Abstract
Salmonella enterica serotype Typhi differs from nontyphoidal Salmonella serotypes by its strict host adaptation to humans and higher primates. Since fimbriae have been implicated in host adaptation, we investigated whether the serotype Typhi genome contains fimbrial operons which are unique to this pathogen or restricted to typhoidal Salmonella serotypes. This study established for the first time the total number of fimbrial operons present in an individual Salmonella serotype. The serotype Typhi CT18 genome, which has been sequenced by the Typhi Sequencing Group at the Sanger Centre, contained a type IV fimbrial operon, an orthologue of the agf operon, and 12 putative fimbrial operons of the chaperone-usher assembly class. In addition to sef, fim, saf, and tcf, which had been described previously in serotype Typhi, we identified eight new putative chaperone-usher-dependent fimbrial operons, which were termed bcf, sta, stb, ste, std, stc, stg, and sth. Hybridization analysis performed with 16 strains of Salmonella reference collection C and 22 strains of Salmonella reference collection B showed that all eight putative fimbrial operons of serotype Typhi were also present in a number of nontyphoidal Salmonella serotypes. Thus, a simple correlation between host range and the presence of a single fimbrial operon seems at present unlikely. However, the serotype Typhi genome differed from that of all other Salmonella serotypes investigated in that it contained a unique combination of putative fimbrial operons.
The genus Salmonella contains pathogens which are closely related genetically but differ in their host range (7). One end of the spectrum is formed by broad-host-range pathogens such as Salmonella enterica serotype Typhimurium, which is frequently associated with cases of disease in a number of animal species, including mice, pigs, poultry, horses, cattle, and sheep (19, 43, 52). At the other end of the spectrum are pathogens whose ability to cause disease is restricted to a single genus or related genera of vertebrate species. Serotype Typhi is a prototypical host-restricted serotype which causes typhoid fever in humans and higher primates but is unable to produce illness in other vertebrate species. Since there is no inexpensive animal model with which to study serotype Typhi pathogenesis, little is known about virulence factors which are responsible for its apparent adaptation to the human host and its ability to cause typhoid fever. With the sequence of the whole serotype Typhi genome now almost complete, we can begin to address these questions using comparative genomic analysis.
One of the virulence factors recently implicated in adaptation of serotype Typhi to the human host is a fimbrial operon termed tcf, for Typhi colonization factor. Serotype Typhi is the only serotype within Salmonella reference collection C (SARC), a strain collection consisting of 16 isolates representing all phylogenetic lineages within the genus Salmonella, which hybridizes with a DNA probe specific to the tcf operon (20). The serotype Typhi tcf operon consists of four genes, tcfABCD, which display sequence homology to genes within the coo operon, encoding CS1 fimbriae of human-adapted enterotoxigenic Escherichia coli. Interestingly, colonization factor antigens such as CS1 confer the species specificity of enterotoxigenic E. coli isolates that are adapted to the human host (14). From the restricted distribution among Salmonella serotypes and its homology with genes encoding a human colonization factor, Folkesson and coworkers concluded that the tcf operon encodes serotype Typhi-specific fimbriae, which may play a role in the strict human specificity observed for this pathogen (20).
The advent of complete genome sequencing allows identification of all putative fimbrial operons present in a bacterial pathogen. The information obtained from shotgun sequencing of the serotype Typhi genome can hence be used to investigate further whether the presence of genes encoding an individual adhesin or a combination of putative fimbrial operons correlates with adaptation of this pathogen to the human host. To cover the spectrum of genetic diversity among Salmonella serotypes, the distribution of putative serotype Typhi fimbrial operons can be determined by hybridization with strains of the SARC collection, which represent isolates of all phylogenetic lineages within the genus Salmonella, including Salmonella bongori and S. enterica subspecies I, II, IIIa, IIIb, IV, VI, and VII (9). Within the SARC collection, one phylogenetic group, S. enterica subspecies I, is of particular interest for public health because it contains approximately 60% of known Salmonella serotypes (38). Furthermore, members of S. enterica subspecies I are frequently isolated from mammals and birds and account for more than 99% of cases of disease reported from humans and domesticated animals (1). In contrast, members of S. bongori and S. enterica subspecies II to VII are rarely isolated from mammals or birds but rather represent reptile-associated Salmonella serotypes (7, 38). The SARC collection contains only two serotypes of S. enterica subspecies I, the host-restricted serotype Typhi and the broad-host-range serotype Typhimurium (9). Thus, to compare the repertoire of putative serotype Typhi fimbrial operons with that of other serotypes adapted to humans, livestock, or domestic fowl, hybridization analysis with fimbrial biosynthesis genes has to be extended to include common S. enterica subspecies I serotypes which are not represented in the SARC collection. For instance, S. enterica subspecies I contains a number of well-characterized host-restricted serotypes, such as the avian-adapted Gallinarum, the bovine-adapted Dublin, the porcine-adapted Choleraesuis, and several human-adapted typhoidal serotypes, including Paratyphi A, Paratyphi B, Paratyphi C, and Sendai. In addition, several broad-host-range serotypes of S. enterica subspecies I, such as Enteritidis, Heidelberg, and Agona, are frequently associated with diarrheal disease in humans and hence should be included in an investigation on the distribution of fimbrial operons within this genus.
In this study, we have identified open reading frames in the serotype Typhi genome which display homology to fimbrial biosynthesis genes. Hybridization analysis was performed to compare the genomic repertoire of putative fimbrial operons present in serotype Typhi with that present in strains of the SARC collection and a representative collection of S. enterica subspecies I serotypes. This analysis provided an impression of the complete makeup of fimbrial biosynthesis genes present in serotype Typhi and its relation to the fimbrial repertoires present in other members of its genus.
MATERIALS AND METHODS
Bacterial strains.
SARC has been described previously (9). Salmonella serotypes of S. enterica subspecies I were isolates from Salmonella reference collection B (SARB), which has been reported recently (8). CT18 is multiple-antibiotic-resistant, Vi-positive serotype Typhi strain carrying two plasmids (pHCM1 and pHCM2) which was isolated in the Mekong Delta (Vietnam) from a child with uncomplicated typhoid fever. The serotype Typhi strain Ty2 was first isolated in 1918 in Cherson, and this laboratory strain was obtained from stock maintained at the American Type Culture Collection (ATCC 19430). An avirulent derivative of strain Ty2, termed Ty21a, which was subsequently licensed for use as a live oral vaccine, was originally described by Germanier and Fürer (21) and received from the American Type Culture Collection (ATCC 33459). H901 is a serotype Typhi strain with reduced virulence which was obtained from the American Type Culture Collection (ATCC 33458) and has been described previously (24). Clinical serotype Typhi strains from the State of California Health Laboratory isolated in different years have been described previously (29). The E. coli strain TA One Shot was purchased from Invitrogen.
DNA analysis.
Nucleotide sequences of serotype Typhi with homology to fimbrial biosynthesis genes were identified by homology search using the programs BlastX and BlastP (2, 3). The sequence data of clinical isolate CT18 were produced by the Typhi Sequencing Group at the Sanger Centre and can be obtained from http://www.sanger.ac.uk/Projects/S_typhi. Comparison of deduced amino acid sequences and design of PCR primers were performed using the program MacVector 6.5 (Oxford Molecular Group Inc.).
Generation of nucleotide probes.
DNA probes specific for the pef, lpf, agf, fim, and sef operons have been described previously (6). The major fimbrial subunits of the tcf operon (tcfA) and the bcf operon (bcfA) were PCR amplified and cloned into vector pCR2.1-TOPO (Invitrogen) to give rise to plasmids pNK37 and pNK38, respectively (Table 1). A fragment of the safC gene was PCR amplified and cloned (pST4) to generate a DNA probe specific for the saf operon. DNA regions containing fragments of staAB, stbA, stcAB, stdA, steB, stgAB, and sthAB were PCR amplified and cloned into plasmid vector pCR2.1-TOPO to give rise to plasmids pNK4, pNK9, pNK29, pNK13, pST11, pNK1, and pST13, respectively. The primers for PCR amplification of fimbrial biosynthesis genes are listed in Table 1. After agarose gel electrophoresis, PCR products were isolated using a QiaexII kit (Qiagen) and the protocols provided by the manufacturer. Subsequently, PCR products were cloned using a TA-TOPO cloning kit from Invitrogen. Plasmid DNA was isolated using ion exchange columns from Qiagen. DNA probes were generated after EcoRI restriction of plasmids and gel purification of insert DNA by labeling the restriction fragments depicted in Fig. 1 using the labeling and detection kit (nonradioactive) from NEN.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Nucleotide sequence (5′ → 3′) |
|---|---|
| tcf-1 | CATTTATTCTCAGGGGGAGCG |
| tcf-2 | CATCCTCTTTATCTGTTGCCACG |
| bcf-1 | TCCCCCGGGGATACTACAACCGTCACTGG |
| bcf-2 | GCGGATAAATCACCCTGGTC |
| saf-1 | TGTTCTGGCTCCTTGTTTGACG |
| saf-2 | TTCTGTTTGACCTCCACCCGAG |
| sta-1 | CCGAATTCAGTTGAAGGCGGCCAGATCG |
| sta-2 | CACCCGGGCGTCGTTATCGTTACCCAGGTC |
| stb-1 | CCGTCGACCGTTTCAATGCCAGACCGTAGC |
| stb-2 | CCGAATTCTGTTTCTGATAACACCATCACC |
| stc-1 | CCGAATTCTGTTGATGAGTATGATTCAGG |
| stc-2 | GACCCGGGGACTTCTTTCTTCTCTGCC |
| std-1 | CCGAATTCAGTCGATCCACCAACAGCAGG |
| std-2 | CACCCGGGGACGGAAAATGTCAGGGTAATCAG |
| ste-1 | ATAGCGTGTGGAGTGGGAATGC |
| ste-2 | ATAGCGTGTGGAGTGGGAATGC |
| stg-1 | CCGAATTCATCTGATGGCACCGTTCACTTCC |
| stg-2 | CACCCGGGATCCTTTGGCAACTCACC |
| sth-1 | GACCCGGGGGTTACGGTTGCGGAACCATCG |
| sth-2 | GTATTGTAATGGGCTGTGGTGTCTC |
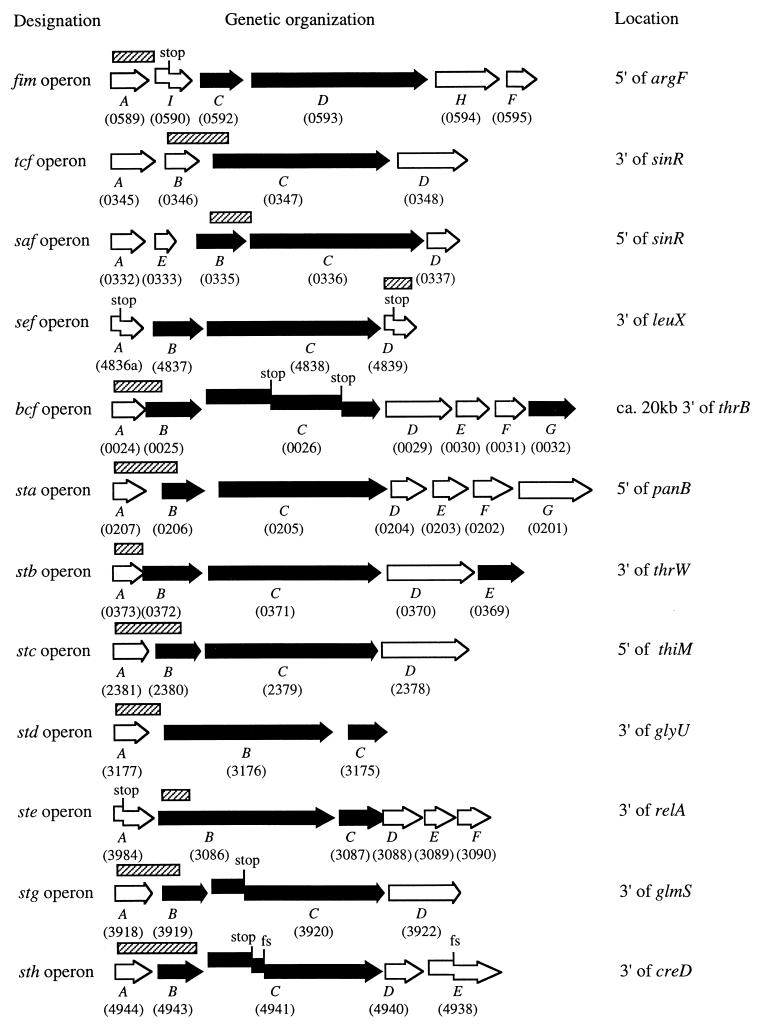
FIG. 1.
Genetic organization of putative serotype Typhi operons of the chaperone-usher-dependent assembly class. The designation of genes is indicated, and the systematic number designation (STY number) of open reading frames annotated in the serotype Typhi genome is given in parentheses. Open arrows indicate the positions of open reading frames with homology to genes encoding fimbrial subunits. Solid arrows indicate the positions of open reading frames with homology to genes involved in transport and assembly of subunits (fimbrial chaperones and ushers). The positions of DNA fragments used as DNA probes for hybridization analysis with strains of the SARC and SARB collections are indicated by hatched bars. stop, stop codon; fs, frameshift mutation.
Southern hybridization.
Isolation of total bacterial DNA was performed as recently described (4). DNA was restricted with EcoRI, and the fragments were separated on a 0.5% agarose gel. Southern transfer of DNA onto a nylon membrane was performed as previously described (4). Hybridization was performed at 65°C in solutions without formamide. One 15-min wash was performed under nonstringent conditions at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS). Subsequently, one 15-min wash was performed under stringent conditions at 65°C in 0.2× SSC–0.1% SDS. Hybrids were detected using the labeling and detection kit (nonradioactive) from NEN.
RESULTS
Identification of putative fimbrial operons.
Three pathways for the assembly of fimbrial adhesins have been described in members of the family Enterobacteriaceae to date. These include the chaperone-usher-dependent assembly pathway (46), the nucleator-dependent assembly pathway (22), and the assembly pathway for type IV fimbriae (25). While exported fimbrial subunits that assemble into fimbrial filaments show little sequence homology, the amino acid sequences of components involved in the export and assembly process do tend to be conserved within each pathway. To identify putative fimbrial operons in the serotype Typhi genome, we therefore performed homology searches using sequences of fimbrial proteins known to be involved in assembly.
Putative serotype Typhi fimbrial operons of the nucleator-dependent assembly class were identified by homology search (BlastP) with amino acid sequences of aggregative (also known as curli) fimbrial proteins from serotype Typhimurium (40). Only one fimbrial operon was identified, which represented the serotype Typhi orthologue of the aggregative fimbrial (agf) operon of serotype Typhimurium.
The presence in some Salmonella serotypes of genes encoding type IV fimbriae is suggested by hybridization analysis using genes encoding the bundle-forming pilus (bfp) of enteropathogenic E. coli as DNA probes. However, this DNA probe does not hybridize with genomic DNA from serotype Typhi (42). The identification of a gene cluster for the production of serotype Typhi type IV pili was described recently (54). This fimbrial operon was located on a large (>135 kb) DNA region which was absent from serotype Typhimurium (Genome Sequencing Center, personal communication) and contained capsule (Vi antigen) biosynthesis genes and a bacteriophage carrying the sopE1 gene. The serotype Typhi genome was scanned for additional putative type IV fimbrial operons using the amino acid sequences of the type IV prepilin peptidases TcpJ of Vibrio cholerae, BfbP of enteropathogenic E. coli, PilC and PilD of Pseudomonas aeruginosa, and PilD of Neisseria gonorrhoeae (26, 28, 35, 53). An operon orthologous to the cryptic E. coli ppdD hopBC operon was detected in the serotype Typhi genome (51). Due to the apparent lack of type IV fimbriae in E. coli K-12 and the lack of phenotype of an E. coli hopB mutant, the function of the ppdD hopBC operon, if any, is presently unclear. It is questionable whether the cryptic ppdD hopBC operon is related to fimbrial biosynthesis, because homologues of these genes encode transfer functions of conjugative plasmids or are found in the genomes of filamentous phages. The ppdD and hopBC genes may hence represent the remnant of an integrated plasmid or bacteriophage, and the serotype Typhi orthologues were not characterized further in this study.
Twelve putative serotype Typhi fimbrial operons of the chaperone-usher-dependent assembly class were identified by NCBI Blast homology searches within the serotype Typhi genome (Fig. 1). Four of these operons, sef, tcf, saf, and fim, had been described previously (16, 20, 41). However, the CT18 genome contained stop codons in sefA, sefD, and fimI (Fig. 1). Furthermore, an additional open reading frame (safE) was detected in the saf operon of strain CT18. The saf and tcf operons were present on an approximately 60-kb DNA region which was absent from the E. coli K-12 genome and flanked on one side by a tRNA gene (aspV). Similarly, the sef operon was located on a DNA region that was absent from E. coli K-12 and flanked on one side by a tRNA gene (leuX). In contrast, an orthologue of the serotype Typhi fimACDHF operon (designated sfmACDHF) was located at the corresponding map position (5′ of the folD gene) in the E. coli K-12 genome. It should be pointed out that the E. coli orthologue of the serotype Typhi fim operon is not identical with the E. coli fim operon. The fim operon of E. coli maps to a different location than that of serotype Typhi, and both gene clusters are distinct in gene order and the number of genes present. Thus, despite the fact that the fim operons of E. coli and serotype Typhi both encode mannose-sensitive adhesins which are termed type I fimbriae, these two operons are clearly paralogous. An orthologue of the E. coli fim operon was not present in the serotype Typhi genome.
One operon that had not previously been described in serotype Typhi represented an orthologue of the putative bcf fimbrial operon recently identified in serotype Typhimurium (48). This operon was located between orthologues of dnaJ and nhaA on a 30-kb DNA region that was absent from the E. coli K-12 genome. The remaining seven putative fimbrial operons had not previously been described in the genus Salmonella and were designated sta, stb, stc, std, ste, stg, and sth. Our subsequent analysis focused on characterizing these putative fimbrial operons of the chaperone-usher-dependent assembly class.
The staABCDEFG operon was located upstream of the panB gene and represented an orthologue of an operon present in the E. coli K-12 genome, which is formed by the genes yadCKLM, htrE, ecpD, and yadN (39). Although the predicted amino acid sequences of fimbrial proteins in serotype Typhi and E. coli had only between 33.9 and 60.3% identity, the gene order was conserved. Furthermore, sequences upstream (filK and pcnB) and downstream (panDCB) of sta were highly conserved between the two organisms (between 82.3 and 96% amino acid sequence identity).
The stbABCDE operon was located on a 26.7-kb DNA region that was absent from the E. coli K-12 genome. The tRNA gene thrW and an attachment site for bacteriophage P22 (ataA) formed one border of this genetic island, while the second border was formed by an orthologue of the E. coli open reading frame yahN. This island also carried foxA, a gene encoding an outer membrane ferrioxamine receptor described recently in serotype Typhimurium (27).
The genes stcABCD were found to be orthologues of open reading frames yehDCBA, respectively, of E. coli K-12. The predicted amino acid sequences of fimbrial proteins in serotype Typhi and E. coli had only between 32 and 75% identity, while proteins encoded upstream (MetG, 95% identity) or downstream of the stc operon were highly conserved.
The stdABC operon was located on a 19.5-kb DNA region that was absent from the E. coli K-12 genome. The tRNA gene glyU and an orthologue of the E. coli open reading frame b2845 formed the borders of this genetic island.
The steABCDEF operon was flanked by mazG and relA and was located on a 9-kb DNA region that was absent from the E. coli K-12 genome. The first gene in the operon, steA, displayed homology to genes encoding fimbrial subunits. However, sequence homology suggested that a stop codon detected at codon 88 of steA would result in truncation of the putative fimbrial subunit (Fig. 1).
The stgABCD operon was located on a 9-kb DNA region that was absent from the E. coli K-12 genome. The insertion, relative to the E. coli K-12 sequence, was located between orthologues of glmS and pstS. A TAA stop was detected at codon 171 of the stgC coding sequence, the gene encoding the putative usher of the stg operon (Fig. 1).
The sthABCDE operon was located on a 6-kb DNA region that was absent from the E. coli K-12 genome. This genetic region was flanked by orthologues of the E. coli creD and arcA genes. DNA sequences with homology to genes encoding fimbrial usher proteins (sthC) and fimbrial subunits (sthE) were pseudogenes (interrupted by a stop codon or a frameshift mutation) (Fig. 1).
Distribution of putative fimbrial operons of the chaperone-usher-dependent assembly pathway within the genus Salmonella.
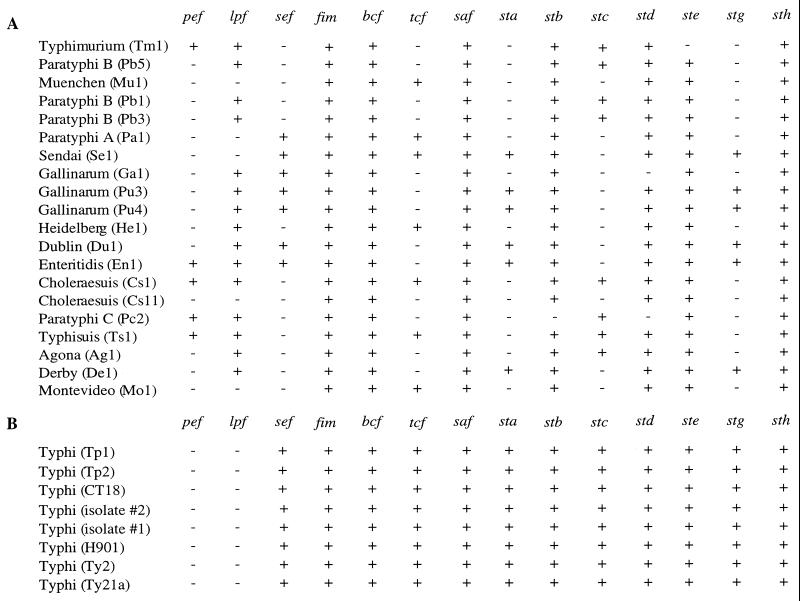
The serotype Typhi genome contained 12 putative fimbrial operons of the chaperone-usher-dependent assembly class (Fig. 1). The distribution among a representative set of S. enterica subspecies I serotypes has been determined for only three of these operons, fim, saf, and sef (5, 16, 20, 45). To determine the distribution of the remaining fimbrial operons of the chaperone-usher-dependent assembly pathway among strains of S. enterica subspecies I, Southern blot analysis was performed with genomic DNA from 20 strains of the SARB collection (Fig. 2A). Each DNA probe gave hybridization signals with several Salmonella serotypes. Furthermore, none of the fimbrial operons was restricted to typhoidal Salmonella serotypes (Typhi and Paratyphi A, B, and C). While hybridization analysis demonstrated that safB, bcfAB, stbA, stdA, steB, and sthAB are well conserved among isolates of S. enterica subspecies I, DNA probes specific for tcfB, staAB, stcAB, and stgAB produced a scattered hybridization pattern within this group. The scattered phylogenetic distribution of tcfBC, staAB, stcAB, and stgAB suggests that deletions and/or horizontal transfer events of the corresponding putative fimbrial operons occurred multiple times within S. enterica subspecies I, generating different combinations of fimbrial operons in different serotypes.
FIG. 2.
Distribution of known fimbrial operons among serotypes of S. enterica subspecies I. (A) Distribution of fimbrial operons among 20 strains of the SARB collection determined by Southern hybridization. The distribution of the fim, lpf, pef, agf, and sef operons among these isolates has been reported previously (6). Serotypes: Ag, Agona; Cs, Choleraesuis; De, Derby; Du, Dublin; En, Enteritidis; Ga, Gallinarum (biotype Gallinarum); He, Heidelberg; Mo, Montevideo; Mu, Muenchen; Pa, Paratyphi A; Pb, Paratyphi B; Pc, Paratyphi C; Pu, Gallinarum (biotype Pullorum); Se, Sendai; Tm, Typhimurium; Ts, Typhisuis. (B) Distribution of fimbrial operons among eight serotype Typhi isolates. The presence of fimbrial operons in strain CT18 was determined by analyzing the genomic sequence determined by the Sanger Centre. Data for the remaining isolates were determined by Southern hybridization. Strains Tp1 and Tp2 are part of the SARB collection. +, operon present; −, operon not present.
To assess whether the repertoire of putative fimbrial operons detected in the genomic sequence of the clinical serotype Typhi isolate CT18 was conserved among other members of this serotype, Southern hybridization was performed with genomic DNA from serotype Typhi strains Ty2, Ty21a, and H901, two clinical isolates from California, and two serotype Typhi isolates (Tp1 and Tp2) present in the SARB collection. DNA probes specific for agf, fim, bcf, sef, tcf, sta, stb, stc, std, ste, stg, and sth hybridized with genomic DNA from all isolates. In contrast, no hybridization signal was obtained with DNA probes specific for the serotype Typhimurium fimbrial operons pef and lpf (Fig. 2B).
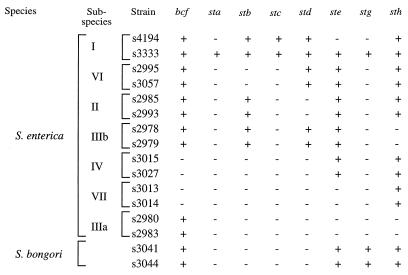
DNA probes specific to newly identified putative fimbrial operons were used for Southern hybridization with total DNA from the 16 strains of the SARC collection, representing S. bongori and S. enterica subspecies I to VII (Fig. 3). DNA probes specific for two fimbrial operons, sta and stc, hybridized only with genomic DNA from S. enterica subspecies I serotypes. Similarly, a previous report indicates that sequences homologous to tcf and saf are restricted to serotypes of S. enterica subspecies I (20). In contrast, in addition to hybridizing with DNA from isolates of S. enterica subspecies I, DNA probes specific for bcf, stb, std, ste, stg, and sth hybridized with genomic DNA from serotypes of S. bongori of other S. enterica subspecies (Fig. 3).
FIG. 3.
Distribution of fimbrial operons among 16 strains of the SARC collection determined by Southern hybridization. The SARC collection contains serotypes of S. bongori (s3041 and s3044), and of S. enterica subspecies I (serotype Typhimurium s4194, serotype Typhi s3333), II (s2985 and s2993), IIIa (s2980 and s2983), IIIb (s2978 and s2979), IV (s3015 and s3027), VI (s2995 and s3057), and VII (s3013 and s3014). +, operon present; −, operon not present.
DISCUSSION
While the distribution among Salmonella serotypes is known for individual fimbrial operons or a selected subset of operons, previous studies fall short of establishing the total number of fimbrial operons present in a particular Salmonella serotype (6, 11, 15, 20, 45, 47). A study showing that a serotype Typhimurium strain carrying mutations in all known fimbrial operons (fim, lpf, pef, and agf) is still able to express several morphologically distinct fimbrial structures suggests that a number of fimbrial operons have not yet been described (50). To obtain a first impression of how many fimbrial operons may be present in a particular Salmonella serotype, we searched the genomic sequence of the serotype Typhi strain CT18 for putative fimbrial genes. The serotype Typhi genome contained one member of the nucleator-dependent assembly class of fimbrial operons which represented an orthologue of the serotype Typhimurium agf operon (40). Furthermore, a type IV pilus operon present in CT18 has been characterized previously in serotype Typhi (54). In addition, the serotype Typhi genome contained 12 putative fimbrial operons of the chaperone-usher-dependent assembly class, 7 of which had not previously been described in the genus Salmonella (Fig. 1). The 12 putative fimbrial operons of the chaperone-usher-dependent assembly class detected in strain CT18 were also present in seven other serotype Typhi strains investigated, including the current oral typhoid vaccine strain Ty21a (Fig. 2B).
A recent study has implicated the tcf fimbrial operon in the adaptation of serotype Typhi to the human host. In support of this idea, it was shown by hybridization analysis using strains of the SARC collection that the tcf operon is a DNA region which is restricted to serotype Typhi (20). Since the 16 strains of the SARC collection include only two serotypes of S. enterica subspecies I (Typhi and Typhimurium) (Fig. 3), we extended this analysis to include other serotypes frequently isolated from humans or domestic animals. Hybridization analysis performed with DNA from 20 representatives of S. enterica subspecies I established that sequences with homology to tcf were not restricted to serotype Typhi (Fig. 2A). While DNA from three typhoidal serotypes, including Typhi, Paratyphi A, and Sendai, hybridized with the tcfA-specific DNA probe, two typhoidal serotypes, Paratyphi B and Paratyphi C, did not contain this DNA region (Fig. 2A and B). Furthermore, DNA from a number of nontyphoidal serotypes, including Heidelberg, Choleraesuis, Montevideo, Typhisuis, and Muenchen, hybridized with the tcfA-specific DNA probe. Thus, the tcf operon is neither specific to serotype Typhi nor restricted to or characteristic of human-adapted typhoidal serotypes. Similarly, none of the other putative fimbrial operons investigated in this study were found to be restricted to serotype Typhi or to human-adapted typhoidal serotypes. This finding may not be surprising, as the detection of fimbrial biosynthesis genes by Southern hybridization does not provide clues about the potential roles of the encoded adhesins during host-pathogen interaction. For instance, the fim operon is well conserved among Salmonella serotypes, as indicated by Southern blot analysis (6, 45). However, the fim operon in serotypes Typhimurium and Typhi encodes an adhesin, termed type 1 fimbriae, which mediates mannose-sensitive hemagglutination (18, 31, 36), whereas in serotype Gallinarum, it encodes surface appendages, termed type 2 fimbriae, which do not mediate mannose-sensitive hemagglutination (13, 37). These differences in the binding specificity of the adhesin encoded by the fim operon are not detected by Southern hybridization (Fig. 2), possibly because the adhesive properties of fimbriae can be altered by point mutations, as recently shown for type 1 fimbriae of E. coli (44). Since the presence of a fimbrial operon in two Salmonella serotypes does not imply that the encoded adhesins possess identical binding properties, it is not possible to make reliable inferences regarding host adaptation using data from Southern hybridization. Although fimbriae may play a role in host adaptation, a simple correlation between host range and the presence of a single fimbrial operon seems at present unlikely.
While alterations in the binding specificity of fimbriae may occur, fimbriae appear to be antigenically conserved (34). Fimbrial antigens are composed of about 1,000 copies of a major fimbrial subunit. The majority of an antibody response against intact whole fimbriae is elicited by the major fimbrial subunit (23, 30). Sequence analyses show that FimA, the major fimbrial subunit of type 1 fimbriae, has 99% sequence identity between serotypes Typhimurium and Enteritidis (17). Similarly, the sequence of serotype Typhimurium lpfA, encoding the major subunit of LP (long polar) fimbriae, differs by only one nonsynonymous change from that of serotype Enteritidis lpfA (33). As a consequence of the high degree of sequence conservation between major fimbrial antigens, serum raised against type 1 fimbriae of serotype Typhimurium or Enteritidis cross-reacts with type 2 fimbriae of serotype Gallinarum and type 1 fimbriae of other Salmonella serotypes (13, 17, 37). Similarly, LpfA of serotypes Enteritidis and Typhimurium, SefA of serotypes Dublin and Enteritidis, and AgfA of various Salmonella serotypes cross-react serologically (15, 16, 33). Furthermore, fimbriae are targeted by the immune response during an infection, as shown by analyzing the serological response to SEF-14 fimbriae during serotype Enteritidis infection in chickens (12), to type 1 fimbriae in human typhoid fever patients (10), and to LpfA during serotype Typhimurium infection in mice (32). Determining the distribution of fimbrial operons is thus a first step in assessing which fimbrial antigens may be expressed by a Salmonella serotype. Interestingly, the repertoire of putative fimbrial operons of the chaperone-usher-dependent assembly class that was detected in the genomic sequence of CT18 was found to be conserved among serotype Typhi isolates (Fig. 2B) but was distinct from that of all other Salmonella serotypes investigated (Fig. 2A). The presence of a unique repertoire of fimbrial biosynthesis genes in serotype Typhi is intriguing and illustrates that our understanding of this aspect in the genetic design of Salmonella serotypes is rather limited. For instance, it is currently an enigma which selective pressures are responsible for generating and maintaining the substantial heterogeneity of fimbrial repertoires found among Salmonella serotypes (Fig. 2).
Evidence for expression of fimbriae assembled by the chaperone-usher-dependent pathway in serotype Typhi is so far only available for the fim operon (18, 31). Thus, with the exception of fim, all operons described in this study should be considered putative. This conclusion is underscored by the finding that several open reading frames in putative fimbrial operons were interrupted by stop codons or frameshift mutations and should thus be considered pseudogenes (Fig. 1). In case these mutations affect genes important for assembly, these operons may not be functional in serotype Typhi. For instance, both fimbrial subunits encoded by the sef gene cluster were pseudogenes in serotype Typhi strain CT18, suggesting that this operon may not encode a functional adhesin. Consistent with this idea, expression of the SefA fimbrial subunit is not detectable by Western blot in serotype Typhi strains hybridizing with a sefA-specific DNA probe (16, 49).
ACKNOWLEDGMENTS
We thank the Genome Sequencing Center, Washington University, St. Louis, for communication of DNA sequence data prior to publication.
Sequence analysis of the serotype Typhi CT18 genome is funded by the Beowulf Genomics Initiative of the Wellcome Trust. Work in A.B.'s laboratory is supported by the Texas Advanced Research (Technology) Program under grant 000089-0051-1999 and Public Health Service grants AI40124 and AI44170.
REFERENCES
- 1.Aleksic S, Heinzerling F, Bockemühl J. Human infection caused by salmonellae of subspecies II to VI in Germany, 1977–1992. Zentbl Bakteriol. 1996;283:391–398. doi: 10.1016/s0934-8840(96)80074-0. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley & Sons; 1994. [Google Scholar]
- 5.Bäumler A J. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 6.Bäumler A J, Gilde A J, Tsolis R M, van der Velden A W M, Ahmer B M M, Heffron F. Contribution of horizontal gene transfer and deletion events to the development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J Bacteriol. 1997;179:317–322. doi: 10.1128/jb.179.2.317-322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bäumler A J, Tsolis R M, Ficht T A, Adams L G. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd E F, Wang F S, Beltran P, Plock S A, Nelson K, Selander R K. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J Gen Microbiol. 1993;139:1125–1132. doi: 10.1099/00221287-139-6-1125. [DOI] [PubMed] [Google Scholar]
- 9.Boyd E F, Wang F S, Whittam T S, Selander R K. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cefalu M, Cutore P. The serological view of typhyoid fever and the anti-fimbriae antibodies. Riv Ist Sieroter Ital. 1967;42:310–322. [PubMed] [Google Scholar]
- 11.Clouthier S C, Collinson S K, Kay W W. Unique fimbriae-like structures encoded by sefD of the SEF14 fimbrial gene cluster of Salmonella enteritidis. Mol Microbiol. 1994;12:893–903. doi: 10.1111/j.1365-2958.1994.tb01077.x. [DOI] [PubMed] [Google Scholar]
- 12.Cooper G L, Thorns C J. Evaluation of SEF14 fimbrial dot blot and flagellar western blot tests as indicators of Salmonella enteritidis infection in chickens. Vet Rec. 1996;138:149–153. doi: 10.1136/vr.138.7.149. [DOI] [PubMed] [Google Scholar]
- 13.Crichton P B, Yakubu D E, Old D C, Clegg S. Immunological and genetical relatedness of type 1 and type 2 fimbriae in salmonellas of serotype Gallinarum, Pullorum and Typhimurium. J Appl Bacteriol. 1989;67:283–291. doi: 10.1111/j.1365-2672.1989.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 14.de Graaf F K, Gaastra W. Fimbriae of enterotoxic Escherichia coli. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 58–83. [Google Scholar]
- 15.Doran J L, Collinson S K, Burian J, Sarlos G, Todd E C, Murno C K, Kay C M, Banser P A, Peterkin P I, Kay W W. DNA-based diagnostic test for Salmonella species targeting agfA, the structural gene for thin, aggregative fimbriae. J Clin Microbiol. 1993;31:2263–2273. doi: 10.1128/jcm.31.9.2263-2273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doran J L, Collinson S K, Clothier S C, Cebula T A, Koch W H, Burian J, Banser P A, Todd E C D, Kay W W. Diagnostic potential of sefA DNA probes to Salmonella enteritidis and certain other O-serogroup D1 Salmonella serovars. Mol Cell Probes. 1996;10:233–246. doi: 10.1006/mcpr.1996.0033. [DOI] [PubMed] [Google Scholar]
- 17.Doran J L, Collinson S K, Kay C M, Banser P A, Burian J, Munro C K, Lee S H, Somers J M, Todd E C, Kay W W. fimA and tctC based DNA diagnostics for Salmonella. Mol Cell Probes. 1994;8:291–310. doi: 10.1006/mcpr.1994.1041. [DOI] [PubMed] [Google Scholar]
- 18.Duguid J P, Anderson E S, Campbell I. Fimbriae and adhesive properties in salmonellae. J Pathol Bacteriol. 1966;92:107–137. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- 19.Edwards P R, Bruner D W. The occurrence and distribution of Salmonella types in the United States. J Infect Dis. 1943;72:58–67. doi: 10.1093/infdis/83.3.220. [DOI] [PubMed] [Google Scholar]
- 20.Folkesson A, Advani A, Sukupolvi S, Pfeifer J D, Normark S, Lofdahl S. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol Microbiol. 1999;33:612–622. doi: 10.1046/j.1365-2958.1999.01508.x. [DOI] [PubMed] [Google Scholar]
- 21.Germanier R, Fürer E. Isolation and characterization of galE mutant Ty21a of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975;131:553–558. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- 22.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson M S, Brinton C C., Jr Identification and characterization of E. coli type-1 pilus tip adhesion protein. Nature. 1988;332:265–268. doi: 10.1038/332265a0. [DOI] [PubMed] [Google Scholar]
- 24.Hickman F W, Rhoden D L, Esaias A O, Baron L S, Brenner D J, Farmer J J., 3rd Evaluation of two Salmonella typhi strains with reduced virulence for use in teaching and proficiency testing. J Clin Microbiol. 1982;15:1085–1091. doi: 10.1128/jcm.15.6.1085-1091.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman M R, Seyer J M, Taylor R K. Processing of TCP pilin by TcpJ typifies a common step intrinsic to a newly recognized pathway of extracellular protein secretion by gram-negative bacteria. Genes Dev. 1991;5:1834–1846. doi: 10.1101/gad.5.10.1834. [DOI] [PubMed] [Google Scholar]
- 27.Kingsley R A, Reissbrodt R, Rabsch W, Ketley J M, Tsolis R M, Everest P, Dougan G, Baumler A J, Roberts M, Williams P H. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl Environ Microbiol. 1999;65:1610–1618. doi: 10.1128/aem.65.4.1610-1618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauer P, Albertson N H, Koomey M. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol Microbiol. 1993;8:357–368. doi: 10.1111/j.1365-2958.1993.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 29.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindberg F, Lund B, Johansson L, Normark S. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature. 1987;328:84–87. doi: 10.1038/328084a0. [DOI] [PubMed] [Google Scholar]
- 31.Muscas P, Rossolini G M, Chiesurin A, Santucci A, Satta G. Purification and characterization of type 1 fimbriae of Salmonella typhi. Microbiol Immunol. 1994;38:353–358. doi: 10.1111/j.1348-0421.1994.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 32.Nicholson T L, Bäumler A J. Salmonella enterica serotype Typhimurium elicits cross-immunity against a Salmonella enterica serotype Enteritidis strain expressing LP fimbriae from the lac promoter. Infect Immun. 2001;69:204–212. doi: 10.1128/IAI.69.1.204-212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norris T L, Baumler A J. Phase variation of the lpf operon is a mechanism to evade cross-immunity between Salmonella serotypes. Proc Natl Acad Sci USA. 1999;96:13393–13398. doi: 10.1073/pnas.96.23.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowotarska M, Mulczyk M. Serologic relationship of fimbriae among Enterobacteriaceae. Arch Immunol Ther Exp. 1977;25:7–16. [PubMed] [Google Scholar]
- 35.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Old D C. Inhibition of the interaction between fimbrial hemagglutinin and erythrocytes by d-mannose and other carbohydrates. J Gen Microbiol. 1972;71:149–157. doi: 10.1099/00221287-71-1-149. [DOI] [PubMed] [Google Scholar]
- 37.Old D C, Payne S B. Antigens of the type 2 fimbriae of salmonellae: “cross reacting material” (CRM) of type 1 fimbriae. J Med Microbiol. 1971;4:215–225. doi: 10.1099/00222615-4-2-215. [DOI] [PubMed] [Google Scholar]
- 38.Popoff M Y, Le Minor L. WHO Collaborating Center for Reference and Research on Salmonella. 5th ed. Paris, France: Institute Pasteur; 1992. Antigenic formulas of the Salmonella serovars. [Google Scholar]
- 39.Raina S, Missiakas D, Baird L, Kumar S, Georgopoulos C. Identification and transcriptional analysis of the Escherichia coli htrE operon, which is homologous to pap and related pilin operons. J Bacteriol. 1993;175:5009–5021. doi: 10.1128/jb.175.16.5009-5021.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Römling U, Bian Z, Hammar M, Sierralta W D, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossolini G M, Muscas P, Chiesurin A, Satta G. Analysis of the Salmonella fim gene cluster: identification of a new gene (fimI) encoding a fimbrin-like protein and located downstream from the fimA gene. FEMS Microbiol Lett. 1993;114:259–265. doi: 10.1111/j.1574-6968.1993.tb06583.x. [DOI] [PubMed] [Google Scholar]
- 42.Sohel I, Puente J L, Murray W J, Vuopio-Varkila J, Schoolnik G K. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 43.Sojka W J, Wray C, Shreeve J, Benson A J. Incidence of Salmonella infection in animals in England and Wales, 1968–1974. J Hyg (London) 1977;78:43–56. doi: 10.1017/s0022172400055923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokurenko E V, Chesnokova V, Dykhuizen D E, Ofek I, Wu X R, Krogfelt K A, Struve C, Schembri M A, Hasty D L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swenson D L, Clegg S, Old D C. The frequency of fim genes among Salmonella serovars. Microb Pathog. 1991;10:487–492. doi: 10.1016/0882-4010(91)90115-q. [DOI] [PubMed] [Google Scholar]
- 46.Thanassi D G, Saulino E T, Hultgren S J. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr Opin Microbiol. 1998;1:223–231. doi: 10.1016/s1369-5274(98)80015-5. [DOI] [PubMed] [Google Scholar]
- 47.Thorns C J, Sojka G M, Mclaren I M, Dibb-Fuller M. Characterization of monoclonal antibodies against a fimbrial structure of Salmonella enteritidis and certain other serogroup D salmonellae and their application as serotyping reagents. Res Vet Sci. 1992;53:300–308. doi: 10.1016/0034-5288(92)90130-t. [DOI] [PubMed] [Google Scholar]
- 48.Tsolis R M, Townsend S M, Miao E A, Miller S I, Ficht T A, Adams L G, Bäumler A J. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turcotte C, Woodward M J. Cloning, DNA nucleotide sequence and distribution of the gene encoding the SEF14 fimbrial antigen of Salmonella enteritidis. J Gen Microbiol. 1993;139:1477–1485. doi: 10.1099/00221287-139-7-1477. [DOI] [PubMed] [Google Scholar]
- 50.van der Velden A W M, Bäumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitchurch C B, Mattick J S. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene. 1994;150:9–15. doi: 10.1016/0378-1119(94)90851-6. [DOI] [PubMed] [Google Scholar]
- 52.Wray C, Sojka W J, Bell J C. Salmonella infection in horses in England and Wales, 1973 to 1979. Vet Rec. 1981;109:398–401. doi: 10.1136/vr.109.18.398. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H Z, Lory S, Donnenberg M S. A plasmid-encoded prepilin peptidase gene from enteropathogenic Escherichia coli. J Bacteriol. 1994;176:6885–6891. doi: 10.1128/jb.176.22.6885-6891.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X L, Tsui I S, Yip C M, Fung A W, Wong D K, Dai X, Yang Y, Hackett J, Morris C. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect Immun. 2000;68:3067–3073. doi: 10.1128/iai.68.6.3067-3073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]