Abstract
Peucedanum franchetii is a famous folk medicinal plant in China. However, the taxonomy of the P. franchetii has not been sufficiently resolved. Due to similar morphological features between P. franchetii and Ligusticopsis members, the World Flora Online (WFO) Plant List suggested that this species transformed into the genus Ligusticopsis and merged with Ligusticopsis likiangensis. However, both species are obviously diverse in leaf shape, bracts, and bracteoles. To check the taxonomic position of P. franchetii, we newly sequenced and assembled the plastome of P. franchetii and compared it with nine other plastomes of the genus Ligusticopsis. Ten plastomes were highly conserved and similar in gene order, codon bias, RNA editing sites, IR borders, and SSRs. Nevertheless, 10 mutation hotspot regions (infA, rps8, matK, ndhF, rps15, psbA-trnH, rps2-rpoC2, psbA-trnK, ycf2-trnL, and ccsA-ndhD) were still detected. In addition, both phylogenetic analyses based on plastome data and ITS sequences robustly supported that P. franchetii was not clustered with members of Peucedanum but nested in Ligusticopsis. P. franchetii was sister to L. likiangensis in the ITS topology but clustered with L. capillacea in the plastome tree. These findings implied that P. franchetii should be transferred to genus Ligusticopsis and not merged with L. likiangensis, but as an independent species, which was further verified by morphological evidences. Therefore, transferring P. franchetii under the genus Ligusticopsis as an independent species was reasonable, and a new combination was presented.
Keywords: Apiaceae, Ligusticopsis, Peucedanum franchetii, new combination, plastome, phylogeny, taxonomy
1. Introduction
The genus Peucedanum, with P. officinale L. as type species [1], is one of the largest taxonomically notorious genera of the Apiaceae family and has a broad circumscription widely distributed in Eurasia and Africa, with Europe and East Asia as the major distribution centers [2,3]. The genus includes about 100–120 species in total, and 40 species (33 species are endemic) occur in China [4,5]. However, the plants of this genus exhibit great diversity in flowers, leaves, bracteoles, and mericarps [3]. Moreover, all previous molecular phylogenetic analysis supported that the genus Peucedanum is not monophyletic [5]. Therefore, some authors have implemented several taxonomic changes about this genus [6,7,8,9]. Reduron et al. confirmed that the restitution of Cervaria, Imperatoria, Oreoselinum, Pteroselinum, Thysselinum, Xanthoselinum, and Holandrea relied on morphological and phytochemical data [10]. Pimenov et al. suggested all members of Peucedanum should be shifted into other genera, excluding 8–10 species in P. sect. Peucedanum [11]. Winter et al. [12] transformed 24 Peucedanum species into Afroligusticum C. Norman, Cynorhiza Eckl. & Zeyh., and Lefebvrea A. Rich. Although numerous taxonomic treatments for Peucedanum members have been achieved, the revision of this genus is still on the way, especially for those taxa endemic to China.
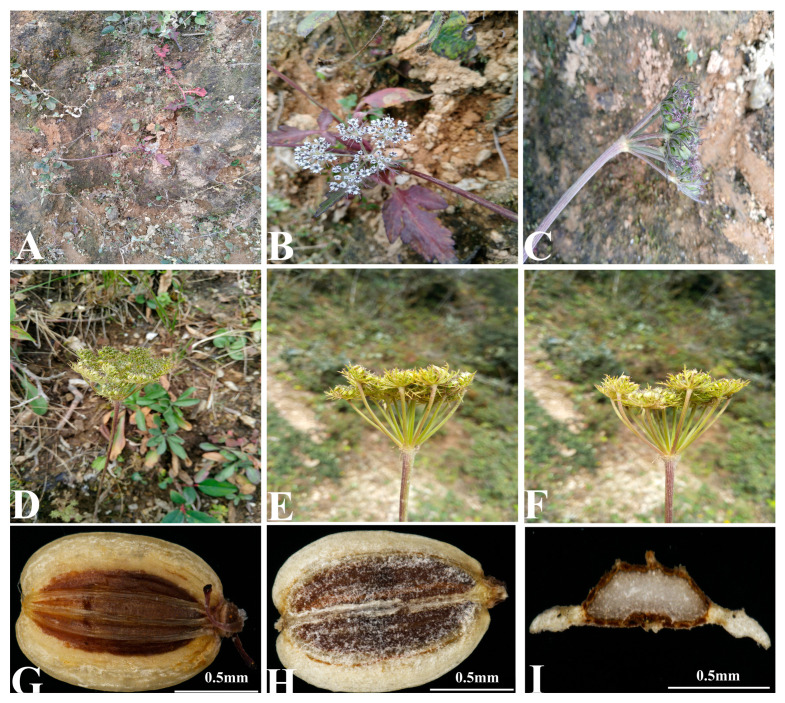
Peucedanum franchetii C.Y.Wu & F.T.Pu is endemic to China, growing on grassland or calcareous hillside at an altitude of about 3000 m. Due to dorsally compressed mericarps with slightly prominent dorsal ribs and narrowly winged lateral ribs appearing in P. franchetii (Figure 1), this species is deemed to be a member of the genus Peucedanum [5]. However, it is similar to the genus Ligusticopsis, which was established by Leute in 1969 with the type species Ligusticopsis rechingeriana (H.Wolff) Leute [13]. Therefore, in the world flora plant list (http://worldfloraonline.org (accessed on 28 November 2022)), P. franchetii was regarded as a synonym of Ligusticopsis likiangensis (H.Wolff) Lavrova & Kljuykov. However, making detailed field investigations and collecting the material of this plant in the type locality, we found P. franchetii was extraordinarily different from L. likiangensis in leaf shape, bracts, and bracteoles. Therefore, we hypothesize that P. franchetii may be not the synonym of L. likiangensis and could be an independent species. Moreover, the phylogenetic position of P. franchetii has not been inferred so far.
Figure 1.
Illustrations of Peucedanum franchetii. (A) Habit; (B) basal leaves; (C) stem leaves; (D) umbel with bracts and bracteoles; (E) flower; (F) young mericarp with calyx teeth; (G) dorsal of mericarp; (H) commissural of mericarp; (I) cross-section of mericarp. Scale bars: 0.5 mm (G,H,I).
In this work, we newly sequenced the complete plastome of P. franchetii. Together with nine previously published Ligusticopsis plastomes, we carried out a comprehensive comparative analysis among the ten plastomes. Then, we conducted phylogenetic analyses based on plastome data and ITS sequences to confirm the position of P. franchetii. Finally, combining with morphological evidence, we made a taxonomic revision for P. franchetii.
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction, and Sequencing
The fresh leaf of P. franchetii used in this experiment was collected from its natural habitat (Figure 1) in Eryuan, Yunnan, China (type locality), and dried in silica gel. The formal identification of P. franchetii plant was performed according to the Flora of China [5]. The voucher specimens are currently stored at the herbarium of Sichuan University (Chengdu, China) under deposition number LCK2020001. Total genomic DNA was extracted from silica gel dried leaves, employing the modified cetyltrimethylammonium bromide (CTAB) method [14], and carried on the Illumina NovaSeq platform at Personalbio (Shanghai, China), applying the paired-end 150 bp reads with an average insert size of 300–400 bp. Then, the obtained raw data were trimmed by removing adaptors and low-quality reads using AdapterRemoval v2 (trimwindows = 5; minlength = 50) [15], and finally, we filtered low-quality reads to gain high-quality reads with fastP v0.15.0 (-n 10 and -q 15) [16].
In addition, total DNA was also employed to amplify ITS sequence. We operated 30 µL amplification system, including 2 µL extracted total DNA, 10 µL ddH2O, 15 µL Taq MasterMix (CWBio, Beijing, China), 1.5 µL of 10 pmol µL−1 forward primers (ITS-4: 5′-TCC TCC GCT TAT TGA TAT GC-3′), and 1.5 µL of 10 pmol µL−1 reverse primers (ITS-5: 5′-GGA AGT AAA AGT CGT AAC AAG G-3′) [17]. PCR cycling profile consisted of an initial denaturation step at 94 °C for 3 min, followed by denaturation step at 94 °C for 45 s, 30 cycles of 45 s at 94 °C, annealing at 55 °C for 45 s and extension at 72 °C for 45 s, a final extension for 7 min at 72 °C, and storage at 4 °C. All PCR products were separated, manipulating a 1.5% (wv−1) agarose TAE gel and delegated Sangon Biotech (Shanghai, China) for sequencing. DNAstar–SeqMan [18] was occupied to edit the newly sequenced DNA and gain consensus sequences.
2.2. Plastome Assembly and Annotation
The plastome of P. franchetii was de novo assembled via GetOrganelle v1.6.2 software [19] considering the plastome of Ligusticopsis brachyloba (Franch.) Leute (NC_065500) as reference. To assess the sequencing depth and overlapping contigs, the cleaned reads were mapped to the reference plastome by Geneious v9.0.2 [20]. The Plastid Genome Annotator (PGA) was applied to annotate the plastome [21], followed by the validation of annotation adopting online GeSeq tool [22] with default parameters. The plastome of L. brachyloba (NC_065500) was also chosen as the reference to predict protein-coding genes (PCGs) and transfer RNA (tRNA) genes and ribosomal RNA (rRNA) genes. Finally, we checked the annotation results with BLAST and DOGMA [23] and corrected the wrong annotation in Geneious v9.0.2 [20] manually. A detailed plastome map was drawn by the online Organellar Genome DRAW tool (OGDRAW: https://chlorobox.mpimp-golm.mpg.de/geseq.html, accessed on 1 October 2022) [24]. The newly yielded ITS sequence and plastome of P. franchetii were submitted into the NCBI under the accession numbers OP740295 and OP745624, respectively. Additionally, we downloaded 54 ITS sequences and 54 plastomes of Apioideae for the phylogenetic analyses. The names of the species and GenBank accession numbers for all samples were listed in Table S1.
2.3. Codon Usage, RNA Editing, and SSRs Analyses
To identify codon usage pattern and relative synonymous codon usage (RSCU) values [25], CodonW v1.4.2 program [26] was used to analyze the codon usage bias, and the heatmap was depicted by TBtools [27]. To acquire the composition and characteristics of RNA editing, 41 shared protein-coding genes for 10 plastomes were carried out through the online PREP-Cp program with a cutoff value of 0.8, and other parameters were default to predict RNA editing sites [28]. Simple sequence repeats (SSRs) of ten plastomes were calculated by MISA web (http://misaweb.ipk-gatersleben.de/, accessed on 2 October 2022) [29] tool with the following criteria: 10, 5, 4, 3, 3, and 3 repeat units for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively.
2.4. Comparative Plastome Analyses
The expansion or contraction of the IR regions may lead to the size variation of the plastome; the boundaries of IR regions in ten plastomes were analyzed by IRscope [30] and manually inspected in Geneious v9.0.2 [20]. To better determine whether any specific pattern of structural variation exists in the ten plastomes, gene arrangement comparison was visualized by the Mauve alignment program [31] with default settings in Geneious v9.0.2 [20]. Furthermore, the sequence identity among the ten species was detected using mVISTA software (https://genome.lbl.gov/vista/mvista/submit.shtml, accessed on 3 October 2022) [32] with the default settings, and P. franchetii was chosen as the reference.
2.5. Identification of Divergence Hotspots
To further discern the mutation hotspot regions, the protein-coding regions, non-coding regions, and intron regions of the ten plastomes were first extracted in Geneious v9.0.2 [20] and aligned employing MAFFT version 7.427 [33] with default parameters. Then, the alignments were manually adjusted. Finally, the nucleotide diversity (Pi) was calculated for alignments with more than 200 bp in length carrying DnaSP version 5.0 [34].
2.6. Phylogenetic Analyses
To pinpoint the phylogenetic position of P. franchetii, 54 ITS and 54 complete plastome sequences (Table S1) were employed to reconstruct the phylogenetic trees. Anthriscus sylvestris (L.) Hoffm and Anthriscus cerefolium (L.) Hoffm were selected as outgroups according to the results of Wen et al. [35]. Then, 54 ITS and 54 plastome sequences were respectively aligned in Geneious prime 2021.1.1 plugin MAFFT 7.427 [33] and modified manually. Alignments were used for maximum likelihood analyses (ML) and Bayesian inference (BI) methods. The best-scoring ML tree was inferred adopting RAxML version 8.2.11 [36] with the GTRGAMMAX model for each partition, and branch support was assessed by the rapid bootstrap algorithm [37] with 1000 replicates. For BI analysis, four parallel Markov chain Monte Carlo (MCMC) runs were executed in MrBayes version 3.2.7 [38] and the best-fitting substitution model (SYM + I + G) for ITS dataset and (GTR + I + G) for plastome dataset determined by Modeltest v3.7 [39]. A total of 1,000,000 generations were run with sampling every 500 generations, and the first 25% of samples were discarded as burn-in and the remainder were maintained to produce the consensus tree. Results of phylogenetic analyses were visualized and exhibited in FigTree v1.4.2 software [40] and beautified by the online tool iTOL (https://itol.embl.de/itol.cgi, accessed on 4 October 2022) [41].
3. Results
3.1. Characteristics of Plastome
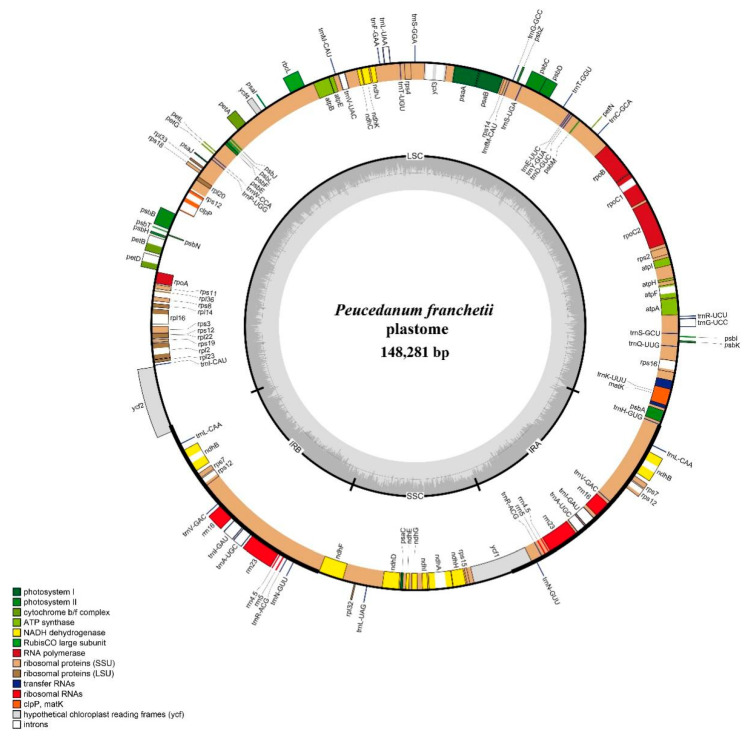
The length of the complete plastome sequence in P. franchetii was 148,281 bp and displayed a typical quadripartite structure (Figure 2), containing one LSC region (92,298 bp) and one SSC region (17,691 bp) separated by a pair of IR regions (19,146 bp). Moreover, the total GC content was 37.5%. The plastome encoded 113 unique genes, and these genes had four categories: self-replication, genes for photosynthesis, other genes, and genes of unknown function. Among them, 15 genes harbored a single intron, and 3 genes (ycf3, clpP, and rps12) had 2 introns (Table S2).
Figure 2.
Gene map of the P. franchetii plastome. The genes exhibited outside of the circle are transcribed clockwise, while those are counterclockwise inside. The genes belonging to different functional groups are color-coded.
The study also compared P. franchetii plastome with nine Ligusticopsis plastomes. For the ten plastomes, the overall size ranged from 147,752 bp (L. involucrata) to 148,633 bp (L. brachyloba). They hold the same unique gene number, comprising 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The overall GC content was corresponding in the ten plastomes (37.3–37.5%). Hence, the structure of the plastome appeared to be largely conserved across the ten species (Table 1).
Table 1.
Comparative analyses of ten plastomes. P. franchetii included.
| Taxon | Length (bp) | Number of Genes (Unique) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Genome | LSC | SSC | IR | Total | CDS | rRNA | tRNA | GC (%) | |
| P. Franchetii C.Y.Wu & F.T.Pu | 148,281 | 92,298 | 17,691 | 19,146 | 113 | 79 | 4 | 30 | 37.50% |
| L. brachyloba (Franch.) Leute | 148,633 | 92,265 | 17,588 | 19,390 | 113 | 79 | 4 | 30 | 37.40% |
| L. capillacea (H.Wolff) Leute | 147,808 | 91,907 | 17,503 | 19,199 | 113 | 79 | 4 | 30 | 37.50% |
| L. hispida Lavrova & Kljuykov | 147,797 | 91,846 | 17,627 | 19,162 | 113 | 79 | 4 | 30 | 37.40% |
| L. involucrata Lavrova | 147,752 | 91,782 | 17,560 | 19,205 | 113 | 79 | 4 | 30 | 37.40% |
| L. likiangensis (H.Wolff) Lavrova & Kljuykov | 148,196 | 92,305 | 17,575 | 19,158 | 113 | 79 | 4 | 30 | 37.50% |
| L. scapiformis (H.Wolff) Leute | 148,107 | 92,214 | 17,581 | 19,156 | 113 | 79 | 4 | 30 | 37.50% |
| L. rechingeriana Leute | 148,525 | 91,813 | 17,654 | 19,529 | 113 | 79 | 4 | 30 | 37.30% |
| L. modesta (Diels) Leute | 148,133 | 92,247 | 17,568 | 19,159 | 113 | 79 | 4 | 30 | 37.50% |
| L. wallichiana Fedde ex H.Wolff | 148,594 | 92,281 | 17,567 | 19,373 | 113 | 79 | 4 | 30 | 37.40% |
3.2. Codon Usage, RNA Editing in Protein-Coding Genes
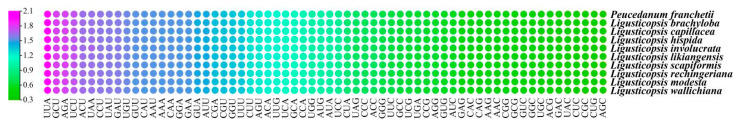
Our study summarized the codon usage frequency of 79 protein-coding genes among the 10 plastomes. The result demonstrated that codon usage bias was similar in them. The 79 protein-coding genes encoded 22,632–22,553 codons. In these codons, the Leu was encoded by the highest number of codons 2399–2410 (10.64%–10.65%), while the Cys was the least 231–237 (1.02–1.05%) (Table S3). Figure 3 showed the relative synonymous codon usage (RSCU) values of all protein-coding genes for the ten plastomes. The RSCU values ranged from 0.34 to 2.00. The UAA codon possessed the highest value (RSCU = 2.0) in the P. franchetii, L. brachyloba, L. capillacea, and L. scapiformis plastomes, while the AGC codon possessed the smallest value (RSCU = 0.34) only in the L. involucrata plastome. The RSCU value of codons AUG and UGG was 1.00 (Table S3). In addition, the RSCU values of 30 codons were greater than 1.00, and these codons were ended with A/U, except UUG in ten plastomes.
Figure 3.
The RSCU values of all concatenated protein-coding genes for ten plastomes. Color key: the purple values represent higher RSCU values while the green values indicate lower RSCU values.
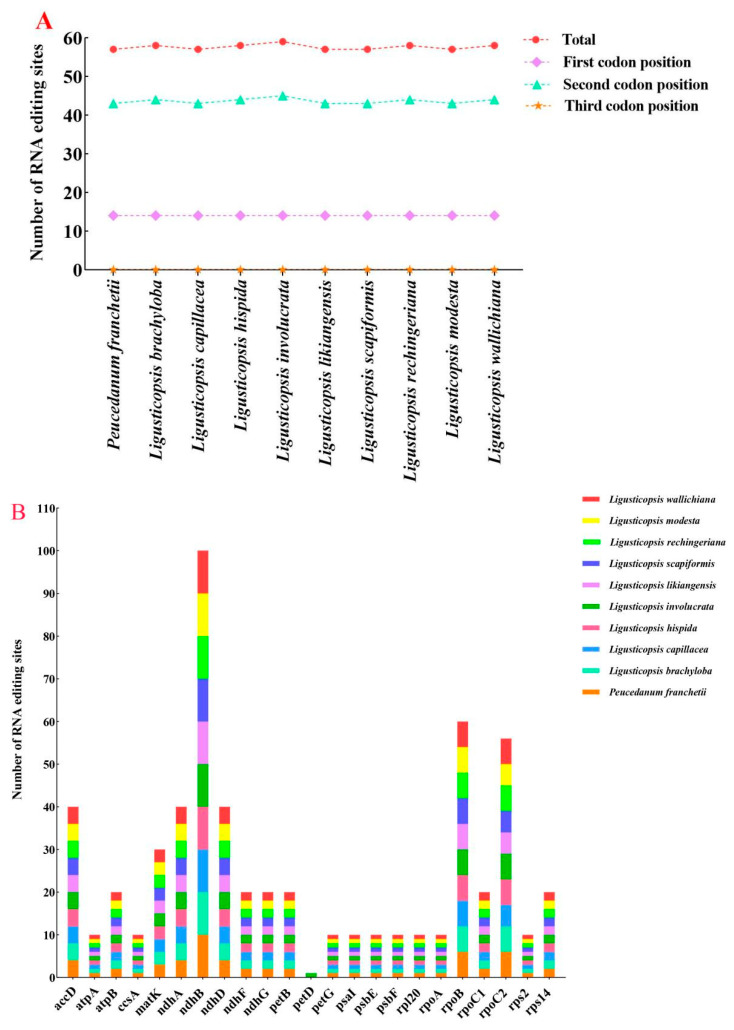
This work also predicted the RNA editing sites of the ten plastomes. We found 22–23 protein-coding genes and identified 57–59 potential RNA editing sites in the 10 plastomes (Table S4, Figure 4). Among these protein-coding genes, the ndh gene had the most editing sites (22), while other genes (atpA, ccsA, petG, psaI, psbE, psbF, rpl20, rpoA, and rps2) only had one editing site. Among all detected RNA editing sites, the second codon position occupied the most editing sites (43–45), followed by the first codon position (14), but no sites detected in the third codon position (Figure 4A). Although ten plastomes appeared to have a similar pattern of RNA editing, one specific editing site had been picked out: petD (one site; only identified in L. involucrata) (Figure 4B). Moreover, the amino acid conversion S to L occurred most frequently, while A to V and R to C occurred least (Table S4).
Figure 4.
The RNA editing sites in ten plastomes. (A) Numbers of RNA editing sites distributed in different codon positions; (B) numbers of RNA editing sites presented in genes.
3.3. Simple Sequence Repeats (SSRs)
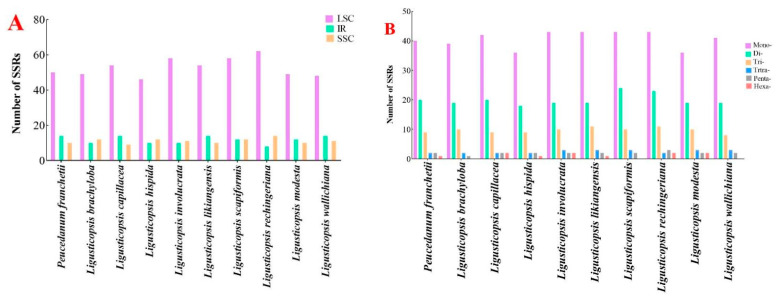
We estimated and compared the SSRs in the ten plastomes. The total number of SSRs altered from 71 to 84, of which the LSC region had the richest repeats (46–62) and the SSC region (9–14) had the poorest following IR regions (8–14) (Table S5, Figure 5A). Figure 5B displayed the number of different repeat types in ten plastomes. Among these SSRs, the mononucleotide repeats were the maximum (36–42), followed by the dinucleotides (18–24) and the hexanucleotide repeats, which were the minimal (0–1).
Figure 5.
Analyses of simple sequence repeats (SSRs) in ten plastomes. (A) Presence of SSRs in LSC, SSC and IR; (B) numbers of different repeat types.
3.4. Plastome Comparison and Hotspots Identification
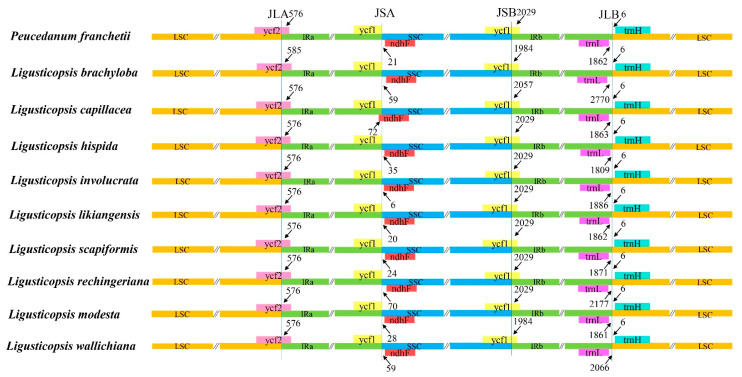
In the current study, we conducted a detailed comparison of the IR boundaries among the ten plastomes. The LSC/IRa junction was located in the ycf2 gene and expanded 576 bp into the ycf2 gene except for L. brachyloba (585 bp). The SSC/IRa junction was located in the intergenic spacer region between the ndhF and ycf1 gene, but expanded 72 bp into ndhF gene in L. capillacea. The SSC/IRb junction was located in the ycf1 gene and expanded 1984–2029 bp into the ycf1 gene. The LSC/IRb junction was located between the trnL and trnH genes, and the trnL gene was 1860–2771 bp away from LSC, while the trnH gene had the same length (6 bp) away from the IRb borders (Figure 6).
Figure 6.
Comparison of the border positions of LSC, SSC, and IR regions across the ten plastomes. Functional genes and truncated fragments are denoted by colored boxes. The sizes of gene fragments located at boundaries are indicated by the base pair lengths.
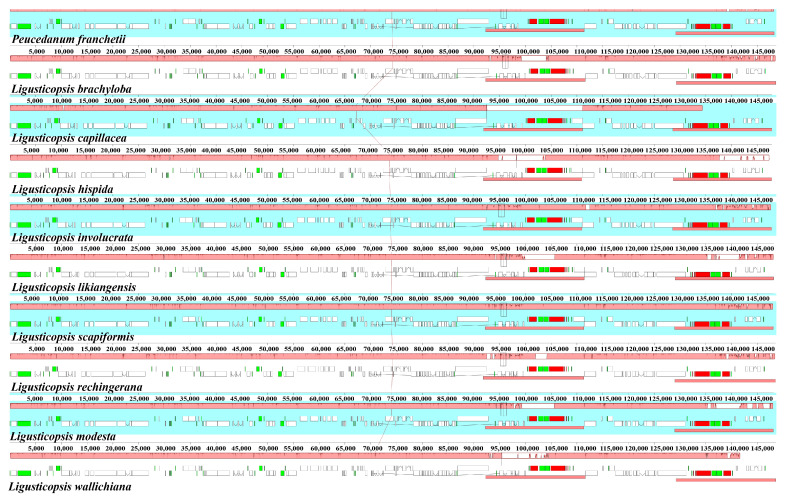
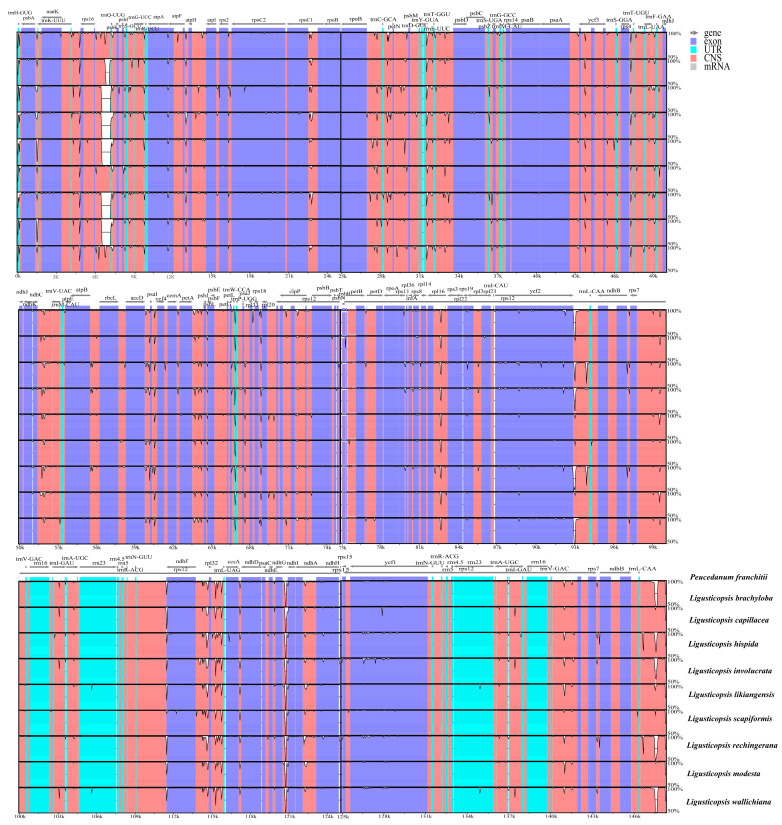
The Mauve result demonstrated that the gene arrangement was highly conservative and, moreover, the gene retained a similar sequence among the ten plastomes (Figure 7 and Figure 8).
Figure 7.
Mauve alignment of ten plastomes. Local collinear blocks within each alignment are represented by blocks of the same color connected with lines.
Figure 8.
Sequence identity plot comparing the ten plastomes using mVISTA. The y-axis corresponds to percentage identity (50–100%), while the x-axis shows the position of each region within the locus. Arrows indicate the transcription of annotated genes in the reference genome.
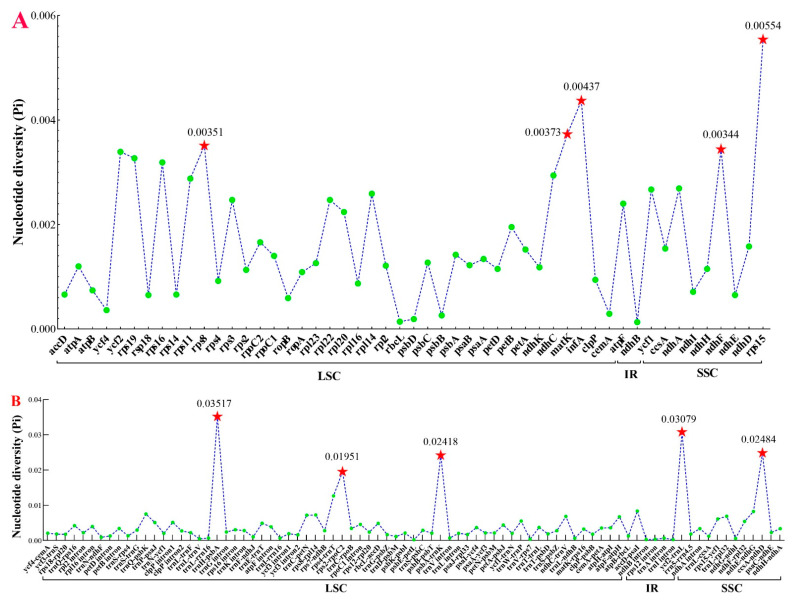
The nucleotide diversity (Pi) was calculated to analyze the sequence divergence level of four regions across the ten plastomes. The results indicated that the divergence level of LSC and SSC regions were higher than the IR regions (Figure 9). We also screened ten mutation hotspot regions as candidate DNA barcodes, including five protein-coding genes (rps8, matK, infA, ndhF, and rps15) with the Pi > 0.00344 (Figure 9A) and five non-coding genes (psbA-trnH, rps2-rpoC2, psbA-trnK, ycf2-trnL, and ccsA-ndhD) with the Pi > 0.01950 (Figure 9B).
Figure 9.
Comparative analysis of the nucleotide diversity (Pi) values among the ten plastomes. (A) protein-coding genes; (B) non-coding and intron regions.
3.5. Phylogenetic Analyses
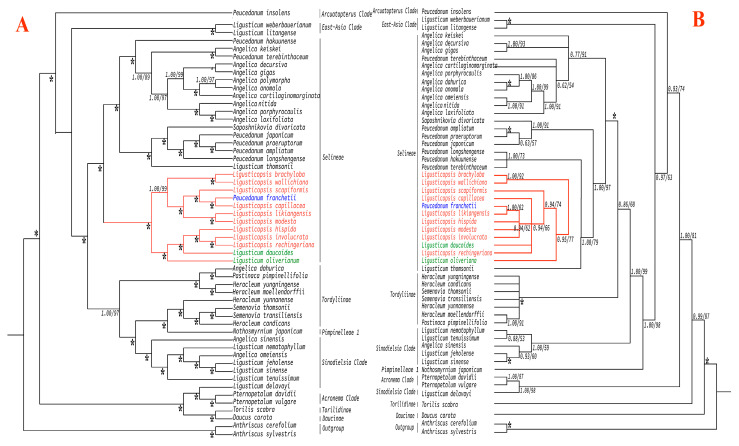
We used 54 ITS sequences and 54 plastome sequences of Apioideae to conduct the phylogenetic analyses (Table S1). Although the topological tree between the ITS and plastome were conflicting, both demonstrate that P. franchetii was nested in the genus Ligusticopsis. In the plastome tree, P. franchetii clustered with L. capillacea and resolved as a sister to other Ligusticopsis taxa with high support (BS = 100%, PP = 1.00) (Figure 10A). However, in the ITS topology, P. franchetii clustered with L. likiangensis also with high support (BS = 92%, PP = 1.0) and resolved it as a sister to other Ligusticopsis taxa (Figure 10B). Moreover, both phylogenetic trees also suggested that Ligusticum daucoides (Franch.) Franch and Ligusticum oliverianum (de Boiss.) Shan are nested in the genus Ligusticopsis (Figure 10).
Figure 10.
Phylogeny of the 54 taxa inferred from maximum likelihood (ML) and Bayesian inference (BI) analyses. Branch support was assessed using ML bootstrap percentage (BP) and Bayesian posterior probability (PP), and internal branches with less than 100 BP/1.0 PP were indicated with corresponding values. (A) Phylogenetic tree of 54 Apioideae taxa based on plastomes. (B) Phylogenetic tree of 54 Apioideae taxa based on ITS sequences. (* means the node with BS = 100 and PP = 1.00).
4. Discussion
4.1. Plastome Characteristics
We reported the newly sequenced and assembled complete plastome of P. franchetii and performed a comparative analysis with nine other Ligusticopsis species. The results indicated that ten plastomes exhibited a typical quadripartite structure, which was in accordance with the plastome structure of Apiaceae published previously [42,43,44,45,46,47,48]. Furthermore, all plastomes were similar in genome size, gene order, and GC content. These results demonstrated that ten plastomes were highly conserved. In addition, higher GC content was detected in IR regions compared to the LSC and SSC regions, for which was present four rRNA (rrn23, rrn16, rrn5, and rrn4.5) genes with high GC content (50–56.4%) in IR regions [49,50].
4.2. Codon Usage Analyses
Codons act as a bridge between nucleic acids and proteins in spreading genetic information [51]. Each amino acid has two or more codons and each has its preferred codon for codon usage bias (CUB). CUB is a common phenomenon that widely exists in organisms [52,53], and it has been hypothesized that it controls translation dynamics such as reliability, accuracy, and protein folding [54]. Recent research has proclaimed that codon usage greatly influences the evolution of the plastome [55,56]. In this investigation, Leu was the more frequent amino acid and Cys was the least. The effects were consistent with those observed in other plant plastome studies [57,58,59,60,61]. The most preferred codons end with A/U (Table S3, Figure 3), which was commonly examined in previously reported genera of flowering plants [62,63,64]. We also counted RSCU values for 64 mutant codons, 30 of which had RSCU values larger than 1.00, and codons AUG and UGG had no bias (RSCU = 1.00). These results correspond to prior plastome studies, demonstrating that usage bias of particular codons was induced by adaptation evolution of the plastome [62].
4.3. RNA Editing Analyses
RNA editing as a post-transcriptional modification process is important for correcting DNA mutations on the RNA level [65,66,67]. In the present study, the editing sites were cytidine (C) to uridine (U) conversions in ten plastomes (Table S4), which was reported in higher plants [43,44,45]. Among these RNA editing sites, changing a nucleotide at the second position was more common than other position shifts. It corresponded to the general characteristics of plastid gene RNA editing in higher plants [59,68,69,70]. Moreover, a multitudinous number was discovered in the ndhB gene, which was line with the previous conclusion that ndh group genes have the largest number of plastome editing sites in flowering plants [70].
4.4. Simple Sequence Repeats (SSRs) Analyses
SSRs are usually small tandem single mononucleotide repeats, which show intraspecific repeat number differences [71,72]. Owing to the characteristics of SSR’s codominant inheritance, high reproducibility, multiallelic composition, richness, and ease of detection, it is widely discussed in plant diversity analysis [73,74]. This work gained a total number of (71–84) SSRs, which had coincident results among other Apiaceae in numbers [44,75]. The results also showed that the SSR loci of the LSC region appeared more usually than in the SSC and IR regions; it may be hypothesized that it was relevant to the longer LSC region [43]. Moreover, the mononucleotide repeats were abundant and mostly composed of A/T motifs. The same results were reported in many other angiosperms plastomes [76,77]. However, two hexanucleotide simple sequence repeats (AATATT/AATATT) were observed in the plastome of L. capillacea, though not in other plastomes, which could be served as specific molecular markers to recognize this species. Such studies implied that SSRs changed the plastome and played an important role in detecting genomic variability [58].
4.5. The IR/SC Boundaries of the Ten Plastomes
IR contraction and expansion are extremely common in plastomes of angiosperm plants [78,79,80,81]. Here, we compared the IR/SC boundaries and found that the crossroads and surrounding genes of four sections IRa/SSC/IRb/LSC were the same. Moreover, the ten plastomes had same gap between the IRb/LSC junctions. These results further justified the conservation of the ten plastomes. Our research supported this hypothesis, demonstrating that the IR region is more conserved and that most substitutions occur in the SSC and LSC regions. The same phenomena have been achieved in other plastid genomes as well [82,83,84,85].
4.6. Candidate DNA Barcodes
DNA barcodes are considered as short DNA sequences with adequate variations to identify species [86]. In plants, three variable loci (matK, rbcL, and trnH-psbA) from plastome were extensively defined as the standard DNA barcodes. But these fragments had been proved to be insufficient for the identification of a taxonomically complex group [87]. For example, Liu et al. operated the rbcL gene as DNA barcodes suffered from weak solutions among Peucedanum [43]. Therefore, additional loci are urgently needed to be developed for specific taxa. In this study, we screened the ten most divergent loci, containing five protein-coding genes (rps8, matK, infA, ndhF, and rps15) and five non-coding protein genes (psbA-trnH, rps2-rpoC2, psbA-trnK, ycf2-trnL, and ccsA-ndhD) based on the complete plastome. These regions could be served as potential DNA barcodes for species identification in Ligusticopsis.
4.7. Phylogenetic Relationships and Taxonomic Implication of P. franchetii
Although plastome is highly conserved in genome structure, gene order, and content, it shows highly variable characters. Therefore, plastome data have been applied to generate highly resolved phylogeny in plant species, especially in taxonomically complex groups [88,89,90]. In this article, we constructed the phylogenetic framework depending on plastome data to make a definite position of P. franchetii. Furthermore, we also performed phylogenetic analysis using ITS sequences, mainly due to the fact that chloroplast genomes only represent maternal inheritance.
Previous phylogenetic analysis based on nrDNA ITS sequences performed by Zhou et al. [91] confirmed that the generitype species Peucedanum officinale was distant from Ligusticopsis (including the type species Ligusticopsis rechingeriana). Although there was little difference between the plastome and ITS topologies, the P. franchetii fell into genus Ligusticopsis with high support value, which suggested that P. franchetii was a member of genus Ligusticopsis. The morphological characteristics of P. franchetii also supported the above phylogenetic results, such as pinnate leaves, pinnate and linear coexisted bracteoles, conspicuous calyx teeth, and numerous vittae in mericarp commissure and each furrow (Figure 1, Table S6). All these morphological traits were in accordance with the genus Ligusticopsis. Thus, P. franchetii should be transferred from genus Peucedanum to Ligusticopsis.
P. franchetii was regarded as a synonym of L. likiangensis in the World Flora plant list (http://worldfloraonline.org, accessed on 10 December 2022). However, our results did not support this treatment. Although P. franchetii was a sister to L. likiangensis in the ITS topology, this species clustered with L. capilliacea in the plastome tree. Moreover, obviously different morphological characters were detected for the both species. The absent bracts and pinnate and linear coexisted bracteoles were observed in P. franchetii, while pinnate and linear coexisting bracts and pinnate bracteoles were found in L. likiangensis (Table S6). Therefore, merging P. franchetii into L. likiangensis was inappropriatem and P. franchetii should be regarded as a dependent species of genus Ligusticopsis.
In addition, our phylogenetic tree also indicated that Ligusticum daucoides and Ligusticum oliverianum nested in the genus Ligusticopsis and belonged to Selineae. Both species were obviously distant from the type species of Ligusticum (Ligusticum scoticum L.), which is located in Acronema Clade [91,92]. These results supported the taxonomic treatments performed by Pimenov [93], who transferred both species into the genus Ligusticopsis. This conclusion is consistent with the definition of Ligusticopsis based on the chloroplast genome and morphological boundary performed by Li et al. [44]. Our results with high supports provided additional evidence to accept the previous treatments.
4.8. Taxonomic Treatment
Ligusticopsis franchetii (C.Y.Wu & F.T.Pu) B.N.Song, C.K.Liu & X.J.He, comb. nov.
≡ Peucedanum franchetii C.Y.Wu & F.T.Pu in Wang Wentsai (ed.), Vasc. Pl. Hengduan Mount. 1349 (1993)
Type: China. Yunnan: in pratis calcareis montis Hee-can-men, 3000 m, 3 October 1884, Delavay 192 (isolectotype: P! P02272009; lectype P! P02272008) [93].
Descriptions: Perennial herbs, 20–30cm, pallid green, often purplish-tinged. Stems several, hollow, puberulous above. Leaf blade long ovate, pinnate, thinly coriaceous, abaxially strongly reticulate, white villous, margins dentate and slightly reflexed; pinnae 1–2 × 0.5–1 cm, 2–3 pairs, lateral pinnae rhombic or oblique ovate, base cuneate or truncate, apical pinnae ovate, base cuneate, decurrent. Umbels terminal, 2–3 cm across; peduncles elongate, straight, apex villous; bracts absent; rays 8–14, 1–2 cm, 4-angled, inner faces white hispid, outer faces glabrous; bracteoles 8–10, pinnate and linear coexisted. Umbellules 12–16-flowered. Calyx teeth conspicuous. Petals white. Styles longer than stylopodium. Fruit ovoid-oblong, ca. 3 × 2 mm, glabrous; dorsal ribs prominent, lateral ribs winged, wings ca. 1 mm, ca. 1/3 width of body, thin; vittae 1–3 in each furrow, 4–8 on commissure.
Distribution and habitat in China: Ligusticopsis franchetii is endemic to south-western China (north-western Yunnan). It grows in alpine meadows in limestone areas, at an elevation of 2900–4000 m.
Additional specimens examined: China—Yunnan: Lijiang County, 2900 m, 4 September 1992, N.H. Wang and C.N. Zhang 92027 (NAS); Dali, 3037 m, 22 August 2021, Z.X. Li 210822-2-1 (SZ); 30 October 2021, C.K. Liu LCK20211030001 (SZ).
5. Conclusions
To summarize, comparative analyses displayed that the genome structure and size, number, gene order, GC content, RNA editing, and codon usage were extremely conservative among the ten plastomes. However, ten highly variable regions (rps8, matK, infA, ndhF, rps15, psbA-trnH, rps2-rpoC2, psbA-trnK, ycf2-trnL, and ccsA-ndhD) were identified, which may be strong potential DNA barcodes for species identification and phylogenetic relationship construction. Furthermore, both morphological characteristics and molecular data robustly supported that P. franchetii should be transferred from the genus Peucedanum to Ligusticopsis as an independent species. In a word, our result will be beneficial to future phylogeny, taxonomy, and evolutionary studies of the genus Ligusticopsis.
Acknowledgments
We are grateful to Ting Ren, Qiu-ping Jiang, and Huan-huan Qin for their valuable discussions. We also thank the Shanghai Personal Biotechnology company for sequencing.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12010097/s1, Table S1: DNA sequences included in phylogenetic analyses with GenBank accession; Table S2: List of unique genes identified in plastomes of P. franchetii; Table S3: Codon usage and relative synonymous codon usage (RSCU) values of protein-coding genes for the ten plastomes; Table S4: RNA editing sites analyses for the ten plastomes; Table S5: Numbers of SSR motifs identified in the ten plastomes; Table S6: Synopsis of the morphological data from 10 species involved in this study. Some data from type specimens and Flora of China.
Author Contributions
S.Z. and X.H. designed the research. B.S., Y.X. and R.T. collected the data. B.S., C.L., D.X. and Z.L. conducted data analysis. B.S., S.Z. and X.H. prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The newly yielded ITS sequence and plastome of P. franchetii was submitted into the NCBI with accession numbers OP740295 and OP745624, respectively.
Conflicts of Interest
The authors declare that the research has no conflict of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (Grant No. 32170209, 32070221, 31872647) and Survey on the Background Resources of Chengdu Area of Giant Panda National Park (Project No.: 510101202200376).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Linnaeus C. Species Plantarum.Laurentii Salvii. Oxford University Press; Stockholm, Sweden: 1753. [Google Scholar]
- 2.Pimenov M.G., Leonov M.V., Constance L. The Genera of the Umbelliferae: A Nomenclator[M] Royal Botanic Gardens; Kew, UK: 1993. [Google Scholar]
- 3.Zhang X. Studies on geographic distribution of Peucedanum L. in southwest of China. Chin. Agric. Sci. Bull. 2011;27:177–180. doi: 10.1093/mp/ssq070. [DOI] [Google Scholar]
- 4.Spalik K., Reduron J.P., Downie S.R. The phylogenetic position of Peucedanum sensu lato and allied genera and their placement in tribe Selineae (Apiaceae, subfamily Apioideae) Plant Syst. Evol. 2004;243:189–210. doi: 10.1007/s00606-003-0066-2. [DOI] [Google Scholar]
- 5.Sheh M.L., Watson M.F. Flora of China. Volume 14. Science Press & Missouri Botanic Garden Press; Beijing, China: St. Louis, MO, USA: 2005. Peucedanum Linnaeus; pp. 182–192. [Google Scholar]
- 6.Calestani V. Conspectus specierum europaearum generis Peucedani. Bull. Soc. Bot. Ital. Ann. 1905;1905:193–201. [Google Scholar]
- 7.Leute G.H. Die Gattungen Imperatoria L. Und Tommasinia Bertol. (Apiaceae) Ann. Des Nat. Mus.Wien. 1965:69–79. [Google Scholar]
- 8.Pimenov M.G., Hedge I.C., Lamond J.M., Rechinger K.H. Cervaria. In: Rechinger K.H., Iranica F., editors. Umbelliferae. Volume 162. Springer; Berlin/Heidelberg, Germany: 1987. pp. 83–110. [Google Scholar]
- 9.Frey R. Taxonomische Revision der Gattung Peucedanum: Sektion Peucedanum und Sektion Palimbioidea (Umbelliferae) Candollea. 1989;44:257–327. [Google Scholar]
- 10.Tutin T.G., Heywood V.H., Burges N.A., Moore D.M., Valentine D.H., Walters S.M., Webb D.A., editors. Flora Europaea. Volume 2 Cambridge University Press; Cambridge, UK: 1968. [Google Scholar]
- 11.Reduron J.-P., Charpin A., Pimenov M.G. Contribution a la nomenclature generique des Apiaceae (Ombelliferes) J. Bot. Soc. Bot. Fr. 1997:91–104. [Google Scholar]
- 12.Winter P.J.D., Magee A.R., Phephu N., Tilney P.M., Downie S.R., van Wyk B.E. A new generic classification for African peucedanoid species (Apiaceae) Taxon. 2008;57:347–364. doi: 10.2307/25066009. [DOI] [Google Scholar]
- 13.Leute G.H. Untersuchungen über den Verwandtschaftkreis der Gattung Ligusticum L. (Umbelliferae) 1. Ann. Des Nat. Mus. Wien. 1969;73:55–98. [Google Scholar]
- 14.Doyle J., Doyle J.L. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 15.Davis J.I., Mcneal J.R., Barrett C.F., Chase M.W., Cohen J.I., Duvall M.R. Contrasting patterns of support among plastid genes and genomes for major clades of the monocotyledons. Early Events Monocot Evol. 2013;83:315–349. [Google Scholar]
- 16.Chen S., Zhou Y., Chen Y., Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White T.J., Bruns S., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 18.Burland T.G. DNASTAR’s Lasergene Sequence Analysis Software. In: Misener S., Krawetz S.A., editors. Bioinformatics Methods and Protocols. Methods in Molecular Biology™. Volume 132. Humana Press; Totowa, NJ, USA: 2000. pp. 71–91. [DOI] [PubMed] [Google Scholar]
- 19.Jin J.J., Yu W.B., Yang J.B., Song Y., Depamphilis C., Yi T.S., Li D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu X.J., Moore M.J., Li D.Z., Yi T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:50. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:6–11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 26.Peden J.F. Ph.D. Thesis. University of Nottingham; Nottingham, UK: 1999. Analysis of Codon Usage. [Google Scholar]
- 27.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Mower J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes, and user-defined alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amiryousefi A., Hyvönen J., Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 31.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32((Suppl. S2)):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 35.Wen J., Xie D.F., Price M., Ren T., Deng Y., Gui L., Guo X.L., He X.J. Backbone phylogeny and evolution of Apioideae (Apiaceae): New insights from phylogenomic analyses of plastome data. Mol. Phylogenet. Evol. 2021;161:107183. doi: 10.1016/j.ympev.2021.107183. [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 38.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Posada D., Crandall K.A. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 40.Rambaut A., Drummond A. FigTree, Version 1.4.2. 2015. [(accessed on 4 October 2021)]. Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 41.Letunic I., Bork P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 42.Kang L., Xie D.F., Xiao Q.Y., Peng C., Yu Y., He X.J. Sequencing and analyses on chloroplast genomes of Tetrataenium candicans and two allies give new insights on structural variants, DNA barcoding and phylogeny in Apiaceae subfamily Apioideae. PeerJ. 2019;7:e8063. doi: 10.7717/peerj.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C.K., Lei J.Q., Jiang Q.P., Zhou S.D., He X.J. The complete plastomes of seven Peucedanum plants: Comparative and phylogenetic analyses for the Peucedanum genus. BMC Plant Biol. 2022;22:101. doi: 10.1186/s12870-022-03488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren T., Li Z.X., Xie D.F., Gui L.J., Peng C., Wen J., He X.J. Plastomes of eight Ligusticum species: Characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 2020;20:519. doi: 10.1186/s12870-020-02696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z.X., Guo X.L., Price M., Zhou S.D., He X.J. Phylogenetic position of Ligusticopsis (Apiaceae, Apioideae): Evidence from molecular data and carpological characters. AoB Plants. 2022;14:plac008. doi: 10.1093/aobpla/plac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Logacheva M.D., Krinitsina A.A., Belenikin M.S., Khafizov K., Konorov E.A., Kuptsov S.V., Speranskaya A.S. Comparative analysis of inverted repeats of polypod fern (Polypodiales) plastomes reveals two hypervariable regions. BMC Plant Biol. 2017;17:61–73. doi: 10.1186/s12870-017-1195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Geng M., Xu Z., Wang Q., Li L., Xu M., Li M.M. The complete plastome of Peucedanum praeruptorum (Apiaceae) Mitochondrial DNA B Resour. 2019;4:3612–3613. doi: 10.1080/23802359.2019.1676180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gou W., Jia S.B., Price M., Guo X.L., Zhou S.D., He X.J. Complete plastid genome sequencing of eight species from Hansenia, Haplosphaera and Sinodielsia (Apiaceae): Comparative analyses and phylogenetic implications. Plants. 2020;9:1523. doi: 10.3390/plants9111523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asaf S., Khan A.L., Khan M.A., Imran Q.M., Kang S.-M., Al-Hosni K., Jeong E.J., Lee K.E., Lee I.-J. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS ONE. 2017;12:e0182281. doi: 10.1371/journal.pone.0182281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Souza U.J.B., Nunes R., Targueta C.P., Diniz-Filho J.A.F., de Campos Telles M.P. The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae-Caesalpinioideae): Comparative analysis with related Mimosoid species. Sci. Rep. 2019;9:14206. doi: 10.1038/s41598-019-50620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lagerkvist U. “Two out of three”: An alternative method for codon reading. Proc. Natl. Acad. Sci. USA. 1978;75:1759–1762. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 53.Plotkin J.B., Kudla G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y. Codon Usage Infuences the Local Rate of Translation Elongation to Regulate Cotranslational Protein Folding. Mol. Cell. 2015;59:744–754. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittal P., Brindle J., Stephen J., Plotkin J.B., Kudla G. Codon usage infuences ftness through RNA toxicity. Proc. Natl. Acad. Sci. USA. 2018;115:8639–8644. doi: 10.1073/pnas.1810022115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen X., Guo S., Yin Y., Zhang J., Yin X., Liang C., Wang Z.W., Huang B.F., Liu Y.H., Xiao S.M., et al. Complete chloroplast genome sequence and phylogenetic analysis of aster tataricus. Molecules. 2018;23:2426. doi: 10.3390/molecules23102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu B., Qian F., Hou Y., Yang W., Cai M., Wu X. Complete chloroplast genome features and phylogenetic analysis of Eruca sativa (Brassicaceae) PLoS ONE. 2021;16:1–19. doi: 10.1371/journal.pone.0248556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du X., Zeng T., Feng Q., Hu L., Luo X., Weng Q., He J.F., Zhu B. The complete chloroplast genome sequence of yellow mustard (Sinapisalba L.) and its phylogenetic relationship to other Brassicaceae species. Genes. 2020;731:144340. doi: 10.1016/j.gene.2020.144340. [DOI] [PubMed] [Google Scholar]
- 59.Tang D., Wei F., Kashif M.H., Munsif F., Zhou R. Identifcation and analysis of RNA editing sites in chloroplast transcripts of kenaf (Hibiscus cannabinus L.) 3 Biotech. 2019;9:361. doi: 10.1007/s13205-019-1893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Redwan R.M., Saidin A., Kumar S.V. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015;15:196. doi: 10.1186/s12870-015-0587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan C., Du J., Gao L., Li Y., Hou X. The complete chloroplast genome sequence of watercress (Nasturtium ofcinale R. Br.): Genome organization, adaptive evolution and phylogenetic relationships in Cardamineae. Gene. 2019;699:24–36. doi: 10.1016/j.gene.2019.02.075. [DOI] [PubMed] [Google Scholar]
- 62.Ren T., Yang Y.C., Zhou T., Liu Z.L. Comparative plastid genomes of Primula species: Sequence divergence and phylogenetic relationships. Int. J. Mol. Sci. 2018;19:1050. doi: 10.3390/ijms19041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong S., Ying Z., Yu S., Wang Q., Liao G., Ge Y., Cheng R. Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: Insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships. BMC Genom. 2021;6:880. doi: 10.1186/s12864-021-08193-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X., Zhou T., Bai G., Zhao Y. Complete chloroplast genome sequence of Fagopyrum dibotrys: Genome features, comparative analysis and phylogenetic relationships. Sci. Rep. 2018;8:12379. doi: 10.1038/s41598-018-30398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castandet B., Araya A. RNA editing in plant organelles. Why make it easy? Biochemistry. 2011;76:924–931. doi: 10.1134/S0006297911080086. [DOI] [PubMed] [Google Scholar]
- 66.Chen H., Deng L., Jiang Y., Lu P., Yu J.N. RNA editing sites exist in protein-coding genes in the chloroplast genome of Cycas taitungensis. J. Integr. Plant Biol. 2011;53:961–970. doi: 10.1111/j.1744-7909.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 67.Wang M., Liu H., Ge L., Xing G.W., Wang M., Song W.N., Nie X.J. Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes. 2016;8:13. doi: 10.3390/genes8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corneille S., Lutz K., Maliga P. Conservation of RNA editing between rice and maize plastids: Are most editing events dispensable? Mol. Gen. Genet. 2000;264:419–424. doi: 10.1007/s004380000295. [DOI] [PubMed] [Google Scholar]
- 69.Hanson M.R., Sutton C.A., Lu B. Plant organelle gene expression: Altered by RNA editing. Trends Plant Sci. 1996;1:57–64. doi: 10.1016/S1360-1385(96)80030-6. [DOI] [Google Scholar]
- 70.He P., Huang S., Xiao G., Zhang Y., Yu J. Abundant RNA editing sites of chloroplast protein-coding genes in Ginkgo biloba and an evolutionary pattern analysis. BMC Plant Biol. 2016;16:257. doi: 10.1186/s12870-016-0944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Provan J., Powell W., Hollingsworth P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001;16:142–147. doi: 10.1016/S0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- 72.Jakobsson M., Säll T., Lind-Halldén C., Halldén C. Evolution of chloroplast mononucleotide microsatellites in Arabidopsis thaliana. Theor. Appl. Genet. 2007;114:223. doi: 10.1007/s00122-006-0425-9. [DOI] [PubMed] [Google Scholar]
- 73.Powell W., Machray G.C., Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996;1:215–222. doi: 10.1016/S1360-1385(96)86898-0. [DOI] [Google Scholar]
- 74.Ranade S.S., Lin Y.-C., Zuccolo A., Van de Peer Y., del Rosario García-Gil M. Comparative in silico analysis of EST-SSRs in angiosperm and gymnosperm tree genera. BMC Plant Biol. 2014;14:220. doi: 10.1186/s12870-014-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J., Xie D.F., Guo X.L., Zheng Z.Y., He X.J., Zhou S.D. Comparative analysis of the complete plastid genome of five Bupleu rum species and new insights into DNA barcoding and phylogenetic relationship. Plants. 2020;9:543. doi: 10.3390/plants9040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tangphatsornruang S., Sangsrakru D., Chanprasert J., Uthaipaisanwong P., Yoocha T., Jomchai N., Tragoonrung S. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: Structural organization and phylogenetic relationships. DNA Res. 2010;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin D., Wang Y., Zhang X., Ma X., He X., Zhang J. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci. Rep. 2017;7:11649. doi: 10.1038/s41598-017-12026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Menezes A.P.A., Resende-Moreira L.C., Buzatti R.S.O., Nazareno A.G., Carlsen M., Lobo F.P., Kalapothakis E., Lovato M.B. Chloroplast genomes of Byrsonima species (Mal pighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018;8:2210. doi: 10.1038/s41598-018-20189-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mehmood F.A., Shahzadi I., Waseem S., Mirza B., Ahmed I., Waheed M.T. Chloroplast genome of Hibiscus rosasinensis (Malvaceae): Comparative analyses and identifcation of mutational hotspots. Genomics. 2020;112:581–591. doi: 10.1016/j.ygeno.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 80.Song Y., Wang S., Ding Y., Xu J., Li M.F., Zhu S., Chen N.J. Chloroplast Genomic Resource of Paris for Species Discrimination. Sci. Rep. 2017;7:3427. doi: 10.1038/s41598-017-02083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim K.J., Lee H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 82.Li Q., Su N., Zhang L., Tong R., Zhang X., Wang J.R., Chnag Z.Y., Zhao L., Potter D. Chloroplast genomes elucidate diversity, phylogeny, and taxonomy of Pulsatilla (Ranunculaceae) Sci. Rep. 2020;10:19781. doi: 10.1038/s41598-020-76699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mehmood F., Abdullah Ubaid Z., Shahzadi I., Ahmed I., Waheed M.T., Poczai P., Mirza B. Plastid genomics of Nicotiana (Solanaceae): Insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica) PeerJ. 2020;8:e9552. doi: 10.7717/peerj.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao J., Jiang D., Zhao Z., Yuan S., Zhang Y., Zhang T., Zhong Yuan Q., Huang L. Development of Chloroplast Genomic Resources in Chinese Yam (Dioscorea polys tachya) Biomed. Res. Int. 2018;2018:6293847. doi: 10.1155/2018/6293847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu L., Wang Y., He P., Li P., Lee J., Soltis D.E., Fu C.X. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018;19:235. doi: 10.1186/s12864-018-4633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kress W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017;55:291–307. doi: 10.1111/jse.12254. [DOI] [Google Scholar]
- 87.Zhang D., Mo X., Xiang J., Zhou N. Molecular identification of original plants of Fritillariae cirrhosae bulbus, a Tradtional Chinese Cedicine (TCM) using plant DNA barcoding. Afr. J. Tradit. Complement. Altern. Med. 2016;13:74–82. doi: 10.21010/ajtcam.v13i6.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jansen R.K., Ruhlman T.A. Plastid Genomes of Seed Plants. In: Bock R., Knoop V., editors. Genomics of Chloroplasts and Mitochondria. 2012. pp. 103–126. [DOI] [Google Scholar]
- 89.Parks M., Cronn R., Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang H., Shi C., Liu Y., Mao S.-Y., Gao L.-Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014;14:151. doi: 10.1186/1471-2148-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou J., Gao Y., Wei J., Liu W., Downie S.R. Molecular phylogenetics of Ligusticum (Apiaceae) based on nrDNA ITS sequences: Rampant polyphyly, placement of the Chinese endemic species, and a much-reduced circumscription of the genus. Int. J. Plant Sci. 2020;181:306–323. doi: 10.1086/706851. [DOI] [Google Scholar]
- 92.Downie S.R., Spalik K., Katz-Downie D.S., Reduron J.P. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Divers Evol. 2010;128:111–136. doi: 10.1127/1869-6155/2010/0128-0005. [DOI] [Google Scholar]
- 93.Pimenov M.G. Updated checklist of Chinese Umbelliferae: Nomenclature, synonymy, typification, distribution. Turczaninowia. 2017;20:106–239. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly yielded ITS sequence and plastome of P. franchetii was submitted into the NCBI with accession numbers OP740295 and OP745624, respectively.