Abstract
Lys-gingipain (KGP), a lysine-specific cysteine proteinase, is one of the major virulence factors of Porphyromonas gingivalis. Here we examined the involvement of the catalytic domain of KGP (KGPcd) in hemoglobin binding by P. gingivalis, using a specific immunoglobulin G (IgG) elicited by the administration of plasmid DNA encoding KGPcd or the catalytic domain of Arg-gingipain (RGPcd). The pSeq2A/kgpcd and pSeq2B/rgpcd plasmids were constructed by the ligation of kgpcd and rgpcd DNA fragments, respectively. Female BALB/c mice were immunized with each of these plasmids. pSeq2A/kgpcd elicited a strong response to recombinant KGPcd (rKGPcd), as well as to comparably produced rRGPcd-reactive antibodies. The serum antibodies elicited by pSecTag2B/rgpcd also cross-reacted with rKGPcd as well as rRGPcd. Anti-KGPcd IgG significantly inhibited hemoglobin binding by P. gingivalis. Furthermore, the inhibition of hemoglobin binding was markedly enhanced by a combination of anti-KGPcd and anti-fimbriae. Anti-RGPcd IgG showed a negligible inhibitory effect, while both anti-KGPcd and anti-RGPcd IgGs showed significant inhibitory effects on Lys- and Arg-specific proteolytic activities and on the growth of P. gingivalis under iron-restricted conditions where supplemented hemoglobin was the sole iron source. Immunized mice were challenged by intraperitoneal inoculation with P. gingivalis. All nonimmunized mice died within 72 h; however, vaccination with pSeq2A/kgpcd and pSeq2B/rgpcd prevented inflammatory responses and prolonged the survival rate of immunized mice by 43 and 27%, respectively. These results suggest that KGPcd acts as a hemoglobin-binding protein and can also be useful as an immunogen inducing a protective response to P. gingivalis infection.
Iron is an essential nutrient for most living organisms and is abundant in the human body. Free iron, however, is kept at an extremely low level, far below that needed for bacterial growth, resulting in a limitation of bacterial infection (25, 44). Iron is bound extracellularly to transferrin and lactoferrin and is contained intracellularly within ferritin, hemosiderin, and such hemin-containing compounds as hemoglobin and myoglobin (25). Many aerobic and facultative anaerobic bacteria have developed a specific iron acquisition system by using siderophores, which are low-molecular-mass iron chelators that remove iron that has been complexed to host iron-carrying proteins (8). However, some bacterial genera can use heme, hemoglobin, transferrin, lactoferrin, and hemopexin iron directly without the involvement of siderophores (16, 32, 46).
Periodontal diseases are infectious and induce inflammation in the supportive tissues of teeth in response to the accumulation of pathogens in the subgingival crevice (24, 45). The black-pigmented obligate anaerobe Porphyromonas gingivalis is considered to be the most important agent of these infections and causes several types of periodontal diseases, including adult periodontitis (15, 24, 45). The availability of iron in gingival crevicular fluid is crucial for the growth and virulence of this organism, which produces no siderophore (5). P. gingivalis can utilize hemin as an iron source and also seems to store hemin on its cell surface, which causes the black pigmentation of its colonies (36). P. gingivalis specifically utilizes several hemin-containing compounds as iron sources (5, 14); of these, hemoglobin supports bacterial growth much more efficiently than do transferrin, hemin, or inorganic iron compounds (38). We previously reported that the 51-kDa catalytic domain of P. gingivalis Lys-gingipain, lysine-specific cysteine proteinase (KGPcd, encoded by kgpcd), has significant binding ability to human hemoglobin (specific association constant, Ka = 6.90 × 107) and that the recombinant polypeptide of KGPcd also has both a hemoglobin-binding activity and a significant inhibitory effect against the binding of whole-cell extracts to hemoglobin (20). It has been also reported that deletion of the P. gingivalis kgpcd gene generates mutants without pigmentation (6, 7, 23, 30, 40). These findings strongly suggest that KGPcd plays a critical role in hemin acquisition within the cells. On the other hand, HGP15, which has a deduced molecular size of 15 kDa and is encoded by hgp15 downstream of kgpcd and rgpcd (catalytic domain of Arg-gingipain [RGP]-encoding gene) and within hagA (hemagglutinin-encoding gene), was shown to have a marked affinity for hemoglobin (Ka = 2.04 × 107) (27). It was also shown that P. gingivalis fimbriae strongly bind to hemoglobin (Ka = 2.43 × 106) (2); however, the fimbriae were demonstrated to have no association with hemin accumulation and storage by P. gingivalis (6). The above findings concluded that KGPcd and/or HGP15 may play important roles for hemin utilization from hemoglobin of P. gingivalis; however, the exact roles of these two molecules in hemin-hemoglobin transport in P. gingivalis remain to be defined.
Antigen-encoding plasmid DNA immunization (DNA vaccine) is considered to be a powerful approach to the generation of needed antigenic proteins by the host cells. This novel strategy can induce cellular and humoral immune responses to a variety of pathogens, including viruses, parasites, bacteria (35), and tumor cells (19). The antibody responses induced by DNA vaccinations were reportedly lower than those induced by classical immunizations of antigens with adjuvants, because of the low level of secretion of the expressed antigens from the transfected cells (10, 41). Recently, plasmid vectors with a strong heterogenous signal sequence, which mediates efficient antigen secretion in vivo, have been shown to induce significantly higher antibody levels than did previous vectors for little-secreted antigens (10, 41). Since plasmid DNA immunization can be used for immunization of the host without purification of antigenic proteins, this strategy is considered useful for eliciting specific antisera in experimental animals (10, 41). Recently, it was shown that humoral responses were effectively induced against P. gingivalis fimbriae by a DNA vaccination using a P. gingivalis fimbrillin-coding plasmid (18); however, no other studies with this organism have been reported.
We examined the role of the KGPcd in hemoglobin binding by P. gingivalis using specific immunoglobulins elicited by plasmid DNA encoding KGPcd or RGPcd. These plasmids were constructed with a signal sequence to secrete antigens for effective antibody responses. Furthermore, the constructed DNA vaccines were used in a genetic immunization strategy against P. gingivalis challenge in a murine model to evaluate the effect of the KGPcd as an immunogen.
MATERIALS AND METHODS
Bacteria.
P. gingivalis strains ATCC 33277 and W50 were grown in Trypticase soy broth (TSB; BBL Microbiology Systems, Cockeysville, Md.) supplemented with yeast extract (1 mg/ml), menadione (1 μg/ml), and hemin (5 μg/ml) in an anaerobic chamber, as described previously (4). Bacterial cells were harvested, washed in prereduced sterile phosphate-buffered saline (PBS; 10 mM phosphate buffer containing 0.15 M sodium chloride [pH 7.4]), and resuspended in the same buffer. The number of bacteria in the suspension was estimated by measuring the optical density at 600 nm and extrapolated from a standard curve, as described previously (26). To prepare the bacterial extracts, washed cells were suspended in ice-cold PBS containing 3% (wt/vol) zwitterionic detergent 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Pierce, Rockford, Ill.) followed by stirring at 4°C for 40 min. The supernatant obtained by centrifugation at 20,000 × g at 4°C for 1 h was thoroughly dialyzed against PBS and then used as the extract. Escherichia coli XL10-Gold (Stratagene, La Jolla, Calif.) was cultured in Luria-Bertani broth or medium containing 1.5% agar supplemented with ampicillin (100 μg/ml).
Construction of plasmid DNA for immunization.
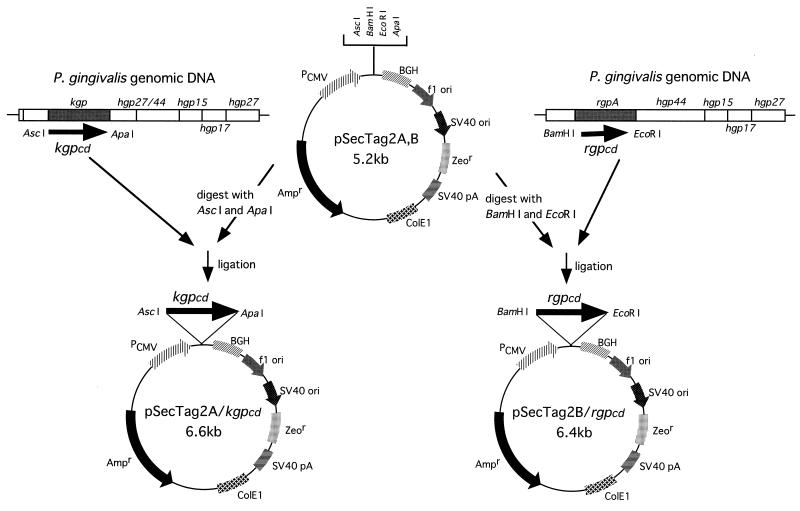
Antigen-encoding plasmids were constructed as illustrated in Fig. 1. P. gingivalis ATCC 33277 genomic DNA was prepared as described previously (3) and used to amplify the kgpcd gene encoding whole amino acid residues of mature KGPcd (489 amino acids (aa) and the rgpcd gene encoding the RGPcd polypeptide corressponding to aa 9 to 431 of whole mature molecule (423 residues), using a PCR method. The PCR primers used were as follows; for kgpcd, the forward primer (5′-TAGGCGCGCCGATGTTTATACAGATCATGGCGAC-3′) and the reverse primer (5′-TAGGGCCCACGGGAAGCTTCTGCCTTCTTTGC-3′) incorporated AscI and ApaI sites; and for rgpcd, the forward primer (5′-TAGGATCCAATGGTCGTATGATCGTCATCG-3′) and the reverse primer (5′-GTGAATTCTCACACTTTCACATCCTTTATC-3′) incorporated BamHI and EcoRI sites (Fig. 1). PCR was performed with a model PCR 2400 thermal cycler (Perkin-Elmer, Norwalk, Conn.), using parameters described previously (20). The resultant kgpcd and rgpcd fragments were inserted into the eukaryotic expression vectors pSecTag2A and pSecTag2B (Invitrogen, Groningen, The Netherlands), respectively. DNA sequencing was performed to confirm the correct in-frame coding alignments of the plasmids with a DNA sequencer (ABI PRISM 310 genetic analyzer; Perkin-Elmer), and the constructed pSecTag2A/kgpcd and pSecTag2B/rgpcd were used for plasmid DNA immunization.
FIG. 1.
Construction of plasmid DNA for immunization. Amplified kgpcd and rgpcd genes were inserted into the eukaryotic expression vectors pSecTag2A and pSecTag2B, respectively (see Materials and Methods). SV40, simian virus 40; PCMV, human cytomegalovirus immediate-early promoter; BGH, bovine growth hormone gene (provides the polyadenylation [pA] signal); Ampr, ampicillin resistance gene; Zeor, Zeocin resistance gene.
Immunization and murine infection protocols.
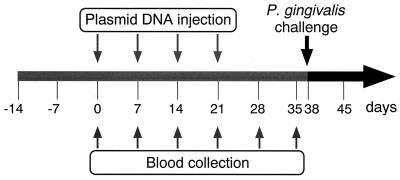
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the Osaka University Graduate School of Dentistry prior to the experiments. Female BALB/c mice (6 weeks old) were maintained in horizontal-flow cabinets and provided with sterile food and water ad libitum. A schematic of the experimental design is shown in Fig. 2. Briefly, mice were immunized by injection of each plasmid DNA dissolved at 1 mg/ml in 50 μl of sterile PBS into the quadriceps muscle. Control mice were immunized with 50 μl of intact pSecTag2A plasmid at 1 mg/ml or PBS. Starting 1 week after primary immunization, the mice were boosted three times at 1-week intervals with the same quantities of each DNA solution. At 0, 7, 14, 21, 28, and 35 days after the primary immunization, blood samples were collected from the orbital sinus or plexus of each mouse and sera were collected by centrifugation following clotting at 4°C. Antibodies elicited by pSecTag2A/kgpcd and pSecTag2B/rgpcd were used as anti-KGPcd and anti-RGPcd antibodies, respectively, for further studies. At 38 days after the first immunization, the mice were intraperitoneally infected with inoculation of 9 × 109 CFU of P. gingivalis W50. The animals were monitored for signs and symptoms of infection and evaluated for (i) the size of eroded skin lesions on the abdomens, (ii) cachexia, and (iii) death. Lesion size was expressed as the average maximum diameter achieved during the 2-week-postinfection period.
FIG. 2.
Protocol for intramuscular immunizations with plasmid DNA containing kgpcd or rgpcd and intraperitoneal challenge in mice. Each plasmid (50 μg) was injected into the quadriceps muscles weekly for a total of four inoculations. At 38 days after the first immunization, mice were intraperitoneally challenged with P. gingivalis W50 (9 × 109 CFU), and their health status was observed over 14 days.
Preparation of rKGPcd and rRGPcd proteins.
The recombinant KGPcd polypeptide (rKGPcd) was expressed in Escherichia coli BL21 (DE3) and purified as described previously (20). To generate the rRGPcd polypeptide, the rgpcd fragments used for pSecTag2B/rgpcd construction were inserted into pGEX2T (Amersham Pharmacia Biotech), which is the vector for expression of the glutathione S-transferase (GST) fusion protein. The construct was then transformed to E. coli XL-10GOLD as described previously (3). The expression of the GST-RGPcd fusion protein was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG) and was purified by affinity chromatography with a glutathione-Sepharose 4B column (Amersham Pharmacia Biotech) as specified by the manufacturer. The rRGPcd was purified using the same column, following enzyme treatment with thrombin (500 NIH units) at 4°C by the procedure specified by the manufacturer.
ELISA.
Serum samples obtained from immunized mice were analyzed for immunoglobulin G (IgG), IgM, and IgA antibodies against rKGPcd and rRGPcd by enzyme-linked immunosorbent assay (ELISA). Individual wells of flat-bottom 96-well plates (Nalge Nunc International, Roskilde, Denmark) were coated with 50 μl of rKGPcd or rRGPcd (10 μg/ml) in bicarbonate-carbonate buffer (pH 9.6) and incubated overnight at 4°C. The wells were blocked overnight at 4°C with PBS containing 10% Block Ace (Dainippon Pharmaceutical Co., Osaka, Japan) and 0.05% Tween 20 (pH 7.4). Sera, in six twofold dilutions from 1:400 to 1:12,800 in PBS containing 0.05% Tween 20 (PBST), were added in triplicate to individual wells, and the mixtures were incubated for 2 h at 37°C. After the wells were washed with PBST, alkaline phosphatase-conjugated goat anti-mouse IgG, IgM, or IgA antibodies (Southern Biotechnology Associates, Birmingham, Ala.) were added and the mixtures were incubated for 2 h at 37°C. All wells were washed, and p-nitrophenyl phosphate in diethanolamine buffer (1 mg/ml; pH 9.4) was added. After 15 min of incubation, color development was stopped by adding 2.5 M NaOH, and the reactions were evaluated at 405 nm with a microplate reader (Titertek MK11; Flow Laboratories, McLean, Va.). The end-point titers for antigen-specific IgG, IgM, and IgA were defined as the last dilution giving an optical density at 405 nm of ≥0.1. The results are expressed as means ± standard deviations of log2 ELISA antibody titers for triplicate wells.
Isolation of specific IgG.
Immune mouse sera whose end-point titers were increased to more than 6,400 were used as anti-KGPcd and anti-RGPcd sera. Antisera against whole cells and fimbriae of P. gingivalis ATCC 33277 were kindly provided by T. Fujiwara (Osaka University). Specific IgG was isolated from the antisera using a HiTrap protein G column (Amersham Pharmacia Biotech).
Immunoblot analysis.
Immunized mouse serum samples were assessed for reactivity with whole-cell extracts of P. gingivalis ATCC 33277, W50, rKGPcd, and rRGPcd. Samples were separated under dissociation conditions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide), and the proteins were transferred onto a nitrocellulose membrane using a Trans-Blot electrophoresis system (Bio-Rad, Hercules, Calif.). The membrane was blocked with 10% Block-Ace in PBST and washed three times with PBST. After overnight incubation with serum IgG (1:500) at 4°C, the membrane was washed three times and incubated with alkaline phosphatase-conjugated goat anti-mouse IgG (1:1,000) for 120 min at 37°C. The membrane was subsequently developed using an alkaline phosphatase substrate.
Hemoglobin-binding activity.
The binding ability of the bacterial extracts to human hemoglobin was determined as described previously (20). Briefly, aliquots of the extracts (200 μl of 150 μg/ml) were immobilized on a nitrocellulose membrane (0.22 μm pore size; Bio-Dot; Bio-Rad) under mild aspiration with a Bio-Dot apparatus (Bio-Rad). The membranes were coated with 10% (vol/vol) Block Ace in PBS and incubated overnight at 4°C with specific IgG antibodies (4 μg/ml) against whole cells, KGPcd, RGPcd, fimbriae, and a combination of these. Nonimmunized mouse IgG and PBS were used as controls. After being washed with PBS, the samples on the membranes were incubated with human hemoglobin in PBS (0.4 mg/ml) (pH 5.5) at 4°C for 3 h. After the membranes were washed with PBS, the binding activity to hemoglobin was quantified to measure the dot intensities as described previously (20). The relative intensities were calculated based on those of the dot of bovine serum albumin BSA (30 μg) as 0% and the dots incubated without inhibitors as 100%. All assays were performed in duplicate on three separate occasions.
Proteinase activity.
The inhibitory effects of anti-KGPcd and anti-RGPcd IgGs against Lys- and Arg-specific proteolytic activities, respectively, were investigated. The bacterial extracts (250 μg/ml) were incubated with five fivefold dilutions of IgG antibodies (100, 20, 4, 0.8, and 0.16 μg/ml) to whole cells, KGPcd, or RGPcd at 4°C for 2 h. Dilutions of nonimmunized mouse IgG and PBS were used as controls. After incubation, aliquots (200 μl) of the complexes were added to 790 μl of a proteinase buffer (20 mM sodium phosphate buffer containing 100 mM NaCl, 10 mM l-cysteine, and 5 mM CaCl2 [pH 8.0]) and 10 μl of the synthetic substrate (10 mM stock solution) was added to a final concentration of 10 μM. Lys- and Arg-specific proteolytic activities were determined as described previously (20). The relative activity was calculated by setting those incubated with PBS as 100%.
Growth inhibition.
P. gingivalis ATCC 33277 cells were grown in enriched TSB under iron-restricted cell conditions, and endogenous stores of iron and hemin were exhausted by two passage of a 10% inoculum into hemin-free medium. These iron-depleted cells were centrifuged and suspended with prereduced PBS. After incubation with IgGs (100 μg/ml) against whole cells, KGPcd, RGPcd, and fimbriae on ice under anaerobic conditions for 2 h, the cells were then inoculated into TSB supplemented with yeast extract (1 mg/ml), menadione (1 μg/ml), and 10 nM human hemoglobin as the sole iron source. Growth was monitored at 600 m in a Bausch & Lomb spectrophotometer (Shimazu Scientific Instruments, Kyoto, Japan) and documented every 6 h.
Statistical analysis.
The multiple comparison was performed by Scheffe's test. The survival curves were calculated by the Kaplan-Meier method and compared by the log rank test.
RESULTS
Immune responses to DNA vaccines.
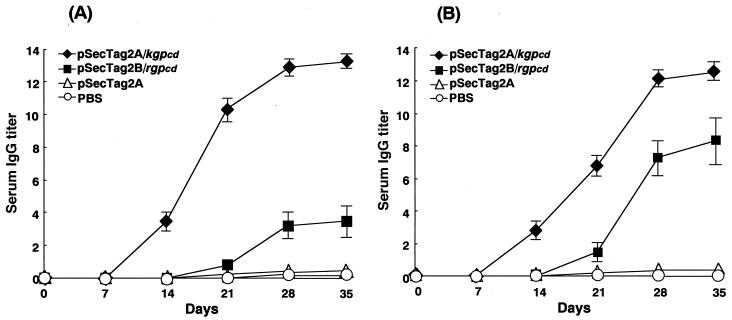
Female BALB/c mice were immunized with pSecTag2A/kgpcd or pSecTag2B/rgpcd. Following the immunization, specific IgG antibodies were clearly induced, and all responses reached a plateau on day 28 (Fig. 3). Injected pSecTag2A/kgpcd elicited a significant antibody response specific to rKGPcd (P < 0.0001) (Fig. 3A), and the induced sera showed reactivities against rRGPcd as well (P < 0.0001) (Fig. 3B). Similarly, specific IgG responses were demonstrated against rRGPcd (P = 0.0029), (Fig. 3B) following immunization with pSecTag2B/rgpcd, and, interestingly, a cross-reaction to rKGPcd was also observed (Fig. 3A). However, the titers were significantly lower than those elicited by pSecTag2A/kgpcd for reactivities with both antigens (P < 0.0001). Serum IgA and IgM responses were only marginal in mice immunized with either pSecTag2A/kgpcd or pSecTag2B/rgpcd. No mice immunized with pSecTag2A or PBS exhibited detectable immune responses to rKGPcd or rRGPcd.
FIG. 3.
Induction of IgG antibodies against rKGPcd and rRGPcd in serum from DNA-immunized mice. Mice were immunized by injection of pSecTag2A/kgpcd (50 μg) and pSecTag2B/rgpcd (50 μg), respectively. At 0, 7, 14, 21, 28, and 35 days after the primary immunization, sera were collected to evaluate specific antibody responses to rKGPcd (A) and rRGPcd (B). Control mice were immunized with intact pSecTag2A plasmid (50 μg) or PBS (50 μl). The values are expressed as means and standard deviations of log2 ELISA antibody titers.
Western blot analysis of antibody responses.
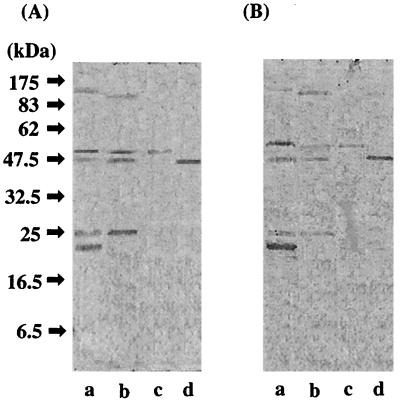
Immunoblotting of serum antibodies elicited by the plasmid DNA immunization was performed with cellular extracts of P. gingivalis. Antibodies elicited by pSecTag2A/kgpcd reacted with KGPcd as well as RGPcd in whole-cell extracts of P. gingivalis ATCC 33277 cells (Fig. 4A, lane a). In addition, bands sized at 110, 25, and 22 kDa were found. In the P. gingivalis W50 preparation, a similar pattern was observed, except that a 95-kDa band was detected instead of the 110-kDa band (lane b). As shown in lanes c and d, both rRGPcd and rKGPcd were recognized by the serum. In the reaction with pSecTag2B/rgpcd-immunized serum, the profiles were similar to those analyzed with pSecTag2A/kgpcd-immunized serum (Fig. 4B). These results were in agreement with the results of the ELISA analysis and point to cross-reactivities between the serum IgG antibodies elicited by pSecTag2B/rgpcd and pSecTag2A/kgpcd.
FIG. 4.
Western blot analysis of serum samples. Immunoblot analyses of serum samples from mice immunized by plasmid DNA are shown. (A) Profiles probed with serum elicited by pSecTag2A/kgpcd. (B) Profiles probed with serum elicited by pSecTag2B/rgpcd. Lanes: a, whole-cell extracts of P. gingivalis ATCC 33277 (40 μg); b, whole-cell extracts of P. gingivalis W50 (40 μg); c, rKGPcd (2 μg); d, rRGPcd (2 μg).
Inhibitory effects of the antibodies on hemoglobin binding by P. gingivalis.
Figure 5 shows the inhibitory effects by the panel of combined IgG antibodies on hemoglobin binding activity by P. gingivalis. Anti-KGPcd, anti-whole cells, and anti-fimbria IgG antibodies significantly inhibited hemoglobin binding by the bacterial extracts (P < 0.0005). Furthermore, the most significant inhibition (93%) was noted in samples incubated with combinations of anti-fimbria and anti-KGPcd antibodies while a negligible inhibitory effect on binding was seen with anti-RGPcd IgG alone (P = 0.8657). These findings suggest that KGPcd and fimbriae are critical molecules for the capture of human hemoglobin by the organism.
FIG. 5.
Inhibitory effects of IgG antibodies on hemoglobin binding by P. gingivalis. Whole-cell extracts of P. gingivalis ATCC 33277 (30 μg), immobilized on the membrane, were incubated with IgG (4 μg/ml) against whole cells of P. gingivalis ATCC 33277, rKGPcd, rRGPcd, fimbriae, and combinations of these and then incubated with human hemoglobin (0.4 mg/ml) at 4°C for 3 h. The densitometric values are expressed as percentages of those obtained with PBS (100%). The experiments were performed twice in triplicate, and the values are expressed as means and standard deviations.
Inhibitory effects of antibodies on the proteinase activity of P. gingivalis.
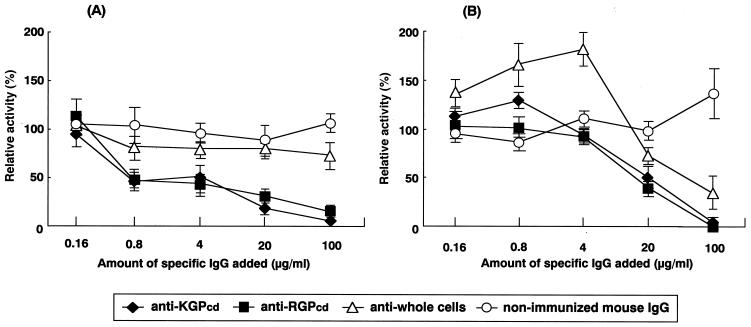
Hydrolytic activities on the Lys-specific substrate by the cell extracts were effectively inhibited in a dose-dependent manner when the extracts were preincubated with anti-KGPcd or anti-RGPcd IgG antibody dilutions (P < 0.0001 and P < 0.0001, respectively) (Fig. 6A). There was no significant difference between the inhibitory effects displayed by anti-KGPcd and anti-RGPcd antibodies. On the Arg-specific substrate, aminolytic activities were unaffected by a 4-μg/ml concentration of anti-KGPcd or anti-RGPcd IgG antibodies but were significantly inhibited by concentrations over 20 μg/ml (P = 0.0126 and P = 0.0026, respectively) (Fig. 6B). The two antibodies showed similar inhibitory effects. However, there was no apparent trend in the inhibition by anti-whole cells or nonimmunized IgG (Fig. 6).
FIG. 6.
Inhibitory effects of the IgG antibodies on proteinase activity of P. gingivalis. Whole-cell extracts of P. gingivalis ATCC 33277 (0.25 μg/μl) were serially incubated with fivefold dilutions of IgG antibodies (×1; 100 μg/ml, ×1/5; 20 μg/ml, ×1/25; 4 μg/ml, ×1/125; 0.8 μg/ml, ×1/625; 0.16 μg/ml) at 4°C for 2 h. The nonimmunized mouse IgG dilution and PBS were used as controls. The Lys-specific (A) and Arg-specific (B) cysteine proteinase activities of the complexes were assayed with Boc-Val-Leu-Lys-MCA and Boc-Gln-Ala-Arg-MCA, respectively. Relative activities were calculated by setting those incubated with PBS as 100%. All assays were performed in triplicate on three separate occasions, and the values are expressed as means and standard deviations.
Growth delay by the antibodies.
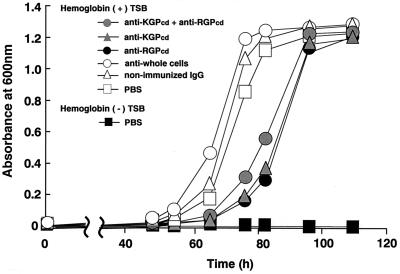
The growth of iron-depleted P. gingivalis cells was evaluated in basal medium supplemented with human hemoglobin as the only iron source, following preincubation with anti-KGPcd and/or anti-RGPcd IgG antibodies. Control cells treated with PBS reached a plateau after 82 h of incubation, while preincubation with anti-KGPcd or anti-RGPcd significantly delayed the growth at 82 h (P = 0.0375 and P = 0.0269, respectively) and caused cell growth to reach the plateau at 110 h of incubation (Fig. 7). In contrast, anti-whole cells or nonimmunized IgG showed no marked effect on the cell growth.
FIG. 7.
Effects of IgG antibodies on the growth of P. gingivalis in medium supplemented with hemoglobin as the sole iron source. Iron-depleted cells of P. gingivalis ATCC 33277 were incubated with various IgG antibodies (100 μg/ml) on ice under anaerobic conditions for 2 h and then inoculated into medium supplemented with human hemoglobin as the sole iron source. Control cells were preincubated with PBS and then inoculated into TSB with or without hemoglobin. All assays were performed in triplicate on three separate occasions.
Effects of plasmid DNA immunization on lethal challenge.
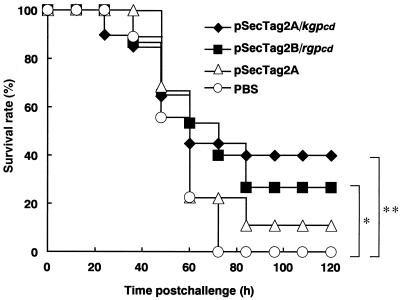
Female BALB/c mice immunized with pSecTag2A/kgpcd or pSecTag2B/rgpcd were intraperitoneally challenged with viable P. gingivalis cells. The immunization of plasmid DNA conferred a significant amount of protection against the lethal challenge compared with that in control animals given PBS. All mice in the control group died within 72 h, while 43% in the pSecTag2A/kgpcd-immunized group and 27% in the pSecTag2B/rgpcd-immunized group survived until the end of the experiment (120 h) (P = 0.0088 and P = 0.0069, respectively) (Fig. 8). Although the degree of inflammatory features varied with the individual mice, the immunization clearly lessened such infectious symptoms as eroded skin lesions on the abdomens, as well as severe cachexia with ruffled hair and hunched bodies (Table 1).
FIG. 8.
Effects of plasmid DNA immunization against lethal challenge with P. gingivalis in mice. Female BALB/c mice immunized with plasmid DNA were intraperitoneally challenged with viable P. gingivalis W50 cells (9 × 109 CFU). Intact pSecTag2A and PBS were used as controls. The log rank test was used for calculation of p values. ∗, P = 0.0069; ∗∗, P = 0.0088.
TABLE 1.
Disease status of P. gingivalis infection in immunized mice
| Mouse group | Total no. of mice | No. of mice with cachexiaa/total no.
|
No. of mice with eroded skin lesionb/total no.
|
||||
|---|---|---|---|---|---|---|---|
| None | Slight | Moderate | Severe | 0–4.0 cm2 | >4.0 cm2 | ||
| pSecTag2A/kgpcd | 21 | 0/21 | 6/21 | 6/21 | 9/21 | 16/21 | 5/21 |
| pSecTag2B/rgpcd | 19 | 0/19 | 2/19 | 5/19 | 12/19 | 14/19 | 5/19 |
| pSeqTag2A | 9 | 0/9 | 0/9 | 0/9 | 9/9 | 6/9 | 3/9 |
| PBS | 10 | 0/10 | 0/10 | 0/10 | 10/10 | 1/10 | 9/10 |
Cachexia was defined as ruffled hair, hunched bodies, and weight loss.
Lesion size was determined and expressed as the average maximum diameter achieved during a 14-day postinfection period.
DISCUSSION
The major proteinase, KGP and RGP are believed to play a critical role in the pathogenesis of P. gingivalis (1, 12, 17, 21, 22, 28, 42, 43). Both are produced as polyprotein moieties and are composed of several functional domains. KGP polyprotein is formed from KGPcd and the hemagglutinin/adhesin domain (HGP27/44, HGP15, HGP17, and HGP27) (31, 33). In our previous study, it was shown that KGPcd binds to human hemoglobin with a significant affinity, which is mediated through an active region(s) distinct from those for proteinase activity. Other studies showed that HGP15 and fimbriae are also capable of binding to hemoglobin (2, 27). Here we attempted to investigate the exact role of KGPcd in hemoglobin binding by P. gingivalis using plasmid DNA-elicited antibodies.
The antibodies elicited by pSecTag2A/kgpcd clearly reacted with KGPcd, RGPcd, and their polyproteins on immunoblotting. Although unknown bands of 25 and 21 kDa were probed by the antibodies, as shown in a previous study using anti-RGP peptide (34), HGP15 was not detected. The KGPcd IgG significantly inhibited the hemoglobin-binding abilities of P. gingivalis. Further, the combination of anti-fimbria and anti-KGPcd antibodies almost completely inhibited hemoglobin binding by the organism. The kgpcd-deficient mutants reportedly increased the expression of fimbriae compared to that caused by the parent strain, but the mutants failed to form black-pigmented colonies (6). These results strongly suggest that the hemoglobin binding for hemin uptake of P. gingivalis is mediated by KGPcd. Fimbriae could have no association with hemin storage but might act as a carrier to pass hemoglobin to KGPcd.
Recent studies by other laboratories support our findings. In the spontaneous mutants of P. gingivalis without pigmentation, the 5′ end of the kgpcd gene was found to be deleted whereas the portion encoding the KGP hemagglutinin domain was innately present (7). These spontaneous kgpcd mutants have markedly decreased binding ability to hemoglobin. It was also shown that inactivation of kgpcd with an IS element generated these mutants, forming a white colony on blood plates as a result of a hemin-hemoglobin defect (6, 23, 40). Chen et al., when speculating that HGP15 is a hemoglobin-binding protein, suggested that proteolytic activity of KGPcd is essential to cleave HGP15 from the its polyproteins and thus the kgpcd mutants are unable to produce HGP15 protein, resulting in the white colonies (6). However, RGPcd reportedly can be used instead of KGPcd to cleave HGP15 and support the expression of its activity (30, 37). Furthermore, it was reported that kgpcd-deficient mutants produced HGP15 slowly after 7 days of incubation but mutants with the mature HGP15 formed white or beige colonies (30). It was also demonstrated that kgpcd mutants exhibited HGP15 on the cell surface after incubation in enriched brain heart infusion broth for 48 h (37). The nonpigmentation of the various mutants seems to be consistent with the absence of KGPcd, regardless of the presence of HGP15. These results strongly suggest that KGPcd acts as a critical receptor in hemin-hemoglobin uptake by P. gingivalis than HGP15. It might be hypothesized that the hemin-hemoglobin uptake by P. gingivalis is mediated by the following pathway: fimbriae and KGPcd bind to hemoglobin, then KGPcd holds the captured hemoglobin for denaturing, and HGP15 and HumR (39) act in the subsequent events such as storage of released hemin. Further study is necessary to understand these details.
The antibodies generated by pSecTag2A/kgpcd and pSecTag2B/rgpcd recognize recombinant and native proteins of KGPcd and RGPcd, respectively. Moreover, both antibodies were found to markedly inhibit Lys-specific and Arg-specific cysteine proteinase activities of P. gingivalis, without significant inefficiencies. These cross-reactivities between the two gingipains were also observed in another study (29). Recently, Curtis et al. analyzed the homology between KGP and RGP by using the diagon plot comparison with a window of 30 amino acids and a stringency of 11 (9). This comparison of the deduced amino acid sequence of the Lys-specific proteinase with that of Arg-specific proteinase reveals considerable conservation in both the propeptide and catalytic domains, suggesting a common ancestral gene for these two loci. The catalytic domains of the two gingipains have only a limited identity (27%): however, both polypeptides have a high similarity of sequences corresponding to a region encompassing the catalytic cysteine residues (10 aa), as well as in the carboxy-terminal region (30 aa) (11, 33). These similar motifs result in cross-reactivities among the antibodies present. However, hemoglobin binding is not influenced by the cross-reactivities among the antibodies; therefore, this interaction would be mediated by a motif distinct from the shared region for proteinase activity. It was reported that immunization of the whole RGPcd molecule failed to elicit a marked immune response (13, 29). This finding could explain why pSecTag2A/kgpcd elicited a stronger response than pSecTag2B/rgpcd as shown in Fig. 3.
Anti-KGPcd IgG was expected to delay the growth of P. gingivalis due to its inhibitory effect on hemin uptake when the cells were grown with human hemoglobin as the only iron source. Therefore, P. gingivalis was grown following preincubation with anti-KGPcd and/or anti-RGPcd IgGs. Cell growth was significantly delayed by anti-KGPcd as anticipated. However, contrary to our expectation, anti-RGPcd also revealed a remarkable effect on cell growth. This may be explained by previous findings that RGP is involved mainly in the proteolysis of nutritive substances for growth (21, 37). The delay could have first been due to the inhibition of hemoglobin uptake by anti-KGPcd, while the prevention of proteolytic abilities by antibodies against KGPcd or RGPcd would also have supported the delay, because of their ability to inhibit the generation of small nutritive peptides by P. gingivalis.
In our preliminary experiments, mice did not die but developed a localized abscess on their abdomen when infected with 1010 CFU of P. gingivalis ATCC 33277, whereas they all died when infected with the same inoculum of strain W50. With respect to the amino acid sequences and functions of Lys- and Arg-specific proteinases, strains ATCC 33277 and W50 were found to be very similar to each other (9). Within the catalytic domain, these strains have a highly conserved identity: the homologies of the amino acid sequences are 94.7% between Lys-specific proteinases (KGP of ATCC 33277 and PrtK of W50), 91.6% between Arg-specific proteinases (RGP and PrtRI), and 98.0% between another kind of Arg-specific proteinases (RgpB and PrtRII). Therefore, we used strain W50 for the challenge experiments. The immunization with pSecTag2A/kgpcd and pSecTag2B/rgpcd showed marked effects on clinical features and survival rates compared to the controls. Immunization with both plasmid DNAs is likely to inhibit the proteolytic abilities of P. gingivalis in vivo. In addition, the inhibitory effects on bacterial hemin uptake may result in effective protection by pSecTag2A/kgpcd immunization. This may explain why pSecTag2A/kgpcd was more effective than pSecTag2B/rgpcd. The intact pSecTag2A without kgpcd or rgpcd also decreased the lesion size, which may be attributable to CpG sequences contained in the plasmid (35). The CpG motifs are known to possess T-helper type 1 immunostimulatory activity and to stimulate monocytes and macrophages to produce a variety of cytokines including interleukin-12, tumor necrosis factor alpha, and alpha/beta interferon. The CpG motifs can also stimulate the production of interleukin-6, which, in turn, promotes B-cell activation and IgM secretion. These immunostimulatory activities of the CpG motifs could have suppressed the lesion formation. Several efforts have been previously made to generate effective vaccines targeting the RGPcd molecule. However, most of those studies suggested that the whole RGPcd molecule was not a suitable immunogen to elicit a marked immune response (13, 29). These findings may also explain the weaker effects of immunization with pSecTag2B/rgpcd. As a result, it is suggested that KGPcd is a more promising candidate for future vaccination than RGPcd.
In summary, the present study found that KGPcd acts as a hemoglobin-binding protein of P. gingivalis and suggests that it may also be a possible immunogen to induce a protective response to P. gingivalis infection.
ACKNOWLEDGMENTS
This work was supported by grants-in-aid for scientific research (C12671994) and encouragement of young scientists (11771154) from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Abe N, Kadowaki T, Okamoto K, Nakayama K, Ohishi M, Yamamoto K. Biochemical and functional properties of lysine-specific cysteine proteinase (Lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J Biochem. 1998;123:305–312. doi: 10.1093/oxfordjournals.jbchem.a021937. [DOI] [PubMed] [Google Scholar]
- 2.Amano A, Nakamura T, Kimura S, Morisaki I, Nakagawa I, Kawabata S, Hamada S. Molecular interactions of Porphyromonas gingivalis fimbriae with host proteins: kinetic analyses based on surface plasmon resonance. Infect Immun. 1999;67:2399–2405. doi: 10.1128/iai.67.5.2399-2405.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Kataoka K, Raj P A, Genco R J, Shizukuishi S. Binding sites of salivary statherin for Porphyromonas gingivalis recombinant fimbrillin. Infect Immun. 1996;64:4249–4254. doi: 10.1128/iai.64.10.4249-4254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano A, Kuboniwa M, Kataoka K, Tazaki K, Inoshita E, Nagata H, Tamagawa H, Shizukuishi S. Binding of hemoglobin by Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;134:63–67. doi: 10.1111/j.1574-6968.1995.tb07915.x. [DOI] [PubMed] [Google Scholar]
- 5.Bramanti T E, Holt S C. Roles of porphyrins and host iron transport proteins in regulation of growth in Porphyromonas gingivalis W50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen T, Dong H, Yong R, Duncan M J. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb Pathog. 2000;28:235–247. doi: 10.1006/mpat.1999.0338. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Kuramitsu H K. Molecular mechanism for the spontaneous generation of pigmentless Porphyromonas gingivalis mutants. Infect Immun. 1999;67:4926–4930. doi: 10.1128/iai.67.9.4926-4930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 10.Drew D R, Lightowlers M, Strugnell R A. Humoral immune responses to DNA vaccines expressing secreted, membrane bound and non-secreted forms of the Taenia ovis 45W antigen. Vaccine. 2000;18:2522–2532. doi: 10.1016/s0264-410x(00)00020-7. [DOI] [PubMed] [Google Scholar]
- 11.Eichinger A, Beisel H G, Jacob U, Huber R, Medrano F J, Banbula A, Potempa J, Travis J, Bode W. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genco C A, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- 13.Genco C A, Odusanya B M, Potempa J, Mikolajczyk-Pawlinska J, Travis J. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect Immun. 1998;66:4108–4114. doi: 10.1128/iai.66.9.4108-4114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genco C A. Regulation of hemin and iron transport in Porphyromonas gingivalis. Adv Dent Res. 1995;9:41–47. doi: 10.1177/08959374950090010801. [DOI] [PubMed] [Google Scholar]
- 15.Grossi S G, Genco R J, Machtei E E, Ho A W, Koch G, Dunford R, Zambon J J, Hausmann E. Assessment of risk for periodontal disease. II. Risk indicators for alveolar bone loss. J Periodontol. 1995;66:23–29. doi: 10.1902/jop.1995.66.1.23. [DOI] [PubMed] [Google Scholar]
- 16.Guerinot M L. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki T, Nakayama K, Yoshimura F, Okamoto K, Abe N, Yamamoto K. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J Biol Chem. 1998;273:29072–29076. doi: 10.1074/jbc.273.44.29072. [DOI] [PubMed] [Google Scholar]
- 18.Kawabata S, Terao Y, Fujiwara T, Nakagawa I, Hamada S. Targeted salivary gland immunization with plasmid DNA elicits specific salivary immunoglobulin A and G antibodies and serum immunoglobulin G antibodies in mice. Infect Immun. 1999;67:5863–5868. doi: 10.1128/iai.67.11.5863-5868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalczyk D W, Ertl H C. Immune responses to DNA vaccines. Cell Mol Life Sci. 1999;55:751–770. doi: 10.1007/s000180050330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys-gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 21.Kuramitsu H K. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol. 1998;13:263–270. doi: 10.1111/j.1399-302x.1998.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 22.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis J P, Dawson J A, Hannis J C, Muddiman D, Macrina F L. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machtei E E, Dunford R, Hausmann E, Grossi S G, Powell J, Cummins D, Zambon J J, Genco R J. Longitudinal study of prognostic factors in established periodontitis patients. J Clin Periodontol. 1997;24:102–109. doi: 10.1111/j.1600-051x.1997.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 25.Martinez J L, Delgado-Iribarren A, Baquero F. Mechanisms of iron acquisition and bacterial virulence. FEMS Microbiol Rev. 1990;75:45–56. doi: 10.1111/j.1574-6968.1990.tb04085.x. [DOI] [PubMed] [Google Scholar]
- 26.Nagata H, Murakami Y, Inoshita E, Shizukuishi S, Tsunemitsu A. Inhibitory effect of human plasma and saliva on co-aggregation between Bacteroides gingivalis and Streptococcus mitis. J Dent Res. 1990;69:1476–1479. doi: 10.1177/00220345900690080501. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien-Simpson N M, Black C L, Bhogal P S, Cleal S M, Slakeski N, Higgins T J, Reynolds E C. Serum immunoglobulin G (IgG) and IgG subclass responses to the RgpA-Kgp proteinase-adhesin complex of Porphyromonas gingivalis in adult periodontitis. Infect Immun. 2000;68:2704–2712. doi: 10.1128/iai.68.5.2704-2712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien-Simpson N M, Paolini R A, Reynolds E C. RgpA-Kgp peptide-based immunogens provide protection against Porphyromonas gingivalis challenge in a murine lesion model. Infect Immun. 2000;68:4055–4063. doi: 10.1128/iai.68.7.4055-4063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain) J Biochem. 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 32.Otto B R, Verweij-van Vught A M J J, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 33.Pavloff N, Pemberton P A, Potempa J, Chen W C, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272:1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 34.Potempa J, Pike R, Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun. 1995;63:1176–1182. doi: 10.1128/iai.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson H L, Torres C A. DNA vaccines. Semin Immunol. 1997;9:271–283. doi: 10.1006/smim.1997.0083. [DOI] [PubMed] [Google Scholar]
- 36.Shah H N, Bonnett R, Mateen B, Williams R A D. The porphyrin pigmentation of subspecies of Bacteroides melaninogenicus. Biochem J. 1979;180:45–50. doi: 10.1042/bj1800045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, Ratnayake D B, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 38.Shizukuishi S, Tazaki K, Inoshita E, Kataoka K, Hanioka T, Amano A. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;131:313–317. doi: 10.1111/j.1574-6968.1995.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 39.Simpson W, Olczak T, Genco C A. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson W, Wang C Y, Mikolajczyk-Pawlinska J, Potempa J, Travis J, Bond V C, Genco C A. Transposition of the endogenous insertion sequence element IS1126 modulates gingipain expression in Porphyromonas gingivalis. Infect Immun. 1999;67:5012–5020. doi: 10.1128/iai.67.10.5012-5020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svanholm C, Bandholtz L, Lobell A, Wigzell H. Enhancement of antibody responses by DNA immunization using expression vectors mediating efficient antigen secretion. J Immunol Methods. 1999;228:121–130. doi: 10.1016/s0022-1759(99)00086-1. [DOI] [PubMed] [Google Scholar]
- 42.Tokuda M, Karunakaran T, Duncan M, Hamada N, Kuramitsu H. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect Immun. 1998;66:1159–1166. doi: 10.1128/iai.66.3.1159-1166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 44.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolff L, Dahlen G, Aeppli D. Bacteria as risk markers for periodontitis. J Periodontol. 1994;65:498–510. doi: 10.1902/jop.1994.65.5s.498. [DOI] [PubMed] [Google Scholar]
- 46.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;64:65–102. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]