Abstract
The 58-kiloDalton mannoprotein (mp58) on the surface of Candida albicans is highly immunogenic, is expressed by all C. albicans isolates tested, and elicits strong antibody responses during candidiasis. It belongs to a family of immunodominant fungal antigens with representatives also in different species of Aspergillus. The amino acid sequence of the protein portion of mp58 as deduced from the DNA sequence of its encoding gene (FBP1/PRA1) was used to synthesize a complete set of overlapping dodecapeptides (overlap, 7; offset, 5) covalently attached to the surface of derivatized polyethylene pins. The pin-coupled peptides were used in a modified enzyme-linked immunosorbent assay (ELISA) to identify continuous epitopes recognized by a number of antiserum preparations containing anti-mp58 antibodies. This comprehensive epitope-scanning study revealed the presence of multiple immunoreactive continuous B-cell epitopes within the protein sequence. Regions of increased reactivity included both the amino and carboxy termini of the mature protein (encompassing amino acid residues 16 to 50 and 286 to 299, respectively) and four internal regions spanning amino acids at positions 66 to 92, 121 to 142, 148 to 192, and 211 to 232. Further delineation of epitopic regions and identification of the boundaries of the antigenic sites was performed upon ELISA testing with a second Pepset consisting of completely overlapping 8-mer peptides spanning these reactive regions in the protein moiety of mp58. The highly reactive epitopic region at the C terminus of the protein was further evaluated using both window net and replacement net analyses. A synthetic peptide corresponding to the last 10 amino acid residues at the C terminus of the protein was immunogenic when injected into mice after being coupled to a carrier protein. Moreover, antibodies in the resulting sera specifically recognized the homologus mp58 in ELISAs and immunoblot assays. Delineation of the antibody responses to mp58 could provide the basis for the development of novel immunity-based prophylactic, therapeutic, and diagnostic techniques for the management of candidiasis.
Candida albicans is both a commensal and an opportunistic pathogen of humans. Depending on the underlying host defect, this microorganism is able to cause a variety of infections that range from mucosal to life threatening disseminated candidiasis. C. albicans pathogenicity also depends on a complex array of microorganism-related virulence factors (reviewed in reference 15). Most of the biological functions related to pathogenicity and virulence reside in the fungal cell wall, since as the outermost part of the cell, the cell wall is the structure that mediates the host-fungus interplay (14). This includes the triggering and modulation of host immune responses, which in the case of C. albicans appears to rely on a complex interplay between natural and adaptive immunity, posing interesting challenges to the host (10, 20). Candidal antigens may stimulate specific cell-mediated and humoral immune responses, and there is a renewed interest in the study of the host antibody response to C. albicans (9, 10, 13, 16–18, 25–27, 41, 43, 47, 51, 55). The identification and characterization of immunodominant antigens eliciting potent immune responses during candidiasis could have important repercussions for developing novel diagnostic, prophylactic, and therapeutic techniques for candidiasis (41).
We have identified a 58-kDa mannoprotein (mp58) in the cell wall (surface) of C. albicans that is also an immunodominant antigen during infection (12, 50, 53). mp58 is present in cell wall extracts of both yeast cells and germ tubes (12) and is heterogeneously distributed at the C. albicans cell surface (42). The mp58 species was initially identified because of its ability to bind fibrinogen in ligand affinity blotting experiments (12) and may represent a specific candidal receptor for fibrinogen, since other mammalian proteins, such as laminin, fibronectin, type IV collagen, and C3d, did not bind in similar experiments (12, 36–38). All C. albicans strains tested so far express this moiety (33, 53). mp58 is also expressed by fungal cells in vivo in infected tissues (12, 39). These properties suggest an active role for mp58 during candidiasis. A cDNA clone for the protein portion of mp58 was isolated by immunoscreening a C. albicans expression library with antibodies generated against the purified molecule (40). Its sequence is almost identical to that of the gene for a C. albicans pH-regulated antigen (PRA1), also identified by using cDNA cloning techniques (52). It also shows homology with a family of immunodominant antigens in different species of Aspergillus (5, 8, 52). Thus, antigenicity rather than binding properties may be the primary role of this cell wall component (1, 52, 53). The gene showed condition-dependent transcription, since the mRNA transcript was found only when both yeast and germ tubes were grown in a minimal medium and was not detected when the cells were incubated in rich medium (1). C. albicans mp58 also contains N- and O-glycosidically linked sugar residues that represent 18 to 20 and 3 to 4%, respectively, of its apparent molecular mass (12), and the carbohydrate component of mp58 may play an important role in the ability of mp58 to bind fibrinogen (12). Since the primary structure of the protein moiety of mp58 can be deduced from its encoding gene, FBP1/PRA1 (40, 52), we have used the method of Geysen et al. for epitope mapping in order to identify antigenic regions in mp58. This method consists of synthesizing overlapping peptides spanning the entire sequence of a given protein and assessing the reactivities of the resulting sets of peptides with antibody preparations, thus allowing the identification of linear (continuous) B-cell epitopes (23).
MATERIALS AND METHODS
Organism, culture conditions, and preparation of cell wall extracts.
C. albicans strains 3153A and ATCC 26555 were used in this work. They were maintained on Sabouraud medium containing 2% (wt/vol) agar. Yeast cells were grown in suspension culture in the medium of Lee et al. (32) at 22°C. Germ tubes were induced from stationary-phase yeast cells by incubating them at 37°C in the same medium for 4 to 6 h. Cell wall extracts were prepared from intact cells (germ tubes) by treatment with β-mercaptoethanol (β-ME) as described before (11, 31). Briefly, germ tubes were resuspended in alkaline buffer containing 1% (vol/vol) β-ME and incubated for 45 min at 37°C with gentle agitation. After treatment, the cells were sedimented, and the supernatant fluid was recovered, dialyzed, and lyophylized (β-ME extract). The total sugar content in the extract was determined colorimetrically with mannose as the standard (19).
Purification of C. albicans mp58.
For the purification of mp58, the components of β-ME were separated by preparative sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis under denaturing conditions as described by Laemmli (30) with minor modifications (11, 12, 30, 32). Briefly, about 10 mg (based on total sugar content) of the β-ME extract was applied to a 13-cm-wide by 20-cm-high 5 to 15% polyacrylamide slab gradient gel. Prestained molecular weight standards (Gibco-BRL, Life Technologies Inc., Gaithersburg, Md.) were run in parallel in a single reference well formed to one side of the resolving gel slab. After electrophoretic separation, the transverse sections of the gels corresponding to mp58 (as identified by Coomassie staining) were excised and crushed, and the polypeptide moieties were electroeluted (32, 49). Alternatively, the materials present in the preparative gels were transferred to nitrocellulose membranes (56), and the transverse section corresponding to mp58 was identified by Ponceau staining, cut out, washed with glass-distilled water to remove the dye, air dried, and stored at −70°C until it was used (12, 35, 56). The purity of the resulting preparations of mp58, either bound to nitrocellulose or in solution, were tested by (i) Coomassie staining in polyacrylamide gels, (ii) immunoblot techniques to confirm their reactivity against anti-mp58 antibodies but lack of reactivity with antibodies against other cell wall and cytosolic antigens, and (iii) ability to bind fibrinogen in ligand-binding assays.
N-terminal sequencing of purified mp58.
N-terminal amino acid sequencing of C. albicans mp58 purified electrophoretically was performed by automated Edman degradation using a Procise cLC492 protein sequencer with on-line high-performance liquid chromatography detection of phenylthiohydantion amino acids and data collection and analysis software.
Predictions of physicochemical parameters and secondary structure of the protein moiety of mp58.
The amino acid sequence of the protein moiety of C. albicans mp58 as deduced from the nucleotide sequence of its encoding gene, FBP1/PRA (40, 52), was subjected to in silico analysis. Computerized algorithms were used to predict the hydrophobicity (29), hydrophilicity (28), surface probability (J. Boger, E. A. Emini, and A. Schmidt, Rep. Sixth Int. Cong. Immunol., p.250, 1986), antigenic index (57), and secondary structure (21). These analyses were performed with the biocomputing software program ANTHEPROT (22).
Polyclonal antisera against purified C. albicans mp58 and mp58-containing cell wall extracts.
Hyperimmune sera from mice (BALB/c) immunized with purified C. albicans mp58 were obtained as follows. One mouse was immunized with nitrocellulose-bound mp58 purified by preparative electrophoresis, subsequently transferred to the nitrocellulose support, and sonicated in the presence of a small amount of phosphate-buffered saline (PBS), pH 7.4, until an emulsion was formed. Two mice were immunized with mp58 purified by preparative electrophoresis and subsequent electroelution from the gel slice (see above). The generation of rabbit (New Zealand White) hyperimmune sera has been described before and included serum from a rabbit immunized with purified mp58 (by electrophoresis and transfer to nitrocellulose supports) (12) and sera from two rabbits immunized with β-ME cell wall extracts (where mp58 is a major antigenic component) from two different C. albicans type strains, ATCC 26555 (35) and 3153A (37). The immunization schedules included an initial subcutaneous injection with the antigen mixed with complete Freund's adjuvant and subsequent booster injections with incomplete Freund's adjuvant, except for the rabbit injected with cell wall extracts of C. albicans 3153A, for which Ribi adjuvant was used (Ribi ImmunoChem Research, Inc., Hamilton, Mont.). Immunizations were performed every 3 weeks, and hyperimmune sera were collected 10 days after the final booster. All antiserum preparations were demonstrated to contain antibodies against C. albicans mp58 in immunoblot assays.
Epitope mapping.
Analysis of continuous B-cell epitopes on C. albicans mp58 was carried out by means of the Multipin Peptide Technology (PepScan) of Chiron Mimotopes (San Diego, Calif.). The amino acid sequence of the protein portion of mp58 was used to synthesize a complete set of overlapping dodecapeptides (overlap, 7; offset, 5) covalently attached to the surfaces of derivatized polyethylene pins in a format compatible with standard enzyme-linked immunosorbent assays (ELISAs). These overlapping peptides covered the entire sequence of the protein, which includes a 15-amino-acid signal peptide and a mature protein containing 284 amino acids. The final Pepset consisted of a total of 59 peptides plus 2 control peptides. The reactivities of various serum preparations containing anti-mp58 antibodies with the pin-bound peptides were detected by a modified enzyme immunoassay. Briefly, pins were precoated for 1 h at room temperature with 200 μl of PBS containing 3% bovine serum albumin (BSA) per well in the wells of a microtiter plate. They were then incubated overnight at 4°C with 200 μl of a 1:1,000 dilution of the primary antiserum in PBS containing 0.05% Tween-20 (PBST) and 1% BSA. The plate containing the primary antibody was discarded, and the pin block was washed four times for 10 min each time in a tray with PBST. Then, the corresponding species-specific peroxidase-conjugated secondary antibody (goat anti-mouse immunoglobulin G [IgG] or goat anti-rabbit IgG [Bio-Rad, Hercules, Calif.] at a 1:2,000 dilution in PBST plus 1% BSA) was added to the wells of a microtiter plate, and the pin block was inserted and incubated for 1 h at room temperature. After being washed as before, the block was inserted in a new microtiter plate containing 200 μl of o-phenylenediamine substrate per well and developed in the dark for 10 min with gentle agitation. Color development was stopped by the addition of 100 μl of 1 M H2SO4 per well, and the plate was read at 490 nm in a Benchmark microplate reader (Bio-Rad). For mouse and rabbit sera, the results from an experiment using the corresponding preimmune sera were subtracted from the experimental values. To reuse the pin block, it was placed in an ultrasonic bath with stripping buffer (100 mM sodium phosphate [pH 7.4] supplied with 1% SDS and 0.1% β-ME heated to 60°C). The pin block was sonicated for 10 min, rinsed, and washed in deionized water with an initial temperature of 60°C (twice for 30 s each time and once for 30 min). When not in use, the pins were air dried after immersion in 60°C methanol and stored desiccated at 4°C.
From results of the scanning studies using the above-mentioned set of dodecapeptides, a second Pepset was synthesized consisting of overlapping 8-mer peptides (offset, 1; overlap, 7) spanning potentially important epitopic regions in the protein moiety of mp58. This allowed further delineation of epitopic regions and identification of the boundaries of the antigenic sites upon subsequent ELISAs with murine and rabbit antibodies using the same procedures as described above.
A third Pepset was constructed that included both a “window net” and a “replacement net” synthesis to further analyze the C-terminal domain within the protein moiety of C. albicans mp58, which was identified as a highly reactive epitope, using the same antibody preparations described above. The window net is performed to identify the precise boundaries of an identified epitope and consists of synthesizing all of the shorter overlapping sequences covering an identified antibody-binding peptide, which in this case were all 4-mers, 5-mers-, 6-mers, and so on. While the general and window net syntheses provide basic information on the location and bundaries of epitopes, the replacement net consisting of synthesizing peptide analogs with single-amino-acid substitutions, provides information on the contribution to antibody binding by individual residues within an epitope. In this case, alanine was substituted for each residue one at a time along the C-terminal reactive peptide, and the alanine in the original sequence was replaced by glycine. Molecular modeling of this epitopic region was originated with Swiss-Pdbviewer (version 3.0).
Synthetic peptides, ELISA, and immunoblot techniques.
On the basis of the epitope-mapping studies described above, a synthetic peptide corresponding to the C-terminal 10 amino acids (HTHADGEVHC) of the protein moiety of C. albicans mp58 was synthesized with 9-fluorenylmethoxycarbonyl chemistry, purified by high-performance liquid chromatography, coupled to keyhole lympet hemocyanin (KLH; Pierce, Rockford, III.), and used to immunize two mice in order to generate anti-peptide antibodies. The immunization schedules were as described above with a dose of 50 μg of peptide in PBS mixed with an equal volume of Freund's adjuvant (complete for the initial dose; incomplete for booster injections). Characterization of the resulting antisera was performed by ELISA and immunoblot techniques. For ELISA, wells of a microtiter plate (Costar High Binding; Corning Costar Corp., Cambridge, Mass.) were coated with different quantities of free (unconjugated) peptide in PBS or with electrophoretically purified mp58 in borate buffer, and then the antisera (at a 1:1,000 dilution in PBST plus 1% BSA for a dose-response ELISA and at serial dilutions in the same buffer for titration experiments) were added to the wells and the ELISA was completed as described above with peroxidase-conjugated anti-mouse IgG as a secondary antibody. A competitive ELISA was developed to assess the ability of soluble linear peptide to inhibit binding of the anti-peptide antiserum to solid-phase-coated peptide. For this purpose, the wells of a microtiter plate (Costar High Binding) were coated with 0.1 μg of unconjugated decapeptide. A 1:5,000 dilution (in PBST with 1% BSA) of anti-peptide antiserum was added to the wells of a second plate and reacted with serial dilutions (also in PBST with 1% BSA) of soluble peptide. This plate was incubated at room temperature for 2 h, the contents of each well was transferred to the plate coated with a fixed amount of peptide, and the ELISA was performed as described above. Values for the uninhibited serum were considered 100% binding. For immunoblotting, materials present in the C. albicans β-ME extract were separated by SDS-polyacrylamide gel electrophoresis using precast 4 to 15% gradient minigels (Bio-Rad) and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, N.H.) (37). After the membranes were blocked in Tris-HCl buffer plus 0.9% (wt/vol) NaCl (TBS) containing 3% (wt/vol) BSA, they were incubated in the presence of the anti-peptide antisera (diluted 1:1,000 in TBS with 0.05% Tween 20 and 1% BSA). Peroxidase-labeled goat anti-mouse IgG (diluted 1:2,000 in TBS with 0.05% Tween 20 and 1% BSA) (Bio-Rad) was used as the secondary antibody. Colored reactive bands were developed with H2O2 and 4-chloro-1-napthol as the chromogenic reagent.
The anti-carboxyl terminus decapeptide antisera were also used in PepScan experiments with both the 12-mer and the 8-mer Pepsets using the procedures described above.
RESULTS
In silico analysis of the protein moiety of mp58.
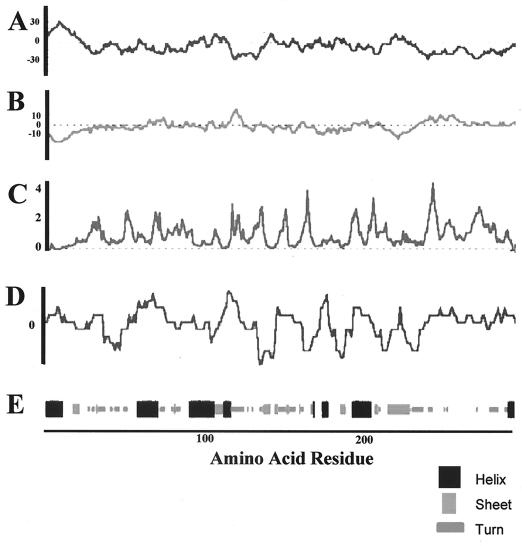
Computer-based analysis of hydrophicility, antigenic index, and surface probability are often used to predict B-cell-reactive continuous epitopes in protein sequences. Using the deduced amino acid sequence of the gene encoding the protein portion of mp58, these predictions revealed the presence of putative antigenic regions randomly distributed throughout the protein, with the exception of the first 15 amino acid residues at the N terminus, which constitute the putative signal peptide (Fig. 1A to D). The predictions of the secondary structure of the protein moiety of mp58 are shown in Fig. 1E.
FIG. 1.
Predictions of hydrophobicity (A), hydrophilicity (B), surface probability (C), antigenic index (D), and secondary structure (E) of the protein moiety of C. albicans mp58.
Determination of the N-terminal amino acid sequence of C. albicans mp58 present at the cell wall.
The N-terminal amino acid sequence of mp58 purified from C. albicans cell wall extracts was determined by automated Edman degradation. At each cycle, a single amino acid was detected; therefore, we concluded that the electrophoretic procedure rendered a pure mp58 preparation. The analysis yielded the following sequence: Ala-Pro-Val-X-Val-X-Arg-Phe-Val. This sequence corresponds to the deduced amino acid sequence (amino acid residues 16 to 24) from its encoding gene, FBP1/PRA1 (40, 52), where X is predicted to be threonine and could be modified (glycosylated) in the native state. This analysis also confirmed cleavage following A1a15 for mp58 in its native state within the C. albicans cell wall, which was correctly predicted using the algorithms described above.
Epitope mapping with murine sera to purified C. albicans mp58.
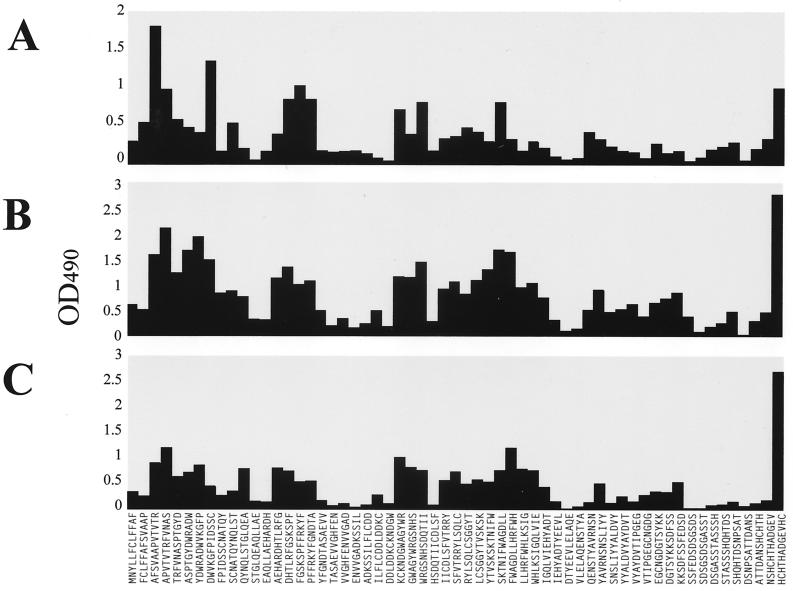
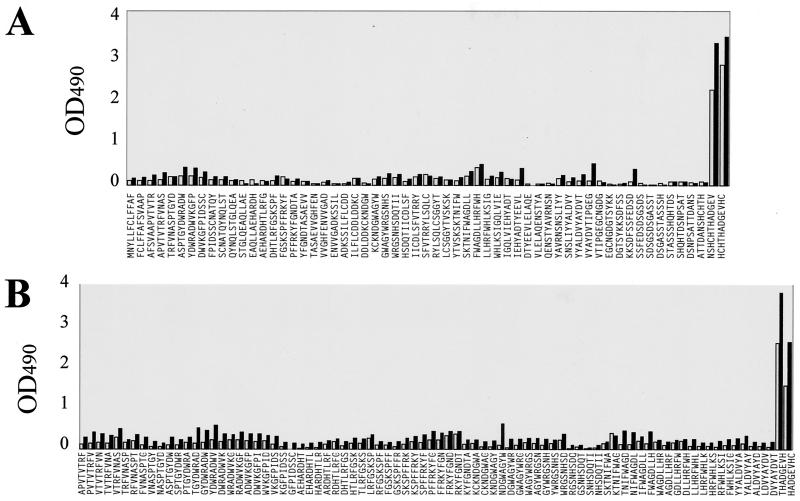
A PepScan analysis was carried out with sera from three mice immunized with electrophoretically purified C. albicans mp58. When tested by an immunoblot assay, these hyperimmune sera recognized C. albicans mp58 with high specificity among other proteinaceous components present in cell wall extracts (not shown). The three sera were tested individually against the pin-bound dodecapeptides spanning the entire amino acid sequence of mp58 to allow determination of continuous B-cell epitopes in its protein moiety. As shown in Fig. 2, the individual murine antisera were capable of recognizing multiple dodecapeptides, thus indicating a complex polyclonal response to the protein moiety of mp58. The results identified areas of increased reactivity corresponding to both the N and C termini of the mature protein (amino acid residues 16 to 50 and 286 to 299, respectively) together with three internal regions which mapped to residues 66 to 87, 121 to 142, and 148 to 192. In the case of sera from the two mice immunized with mp58 purified by electroelution (Fig. 2B and C), the highest reactivity was detected with the C-terminal dodecapeptide, whereas increased reactivity against the N terminus was detected with the serum from the mouse immunized with nitrocellulose-bound mp58 (Fig. 2A).
FIG. 2.
IgG reactivity of hyperimmune sera from mice immunized with electrophoretically purified mp58 with dodecapeptides spanning the entire sequence of the protein portion of C. albicans mp58. (A) Results with serum from a mouse immunized with nitrocellulose-bound mp58; (B and C) results with sera from two mice immunized with electroeluted mp58. OD490, optical density at 490 nm.
Epitope mapping with rabbit antibodies to C. albicans mp58.
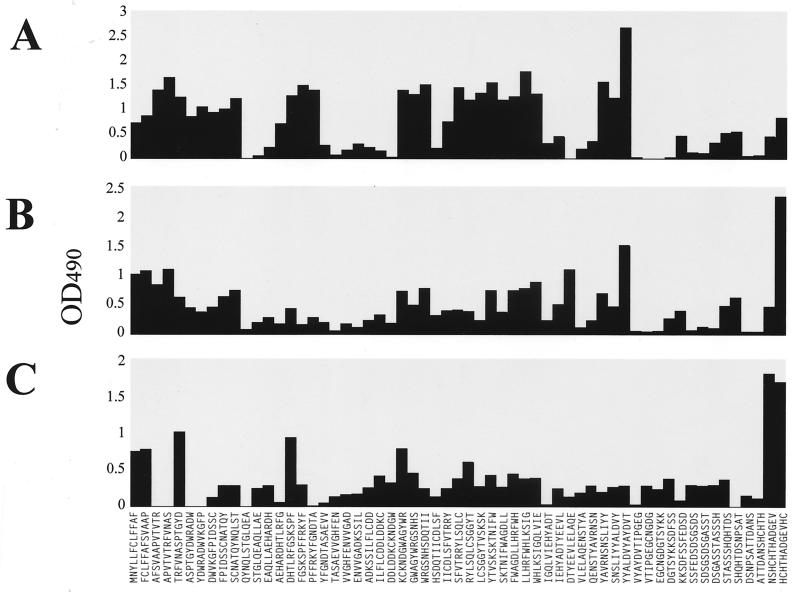
Anti-mp58 antibodies were present in serum samples from a rabbit immunized with purified mp58 and in those from two rabbits immunized with cell wall extracts of C. albicans, where mp58 is an immunodominant antigen (12, 35, 37). These sera were used to further analyze the immunogenicity of C. albicans mp58 by using the same Pepset with dodecapeptides spanning the entire sequence of the protein moiety of mp58. Figure 3 shows the results from this comprehensive scanning. This analysis identified immunogenic domains within mp58 similar to the ones revealed by epitope-mapping experiments using murine sera. One additional epitopic region spanning amino acid residues 211 to 232 was also detected (Fig. 3A and B). This was the region which showed the highest levels of reactivity with the serum from the rabbit injected with purified mp58 (Fig. 3A), whereas antibodies present in sera from rabbits injected with C. albicans cell wall extracts exhibited higher levels of reactivity against the C terminus of the protein (Fig. 3B and C).
FIG. 3.
IgG reactivity of serum samples from rabbits immunized with purified C. albicans mp58 (A) and with mp58-containing cell wall extracts from C. albicans type strains ATCC 26555 (B) and 3153A (C) with the dodecapeptides spanning the entire sequence of the protein portion of mp58. OD490, optical density at 490 nm.
Identification of discrete continuous epitopes in the protein moiety of mp58.
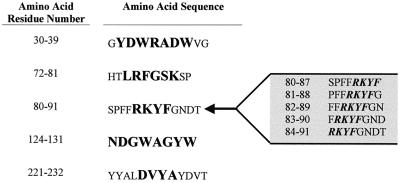
Upon completion of the epitope-mapping analysis with the dodecapeptide Pepset, a second array of peptides was synthesized consisting of completely overlapping octapeptides spanning the immunoreactive domains identified in the previous experiments. Epitope-mapping analysis of the 8-mer set with the same serum samples containing anti-mp58 antibodies allowed further delineation of epitopic regions and identification of the boundaries of the antigenic sites. As shown in Fig. 4, discrete linear continuous epitopes identified in these studies, ranging from 4 to 8 amino acids long, mapped to residues 31 to 37 (YDWRADW) , 74 to 79 (LRFGSK) , 84 to 87 (RKYF), 124 to 131 (NDGWAGYW) , and 225 to 228 (DVYA). Interestingly, most of the antibody-binding activity at the C-terminal epitopic region (identified previously by using murine sera and the 12-mer set [see above] was lost when the same antiserum preparations were used with the set consisting of overlapping octapeptides.
FIG. 4.
Amino acid sequences representing IgG epitopes as identified by reactivity of antibodies against completely overlapping octapeptides spanning reactive segments in the protein sequence of C. albicans mp58. The identified epitopes are shown in boldface. Several reactive sequences containing the RKYF epitope are shown to the right as an example of how the boundaries of this epitope were delineated by synthesizing several overlapping octapeptides.
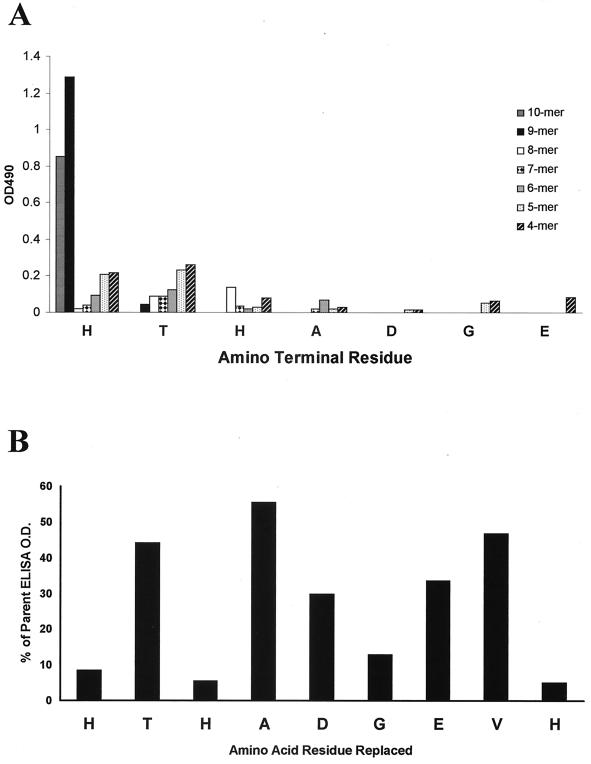
Analysis of the epitopic region at the C-terminal domain of the protein moiety of C. albicans mp58.
On the basis of antibody reactivity with pepsets consisting of overlapping peptides, the C-terminal domain of mp58 was identified as a highly reactive region. In order to further delineate the specific boundaries of this epitopic region and to study the contribution of individual amino acid residues to the antibody-binding properties of this domain, a combined window net and replacement net approach was used. The window net analysis identified the nonapeptide 290HTHADGEVH298 as the minimal region that retained most of the antibody-binding activity, whereas a sharp decrease in binding was observed for all derived octapeptides (Fig. 5A), also in accordance with results obtained using the Pepset consisting of overlapping octapeptides (see above). The replacement net analysis revealed the important role of the histidines (residues 290, 292, and 298) in recognition by antibodies, since single substitutions for each of these histidine residues resulted in negligible levels of reactivity (less than 10% compared to the reactivity with the parent peptide). Also, single substitution of Gly295 resulted in an abrupt drop in reactivity (to about 13%). However, all other single substitutions maintained moderate levels of reactivity, ranging from 40 to 55% of the optical density observed with the parent peptide (Fig. 5B).
FIG. 5.
(A) Window net analysis of the C-terminal antibody-binding region. The results show the IgG reactivities of hyperimmune sera from mice immunized with electrophoretically purified mp58 against all overlapping tetra-, penta-, hexa-, hepta-, octa-, nona-, and decapeptides spanning the C terminus of mp58. The results represent anti-IgG reactivity (measured as optical density at 490 nm [OD=490]; average values of two experiments using hyperimmune sera from mice immunized with electrophoretically purified mp58). (B) Results of the replacement net experiment. The epitopic region HTHADGEVH identified in previous analysis was the basis for synthesis of single-residue replacement analogs. Alanine was substituted for each residue one at a time along the C-terminal reactive peptide, and the alanine in the original sequence was replaced by glycine. The results represent the percentage of reactivity (anti-IgG; average of two experiments using hyperimmune sera from mice immunized with electrophoretically purified mp58) retained by peptides with the indicated residue replaced compared to the reactivity observed with the native (unsubstituted) peptide.
Murine antibody responses to a synthetic peptide corresponding to the last 10 amino acid residues at the C terminus of the protein moiety of C. albicans mp58.
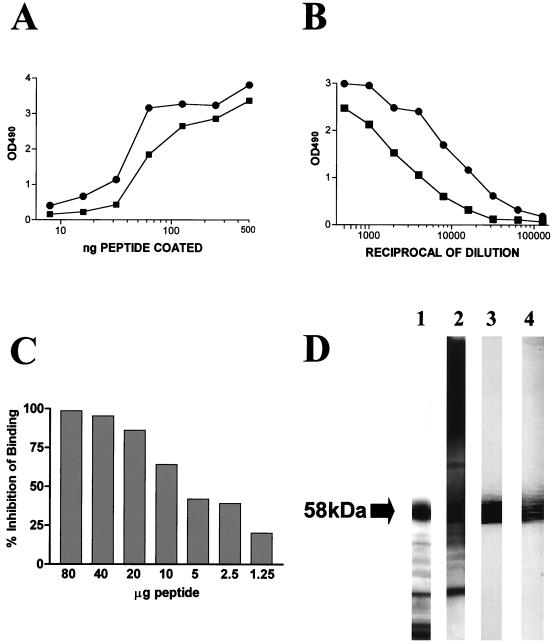
To determine the potential relevance of the epitopic region at the C terminus of the protein and based on the epitope-mapping results above, a 10-mer synthetic peptide encompassing the last 10 amino acid residues at the C terminus of the mp58 sequence (the 290HTHADGEVH298 epitope plus the C-terminal cysteine residue, which is already present in the native protein and served as a spacer arm for conjugation to the carrier protein) was synthesized. This decapeptide was coupled to KLH and used to immunize two mice. Serum samples obtained from the immunized animals 10 days after the last booster were examined by a variety of immunological procedures. In a dose-response ELISA, the two antisera were able to recognize both the free (unconjugated) decapeptide and the purified mp58 coating the surfaces of microtiter wells in a saturable and dose-dependent fashion (Fig. 6A). An endpoint dilution ELISA revealed high titers of anti-peptide antibodies (Fig. 6B). The specificity of the reaction was demonstrated by the fact that soluble peptide was an effective competitor of binding of the anti-peptide antiserum to the same peptide immobilized in the wells of a microtiter plate and was able to completely abolish binding at high concentrations (Fig. 6C). In an immunoblot assay with cell wall extracts of the fungus, sera from both animals recognized C. albicans mp58 with high specificity (Fig. 6D). Finally, in PepScan analyses with the dodecapeptide and octapeptide sets derived from the protein sequence of mp58, reactivity with both anti-synthetic-peptide antisera was limited to peptides corresponding to the C-terminal region of the amino acid sequence (Fig. 7).
FIG. 6.
Characterization of the two antisera generated in mice against the KLH-conjugated synthetic peptide encompassing the 10 amino acid residues (HTHADGEVHC) at the C terminus of C. albicans mp58. (A) Results of an experiment in which the wells of a microtiter plate were coated with decreasing amounts of free (unconjugated) decapeptide and incubated with a 1:1,000 dilution of anti-peptide antisera (dose-response ELISA). (B) Titration curves of the resulting antisera (endpoint ELISA), for which wells of a microtiter plate were coated with a fixed amount of 1 μg of free peptide per well. (C) Results of a competitive-inhibition ELISA in which different quantities of free soluble decapeptide inhibited binding of the anti-peptide antiserum to solid-phase-coated peptide in a dose-dependent manner. Binding of the uninhibited antibody was considered 100% (0% inhibition). The values shown represent the results from a single experiment. The experiment was repeated with similar results. (D) Results of an immunoblot experiment where the anti-peptide antisera (lanes 3 and 4) were able to recognize mp58 with high specificity among other cell wall components present in the nitrocellulose membrane as revealed by Coomassie staining (lane 1) and immunoblot analysis with rabbit polyclonal antiserum generated against cell wall extracts of the fungus (lane 2). OD490, optical density at 490 nm.
FIG. 7.
IgG reactivity of hyperimmune sera from mice immunized with KLH-conjugated Cterminal decapeptide of C. albicans mp58 against 12-mer (A) and 8-mer (B) Pepsets. OD490, optical density at 490 nm.
DISCUSSION
The lack of an early and accurate diagnostic procedure, the limited arsenal of therapeutic weapons to combat infection, and the emergence of resistant strains due to empirical prophylactic treatment are responsible for the high morbidity and mortality associated with infections caused by C. albicans. For these reasons, there is an increasing interest in the search for new or alternative therapies to enhance or complement a multicomponent approach to the management (diagnosis, prevention, and treatment) of candidiasis (13, 17, 18, 41). Although the role of antibody response in host defense against disseminated candidiasis is still controversial, there is a renewed interest in the study of the host antibody response to Candida and the possibility of immunointervention as a feasible prophylactic or therapeutic approach for the management of candidiasis (9, 10, 13, 17, 18, 25–27, 41, 43, 45, 47, 48, 51, 55). The cell wall of C. albicans is a significant source of antigens (41), and several immunodominant antigens in candidiasis, including the 58-kDa mannoprotein (mp58), are found in the cell wall (2, 12, 33, 34, 46, 54).
The goal of this study was to define continuous B-cell epitopes on C. albicans mp58. The gene encoding the protein portion of mp58 has been identified by using cDNA cloning techniques that take advantage of its antigenic nature (40, 52). Here, its deduced amino acid sequence was used to synthesize a set of overlapping dodecapeptides spanning the entire sequence of the protein. By using the resulting Pepset in epitope-mapping experiments with a variety of antiserum preparations containing anti-mp58 antibodies, we were able to identify six different reactive regions throughout the protein sequence (Fig. 2 and 3). Interestingly, these same regions appear to be immunoreactive in preliminary experiments with sera from patients with disseminated candidiasis (not shown). Overall, the results indicated the complexity of the polyclonal IgG response to C. albicans mp58 and supported the validity of the PepScan approach to defining linear peptide epitopes on fungal antigens (24, 44). The immunoreactive regions included both the N and the C termini of the mature protein (as expected, since terminal regions are frequently located on the surfaces of most proteins), together with four internal regions. It must be noted that the epitope-mapping techniques used in the present study are limited to the identification of linear (continuous) epitopes defined by contiguous amino acid residues in the primary structure of the protein. The use in these experiments of antibody preparations generated against electrophoretically purified mp58 (with electrophoresis performed under denaturing conditions) may maximize the detection of linear epitopes detected by the PepScan technique compared to other, discontinuous epitopes. A significant number of linear B-cell epitopes have also been identified in the case of the Asp f 2 protein of Aspergillus fumigatus, belonging to the same family of immunodominant fungal antigens (3). Also, multiple linear epitopes have been detected on two other important antigens of C. albicans, the 47-kDa fragment of hsp90 and secretory proteinase 2 (24, 44). In any case, other discontinuous epitopes, defined by the interactions between amino acid residues that are distant in the protein sequence but brought together as a result of the natural folding of the protein, may be found on mp58 in its native conformation. Also, since C. albicans mp58 is a glycoprotein, the carbohydrate portion of the molecule is likely to contain antigenic determinants that cannot be identified using this methodology. As noted by other investigators (6, 58), in the case of C. albicans mp58, comparisons between data from epitope-mapping experiments and computer-based algorithms used to predict antigenic regions (Fig. 1) showed limited correlation. Most antigenicity prediction methods are based on identification of short stretches of amino acids that are likely to be located at the protein surface (58). However, confirmation of their antigenicity can only be provided by an empirical approach such as the one used in the present study.
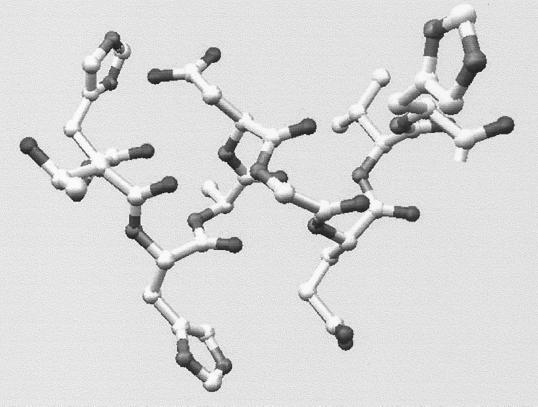
The immunoreactive segments identified using the 12-mer Pepset were further analyzed by synthesizing these sequences as a set of completely overlapping octapeptides. Epitope-scanning experiments using the second Pepset allowed further delineation of immunoreactive regions and identification of discrete epitopes responsible for IgG binding (Fig. 4). In the case of the highly reactive region at the C terminus of the protein moiety of C. albicans mp58, the specificity of antibody binding was further analyzed by using a window net and a replacement net. The results identified a 9-mer (290HTHADGEVH298) as the shortest peptide retaining significant antibody-binding activity and demonstrated the importance of the histidine residues for binding. Both of these observations suggest that antibody recognition by this quite long linear epitope may still be dependent upon certain conformational constraints. In fact, a very apparent three-dimensional structure can be observed in a computer molecular model of this epitope (Fig. 8).
FIG. 8.
Molecular modeling of the epitopic region at the C-terminal domain of C. albicans mp58. Three-dimensional structure of the nonapeptide at the C terminus of C. albicans mp58 identified as an immunodominant B-cell epitope.
Interestingly, as shown in Table 1, some of the sequences representing the IgG-binding linear epitopes identified using the different Pepsets showed significant levels of homology with two other immunodominant fungal proteins, ASPND1 from Aspergillus nidulans (7, 8) and Asp f 2 from A. fumigatus (4, 5). These fungal proteins seem to belong to a family of immunodominant antigens, suggesting an important role for members of this family in the host-parasite interaction during different types of fungal infections. Of note, some of the homologous sequences on Asp f 2 have been reported to represent IgE-binding epitopes (3). It is also important to note the absolute conservation across species, particularly of the histidines within the epitopic regions at the C-terminal domains of each of these proteins, but also the glycine (Table 1 last row), which seems to confirm their contribution to antibody binding, as suggested by the replacement net analysis of this epitope within C. albicans mp58, which demonstrated that these residues are essentially unreplaceable.
TABLE 1.
Sequence homology of five of the identified IgG epitopes on C. albicans mp58 with two immunodominant antigens from A. nidulans and A. fumigatus
| mp58 epitope sequence (C. albicans) | Antigen sequence
|
|
|---|---|---|
| ASPND1 (A. nidulans) | Asp f 2 (A. fumigatus) | |
| LRFGSK | LRWGNE | LRWGNE |
| RKYF | RKYF | RKYFa |
| NDGWAGYW | LEGWGGHW | LEGWGGHW |
| DVYA | EVYA | EAYAa |
| HTHADGEVH | HTHEGGELH | HTHEGGQLHa |
A similar sequence has been detected as an IgE epitope in Asp f 2.
A synthetic peptide corresponding to the C-terminal decapeptide of mp58 (HTHADGEVHC, the identified nonapeptidic epitope plus the native terminal cysteine) was immunogenic when conjugated to a carrier protein (KLH) and injected into mice. The resulting sera were characterized by a variety of immunological procedures (Fig. 6). Hyperimmune sera recognized the free peptide bound to the wells of microtiter plates in a dose-dependent and saturable manner (Fig. 6A). Both antisera generated contained high titers of anti-peptide antibodies as determined by an endpoint ELISA (Fig. 6B). Free soluble peptide was an effective competitor of antibody binding to bound peptide (Fig. 6C) and mp58 (not shown), indicating the specificity of the reaction. The anti-peptide antisera recognized the homologous C. albicans mp58, and no cross-reactivity with other antigens present in cell wall extracts was detected (Fig. 6D). This result suggested that immunization with a single peptide motif led to a highly specific antibody response and that this decapeptide lacks significant sequence homology with other candidal antigens. As expected, the anti-synthetic-peptide antisera selectively recognized peptides corresponding to the C-terminal domain of the protein when used in PepScan experiments with both the 12- and 8-mer sets (Fig. 7). Interestingly, current experiments in our laboratory suggest that the C-terminal epitopic region of mp58 may elicit protective antibody responses, since vaccination with this peptide partially protects mice in a lethal model of hematogenously disseminated candidiasis, and also, patients surviving systemic candidiasis show increased levels of antibodies against this epitope compared to those succumbing to infection (S. Perea, J. Pemán, W. R. Kirkpatrick, T. F. Patterson, and J. L. López-Ribot, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 695, p. 554, 1999; W. R. Kirkpatrick and J. L. López-Ribot, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1111, p. 373, 2000).
In conclusion, we have identified continuous B-cell epitopes on the protein moiety of C. albicans mp58. Our results revealed a complex polyclonal response to this protein. The delineation of antibody responses to immunodominant antigens during candidiasis, including mp58, may provide the basis for the development of new approaches to the management of candidiasis that are urgently needed.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant 1 R29 AI42401 (to J.L.L.R.). A.V. was the recipient of postdoctoral fellowships from the Sociedad Española de Quimioterapia (SEQ) and the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC-Fundación Welcome). S.P. acknowledges the receipt of a NATO postdoctoral fellowship.
Amino acid sequencing and synthesis of the C-terminal synthetic decapeptide were performed at the Protein Core Facility at UTHSCSA.
REFERENCES
- 1.Alloush H M, Lopez-Ribot J L, Chaffin W L. Dynamic expression of cell wall proteins of Candida albicans revealed by probes from cDNA clones. J Med Vet Mycol. 1996;34:91–97. [PubMed] [Google Scholar]
- 2.Angiolella L, Facchin M, Stringaro A, Maras B, Simonetti N, Cassone A. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J Infect Dis. 1996;173:684–690. doi: 10.1093/infdis/173.3.684. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee B, Greenberger P A, Fink J N, Kurup V P. Conformational and linear B-cell epitopes of Asp f 2, a major allergen of Aspergillus fumigatus, bind differently to immunoglobulin E antibody in the sera of allergic bronchopulmonary aspergillosis patients. Infect Immun. 1999;67:2284–2291. doi: 10.1128/iai.67.5.2284-2291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee B, Greenberger P A, Fink J N, Kurup V P. Immunological characterization of Asp f 2, a major allergen from Aspergillus fumigatus associated with allergic bronchopulmonary aspergillosis. Infect Immun. 1998;66:5175–5182. doi: 10.1128/iai.66.11.5175-5182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee B, Kurup V P, Phadnis S, Greenberger P A, Fink J N. Molecular cloning and expression of a recombinant Aspergillus fumigatus protein Asp f II with significant immunoglobulin E reactivity in allergic bronchopulmonary aspergillosis. J Lab Clin Med. 1996;127:253–262. doi: 10.1016/s0022-2143(96)90093-1. [DOI] [PubMed] [Google Scholar]
- 6.Baughn R E, Demecs M, Taber L H, Musher D M. Epitope mapping of B-cell determinants on the 15-kilodalton lipoprotein of Treponema pallidum (Tpp15) with synthetic peptides. Infect Immun. 1996;64:2457–2466. doi: 10.1128/iai.64.7.2457-2466.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calera J A, Ovejero M C, Lopez-Medrano R, Lopez-Aragon R, Puente P, Leal F. The recombinant antigen ASPND1r from Aspergillus nidulans is specifically recognized by sera from patients with aspergilloma. Microbiology. 1998;144:561–567. doi: 10.1099/00221287-144-2-561. [DOI] [PubMed] [Google Scholar]
- 8.Calera J A, Ovejero M C, Lopez-Medrano R, Segurado M, Puente P, Leal F. Characterization of the Aspergillus nidulans aspnd1 gene demonstrates that the ASPND1 antigen, which it encodes, and several Aspergillus fumigatus immunodominant antigens belong to the same family. Infect Immun. 1997;65:1335–1344. doi: 10.1128/iai.65.4.1335-1344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall A. Antibody immunity and invasive fungal infections. Infect Immun. 1995;63:4211–4218. doi: 10.1128/iai.63.11.4211-4218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadevall A, Cassone A, Bistoni F, Cutler J E, Magliani W, Murphy J W, Polonelli L, Romani L. Antibody and/or cell-mediated immunity, protective mechanisms in fungal disease: and ongoing dilemma or an unnecessary dispute? Med Mycol. 1998;36(Suppl. 1):95–105. [PubMed] [Google Scholar]
- 11.Casanova M, Lopez-Ribot J L, Martinez J P, Sentandreu R. Characterization of cell wall proteins from yeast and mycelial cells of Candida albicans by labelling with biotin: comparison with other techniques. Infect Immun. 1992;60:4898–4906. doi: 10.1128/iai.60.11.4898-4906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casanova M, Lopez-Ribot J L, Monteagudo C, Llombart-Bosch A, Sentandreu R, Martinez J P. Identification of a 58-kilodalton cell surface fibrinogen-binding mannoprotein from Candida albicans. Infect Immun. 1992;60:4221–4229. doi: 10.1128/iai.60.10.4221-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassone A, De Bernardis F, Ausiello C M, Gomez M J, Boccanera M, La Valle R, Torosantucci A. Immunogenic and protective Candida albicans constituents. Res Immunol. 1998;149:289–299. doi: 10.1016/s0923-2494(98)80753-0. [DOI] [PubMed] [Google Scholar]
- 14.Chaffin W L, Lopez-Ribot J L, Casanova M, Gozalbo D, Martinez J P. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev. 1998;62:130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 16.De Bernardis F, Boccanera M, Adriani D, Spreghini E, Santoni G, Cassone A. Protective role of antimannan and anti-aspartyl proteinase antibodies in an experimental model of Candida albicans vaginitis in rats. Infect Immun. 1997;65:3399–3405. doi: 10.1128/iai.65.8.3399-3405.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deepe G S., Jr Prospects for the development of fungal vaccines. Clin Microbiol Rev. 1997;10:585–596. doi: 10.1128/cmr.10.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon D M, Casadevall A, Klein B, Mendoza L, Travassos L, Deepe G S., Jr Development of vaccines and their use in the prevention of fungal infections. Med Mycol. 1998;36Suppl. 1:57–67. [PubMed] [Google Scholar]
- 19.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 20.Fukazawa Y, Cassone A, Bistoni F, Howard D H, Kagaya K, Murphy J W, Cenci E, Lane T E, Mencacci A, Puccetti P, et al. Mechanisms of cell-mediated immunity in fungal infection. J Med Vet Mycol. 1994;32(Suppl. 1):123–131. doi: 10.1080/02681219480000781. [DOI] [PubMed] [Google Scholar]
- 21.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 22.Geourjon C, Deleage G. ANTHEPROT 2.0: a three-dimensional module fully coupled with protein sequence analysis methods. J Mol Graph. 1995;13:209–212. doi: 10.1016/0263-7855(95)00035-5. [DOI] [PubMed] [Google Scholar]
- 23.Geysen H M, Rodda S J, Mason T J, Tribbick G, Schoofs P G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 24.Ghadjari A, Matthews R C, Burnie J P. Epitope mapping Candida albicans proteinase (SAP 2) FEMS Immunol Med Microbiol. 1997;19:115–123. doi: 10.1111/j.1574-695X.1997.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 25.Han Y, Cutler J E. Antibody response that protects against disseminated candidiasis. Infect Immun. 1995;63:2714–2719. doi: 10.1128/iai.63.7.2714-2719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y, Morrison R P, Cutler J E. A vaccine and monoclonal antibodies that enhance mouse resistance to Candida albicans vaginal infection. Infect Immun. 1998;66:5771–5776. doi: 10.1128/iai.66.12.5771-5776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y, Ulrich M A, Cutler J E. Candida albicans mannan extract-protein conjugates induce a protective immune response against experimental candidiasis. J Infect Dis. 1999;179:1477–1484. doi: 10.1086/314779. [DOI] [PubMed] [Google Scholar]
- 28.Hopp T P, Woods K R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Ribot J L, Martinez J P, Monteagudo C, Alloush H M, Mattioli N V, Chaffin W L. Evidence for the presence of complex carbohydrates in Candida albicans cell wall glycoproteins. Rev Iberoam Micol. 1999;16:23–26. [PubMed] [Google Scholar]
- 33.Lopez-Ribot J L, Kirkpatrick W R, McAtee R K, LaValle R, Patterson T F. Low levels of antigenic variability in fluconazole-susceptible and -resistant Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Clin Diagn Lab Immunol. 1999;6:665–670. doi: 10.1128/cdli.6.5.665-670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Ribot J L, Alloush H M, Masten B J, Chaffin W L. Evidence for presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect Immun. 1996;64:3333–3340. doi: 10.1128/iai.64.8.3333-3340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Ribot J L, Casanova M, Gil M L, Martinez J P. Common and form-specific cell wall antigens of Candida albicans as released by chemical and enzymatic treatments. Mycopathologia. 1996;134:13–20. doi: 10.1007/BF00437047. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Ribot J L, Casanova M, Monteagudo C, Sepulveda P, Martinez J P. Evidence for the presence of a high-affinity laminin receptor-like molecule on the surface of Candida albicans yeast cells. Infect Immun. 1994;62:742–746. doi: 10.1128/iai.62.2.742-746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Ribot J L, Chaffin W L. Binding of the extracellular matrix component entactin to Candida albicans. Infect Immun. 1994;62:4564–4571. doi: 10.1128/iai.62.10.4564-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Ribot J L, Martinez J P, Chaffin W L. Comparative study of the C3d receptor and 58-kilodalton fibrinogen-binding mannoproteins of Candida albicans. Infect Immun. 1995;63:2126–2132. doi: 10.1128/iai.63.6.2126-2132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Ribot J L, Monteagudo C, Sepulveda P, Casanova M, Martinez J P, Chaffin W L. Expression of the fibrinogen binding mannoprotein and the laminin receptor of Candida albicans in vitro and in infected tissues. FEMS Microbiol Lett. 1996;142:117–122. doi: 10.1111/j.1574-6968.1996.tb08417.x. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Ribot J L, Sepulveda P, Cervera A M, Roig P, Gozalbo D, Martinez J P. Cloning of a cDNA fragment encoding part of the protein moiety of the 58-kDa fibrinogen-binding mannoprotein of Candida albicans. FEMS Microbiol Lett. 1997;157:273–278. doi: 10.1111/j.1574-6968.1997.tb12784.x. [DOI] [PubMed] [Google Scholar]
- 41.Martinez J P, Gil M L, Lopez-Ribot J L, Chaffin W L. Serologic response to cell wall mannoproteins and proteins of Candida albicans. Clin Microbiol Rev. 1998;11:121–141. doi: 10.1128/cmr.11.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez J P, Lopez-Ribot J L, Chaffin W L. Heterogeneous surface distribution of the fibrinogen-binding protein on Candida albicans. Infect Immun. 1994;62:709–712. doi: 10.1128/iai.62.2.709-712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews R, Burnie J. Antibodies against Candida: potential therapeutics? Trends Microbiol. 1996;4:354–358. doi: 10.1016/0966-842x(96)10056-1. [DOI] [PubMed] [Google Scholar]
- 44.Matthews R, Burnie J P, Lee W. The application of epitope mapping in the development of a new serological test for systemic candidosis. J Immunol Methods. 1991;143:73–79. doi: 10.1016/0022-1759(91)90274-j. [DOI] [PubMed] [Google Scholar]
- 45.Matthews R, Hodgetts S, Burnie J. Preliminary assessment of a human recombinant antibody fragment to hsp90 in murine invasive candidiasis. J Infect Dis. 1995;171:1668–1671. doi: 10.1093/infdis/171.6.1668. [DOI] [PubMed] [Google Scholar]
- 46.Matthews R, Wells C, Burnie J P. Characterisation and cellular localisation of the immunodominant 47-Kda antigen of Candida albicans. J Med Microbiol. 1988;27:227–232. doi: 10.1099/00222615-27-4-227. [DOI] [PubMed] [Google Scholar]
- 47.Matthews R C. Pathogenicity determinants of Candida albicans: potential targets for immunotherapy? Microbiology. 1994;140:1505–1511. doi: 10.1099/13500872-140-7-1505. [DOI] [PubMed] [Google Scholar]
- 48.Matthews R C, Burnie J P, Howat D, Rowland T, Walton F. Autoantibody to heat-shock protein 90 can mediate protection against systemic candidosis. Immunology. 1991;74:20–24. [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald C S F, Pappin D, Higgins S. Electroelution of proteins from SDS gels. Trends Genet. 1986;2:35. [Google Scholar]
- 50.Navarro D, Monzonis E, Lopez-Ribot J L, Sepulveda P, Casanova M, Nogueira J M, Martinez J P. Diagnosis of systemic candidiasis by enzyme immunoassay detection of specific antibodies to mycelial phase cell wall and cytoplasmic candidal antigens. Eur J Clin Microbiol Infect Dis. 1993;12:839–846. doi: 10.1007/BF02000404. [DOI] [PubMed] [Google Scholar]
- 51.Polonelli L, De Bernardis F, Conti S, Boccanera M, Gerloni M, Morace G, Magliani W, Chezzi C, Cassone A. Idiotypic intravaginal vaccination to protect against candidal vaginitis by secretory, yeast killer toxin-like anti-idiotypic antibodies. J Immunol. 1994;152:3175–3182. [PubMed] [Google Scholar]
- 52.Sentandreu M, Elorza M V, Sentandreu R, Fonzi W A. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J Bacteriol. 1998;180:282–289. doi: 10.1128/jb.180.2.282-289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sepúlveda P, López-Ribot J L, Murgui A, Canton E, Navarro D, Martínez J P. Candida albicans fibrinogen binding protein: expression in clinical strains and immunogenicity in patients with systemic candidiasis. Int Microbiol. 1998;1:209–216. [PubMed] [Google Scholar]
- 54.Sundstrom P, Aliaga G R. A subset of proteins found in culture supernatants of Candida albicans includes the abundant, immunodominant, glycolytic enzyme enolase. J Infect Dis. 1994;169:452–456. doi: 10.1093/infdis/169.2.452. [DOI] [PubMed] [Google Scholar]
- 55.Tavares D, Ferreira P, Vilanova M, Videira A, Arala-Chaves M. Immunoprotection against systemic candidiasis in mice. Int Immunol. 1995;7:785–796. doi: 10.1093/intimm/7.5.785. [DOI] [PubMed] [Google Scholar]
- 56.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welling G W, Weijer W J, van der Zee R, Welling-Wester S. Prediction of sequential antigenic regions in proteins. FEBS Lett. 1985;188:215–218. doi: 10.1016/0014-5793(85)80374-4. [DOI] [PubMed] [Google Scholar]
- 58.Westhof E, Altschuh D, Moras D, Bloomer A C, Mondragon A, Klug A, Van Regenmortel M H. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature. 1984;311:123–126. doi: 10.1038/311123a0. [DOI] [PubMed] [Google Scholar]