Abstract
Background
Crimean-Congo haemorrhagic fever (CCHF) is a tick-borne viral infection, characterized by haemorrhagic fever in humans and transient asymptomatic infection in animals. It is an emerging human health threat causing sporadic outbreaks in Uganda. We conducted a detailed outbreak investigation in the animal population following the death from CCHF of a 42-year-old male cattle trader in Lyantonde district, Uganda. This was to ascertain the extent of CCHF virus (CCHFV) circulation among cattle and goats and to identify affected farms and ongoing increased environmental risk for future human infections.
Methods
We collected blood and tick samples from 117 cattle and 93 goats, and tested these for anti-CCHFV antibodies and antigen using an enzyme-linked immunosorbent assay (ELISA), quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and target enrichment next generation sequencing.
Results
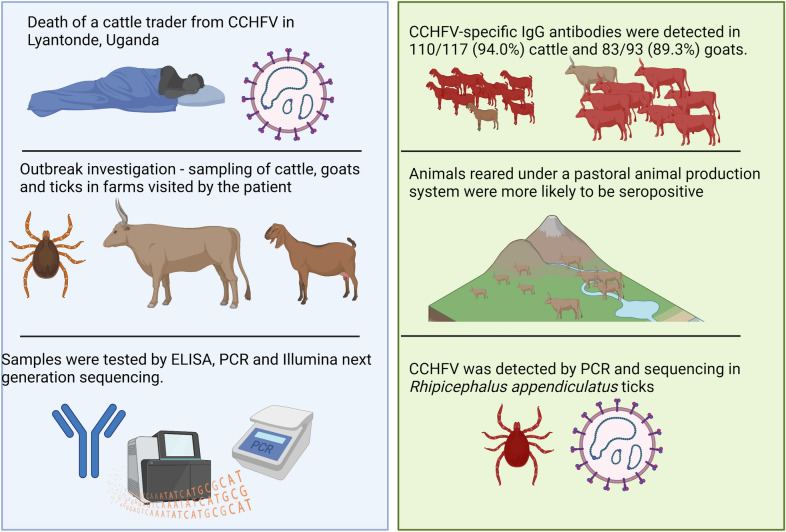
CCHFV-specific IgG antibodies were detected in 110/117 (94.0%) cattle and 83/93 (89.3%) goats. Animal seropositivity was independently associated with female animals (AOR = 9.42, P = 0.002), and animals reared under a pastoral animal production system (AOR = 6.02, P = 0.019] were more likely to be seropositive than tethered or communally grazed animals. CCHFV was detected by sequencing in Rhipicephalus appendiculatus ticks but not in domestic animals.
Conclusion
This investigation demonstrated very high seroprevalence of CCHFV antibodies in both cattle and goats in farms associated with a human case of CCHF in Lyantonde. Therefore, building surveillance programs for CCHF around farms in this area and the Ugandan cattle corridor is indicated, in order to identify opportunities for case prevention and control.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-022-05588-x.
Keywords: Crimean-Congo haemorrhagic fever virus, CCHF antibodies, Tick-borne viral infections, CCHF outbreak, Livestock, Animals, Zoonotic disease, CCHF seroprevalence, Uganda
Background
Crimean-Congo haemorrhagic fever (CCHF) is a severe tick-borne zoonotic disease with a high case fatality rate in humans [1]. Overall, the fatality rate has gradually increased during past decades, with important differences across geographical regions and occupations [2]. The aetiological agent has a wide geographical distribution in parts of Africa, Asia, the Middle East and Eastern Europe [3]. Evidence of exposure to CCHF virus (CCHFV) has been reported in non-endemic countries in southern and western Europe [4–6], suggesting that the distribution of endemic countries might further expand within the next years [7].
CCHFV belongs to the genus Orthonairovirus and family Nairoviridae [8], and displays the typical rapid mutation rate of single-stranded RNA viruses. CCHFV circulates in ticks and is amplified in wild mammalian hosts and livestock [9]. People may become infected with CCHFV from a bite by an infected tick or via blood or fluids of viraemic animals, including humans [1]. Vertebrates, including domestic animals, become infected when bitten by infected ticks. Although domestic animals develop a transient viraemia (7–15 days), they usually remain asymptomatic [7, 10]. Ixodid ticks are infected by vertical transmission or by horizontal transmission when larvae, nymphs or adults take blood meals from viraemic animals or when feeding close to an infected tick (co-feeding infection). The virus persists throughout the tick’s lifespan, leading to repeated opportunities to infect susceptible animals and maintain the virus in the environment [10]
Detection of the virus in animals or in ticks indicates an increased risk of human infection [9]. This study aimed to investigate the seroprevalence of CCHFV exposure in domestic animals following an outbreak involving human cases to ascertain the extent of CCHFV circulation in livestock and to assess the risk for subsequent human infections.
Methods
Ethical clearance
This study was undertaken as part of the arboviral infection study (AVI) approved by the Ethics Review Committee at the College of Veterinary Medicine, Animal Resources and Biosecurity of Makerere University, Kampala, Uganda (Reference Number: SVARREC/20/20l8) and by the Uganda National Council for Science and Technology (Reference Number: HS 2485). Informed written consent was obtained from all study participants before they, and their animals, were enrolled in the study.
Outbreak area
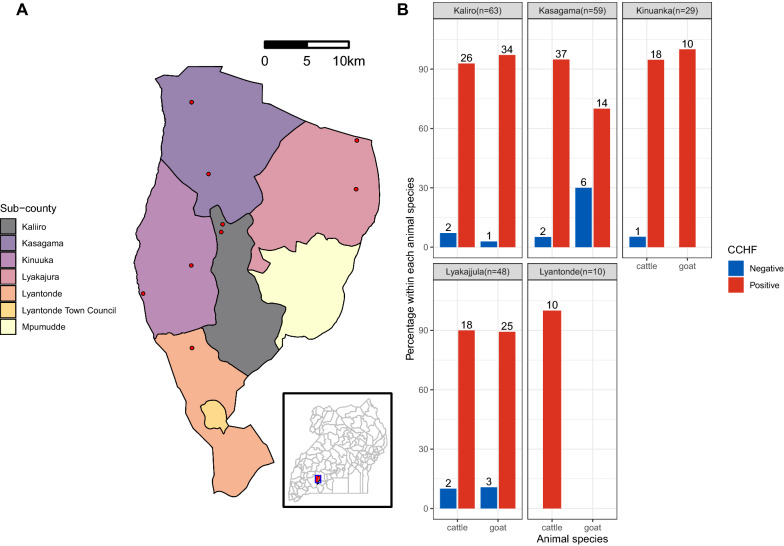
Lyantonde district is located in the Ankole sub-region in Western Uganda (Fig. 1), bordered by Kiruhura, Sembabule, Rakai and Lwengo districts. Lyantonde occupies 888.1 km2, with an estimated population of 93,753 people [11]. Livestock forms the backbone of the economic activity of people in Lyantonde, with the majority (66.1%) rearing cattle and goats of indigenous and exotic crosses under a semi-intensive production system. Kasagama Sub-county (CCHF outbreak site) houses 80% of Lyantonde’s livestock population.
Fig. 1.
Sample sites of the outbreak study. a A map of Lyantonde district showing the sample sites in red and sub-counties as indicated in the legend, and b bar charts of seroprevalence by sub-county and animal species. Positive results are shown in red, negative in blue
CCHF case description
On Monday, 29 July 2019, the Kasagama Sub-county One Health team reported the death from suspected viral haemorrhagic fever (VHF) of a 42-year-old male cattle trader, a resident of Kirindimula village, Kisaruwoko Parish Kasagama Sub-county, in Lyantonde district. He had been treated for malaria in Kasagama on 28 July 2019 following a history of fever, headache and vomiting, abdominal pain and general body pains. On Monday, 29 July 2019, he developed haematemesis and epistaxis. He died on Tuesday, 30 July 2019. Samples taken to the Uganda Virus Research Institute (UVRI) Entebbe for analysis and testing on 31 July 2019 indicated that the patient was positive for CCHF and negative for Ebola, Marburg and Rift Valley fever viruses.
Field investigation
A One Health team comprising the UVRI Ministry of Health, Ministry of Agriculture, Animal Industry and Fisheries, and Lyantonde district local government was convened to undertake the CCHF outbreak investigation in Lyantonde. In order to identify the likely source of the outbreak, an outbreak investigation was carried out on farms purposively selected based on the prior-14-day history of farm visitation by the victim before death from CCHF. A total of 10 farms were identified in this category, and the owners consented to participate in the study. All animals on each farm were placed in a restraint crush and one in four of these were sampled based on random selection and farmers’ choice. For this study, we aimed to collect a total of 100 or more animal samples, based on the numbers used in previous CCHF studies in similar settings in Uganda [12, 13].
A semi-structured herd questionnaire was administered to each farm owner to obtain animal demographic data including age, sex, breed, body temperature and tick infestation number. CCHFV herd level risk factors including herd size, animal production system, tick control practices and history of tick-borne infection were also collected.
Farm animals were selected and blood drawn into sterile EDTA and plain Vacutainer tubes (Becton Dickinson, Plymouth, UK) by veterinary professionals, and transported under cold chain for processing at Mbarara Regional Veterinary Laboratory. Samples were then centrifuged and the serum and plasma aliquoted into 2 ml sterile storage vials (Sarstedt, Inc., Newton, NC, USA). Animal sera were heat-inactivated at 56 °C for 2 h and stored at −80 °C until further laboratory investigation was carried out at the arbovirology laboratory based at UVRI, Entebbe, Uganda.
Anti-CCHFV immunoglobulin (IgG) detection
CCHFV IgG antibodies were tested in duplicate using the commercial ID Screen® CCHF double-antigen multi-species enzyme-linked immunosorbent assay (ELISA) kit (IDVet Innovative Diagnostics, France) following the manufacturer’s protocol [14]. This kit correlates highly with other assays that are routinely used in CCHF diagnostics [12]. Briefly, the test sera were diluted and incubated at 25 °C for 45 min. The conjugate and substrate steps were all conducted at 25 °C for 30 and 15 min, respectively, before the reaction was stopped. Absorbance was read at 450 nm on an automated ELISA reader (BioTek ELx800, USA) using Gen5 version 2.06 software. The sample positivity percentage (S/P%) for each sample was calculated by dividing the optical density (OD) value of the sample (ODS) by the OD of the positive control (ODPC), expressed as a percentage. Serum samples were considered positive if the value for their S/P% was greater than 30%.
CCHFV RNA extraction and quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) testing
Nucleic acid was extracted from plasma using the Beckman Coulter RNA isolation procedure (Brea, CA, USA), following the manufacturer’s instructions. This kit has been found to yield high-quality RNA, suitable for CCHFV nucleic acid testing [15]. Briefly, equal volumes of 200 µl of phosphate-buffered saline (PBS) and test plasma were added to 330 µl lysis buffer and incubated in a water bath at 56 °C for 15 min. After cooling, 410 µl of bind 1/isopropanol solution was added, pipette-mixed and placed on magnetic beads to separate. The supernatant was removed and the beads washed in two subsequent steps using 800 µl of wash buffer/isopropanol and 80% ethanol. This was followed by a DNase treatment step, and the nucleic acid was eluted in 25 µl of nuclease-free water.
The RT-PCR assay was run using the Applied Biosystems 7500 Fast platform and SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen) according to methods previously described by Atkinson’s assay [16] with slight modifications. The 20 µl reaction volume comprised 10 µL of 2× reaction mix, 1.7 µL of PCR-grade water, 1 µL each of CCHFV reverse and forward primer (at 18 µM working concentration), 0.5 µL of probe (25 µM working concentration) and 5 µL of RNA template. The assay was set to run under the following cycling conditions: 50 °C for 10 min, 95 °C for 2 min, followed by 45 cycles of 95 °C for 10 s and 55 °C for 40 s. A cycle threshold (CT) value greater than 40 was considered negative.
Livestock ticks were collected as described previously [17] from half of the body of domestic animals, while environmental ticks were collected by both dragging and flagging methods. Briefly, ticks were transported in 70% ethanol for identification at the species level using morphological keys [18, 19]. Tick pools were created by collection site, species, sex, and the host animal. All tick pools were then crushed in 0.5 ml of Agencourt lysis buffer in a Geno/Grinder 2000 (OPS Diagnostics, Lebanon, NJ, USA), followed by downstream RNA extraction as described above for plasma (Beckman Coulter).
Tick pools were investigated for the presence of CCHFV, Nairobi sheep disease virus (NSDV) and Dugbe virus (DUGV) genomes using target enrichment next-generation sequencing (NGS), as described previously, using a probe library (Arbocap) targeting all arboviruses including nairoviruses [20]. Viral genomes were detected by de novo assembly using dipSPAdes and IDBA, followed by BLASTn and mapping to relevant nairovirus reference sequences using Tanoti. Maximum likelihood phylogenetic analysis was carried out using IQ-TREE and 1000 ultrafast bootstrap replicates [21].
Data analysis
Sociodemographic, epidemiological and laboratory data were analysed using Stata software (v15 StataCorp LP, College Station, TX, USA). Demographic and epidemiological characteristics were summarized using frequencies and percentages, stratified by animal species. We estimated the seropositivity of CCHF as the number of samples that tested positive divided by the total tested, expressed as a percentage. We performed both unadjusted and adjusted regression analysis to determine factors associated with CCHFV exposure. For the unadjusted analysis, the association between CCHFV seropositivity in animals and potential risk factors was first assessed using univariate logistic regression analysis. Multicollinearity was examined among different combinations of variables, and where there was a correlation greater than 0.5, we chose a factor most likely to be associated with CCHF exposure. A backward elimination approach was used to remove factors that were not associated with the outcome in the adjusted analysis (P > 0.1).
Results
The demographic characteristics of cattle and goats sampled are presented in Table 1. Briefly, we collected samples from 117 cattle and 93 goats. A total of 89.7% of the cattle were Friesian crossbreeds, aged 2 years or more (73.5%) and predominantly females (95.7%). The average herd size was 124 cattle (standard deviation [SD] = 0.089, 95% confidence intervals [CI] 2.83–3.18). Most cattle had low to moderate tick infestation. A total of 88.2% of the goats sampled were females, 61.3% were Boer crosses, and the average herd size was 63 goats (SD = 0.062, 95% CI 1.76–2.00).
Table 1.
Sociodemographic and seroprevalence of CCHF in cattle and goats
| Characteristics | Cattle (n = 117) | Goats (n = 93) | ||||||
|---|---|---|---|---|---|---|---|---|
| Total n (%) | Negative n (%) | Positive n (%) | P-value | Total n (%) | Negative n (%) | Positive n (%) | P-value | |
| Overall seropositivity | 7 (6.0) | 110 (94.0) | 10 (10.7) | 83 (89.3) | ||||
| Sub-county | ||||||||
| Kaliro | 29 (24.8) | 2 (6.9) | 27 (93.1) | 0.949 | 35 (37.6) | 1 (2.9) | 34 (97.1) | 0.014 |
| Kasagama | 39 (33.3) | 2 (5.1) | 37 (94.9) | 20 (21.5) | 6 (30.0) | 14 (97.1) | ||
| Kinuanka | 19 (16.2) | 1 (5.3) | 18 (94.7) | 10 (10.8) | 0 (0.0) | 10 (100.0) | ||
| Lyakajjula | 20 (17.1) | 2 (10.0) | 18 (90.0) | 28 (30.1) | 3 (10.2) | 25 (89.3) | ||
| Lyantonde | 10 (8.6) | 0 (0) | 10 (100.0) | – | – | – | ||
| Animal sex | ||||||||
| Male | 5 (4.3) | 2 (33.3) | 4 (66.7) | 0.180 | 11 (11.8) | 5 (45.5) | 6 (54.5) | P < 0.000 |
| Female | 112 (95.7) | 6 (5.4) | 105 (94.6) | 82 (88.2) | 6 (6.1) | 77 (93.9) | ||
| Animal breed | ||||||||
| Indigenous | 12 (10.3) | 1 (8.3) | 11 (91.7) | 0.717 | 36 (38.7) | 7 (19.4) | 29 (80.6) | 0.032 |
| Crossbreed | 105 (89.7) | 6 (5.7) | 99 (94.3) | 57 (61.3) | 3 (61.3) | 54 (94.7) | ||
| Animal age | ||||||||
| < 2 years | 31 (26.5) | 4 (12.9) | 27 (89.1) | 0.164 | 43 (46.2) | 6 (14.0) | 37 (86.0) | 0.289 |
| 2–4 years | 21 (17.9) | 1 (4.8) | 20 (95.2) | 22 (23.7) | 3 (13.6) | 19 (86.4) | ||
| > 4 years | 65 (55.6) | 2 (3.1) | 63 (96.9) | 28 (30.1) | 1 (3.6) | 27 (96.4) | ||
| Body temperature | ||||||||
| ≤ 38 °C | 24 (20.5) | 1 (4.2) | 23 (93.2) | 0.888 | 41 (44.1) | 5 (12.2) | 36 (87.8) | 0.095 |
| 38.1–39.2 °C | 74 (63.3) | 5(6.8) | 69 (93.2) | 29 (31.2) | 5 (17.2) | 24 (82 .8) | ||
| ≥ 39.3 °C | 19 (16.2) | 1 (5.3) | 18 (94.7) | 23 (24.7) | 0 (0.0) | 23 (100.0) | ||
| Herd size | ||||||||
| < 50 | 20 (17.1) | 0 (0.0) | 20 (100.0) | 0.194 | 21 (22.6) | 3 (14.3) | 18 (85.7) | 0.764 |
| 50–99 | 20 (17.1) | 1 (5.0) | 19 (95.0) | 57 (61.3) | 6 (10.5) | 51 (89.5) | ||
| 100–149 | 49 (41.9) | 2 (4.1) | 47 (95.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| 150–199 | 9 (7.7) | 1 (11.1) | 8 (88.9) | 15 (16.1) | 1 (6.8) | 14 (93.3) | ||
| > 200 | 19 (16.2) | 3 (15.8) | 16 (84.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Tick count | ||||||||
| No ticks | 12 (10.3) | 1 (8.3) | 11(91.7) | 0.540 | 13 (14.0) | 0 (0.0) | 13 (100.0) | 0.537 |
| < 50 | 86 (73.5) | 6 (7.0) | 80 (93.0) | 77 (82.8) | 10 (13.0) | 67 (87.0) | ||
| > 50 | 19 (16.2) | 0 (0.0) | 19 (100.0) | 3 (3.2) | 0 (0.0) | 3 (100.0) | ||
| Production system | ||||||||
| Tethering | 14 (12.0) | 2(14.3) | 12 (85.7) | 0.168 | 20 (21.5) | 6 (30.0) | 14 (70.0) | 0.011 |
| Fence/paddocks | 39 (33.3) | 3 (7.7) | 36 (92.3) | 38 (40.9) | 3 (7.8) | 35 (92.1) | ||
| Pastoralism | 64 (54.7) | 2 (3.1) | 62 (96.8) | 35 (37.6) | 1 (2.9) | 34 (97.1) | ||
| Acaricide application | ||||||||
| Once a week | 117 (100.0) | 7 (6.0) | 110 (94.0) | – | 93 (100.0) | 10 (10.8) | 83 (89.2) | – |
| History of tick-borne diseases | ||||||||
| Anaplasmosis | 20 (17.1) | 1 (5.0) | 19 (95.0) | 0.658 | 15 (100.0) | 1 (6.7) | 14 (93.3) | – |
| East Coast fever | 97 (82.9) | 6 (6.2) | 91 (93.0) | – | – | – | ||
Demographic characteristics and CCHF seropositivity clustered by animal species
Temperature (°C) is rectal temperature of the animals measured in degrees Celsius, N is the total number of samples tested for the respective animal species, n is CCHF seroprevalence in numbers and % is seroprevalence expressed as percentages for the respective characteristic
CCHFV antibodies were detected in 110 (94.0%) out of 117 cattle and 83 (89.3%) out of 93 goats tested, with no statistical difference in CCHF seropositivity between cattle and goats (P = 0.208) (Table 1). For the unadjusted model (Table 2), CCHF seropositivity was significantly associated with animal production system [(fence/paddocks: unadjusted odds ratio [UOR] = 3.64, 95% CI 1.15–11.50, P = 0.028), (pastoralism: UOR = 9.85, 95% CI 2.43–39.76, P = 0.001)], all compared with tethered or communally grazed animals. Mature animals over 4 years of age (UOR = 4.69, 95% CI 1.24–17.71, P = 0.023) and female animals (UOR = 9.93, 95% CI 3.05–32.34, P = 0.0001) were also associated with seropositivity.
Table 2.
Risk factors associated with CCHF exposure in animals
| Risk factor | Attribute | Univariate analysis | Multivariate analysis | P-value | |
|---|---|---|---|---|---|
| Odds ratio (95% CI) | P-value | Odds ratio (95% CI) | |||
| Sub-county | Kaliro | Ref | Ref | ||
| Kasagama | 0.31 (0.08–1.24) | 0.099 | – | – | |
| Kinuanka | 1.38 (0.14–13.83) | – | – | ||
| Lyakajjula | 0.42 (0.09–1.86) | – | – | ||
| Lyantonde TC | – | – | – | ||
| Livestock production system | Tethering/communal | Ref | Ref | ||
| Fence/paddock | 3.64 (1.15–11.50) | 0.028 | 2.89 (0.81–10.32) | 0.102 | |
| Pastoralism | 9.85(2.43–39.76) | 0.001 | 6.02 (1.34–27.09) | 0.019 | |
| Animal species | Goats | Ref | Ref | ||
| Cattle | 1.89 (0.69–5.18) | 0.214 | – | – | |
| Animal breed | Indigenous | Ref | Ref | ||
| Crossbreed | 1.03 (0.12–8.53) | 0.975 | – | – | |
| Animal sex | Male | Ref | |||
| Female | 9.93 (3.05–32.34) | < 0.0001 | 9.42 (2.29–38.71) | 0.002 | |
| Animal age | < 2 years | Ref | Ref | ||
| 2–4 years | 1.52 (0.46–5.19) | 0.501 | – | – | |
| > 4 years | 4.69 (1.24–17.71) | 0.023 | – | – | |
| Herd size | < 100 | Ref | |||
| 100–200 | 1.98 (0.48–5.29) | 0.444 | |||
| > 200 | 0.49 (0.12–1.98) | 0.321 | |||
| Animal body temperature | ≤ 38 °C | Ref | |||
| 38.1–39.2 °C | 0.95 (0.33–2.74) | 0.918 | 1.20 (0.37–3.86) | 0.763 | |
| ≥ 39.3 °C | 4.17 (0.48–35.94) | 0.194 | 10.82 (0.93–125.82) | 0.057 | |
| Tick infestation | No tick | Ref | |||
| < 50 ticks | 1.21 (0.13–11.63) | 0.866 | – | – | |
| > 50 ticks | 0.57 (0.05–6.61) | 0.654 | – | – | |
Logistic regression results for risk factors associated with CCHF seropositivity in animals (significant associations at P < 0.05)
Ref indicates the reference variable for the characteristics listed
Dash (–) denotes variables dropped out of the final model because their probability entries were higher than 0.1
In the multivariable regression model, CCHF seropositivity was independently associated with female animals (adjusted odds ratio (AOR) = 9.42, 95% CI 2.29–38.71, P = 0.002) and animals reared under the pastoral production system (AOR = 6.02, 95% CI 1.34–27.09, P = 0.019] compared with those tethered or communally grazed.
All domestic animal samples tested negative for CCHFV on RT-PCR. However, we detected CCHFV in Rhipicephalus appendiculatus in ticks collected from cattle on one farm area (Kaliro Sub-county) visited by the affected patient. Phylogenetic analysis is shown in Fig. 2. The TickP143 Lyantonde 2019 sequence clusters closely with isolates derived from human CCHFV infections between 2013 and 2019.
Fig. 2.
Maximum likelihood tree for CCHFV. a L and b S segments of Ugandan tick isolate 2019, constructed using IQ-TREE with 1000 ultrafast bootstraps and substitution model GTR+F+I+G4 for nucleotide sequences representative of different lineages. Tree scale indicates substitution events. Sequence sampled in this study is shown in red, the Africa II lineage is highlighted in red; Ugandan sequences show region and year of sampling
Discussion
In this outbreak investigation study, we aimed to investigate the source of CCHF infection in a human cattle trader and associated risk factors in the area. Livestock are infected when bitten by infected ticks, and develop asymptomatic transient viraemia lasting 7–15 days. Humans may become infected via contact with the blood or fluids of these viraemic animals or by tick bites. Ticks may remain infected with CCHF throughout their lifespan and act as the main reservoir of CCHF. In keeping with this, we detected CCHF in R. appendiculatus ticks as part of the outbreak investigation, as shown in the phylogenetic analysis with our tick-derived sequence clustering with recently sequenced human cases from Uganda. CCHFV RT-PCR did not reveal PCR-positive animals 9 days after the diagnosis of the human case. This may suggest that the individual was infected by a tick bite rather than direct contact with viraemic animals or that affected animals had become aviraemic after an initial viraemic phase. Vertebrates, including domestic animals, when infected with CCHFV develop transient viraemia lasting for about 2 weeks [9]. Trading of livestock was previously implicated in a CCHF outbreak in India and may be more common than previously reported in other parts of the world, including Uganda [22]. In the farms visited by the cattle trader in this study, the overall seropositivity was extremely high in both cattle (94.0%) and goats (89.3%). We used a commercial CCHF ELISA previously optimized to reduce cross-specific results. Nevertheless, we have previously shown this assay to exhibit cross-reactivity with antibodies targeting the related Dugbe and Nairobi sheep disease orthonairoviruses [17]. We have also detected these viruses in Uganda by sequencing of ticks (less frequently than CCHFV) but did not detect them in our study in Lyantonde. The absence of other orthonairoviruses in ticks in Lyantonde suggests that seropositivity is more likely to relate to CCHFV in this area, but further, more extensive studies are indicated. We also noted the detection of other RNA viruses in the domestic animal samples (Additional file 1: Table S1) using methods that we have developed for NGS over a number of years [20, 23, 24].
In Uganda, previous reports have indicated that over 65% of human cases in the country occur among animal handlers [25]. Evidence of CCHFV prevalence in humans and ticks has been documented in this and other studies [26–28]; however, serosurvey of CCHF in livestock either as part of a viral haemorrhagic fever surveillance program or during outbreaks has not been well described. We report a very high CCHFV seroprevalence in both cattle and goats during an ongoing outbreak, with no interspecies statistical differences, in an area associated with a fatal human infection. A higher seroprevalence in cattle (94.0%) was noted compared with other studies previously conducted in Uganda [12] and nearby countries such as Kenya, Sudan, the Democratic Republic of the Congo, Zambia, Malawi and South Africa [29–36]. Similarly, a higher seroprevalence was noted in goats (89.3%) in comparison with results obtained in different regions of Africa, including Mauritania [37], Nigeria [38] and Senegal [39].
The occurrence of CCHFV antibodies in livestock has been shown to vary depending on serological methods used and host factors including age, sex, breed, livestock management system, vector abundance and competence [12, 31, 34]. Similar to livestock serosurvey conducted in Cameroon [40], our independent regression analysis showed an increased risk of CCHFV seropositivity among animals reared under a pastoral production system. Up to 50.9% of the animals surveyed were occasionally moved to neighbouring districts and wildlife-protected areas of Lake Mburo National Park in search of water and pasture. Domestic animals may acquire and transmit ticks within wildlife grazing grounds, with the risk of inter-district spread, thereby increasing the risk of tick-borne diseases in the area. We also found a significant association between female animals and CCHF seropositivity. Additionally, 94% of the animals reared were hybrid animals and would most likely be kept for a longer time because of their high production value; therefore, they may be more exposed to tick bites.
Conclusion
This investigation demonstrated an extremely high seroprevalence of CCHFV antibodies in both cattle and goats following the occurrence of a fatal human CCHF case in Lyantonde, Uganda. Further research and efforts to improve case prevention and control are indicated, including building surveillance programs for CCHF around farms and the interface between wild hosts, livestock and humans in the cattle corridor in Uganda. Sampling of domestic animals across the country may provide vital information on the ongoing risk to humans in different geographical areas.
Supplementary Information
Additional file 1: Table S1. Viruses detected by NGS following RNA extraction in domestic animal samples.
Acknowledgements
We thank the study participants for making this study possible by providing samples and answering the questionnaire. We extend our gratitude to Lyantonde district medical and veterinary officers and extension workers for their contribution in recruiting study participants. We acknowledge support from the Department of Arbovirology, Emerging and Re-emerging infectious Diseases (UVRI), National Animal Diseases Diagnostics and Epidemiology Centre (MAAIF) and Centre for Virus Research (UoG).
Author contributions
SAA, CM, RT and ECT* conceived the idea and designed the study, collected data, drafted the manuscript. PV, SO, SB, RB and ARA carried out field collection and laboratory testing of the samples, and MN, SA and PA supported with data analysis and reporting. All authors had full access to all the data of the study, discussed and had final responsibility for the decision to submit the final manuscript for publication. All authors read and approved the final manuscript.
Funding
This study was co-funded by the Makerere University-Uganda Virus Research Institute Centre of Excellence for Infection and Immunity Research and Training (MUII) supported by the DELTAS Africa Initiative (Grant Number 107743) of the African Academy of Sciences (AAS), Alliance for Accelerating Excellence in Science in Africa (AESA), and the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (Grant No. 107743), the Wellcome Trust Intermediate Clinical Fellowship fund (102789/Z/13/A) and by a core award from the Medical Research Council (MC_UU_12014/12).
Availability of data and materials
All data collected during the outbreak investigation were analysed, and included in the manuscript for publication. Raw data are available on request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Robert Tweyongyere and Emma C. Thomson have contributed equally to this study
References
- 1.Bente DA, et al. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Nasirian H. New aspects about Crimean-Congo hemorrhagic fever (CCHF) cases and associated fatality trends: a global systematic review and meta-analysis. Comp Immunol Microbiol Infect Dis. 2020;69:101429. doi: 10.1016/j.cimid.2020.101429. [DOI] [PubMed] [Google Scholar]
- 3.Ergönül Ö. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fillatre P, Revest M, Tattevin P. Crimean-Congo hemorrhagic fever: an update. Medecine et maladies infectieuses. 2019;49:574–585. doi: 10.1016/j.medmal.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Monsalve-Arteaga L, et al. Seroprevalence of Crimean-Congo hemorrhagic fever in humans in the World Health Organization European region: a systematic review. PLoS Negl Trop Dis. 2020;14:e0008094. doi: 10.1371/journal.pntd.0008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negredo A, et al. Autochthonous Crimean-Congo hemorrhagic fever in Spain. N Engl J Med. 2017;377:154–161. doi: 10.1056/NEJMoa1615162. [DOI] [PubMed] [Google Scholar]
- 7.Spengler JR, Bergeron É, Spiropoulou CF. Crimean-Congo hemorrhagic fever and expansion from endemic regions. Curr Opin Virol. 2019;34:70–78. doi: 10.1016/j.coviro.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrison AR, et al. ICTV virus taxonomy profile: Nairoviridae. J Gen Virol. 2020;101:798–799. doi: 10.1099/jgv.0.001485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spengler JR, Bergeron É, Rollin PE. Seroepidemiological studies of Crimean-Congo hemorrhagic fever virus in domestic and wild animals. PLoS Negl Trop Dis. 2016;10:e0004210. doi: 10.1371/journal.pntd.0004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spengler JR, et al. A chronological review of experimental infection studies of the role of wild animals and livestock in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus. Antiviral Res. 2016;135:31–47. doi: 10.1016/j.antiviral.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uganda Bureau of Statistics . The national population and housing census 2014 in main report. Kampala, Uganda: Uganda Bureau of Statistics; 2016. [Google Scholar]
- 12.Balinandi S, et al. Serological and molecular study of Crimean-Congo hemorrhagic fever virus in cattle from selected districts in Uganda. J Virol Methods. 2021;290:114075. doi: 10.1016/j.jviromet.2021.114075. [DOI] [PubMed] [Google Scholar]
- 13.Kizito S, et al. Notes from the field: Crimean-Congo hemorrhagic fever outbreak—Central Uganda, August-September 2017. MMWR Morb Mortal Wkly Rep. 2018;67:646–647. doi: 10.15585/mmwr.mm6722a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sas MA, et al. A novel double-antigen sandwich ELISA for the species-independent detection of Crimean-Congo hemorrhagic fever virus-specific antibodies. Antiviral Res. 2018;151:24–26. doi: 10.1016/j.antiviral.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Lista MJ, et al. Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation. PLoS ONE. 2021;16:e0256813. doi: 10.1371/journal.pone.0256813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkinson B, et al. Development of a real-time RT-PCR assay for the detection of Crimean-Congo hemorrhagic fever virus. Vector Borne Zoonotic Dis. 2012;12:786–793. doi: 10.1089/vbz.2011.0770. [DOI] [PubMed] [Google Scholar]
- 17.Atim SA, et al. Risk factors for Crimean-Congo Haemorrhagic Fever (CCHF) virus exposure in farming communities in Uganda. J Infect. 2022 doi: 10.1016/j.jinf.2022.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volcit OV, Keirans JE. A review of African Amblyomma species (Acari, Ixodida, Ixodidae) Acarina. 2009;11:135–214. [Google Scholar]
- 19.Walker AR. Ticks of domestic animals in Africa: a guide to identification of species. Edinburgh: Bioscience Reports Edinburgh; 2003. [Google Scholar]
- 20.Thomson E, et al. Comparison of next-generation sequencing technologies for comprehensive assessment of full-length hepatitis C viral genomes. J Clin Microbiol. 2016;54:2470–2484. doi: 10.1128/JCM.00330-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen LT, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mourya DT, et al. Detection, isolation and confirmation of Crimean-Congo hemorrhagic fever virus in human, ticks and animals in Ahmadabad, India, 2010–2011. PLoS Negl Trop Dis. 2012;6:e1653. doi: 10.1371/journal.pntd.0001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerome H, et al. Metagenomic next-generation sequencing aids the diagnosis of viral infections in febrile returning travellers. J Infect. 2019;79:383–388. doi: 10.1016/j.jinf.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Childs K, et al. Suboptimal SVR rates in African patients with atypical genotype 1 subtypes: implications for global elimination of hepatitis C. J Hepatol. 2019;71:1099–1105. doi: 10.1016/j.jhep.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balinandi S, et al. Clinical and molecular epidemiology of Crimean-Congo hemorrhagic fever in humans in Uganda, 2013–2019. Am J Trop Med Hyg. 2021 doi: 10.4269/ajtmh.21-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoogstraal H. Review article1: the epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J Med Entomol. 1979;15:307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- 27.Balinandi S, et al. Investigation of an isolated case of human Crimean-Congo hemorrhagic fever in Central Uganda, 2015. Int J Infect Dis. 2018;68:88–93. doi: 10.1016/j.ijid.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wampande EM, et al. Phylogenetic characterization of Crimean-Congo hemorrhagic fever virus detected in African blue ticks feeding on cattle in a Ugandan Abattoir. Microorganisms. 2021;9:438. doi: 10.3390/microorganisms9020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obanda V, et al. Livestock presence influences the seroprevalence of Crimean Congo hemorrhagic fever virus on sympatric wildlife in Kenya. Vector Borne Zoonotic Dis. 2021;21:809–816. doi: 10.1089/vbz.2021.0024. [DOI] [PubMed] [Google Scholar]
- 30.Blanco-Penedo I, et al. Seroepidemiology of Crimean-Congo Hemorrhagic Fever Virus (CCHFV) in cattle across three livestock Pastoral regions in Kenya. Dairy. 2021;2:425–434. doi: 10.3390/dairy2030034. [DOI] [Google Scholar]
- 31.Adam IA, Mahmoud MAM, Aradaib IE. A seroepidemiological survey of Crimean Congo hemorrhagic fever among Cattle in North Kordufan State, Sudan. Virol J. 2013;10:178. doi: 10.1186/1743-422X-10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim AM, et al. Epidemiological survey of Crimean Congo hemorrhagic fever virus in cattle in East Darfur State, Sudan. Ticks Tick Borne Dis. 2015;6:439–444. doi: 10.1016/j.ttbdis.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Sas MA, et al. Serosurvey for Crimean-Congo hemorrhagic fever virus infections in ruminants in Katanga province, Democratic Republic of the Congo. Ticks Tick-borne Dis. 2017;8:858–861. doi: 10.1016/j.ttbdis.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Kajihara M, et al. Serologic and molecular evidence for circulation of Crimean-Congo hemorrhagic fever virus in ticks and cattle in Zambia. PLoS Negl Trop Dis. 2021;15:e0009452. doi: 10.1371/journal.pntd.0009452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phonera MC, et al. Seroprevalence and risk factors of Crimean-Congo hemorrhagic fever in cattle of smallholder farmers in Central Malawi. Pathogens. 2021;10:1613. doi: 10.3390/pathogens10121613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Msimang V, et al. Risk factors associated with exposure to Crimean-Congo haemorrhagic fever virus in animal workers and cattle, and molecular detection in ticks, South Africa. PLoS Negl Trop Dis. 2021;15:e0009384. doi: 10.1371/journal.pntd.0009384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz A, et al. Crimean-Congo hemorrhagic fever virus antibody prevalence in Mauritanian livestock (cattle, goats, sheep and camels) is stratified by the animal's age. PLoS Negl Trop Dis. 2021;15:e0009228. doi: 10.1371/journal.pntd.0009228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oluwayelu D, et al. Prevalence of antibodies to Crimean-Congo hemorrhagic fever virus in ruminants, Nigeria, 2015. Emerg Infect Dis. 2020;26:744–747. doi: 10.3201/eid2604.190354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangombi JB, et al. Seroprevalence of Crimean-Congo hemorrhagic fever in domesticated animals in Northwestern Senegal. Vector Borne Zoonotic Dis. 2020;20:797–799. doi: 10.1089/vbz.2019.2592. [DOI] [PubMed] [Google Scholar]
- 40.González Gordon L, et al. Seroepidemiology of Crimean-Congo haemorrhagic fever among cattle in Cameroon: implications from a one health perspective. PLoS Negl Trop Dis. 2022;16:e0010217. doi: 10.1371/journal.pntd.0010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Viruses detected by NGS following RNA extraction in domestic animal samples.
Data Availability Statement
All data collected during the outbreak investigation were analysed, and included in the manuscript for publication. Raw data are available on request.