Abstract
Viral pathogens are the primary cause of canine gastroenteritis. However, few structured comprehensive studies on the viral etiology of canine gastroenteritis have been conducted. In this study, 475 rectal swabs collected over three years (2018-2021) from clinical canine gastroenteritis cases were screened for the presence of six major enteric viruses – canine parvovirus 2 (CPV-2), canine distemper virus (CDV), canine adenovirus 2 (CAdV-2), canine coronavirus (CCoV), canine astrovirus (CaAstV), and canine rotavirus (CRV) – by real-time PCR. The most frequently detected virus was CPV-2, which was present in 64.8% of the samples (subtype 2a, 21.1%; 2b, 77.4%; 2c, 1.5%), followed by CDV (8%), CaAstV (7.2%), CCoV (5.9%), and CAdV-2 (4.6%). Two to four of these viruses in different combinations were found in 16.8% of the samples, and CRV was not detected. The complete genome sequences of Indian isolates of CDV, CCoV, and CaAstV were determined for the first time, and phylogenetic analysis was performed. This study highlights the need for routine prophylactic vaccination with the appropriate vaccines. Notably, 70.3% of animals vaccinated with DHPPiL were found to be positive for at least one virus. Hence, regular molecular analysis of the prevalent viruses is crucial for addressing vaccination failures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-022-05674-6.
Introduction
Dogs (Canis familiaris) are among the most popular companion animals, and like humans, they are prone to diseases such as gastroenteritis (irritation of the stomach and intestines), which can result in vomiting and diarrhoea. The disease can progress quickly and, if left untreated, can be fatal, especially in young animals. Gastroenteritis can occur for a variety of reasons, including dietary indiscretion, tumors, metabolic disorders, toxins, and, most frequently, infectious agents such as bacteria [1], parasites [2, 3], and viruses [4]. Viruses have been reported to be detected in up to 60% of diarrhoeic fecal samples [5].
Canine parvovirus (CPV) [6], canine distemper virus (CDV) [7], canine adenovirus (CAdV) [8], canine coronavirus (CCoV) [9], canine astrovirus (CaAstV) [10], and canine rotavirus (CRV) [11] have frequently been reported as a cause of viral gastroenteritis in dogs. CPV, a member of the family Parvoviridae, is highly contagious, and the CPV-2 variant was first identified in dogs suffering from severe hemorrhagic gastroenteritis and myocarditis [12, 13]. This was later replaced by three antigenic variants, namely CPV-2a, CPV-2b, and CPV-2c [14–17]. CDV belongs to the family Paramyxoviridae and is responsible for high mortality rates in dogs worldwide [7]. The haemagglutinin gene of CDV undergoes genetic drift, and this has led to the introduction of genetically distinct CDV strains [18], and at least 15 different lineages of CDV are circulating worldwide [19, 20]. CAdV belongs to the family Adenoviridae and circulates as two distinct serotypes, CAdV-1 and CAdV-2 [8, 21]. Despite routine vaccination, the re-emergence of CAdV-2 has been documented in various regions of the world [22]. CCoV belongs to the genus Alphacoronavirus of the family Coronaviridae and causes mild to moderate enteritis in dogs, characterized by high morbidity and low mortality [9]. The genus Alphacoronavirus also includes transmissible gastroenteritis virus of swine, porcine epidemic diarrhea virus, feline coronaviruses, and human coronavirus 229E [23]. CaAstV, a member of the genus Mamastrovirus of the family Astroviridae [10], has been detected in diarrheic dogs in multiple countries [24–28]. It is frequently associated with mixed viral enteric infections, especially in young dogs [28, 29]. CRV belongs to the family Sedoreoviridae and causes neonatal diarrhea [11].
In this study, we investigated the prevalence of the major enteric viruses in stool samples from symptomatic dogs, and the first genome sequences of Indian isolates of CDV, CCoV, and CaAstV were determined and used for phylogenetic analysis.
Materials and methods
Collection and processing of clinical samples
Rectal swabs were collected from clinically ill dogs with gastroenteritis at the Teaching Veterinary Clinical Complex, College of Veterinary Science, Hyderabad, and various private clinics in the region. The case histories of 475 samples screened in this study are presented in Supplementary Table S1. Rectal swabs were collected over a period of three years (2018-2021). The samples were homogenized in 3 ml of 0.1 M PBS (pH 7.4) containing antibiotics (100 IU of benzyl penicillin, 100 μg of streptomycin sulfate) per ml and centrifuged at 6000 rpm for 10 min at 4°C, and the supernatant was filtered through a 0.22-µm syringe filter and used for further analysis.
Viral nucleic acid extraction and reverse transcription
Viral RNA was extracted using TRIzol Reagent (Ref: 15596018; Ambion), and cDNA was synthesized using a PrimeScript First-Strand cDNA Synthesis Kit (catalog no. 6110A; TaKaRa), following the manufacturer’s protocol. Viral DNA was extracted using the phenol-chloroform and isoamyl alcohol method as described by Sambrook and Russel [30].
Screening of clinical samples for viruses by real-time PCR
cDNA or viral DNA was used as a template for PCR. Real-time PCR was carried out using SYBR Premix Ex Taq PCR Master Mix (catalog no. RR420A; TaKaRa) and a StepOnePlus Real-Time PCR System (Applied Biosystems). The primers used for the initial screening of six viruses in individual reactions are listed in Table 1 [29, 31–35] along with the real-time PCR reaction conditions. Typing of CAdV isolates as CAdV-1 or CAdV-2 was done based on differences in the melting temperature of the PCR product [32]. Typing for CPV-2/2a/2b/2c was done using a modification of a previously reported TARMS-PCR procedure [36, 37].
Table 1.
Primers used for screening of viruses by real-time PCR
| Primer name | Primer sequence | Target gene | Virus | Amplicon size | PCR conditions | Reference |
|---|---|---|---|---|---|---|
|
CPV-F CPV-R |
5’AAACAGGAATTAACTATACTAATATATTTA3’ 5’AAATTTGACCATTTGGAT AAACT3’ |
VP-2 | CPV | 90 | Initial denaturation at 95°C for 5 min followed by 4 cycles of 95°C for 30 sec and 60°C for 1 min. Melt curve was set starting at 50°C to 95°C with a ramp speed of 1%. | [31] |
|
CAdV-F CAdV-R |
5’AGTAATGGAAACCTAGGGG3’ 5’TCTGTGTTTCTGTCTTGC3’ |
E3 | CAdV | 166 | [32] | |
|
CDV-F CDV-R |
5’AGCTAGTTTCATCTTAACTATCAAATT3’ 5’TTAACTCTCCAGAAAACTCATGC3’ |
N | CDV | 87 | [33] | |
|
CaAstV-F CaAstV-R |
5’GTACTATACCRTCTGATTTAATT3’ 5’AGACCAARGTGTCATAGTTCAG3’ |
ORF1b | CaAstV | 293 | [29] | |
|
CCoV-F CCoV-R |
5’TTGATCGTTTTTATAACGGTTCTACAA3’ 5’AATGGGCCATAATAGCCACATAAT3’ |
M | CCoV | 99 | [34] | |
|
CRV-F CRV-R |
5’TTAGATACTACAAGTAATGGAATCGGATGT3’ 5’TGGGTGTCATTTGATACAACTTCA3’ |
VP7 | CRV | 76 | [35] |
Isolation of virus in cell lines
Dulbecco's modified Eagle medium (DMEM) containing 1% fetal bovine serum (FBS) was used for maintenance of Madin-Darby canine kidney (MDCK), A-72 (canine fibroblast), or Epstein-Barr-virus-transformed marmoset B lymphoblastoid (B95a) cells at 37°C with 5% CO2. The cells were kept in maintenance medium during virus propagation. For virus isolation, fecal samples were emulsified in PBS, and the clarified supernatant was used as a seed for virus culture. Isolation of CAdV-2, CPV-2/2a/2b/2c, and CaAstV was carried out in MDCK cells, whereas B95a cells were used for CDV and A-72 cells were used for CCoV. The cells were incubated at 37°C with 5% CO2 and were observed for CPE, such as granulation, rounding or detachment of cells in clusters, disturbing the confluent monolayer. The cells were frozen and thawed three times, and the viral supernatant was used as a seed for subsequent passages.
Whole-genome sequence analysis
Viral RNA isolated from the cell culture isolates was sent to the sequencing facility MedGenome Labs Ltd., Karnataka, India, for whole-genome sequencing. Briefly, the whole genome was sequenced using a HiSeqX System (Illumina). Around 12.3 Gb of data were generated, with 81 million reads, for CaAstV, and 14 Gb of data were generated, with 95 million reads, for CDV and CCoV. The average Q30% value was above 80. The reads were first aligned to the canine genome sequence (GCF_000002285.3_CanFam3.1), and the unaligned reads were then aligned to the corresponding reference viral genome sequence. De novo assembly was performed using metaSPAades to obtain scaffolds. The scaffolds were used for gene prediction using Prodigal, and the predicted open reading frames (ORFs) were subjected to a BLASTx search. The sequences were deposited in the NCBI database using an online BankIt submission form.
Phylogenetic analysis
The predicted viral genome was subjected to a BLASTn search. At least 15 sequences from the BLASTn results were selected randomly based on percentage identity. A multiple sequence alignment was made using the MUSCLE algorithm in MEGA X software and was exported in MEGA file format. A phylogenetic tree was constructed by the neighbour-joining method in MEGA X software with 1000 bootstrap replicates [38]. The clades were divided according to their geographical distribution or genetic relationships into different lineages/groups.
Results and discussion
In the present study, we screened rectal swabs from 475 dogs with clinically suspected gastroenteritis for the presence of CPV-2/2a/2b/2c, CDV, CAdV-2, CCoV, CRV, and CaAstV, which are the most common etiological agents of viral gastroenteritis.
Prevalence of gastroenteritis-causing viruses
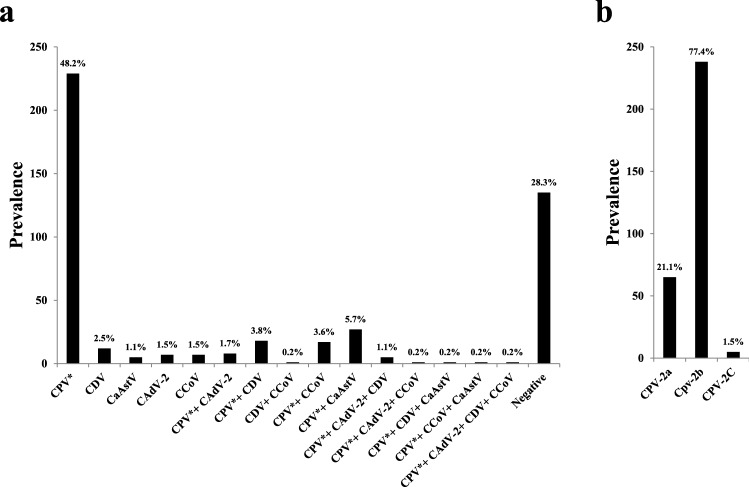
Real-time PCR is a highly sensitive, rapid, and specific technique that can be used to detect low-titer viruses and has been used successfully to screen clinical samples for viruses [31]. Initial screening of the samples showed that 71.6% contained one or more of the viruses tested, but none of them tested positive for CRV (Supplementary Table S1). According to a previous molecular survey of symptomatic dogs, up to 93% of gastroenteritis cases have a viral etiology [39]. Fig. 1a and Supplementary Table S2 show the prevalence of each of the viruses tested by real-time PCR. The most frequently detected virus was CPV-2, which was present in 64.8% of the samples, followed by CDV (8%), CaAstV (7.2%), CCoV (5.9%), and CAdV-2 (4.6%). Antigenic typing of CPV-2 by TARMS-PCR revealed that CPV-2b was the most prevalent antigenic type, followed by CPV-2a and CPV-2c (Fig. 1b and Supplementary Table S3). Based on previous reports, we expected the prevalence rate for CPV-2/2a/2b/2c to be in the range of 50-70% [40, 41], that for CDV to be around 2% [42, 43], that for CaAstV to be in the range of 9-40% [28, 44, 45], that for CCoV to be in the range of 8-65% [43, 46, 47], and that for CAdV to be around 60% [4, 22]. These rates may depend on the age of the dogs (1 month to 5 years), their geographical distribution, and environmental factors. We observed that apart from CPV-2/2a/2b/2c, these viruses were usually found in combination with other viruses, especially CPV-2. Coinfections with two to four viruses in different combinations accounted for 16.8% of the cases. It was reported previously that mixed infections with different viruses are common in dogs with gastroenteritis [3, 39, 48–50]. We therefore suggest that samples from gastroenteritis cases be screened for mixed infections. Most of the pet owners in this study failed to provide a health record of their pet. Of the animals for which a vaccination history was available, it was interesting to note that 70.3% of the animals vaccinated with DHPPiL were positive for at least one of the viruses in question. The possibility that the viruses detected in this study were actually attenuated virus strains originating from the vaccine formulation that were shed in the faeces can be ruled out because (a) rectal swabs were collected from clinical cases in which the animals showed evident signs of severe gastroenteritis and (b) phylogenetic analysis of the virus isolates showed a clear genetic separation from the vaccine virus strains (Fig. 3). These observations indicate the need to focus on updating the vaccine to include currently circulating strains of these viruses. Moreover, it highlights the need for regular molecular and serological screening of prevalent viruses to address vaccination failures.
Fig. 1.
Prevalence of canine enteric viruses. (a) The prevalence of different canine enteric viruses during the years 2018-21 in the Hyderabad region of Telangana state in India. The length of the bar indicates the number of samples that tested positive (y-axis) for each virus either individually or in mixed infections (x-axis). The percentage above the bar indicates the prevalence of that particular virus. CPV* indicates CPV-2/2a/2b/2c. (b) The prevalence of antigenic variants of CPV-2 during the years 2018-21 in the Hyderabad region of Telangana state in India. The length of the bar indicates the number of samples that tested positive (y-axis) for the respective antigenic variant (x-axis). The percentage above the bar indicates the prevalence of that particular antigenic variant.
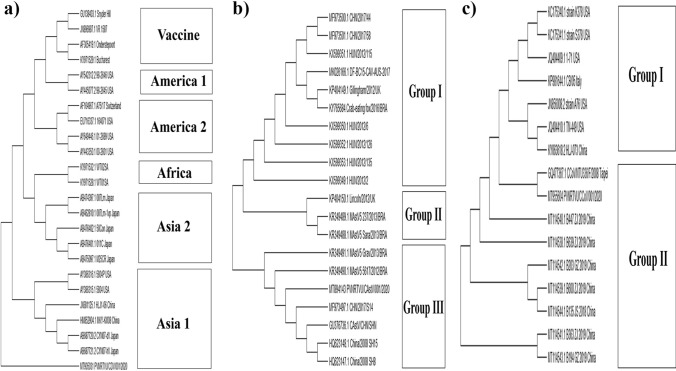
Fig. 3.
Phylogenetic analysis based on whole genome sequences of the isolates from this study and previously published sequences from the NCBI database. The CDV isolates from the current study clustered with members of the Asia 1 lineage (a). The CaAstV isolates from the current study clustered with members of group III, consisting of isolates from China and Brazil (b). The CCoV isolates from the current study clustered with members of group II, consisting of isolates from China and Taipei.
Recovery of virus isolates in cell lines
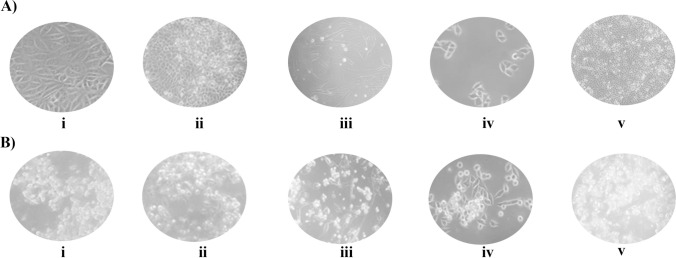
Because of the presence of mixed infections, especially with CPV-2, a highly virulent virus [51], it was difficult to obtain virus isolates from MDCK cells alone. Hence, for the isolation of CDV and CCoV, we used the specialized cell lines B95a [52] and A-72 [53], respectively. The virus isolates were recovered by infecting the respective cell lines as described above. We observed CPE after 5 dpi for CDV in B95a cells, 3 dpi for CPV-2/2a/2b/2c and CaAstV and 5 dpi for CAdV-2 in MDCK cells, and 5 dpi for CCoV in A-72 cells (Fig. 2).
Fig. 2.
Isolation of canine enteric viruses. Panel A shows images of an uninfected cell culture, and panel B shows images of a virus-infected cell culture, both taken at 20X (total of 200X) magnification. From left to right: (i) MDCK/CPV (ii) MDCK/CAdV-2, (iii) A-72/CCoV, (iv) B95a/CDV, (v) MDCK/CaAstV
Whole-genome sequencing-based evolutionary dynamics
Worldwide, there have been a limited number of reports of whole-genome sequences of CDV, CCoV, and CaAstV isolated from dogs [54–56], and we found no reports from India to date. Hence, the whole genome sequences of these three viruses were determined, and ORFs were predicted. The genome length was found to be around 15.6 kb for CDV, 6.5 kb for CaAstV, and 29 kb for CCoV, which is in agreement with previous reports [57–59]. We found six ORFs (N, M, F, H, L, P) in the CDV genome, three ORFs in the CaAstV genome (ORF-1a, 1b, 2), and two overlapping ORFs, encoding ORF-1a and RdRp, along with nine other genes (M, S, E, N, ORF-3a, 3b, 3c and ORF-7a, 7b) in the CCoV genome, as reported previously [29, 59, 60]. The complete sequences and those of their protein-coding regions were deposited in the NCBI GenBank database, and the accession numbers are MT905031 for CDV, MT894143 for CaAstV, and MT955604 for CCoV. Phylogenetic analysis based on whole-genome sequences showed that CDV clustered with members of the Asia-1 lineage, which is distant from the vaccine lineage (Fig. 3a). CaAstV clustered with group III, which consists of Chinese and Brazilian isolates (Fig. 3b). CCoV clustered with group II, which consists of isolates from China and Taipei (Fig. 3c). A previous study showed that all wild-type strains of CDV clustered in groups corresponding to those obtained by H gene analysis, which is routinely used to identify geographically distinct CDV lineages [56]. We observed a similar grouping in this study. Furthermore, we also observed that all of the clinical isolates were genetically distant from the vaccine strains [61, 62]. For CaAstV and CCoV, we found only a few reports of whole-genome-based phylogenetic analysis based on geographical distribution. In one study, a CaAstV isolate from the UK clustered with Chinese isolates [54]. In another study, CCoV isolates from five provinces of China during 2018-19 clustered with earlier isolates from China, similar to our observations [55].
Conclusions
From the present study, we conclude that all three antigenic variants of CPV-2 (CPV-2a/2b/2c), CDV, CaAstV, CAdV-2, and CCoV were involved in cases of canine gastroenteritis during the period 2018-21. CPV-2b was found to be the most prevalent antigenic variant of all of the gastroenteritis viruses surveyed in this study. The majority of the gastroenteritis cases were associated with infection with at least one virus, and the number of viruses infecting each dog ranged from one to four. To our knowledge, this is the first report of complete genome sequences of CDV, CAstV, and CCoV isolates from dogs in India. Phylogenetic analysis revealed that these isolates were genetically distant from the current vaccine strains. We strongly emphasize the need to develop an updated vaccine against the currently circulating variants of viruses that cause canine gastroenteritis.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
The authors acknowledge PV Narsimha Rao Telangana Veterinary University, Hyderabad, for providing a research platform, and DBT, India, for financial support.
Author contributions
AD and MRT performed the experiments and were involved in whole-genome sequence data analysis and submission. BB, HK, and AR performed the sample collection and molecular screening. VKG was involved in whole-genome sequence data analysis and submission and drafting the manuscript. YNR was involved in the conceptualization of the study and mentored the work. KP was involved in the conceptualization of the study, mentored the work, analyzed the data, and drafted the manuscript. All authors proofread the manuscript and approved the final version of the draft.
Funding
This work was supported by the DBT, India (No. BT/ADV/Canine Health/TANUVAS).
Data availability
This manuscript has data included as electronic supplementary material.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This is an observational study. The Institutional Animal Ethics Committee has confirmed that no ethical approval is required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Anusha Dema and Mounika Reddy Tallapally have contributed equally.
References
- 1.Enany M, Wahdan A, El-Metwaly M, et al. Bacterial causes of hemorrhagic gastroenteritis in dogs and cats with detection of some virulence and β-lactamase resistance genes in Escherichia coli and salmonella by multiplex PCR. Suez Canal Vet Med Journal SCVMJ. 2021 doi: 10.21608/scvmj.2021.62502.1030. [DOI] [Google Scholar]
- 2.Gerardi F, Santaniello A, Del Prete L, et al. Parasitic infections in dogs involved in animal-assisted interventions. Ital J Anim Sci. 2018;17:269–272. doi: 10.1080/1828051X.2017.1344937. [DOI] [Google Scholar]
- 3.Headley SA, Alfieri AA, Fritzen JTT, et al. Concomitant canine distemper, infectious canine hepatitis, canine parvoviral enteritis, canine infectious tracheobronchitis, and toxoplasmosis in a puppy. J Vet Diagnostic Investig. 2013;25:129–135. doi: 10.1177/1040638712471344. [DOI] [PubMed] [Google Scholar]
- 4.DiGangi BA, Dingman PA, Grijalva CJ, et al. Prevalence and risk factors for the presence of serum antibodies against canine distemper, canine parvovirus, and canine adenovirus in communities in mainland Ecuador. Vet Immunol Immunopathol. 2019 doi: 10.1016/j.vetimm.2019.109933. [DOI] [PubMed] [Google Scholar]
- 5.Gizzi ABDR, Oliveira ST, Leutenegger CM, et al. Presence of infectious agents and co-infections in diarrheic dogs determined with a real-time polymerase chain reaction-based panel. BMC Vet Res. 2014;10:23. doi: 10.1186/1746-6148-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caddy SL. New viruses associated with canine gastroenteritis. Vet J. 2018;232:57–64. doi: 10.1016/j.tvjl.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quintero-Gil C, Rendon-Marin S, Martinez-Gutierrez M, Ruiz-Saenz J. Origin of canine distemper virus: consolidating evidence to understand potential zoonoses. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramidi A, Ganji VK, Buddala B, et al. E3 gene-based genetic characterization of canine adenovirus-2 isolated from cases of canine gastroenteritis in india revealed a novel group of the virus. Intervirology. 2020;62:216–221. doi: 10.1159/000507329. [DOI] [PubMed] [Google Scholar]
- 9.Buonavoglia C, Decaro N, Martella V, et al. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fauquet CM, Fargette D. International committee on taxonomy of viruses and the 3,142 unassigned species. Virol J. 2005;2:64. doi: 10.1186/1743-422X-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortega AF, Martínez-Castañeda JS, Bautista-Gómez LG, et al. Identification of co-infection by rotavirus and parvovirus in dogs with gastroenteritis in Mexico. Braz J Microbiol. 2017;48:769. doi: 10.1016/J.BJM.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoang M, Lin WH, Le VP, et al. Molecular epidemiology of canine parvovirus type 2 in Vietnam from November 2016 to February 2018. Virol J. 2019;16:52. doi: 10.1186/s12985-019-1159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appel MJ, Scott FW, Carmichael LE. Isolation and immunisation studies of a canine parco-like virus from dogs with haemorrhagic enteritis. Vet Rec. 1979;105:156–159. doi: 10.1136/vr.105.8.156. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, Dhar P, Thakur A, et al. First detection of canine parvovirus type 2b from diarrheic dogs in Himachal Pradesh. Vet World. 2016;9:964–969. doi: 10.14202/vetworld.2016.964-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong Z, Liang L, Zhao J, et al. First isolation of new canine parvovirus 2a from Tibetan Mastiff and global analysis of the full-length VP2 gene of canine parvoviruses 2 in China. Int J Mol Sci. 2014;15:12166–12187. doi: 10.3390/ijms150712166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P, Zeng W, Zhang X, Li S. The genetic evolution of canine parvovirus—a new perspective. PLoS ONE. 2017 doi: 10.1371/journal.pone.0175035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang SY, Wu HY, Chiou MT, et al. Identification of a novel canine parvovirus type 2c in Taiwan. Virol J. 2016;13:160. doi: 10.1186/s12985-016-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glardon O, Stöckli R. Staupeepidemie in der Schweiz: Epidemiologie und Impfanamnese. Schweiz Arch Tierheilkd. 1985;127:707–716. [PubMed] [Google Scholar]
- 19.Bhatt M, Rajak KK, Chakravarti S, et al. Phylogenetic analysis of haemagglutinin gene deciphering a new genetically distinct lineage of canine distemper virus circulating among domestic dogs in India. Transbound Emerg Dis. 2019;66:1252–1267. doi: 10.1111/tbed.13142. [DOI] [PubMed] [Google Scholar]
- 20.Kodi H, Ganji VK, Bhagyalakshmi B, et al. H gene-based molecular characterization of field isolates of canine distemper virus from cases of canine gastroenteritis. Indian J Anim Res. 2020 doi: 10.18805/ijar.b-3989. [DOI] [Google Scholar]
- 21.Mohammadi A, Masoudian M, Nemati Y. Evaluation of PCR techniques for detection and differentiation of canine adenoviruses in faecal samples in Shiraz. Iran Bulg J Vet Med. 2011;14:251. [Google Scholar]
- 22.Balboni A, Mollace C, Giunti M, et al. Investigation of the presence of canine adenovirus (CAdV) in owned dogs in Northern Italy. Res Vet Sci. 2014;97:631–636. doi: 10.1016/j.rvsc.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Decaro N, Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet Microbiol. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams FP. Astrovirus-like, coronavirus-like, and parvovirus-like particles detected in the diarrheal stools of beagle pups. Arch Virol. 1980;66:215–226. doi: 10.1007/BF01314735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi S, Lim SI, Kim YK, et al. Phylogenetic analysis of astrovirus and kobuvirus in Korean dogs. J Vet Med Sci. 2014;76:1141–1145. doi: 10.1292/jvms.13-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martella V, Moschidou P, Catella C, et al. Enteric disease in dogs naturally infected by a novel canine astrovirus. J Clin Microbiol. 2012;50:1066–1069. doi: 10.1128/JCM.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toffan A, Jonassen CM, De Battisti C, et al. Genetic characterization of a new astrovirus detected in dogs suffering from diarrhoea. Vet Microbiol. 2009;139:147–152. doi: 10.1016/j.vetmic.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H, Liu L, Li R, et al. Detection and genetic characterization of canine astroviruses in pet dogs in Guangxi, China. Virol J. 2017;14:156. doi: 10.1186/s12985-017-0823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martella V, Moschidou P, Lorusso E, et al. Detection and characterization of canine astroviruses. J Gen Virol. 2011;92:1880–1887. doi: 10.1099/vir.0.029025-0. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 31.Decaro N, Elia G, Martella V, et al. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Balboni A, Dondi F, Prosperi S, Battilani M. Development of a SYBR Green real-time PCR assay with melting curve analysis for simultaneous detection and differentiation of canine adenovirus type 1 and type 2. J Virol Methods. 2015;222:34–40. doi: 10.1016/j.jviromet.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Elia G, Decaro N, Martella V, et al. Detection of canine distemper virus in dogs by real-time RT-PCR. J Virol Methods. 2006;136:171–176. doi: 10.1016/j.jviromet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Decaro N, Pratelli A, Campolo M, et al. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J Virol Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Logan C, O’Leary JJ, O’Sullivan N. Real-time reverse transcription-PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children. J Clin Microbiol. 2006;44:3189–3195. doi: 10.1128/JCM.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chander V, Chakravarti S, Gupta V, et al. Multiplex amplification refractory mutation system PCR (ARMS-PCR) provides sequencing independent typing of canine parvovirus. Infect Genet Evol. 2016;46:59–64. doi: 10.1016/j.meegid.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 37.Dema A, Ganji VK, Yella NR, Putty K. A novel one-step amplification refractory mutation system PCR (ARMS-PCR) for differentiation of canine parvovirus-2 variants. Virus Genes. 2021;57:426–433. doi: 10.1007/S11262-021-01861-W. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zobba R, Visco S, Sotgiu F, et al. Molecular survey of parvovirus, astrovirus, coronavirus, and calicivirus in symptomatic dogs. Vet Res Commun. 2021;45:31–40. doi: 10.1007/s11259-020-09785-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar M, Nandi S. Molecular typing of canine parvovirus variants by polymerase chain reaction and restriction enzyme analysis. Transbound Emerg Dis. 2010;57:458–463. doi: 10.1111/j.1865-1682.2010.01167.x. [DOI] [PubMed] [Google Scholar]
- 41.Abedi N, Staji H, Shahroozian E. Frequency of canine parvovirus type-2 variants by PCR in enteric and healthy dogs and genotyping of variants by restriction enzyme (RE) mapping by DdeI endonuclease based on partial VP-2 gene sequence. Mol Genet Microbiol Virol. 2018;33:151–155. doi: 10.3103/S0891416818020027. [DOI] [Google Scholar]
- 42.Ashmi JM, Thangavelu A, Senthilkumar TMA, Manimaran K. Molecular characterization of canine distemper virus from Tamil Nadu, India. Indian J Anim Sci. 2017;87:1062–1067. [Google Scholar]
- 43.Agnihotri D, Maan S, Batra K, Jain VK. Comparative evaluation of immunochromatographic and reverse transcriptase polymerase chain reaction based tests for diagnosis of canine distemper. Intas Polivet. 2018;19:414–419. [Google Scholar]
- 44.Takano T, Takashina M, Doki T, Hohdatsu T. Detection of canine astrovirus in dogs with diarrhea in Japan. Arch Virol. 2015;160:1549–1553. doi: 10.1007/s00705-015-2405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Yan N, Ji C, et al. Prevalence and genome characteristics of canine astrovirus in southwest China. J Gen Virol. 2018;99:880–889. doi: 10.1099/jgv.0.001077. [DOI] [PubMed] [Google Scholar]
- 46.Ntafis V, Mari V, Decaro N, et al. Canine coronavirus, Greece. Molecular analysis and genetic diversity characterization. Infect Genet Evol. 2013;16:129–136. doi: 10.1016/j.meegid.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Li C, Guo D, et al. Co-circulation of canine coronavirus I and IIa/b with high prevalence and genetic diversity in Heilongjiang Province, Northeast China. PLoS ONE. 2016;11:e0146975. doi: 10.1371/journal.pone.0146975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratelli A, Martella V, Elia G, et al. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. J Vet Med Ser B. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damián M, Morales E, Salas G, Trigo FJ. Immunohistochemical detection of antigens of distemper, adenovirus and parainfluenza viruses in domestic dogs with pneumonia. J Comp Pathol. 2005;133:289–293. doi: 10.1016/j.jcpa.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Chvala S, Benetka V, Möstl K, et al. Simultaneous canine distemper virus, canine adenovirus type 2, and Mycoplasma cynos infection in a dog with pneumonia. Vet Pathol. 2007;44:508–512. doi: 10.1354/vp.44-4-508. [DOI] [PubMed] [Google Scholar]
- 51.Nandi S, Kumar M. Canine parvovirus: current perspective. Indian J Virol. 2010;21:31–44. doi: 10.1007/s13337-010-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pawar RM, Raj GD, Gopinath VP, et al. Isolation and molecular characterization of canine distemper virus from India. Trop Anim Health Prod. 2011;43:1617–1622. doi: 10.1007/s11250-011-9880-7. [DOI] [PubMed] [Google Scholar]
- 53.Pratelli A, Martella V, Decaro N, et al. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J Virol Methods. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caddy SL, Goodfellow I. Complete genome sequence of canine astrovirus with molecular and epidemiological characterisation of UK strains. Vet Microbiol. 2015;177:206–213. doi: 10.1016/j.vetmic.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He HJ, Zhang W, Liang J, et al. Etiology and genetic evolution of canine coronavirus circulating in five provinces of China, during 2018–2019. Microb Pathog. 2020;145:104209. doi: 10.1016/j.micpath.2020.104209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romanutti C, Keller L, La Torre J, et al. Virus isolation and full-length genome sequencing of a representative canine distemper virus wild type strain of the South America 2 clade. J Virol Methods. 2020 doi: 10.1016/j.jviromet.2020.113857. [DOI] [PubMed] [Google Scholar]
- 57.Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Arch Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mihalov-Kovács E, Martella V, Lanave G, et al. Genome analysis of canine astroviruses reveals genetic heterogeneity and suggests possible inter-species transmission. Virus Res. 2017;232:162–170. doi: 10.1016/j.virusres.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Von Messling V, Zimmer G, Herrler G, et al. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J Virol. 2001;75:6418–6427. doi: 10.1128/JVI.75.14.6418-6427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia RDCNC (2016) Molecular characterization of canine coronavirus. In: Animal coronaviruses. Nature Publishing Group, pp 189–198
- 61.Martella V, Cirone F, Elia G, et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet Microbiol. 2006;116:301–309. doi: 10.1016/j.vetmic.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 62.Zhao JJ, Yan XJ, Chai XL, et al. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet Microbiol. 2010;140:34–42. doi: 10.1016/j.vetmic.2009.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This manuscript has data included as electronic supplementary material.