the recombinant proteins (rHSP60 and rURE) and BSA plus the immunoadjuvant (CpG ODN) in BALB/c mice challenged by the i.p. route with 100 arthroconidia. Mice were immunized by the s.c. route. Mortality was determined at days 1 through 40 postchallenge for 12 mice in each group.
Confirmation of expression of rURE by pSecTag2A.URE-transfected mammalian cells.
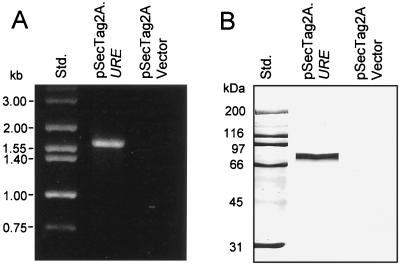
The mammalian plasmid vector construct pSecTag2A.URE was used to transfect murine fibroblast cells by the biolistic method. Transfected clones were selected in growth medium containing Zeocin, and the cell isolates were used as the source of total RNA for RT-PCR analysis of expression of the URE gene. A 1.6-kb band was amplified by PCR using the cDNA template derived from pSecTag2A.URE-transfected cells and the same pair of oligonucleotide primers as used to amplify the 1.6-kb URE gene fragment (Fig. 6A). Total RNA from fibroblasts transfected with the vector alone showed no RT-PCR product with the same primer pair. Immunoblot analysis of fibroblast cell lysates from clones transfected either with pSecTag2A.URE or the vector alone were used to test for production of the urease protein. A single prominent band of 80 kDa was detected by anti-urease antibody in the immunoblot of the lysate derived from the pSecTag2A.URE transfected cells (Fig. 6B). No protein bands were visible in the immunoblot of the control cell lysate. The predicted size of the urease protein expressed by the pSecTag2A.URE construct is 65.2 kDa. The larger molecular size of the actual mammalian cell product is probably the result of posttranslational modification.
FIG. 6.
Expression of URE mRNA and production of recombinant urease protein by pSecTag2A.URE-transfected murine fibroblast cells as determined by RT-PCR (A) and immunoblot analysis (B). The URE gene-specific primers yielded a predicted 1.6-kb PCR product (A), and the urease-specific antiserum detected a single 80-kDa protein band (B). Analyses of control cells transfected with only the pSecTag2A vector showed no products detectable by RT-PCR or immunoblot assay. Std., size standards.
T-cell proliferation and cytokine expression assays of DNA-immunized mice.
T lymphocytes obtained from pSecTag2A.URE-immunized mice and stimulated in vitro with rURE (1.25 to 10 μg) showed high levels of proliferative response compared to control T cells (Fig. 7A). Comparative RT-PCR analysis of cytokine mRNA expression in these same in vitro-stimulated T cells derived from URE DNA-immunized mice revealed distinct IL-2 and IFN-γ gel bands (Fig. 7B). No IL-4 or IL-5 mRNA expression was detected, and IL-10 gene expression was relatively low. The rURE-stimulated immune cells demonstrated a Th1 immune response, and their pattern of cytokine mRNA expression was clearly distinguished from that of the control T cells.
FIG. 7.
(A) Proliferative response of T cells derived from DNA-immunized mice, as indicated, to in vitro stimulation with different concentrations of rURE. Means and standard deviations of three determinations are reported. (B) In vitro expression of selected cytokines and HPRT mRNA in T cells isolated from lymph nodes and spleens of DNA-immunized mice as indicated. The T-cell isolates were stimulated in vitro with rURE (10 μg/ml) or culture medium, and RT-PCR assays were performed as described for Fig. 2.
Survival of mice immunized with DNA.
Mice subjected to biolistic immunization with the pSecTag2A.URE construct revealed 83% survival (10 of 12) at 40 days after i.p. challenge with a lethal inoculum of C. immitis, compared to 17% survival (2 of 12) of mice immunized with the vector alone (Fig. 8). This difference in survival between the two groups of DNA-immunized mice was statistically significant (P = 0.0004). Mice which survived to 42 days postchallenge were sacrificed; their lungs and spleen were homogenized and plated as described above. In the case of the pSecTag2A.URE-immunized mice, 8 of the 10 survivors showed sterile lungs and spleens. The two survivors immunized with the vector alone showed C. immitis-infected organs (>103 CFU/organ).
FIG. 8.
Vaccine efficacy of the pSecTag2A.URE construct in BALB/c mice challenged by the i.p. route with 100 arthroconidia. Mice were immunized intradermally by biolistic injection of DNA using a gene gun. Mortality was determined at days 1 through 40 postchallenge for 12 mice in each group. Control mice were immunized with the pSecTag2A vector alone.
DISCUSSION
We have presented evidence that immunization of BALB/c mice with either the truncated 63-kDa recombinant urease protein (43) or a mammalian plasmid expression vector containing the URE gene which encodes this protein provides protection against a lethal challenge of C. immitis. Our initial selection of urease as a vaccine candidate was based on the results of comparative in vitro immunoassays of rURE and the 66-kDa rHSP60 of C. immitis (40). The heat shock protein was chosen for this study because of its high homology (78%) to the HSP60 protein of Histoplasma capsulatum, an antigen which had been shown to confer protection in BALB/c mice against a lethal intranasal challenge with yeast cells of that fungus (17). H. capsulatum and C. immitis are phylogenetically related fungal pathogens (33), which suggested that rHSP60 protein of C. immitis may also be immunogenic in mice. In fact, we had previously shown that T cells from BALB/c mice immunized with the 66-kDa rHSP60 responded positively to the homologous antigen in an in vitro proliferation assay (40). Comparison of the results of cellular immunoassays of the purified rURE and rHSP60 proteins in the present study, however, demonstrated that rURE is the more potent antigen. This was well illustrated when FKES of C. immitis were used either as the in vitro test antigen to stimulate an immune T-cell response or as the reagent to immunize BALB/c mice. As a test antigen, FKES stimulated responses of both rHSP60- and rURE-immune T cells. However, T lymphocytes isolated from mice which had been immunized with rURE showed the greater in vitro stimulation. The FKES-immune T cells, on the other hand, responded only to rURE at all antigen concentrations tested in the cellular immunoassay. On the basis of these observations, it can be argued that both the native HSP60 and native urease of C. immitis are presented to the host during the course of a natural infection. In fact, HSP60 of C. immitis has been shown to be present in the parasitic cell wall (40), as is also the case for H. capsulatum (16). The mechanism by which this cytosolic protein is transported to the fungal cell wall is unknown. We propose that the native urease, which is also a cytosolic protein, is released with the contents of the mature spherules during the endosporulation phase of the parasitic cycle (18). The response of the murine FKES-immune T cells to the purified rURE in the proliferation assay suggests that under the conditions of immunization used, urease is highly immunogenic and presented to the host in sufficient amounts to establish acquired immunity.
The immunization protocol described in this study employed an ODN which contains immunostimulatory, unmethylated CpG motifs as the vaccine adjuvant (32). Evidence has been presented that unmethylated CpG dinucleotides contained in ODN induce macrophages to secrete IL-12, which stimulates IFN-γ secretion by natural killer cells (7). These and other specific cytokines (e.g., IL-2) can, in turn, induce Th1-dependent delayed-type hypersensitivity (DTH) reactions in the host (38). It has been suggested that T helper cells can be divided into two subsets (21). Th1 cells in essence secrete cytokines that activate macrophages, while Th2 cells secrete cytokines (e.g., IL-4, IL-5, and IL-10) which largely stimulate B cells to produce antibody and do not participate in the DTH response. Cytokine production by T helper cells also plays a role in regulation of Ig isotype expression. Although Th1 cells are poor initiators of antibody response, they participate in isotype switching by release of IFN-γ, which induces expression of IgG2a and IgG3 in mice. IL-4 production in mice, on the other hand, preferentially induces switching to IgG1 and IgE (21). Control of pulmonary infection by H. capsulatum and C. immitis is critically dependent on the release of cytokines from T cells (2, 30). DBA/2 mice, which demonstrate natural resistance to C. immitis infection, preferentially mount a Th1 response to the pathogen, while susceptible BALB/c respond to infection by activation of the Th2 immune pathway (30). It is reasonable to speculate that C. immitis antigens which stimulate expression of Th1 cytokines genes and production of antigen-specific IgG2a would be worthy of further evaluation in protection experiments.
We expanded our comparative evaluation of rHSP60 and rURE as vaccine candidates by examination of expression of selected cytokine genes by in vitro-stimulated immune T cells. Elevated levels of expression of these cytokine mRNAs were used as indicators of Th1 and Th2 pathways of immune response (23). Analysis of expression of the constitutive HPRT gene was used to compare relative amounts of mRNA in each RT-PCR. The results showed that rURE-immune T cells after stimulation with the homologous antigen expressed both IFN-γ and IL-2 mRNAs. In contrast, in vitro-stimulated rHSP60-immune cells expressed relatively low levels of Th1 but elevated levels of Th2 cytokine mRNAs (IL-4, IL-5, and IL-10). Subsequent examinations of cytokine gene expression in lung tissue of immunized and infected mice revealed essentially the same results. Lung tissue from rURE-immunized mice showed distinctly elevated levels of IFN-γ mRNA and no detectable IL-5 gene expression. Lung preparations from rHSP60-immunized and control mice, on the other hand, showed no detectable IFN-γ mRNA but did reveal expression of the IL-5 cytokine gene. IL-2 gene expression was detected in lungs of the rHSP60-immunized mice, but at a lower level than in lungs of rURE-immune mice relative to HPRT gene expression. IL-10 mRNA expression appeared to be slightly elevated in the rURE-immunized mice. Susceptible strains of mice (e.g., BALB/c, C57BL6, and CAST/Ei) infected with C. immitis express more IL-10 and IL-4 mRNA than resistant strains (12). IL-10 is known to inhibit stimulation of the Th1 immune response pathway in mice and blocks cytokine release from macrophages, but it also plays a regulatory role in the production of IL-12 (21). Although we did not examine IL-12 mRNA expression, it is known that an increased level of expression of this cytokine in BALB/c mice correlates with elevated expression of IFN-γ (31).
Evaluations of the protective capacity of rHSP60 and rURE against a lethal C. immitis infection in BALB/c mice were conducted using the same immunization protocol that elicited the in vitro T-cell proliferative response and cytokine mRNA expression. Comparative studies of the fungal burden in different groups of immune mice showed that addition of CpG ODN to the control (BSA) immunization protocol had no significant effect on the CFU of organ homogenates obtained from infected animals. In three separate experiments, mice immunized with rURE plus CpG ODN had about 104- to 105-fold fewer organisms in their lungs and spleens than did the control animals. In contrast, the rHSP60-immune mice showed less than 1 log10 unit reduction in the number of CFU in these same body organs compared to the control animals. The results of survival studies with rHSP60 or rURE plus CpG ODN as the immunogen also revealed that rURE was more protective than rHSP60. In summary, results of the T-cell proliferation assays, cytokine expression studies, fungal burden, and survival experiments identified C. immitis urease protein as a candidate vaccine against coccidioidomycosis. However, its protective efficacy in mice was not equivalent to that of the formalin-killed spherule vaccine, which resulted in sterile lungs in BALB/c mice (29, 35), or to the level of protection provided by a water-soluble, multicomponent fraction of the formalin-killed spherule homogenate (44). Nevertheless, results of protection experiments in BALB/c mice vaccinated with rURE showed better outcome than results of immunization with the previously reported T-cell-reactive antigen (26) or proline-rich antigen (25).
In a recent investigation of the protective capacity of the proline-rich antigen (Ag2/PRA) (22), it was shown that DNA vaccination protected BALB/c mice against i.p. challenge with C. immitis much better than the recombinant protein. The pVR1012 mammalian expression vector (Vical, Inc., San Diego, Calif.) was used for delivery of the gene by the intramuscular route. The authors demonstrated that the protective effects of the genetic vaccine correlated with acquisition of a footpad DTH response and production of IFN-γ. In a repetition of essentially this same experiment in another laboratory, the authors found that the DNA vaccine was protective but was not significantly better than the recombinant Ag2/PRA vaccine (1). On the basis of these studies, we decided to evaluate immunization of BALB/c mice with the URE gene fragment which encodes the 63-kDa recombinant protein. We initially confirmed that murine fibroblasts grown in vitro and transfected by biolistic bombardment (42) with the pSecTag2A.URE mammalian plasmid construct expressed URE mRNA and produced the urease protein. The gene gun was then used to immunize mice by the i.d. route, with either the plasmid vector construct or the vector alone. In vitro assays of proliferative response and cytokine mRNA expression by T cells isolated from URE DNA-immunized mice indicated the production of antigen-specific immune lymphocytes which demonstrated a Th1 response to stimulation. Genetic immunization of BALB/c mice with pSecTag2A.URE resulted in a striking improvement in protection against a lethal i.p. challenge with C. immitis compared to immunization with rURE. While mice immunized with the pSecTag2A vector that survived to 42 days postchallenge still contained C. immitis in their lungs and spleens, 87% of the pSecTag2A.URE-immunized survivors had sterile lungs and spleens.
Although the results of genetic immunization of mice with the urease gene are intriguing, several issues need to be addressed in future studies. We have not evaluated the protective capacity of either the recombinant protein or DNA vaccine against pulmonary challenge. The pulmonary route is the natural route of C. immitis infection. However, i.p. inoculation has been shown to yield reproducible results and is a useful method to initially evaluate candidate antigens (22, 25, 26). DNA delivery into the skin by the biolistic method has been reported to elicit a dominant Th2 response, whereas DNA delivery by intramuscular injection induces a Th1 response (11). However, both in vitro RT-PCR analysis and preliminary studies of cytokine gene expression in lung tissue of pSecTag2A.URE-immunized BALB/c mice revealed a markedly elevated level of IFN-γ mRNA compared to mice immunized with pSecTag2A alone. Current studies are focused on characterization of the nature of the host protective response following pSecTag2A.URE immunization and C. immitis infection.
ACKNOWLEDGMENTS
Support for this study was provided by Public Health Service grants AI19149 and AI37232 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and a grant from the California HealthCare Foundation awarded to G.T.C.
REFERENCES
- 1.Abuodeh R O, Shubitz L F, Siegel E, Snyder S, Peng T, Orsborn K I, Brummer E, Stevens D A, Galgiani J N. Resistance to Coccidioides immitis in mice after immunization with recombinant protein or a DNA vaccine of a proline-rich antigen. Infect Immun. 1999;67:2935–2940. doi: 10.1128/iai.67.6.2935-2940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allendoerfer R, Deepe G S. Regulation of infection with Histoplasma capsulatum by TNFR1 and -21. J Immunol. 2000;165:2657–2664. doi: 10.4049/jimmunol.165.5.2657. [DOI] [PubMed] [Google Scholar]
- 3.Ampel N M, Dols C L, Galgiani J N. Coccidioidomycosis during human immunodeficiency virus infection: results of a prospective study in a coccidioidal endemic area. Am J Med. 1993;94:235–240. doi: 10.1016/0002-9343(93)90054-s. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley; 1989. [Google Scholar]
- 5.Beaman L, Pappagianis D, Benjamini E. Significance of T cells in resistance to experimental murine coccidioidomycosis. Infect Immun. 1977;17:580–585. doi: 10.1128/iai.17.3.580-585.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaman L, Pappagianis D, Benjamini E. Mechanisms of resistance to infection with Coccidioides immitis in mice. Infect Immun. 1979;23:681–685. doi: 10.1128/iai.23.3.681-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu R S, Targoni O S, Krieg A M, Lehmann P V, Harding C V. CpG oligodeoxynucleotides act as adjuvants that switch on T helper 1 (Th 1) immunity. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole G T, Kirkland T N. Conidia of Coccidioides immitis: their significance in disease initiation. In: Cole G T, Hoch H C, editors. The fungal spore and disease initiation in plants and animals. New York, N.Y: Plenum Press; 1991. pp. 403–443. [Google Scholar]
- 9.Cole G T, Kirkland T N, Sun S H. An immunoreactive, water-soluble conidial wall fraction of Coccidioides immitis. Infect Immun. 1987;55:657–667. doi: 10.1128/iai.55.3.657-667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current protocols in immunology. New York, N.Y: John Wiley; 1998. [Google Scholar]
- 11.Feltquate D M, Heaney S, Webster R G, Robinson H L. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunizations. J Immunol. 1997;158:2278–2284. [PubMed] [Google Scholar]
- 12.Fierer J, Walls L, Eckmann L, Yamamoto T, Kirkland T N. Importance of interleukin-10 in genetic susceptibility of mice to Coccidioides immitis. Infect Immun. 1998;66:4397–4402. doi: 10.1128/iai.66.9.4397-4402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frohman M A. RACE: rapid amplification of cDNA ends. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. New York, N.Y: Academic Press; 1990. pp. 28–38. [Google Scholar]
- 14.Galgiani J N. Coccidioidomycosis. West J Med. 1993;159:153–171. [PMC free article] [PubMed] [Google Scholar]
- 15.Galgiani J N. Coccidioidomycosis: a regional disease of national importance. Rethinking approaches for control. Ann Intern Med. 1999;130:293–300. doi: 10.7326/0003-4819-130-4-199902160-00015. [DOI] [PubMed] [Google Scholar]
- 16.Gomez A M, Rhodes J C, Deepe G S. Antigenicity and immunogenicity of an extract from the cell wall and cell membrane of Histoplasma capsulatum yeast cells. Infect Immun. 1991;59:330–336. doi: 10.1128/iai.59.1.330-336.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez F J, Allendoerfer R, Deepe G S. Vaccination with recombinant heat shock protein 60 from Histoplasma capsulatum protects mice against pulmonary histoplasmosis. Infect Immun. 1995;63:2587–2595. doi: 10.1128/iai.63.7.2587-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guevara-Olvera L, Hung C-Y, Yu J-J, Cole G T. Sequence, expression and functional analysis of the Coccidioides immitis ODC (ornithine decarboxylase) gene. Gene. 2000;242:437–448. doi: 10.1016/s0378-1119(99)00496-5. [DOI] [PubMed] [Google Scholar]
- 19.Hung C-Y, Ampel N M, Christian L, Seshan K R, Cole G T. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect Immun. 2000;68:584–593. doi: 10.1128/iai.68.2.584-593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huppert M. Antigens used for measuring immunological reactivity. In: Howard D H, editor. Fungi pathogenic for humans and animals. Part B. Pathogenicity and detection. New York, N.Y: Marcel Dekker; 1983. pp. 219–302. [Google Scholar]
- 21.Janeway C A, Travers P, Walport M, Capra J D. Immunobiology: the immune system in health and disease. 4th ed. London, United Kingdom: Current Biology Publications; 1999. [Google Scholar]
- 22.Jiang C, Magee D M, Quitugua T N, Cox R A. Genetic vaccination against Coccidioides immitis: comparison of vaccine efficacy of recombinant antigen 2 and antigen 2 cDNA. Infect Immun. 1999;67:630–635. doi: 10.1128/iai.67.2.630-635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami K, Tohyama M, Qifeng X, Saito A. Expression of cytokines and inducible nitric oxide synthase mRNA in the lungs of mice infected with Cryptococcus neoformans: effects of interleukin-12. Infect Immun. 1997;65:1307–1312. doi: 10.1128/iai.65.4.1307-1312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkland T N, Fierer J. Inbred mouse strains differ in resistance to lethal Coccidioides immitis infection. Infect Immun. 1983;40:912–916. doi: 10.1128/iai.40.3.912-916.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkland T N, Finley F, Orsborn K I, Galgiani J N. Evaluation of the proline-rich antigen of Coccidioides immitis as a vaccine candidate in mice. Infect Immun. 1998;66:3519–3522. doi: 10.1128/iai.66.8.3519-3522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkland T N, Thomas P W, Finley F, Cole G T. Immunogenicity of a 48-kilodalton recombinant T-cell-reactive protein of Coccidioides immitis. Infect Immun. 1998;66:424–431. doi: 10.1128/iai.66.2.424-431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirkland T N, Zhu S W, Kruse D, Hsu L L, Seshan K R, Cole G T. Coccidioides immitis fractions which are antigenic for immune T lymphocytes. Infect Immun. 1991;59:3952–3961. doi: 10.1128/iai.59.11.3952-3961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine H B. Purification of the spherule-endospore phase of Coccidioides immitis. Sabourandia. 1961;1:112–115. doi: 10.1080/00362176285190231. [DOI] [PubMed] [Google Scholar]
- 29.Levine H B, Kong Y C, Smith C E. Immunization of mice to Coccidioides immitis: dose, regimen and spherulation stage of killed spherule vaccines. J Immunol. 1965;94:132–142. [PubMed] [Google Scholar]
- 30.Magee D M, Cox R A. Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infect Immun. 1995;63:3514–3519. doi: 10.1128/iai.63.9.3514-3519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magee D M, Cox R A. Interleukin-12 regulation of host defenses against Coccidioides immitis. Infect Immun. 1996;64:3609–3613. doi: 10.1128/iai.64.9.3609-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCluskie M J, Davis H L. Oral, intrarectal and intranasal immunizations using CpG and non-CpG oligodeoxynucleotides as adjuvants. Vaccine. 2000;19:413–422. doi: 10.1016/s0264-410x(00)00208-5. [DOI] [PubMed] [Google Scholar]
- 33.Pan S, Sigler L, Cole G T. Evidence for a phylogenetic connection between Coccidioides immitis and Uncinocarpus reesii (Onygenaceae) Microbiology. 1994;140:1481–1494. doi: 10.1099/00221287-140-6-1481. [DOI] [PubMed] [Google Scholar]
- 34.Pappagianis D. Epidemiology of coccidioidomycosis. Curr Top Med Mycol. 1988;2:199–238. doi: 10.1007/978-1-4612-3730-3_6. [DOI] [PubMed] [Google Scholar]
- 35.Pappagianis D the Valley Fever Vaccine Study Group. Evaluation of the protective efficacy of the killed Coccidioides immitis spherule vaccine in humans. Am Rev Respir Dis. 1993;148:656–660. doi: 10.1164/ajrccm/148.3.656. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Smith C E. Epidemiology of acute coccidioidomycosis with erythema nodosum (“San Joaquin” or “Valley Fever”) Am J Public Health. 1940;30:600–611. doi: 10.2105/ajph.30.6.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stacey K J, Blackwell J M. Immunostimulatory DNA as an adjuvant in vaccination against Leishmania major. Infect Immun. 1999;67:3719–3726. doi: 10.1128/iai.67.8.3719-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svetic A, Finkelman F D, Jian Y C, Dieffenbach C W, Scott D E, McCarthy K F, Steinberg A D, Gause W C. Cytokine gene expression after in vivo primary immunization with goat antibody to mouse IgD antibody. J Immunol. 1991;147:2391–2397. [PubMed] [Google Scholar]
- 40.Thomas P W, Wyckoff E E, Pishko E J, Yu J-J, Kirkland T N, Cole G T. The hsp60 gene of the human pathogenic fungus Coccidioides immitis encodes a T-cell reactive protein. Gene. 1997;199:83–91. doi: 10.1016/s0378-1119(97)00351-x. [DOI] [PubMed] [Google Scholar]
- 41.Vanderzanden L, Bray M, Fuller D, Roberts T, Custer D, Spik K, Jahrling P, Huggins J, Schmaljohn A, Schmaljohn C. DNA vaccines expressing either the GP or NP genes of Ebola virus protect mice from lethal challenge. Virology. 1998;246:134–144. doi: 10.1006/viro.1998.9176. [DOI] [PubMed] [Google Scholar]
- 42.Yu J-J, Cole G T. Biolistic transformation of the human pathogenic fungus Coccidioides immitis. J Microbiol Methods. 1998;33:129–141. [Google Scholar]
- 43.Yu J-J, Smithson S L, Thomas P W, Kirkland T N, Cole G T. Isolation and characterization of the urease gene (URE) from the pathogenic fungus Coccidioides immitis. Gene. 1997;198:387–391. doi: 10.1016/s0378-1119(97)00342-9. [DOI] [PubMed] [Google Scholar]
- 44.Zimmermann C R, Johnson S M, Martens G W, White A G, Zimmer B L, Pappagianis D. Protection against lethal murine coccidioidomycosis by a soluble vaccine from spherules. Infect Immun. 1998;66:2342–2345. doi: 10.1128/iai.66.5.2342-2345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]