Graphical Abstract
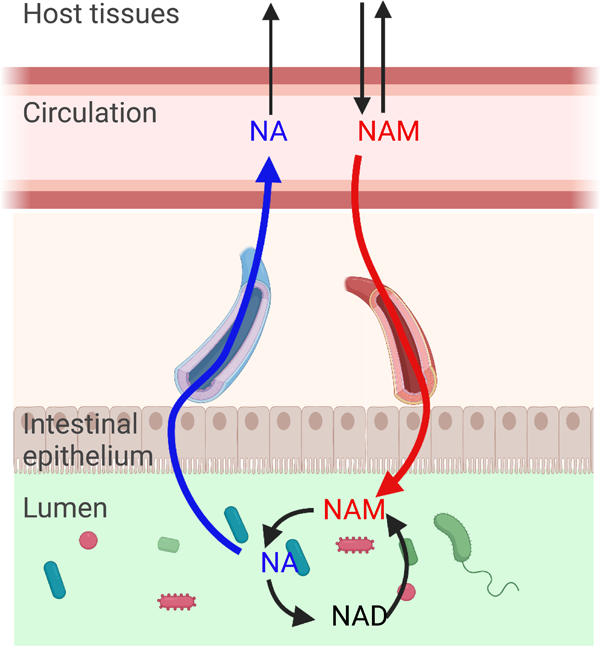
SUMMARY
Nicotinamide adenine dinucleotide (NAD) is an essential redox cofactor in mammals and microbes. Here we use isotope tracing to investigate the precursors supporting NAD synthesis in the gut microbiome of mice. We find that dietary NAD precursors are absorbed in the proximal part of the gastrointestinal tract and not available to microbes in the distal gut. Instead, circulating host nicotinamide enters the gut lumen and supports microbial NAD synthesis. The microbiome converts host-derived nicotinamide into nicotinic acid, which is used for NAD synthesis in host tissues and maintains circulating nicotinic acid levels even in the absence of dietary consumption. Moreover, the main route from oral nicotinamide riboside, a widely used nutraceutical, to host NAD is via conversion into nicotinic acid by the gut microbiome. Thus, we establish the capacity for circulating host micronutrients to feed the gut microbiome, and in turn be transformed in a manner that enhances host metabolic flexibility.
eTOC Blurb
Chellappa et al. show that the gut microbiome primarily obtains its NAD from host-derived circulating nicotinamide. Deamidation of nicotinamide to nicotinic acid by microbes maintains circulating nicotinic acid concentration regardless of dietary intake, providing metabolic flexibility for NAD synthesis. Oral nicotinamide riboside increases systemic nicotinic acid levels through this route.
INTRODUCTION
Nicotinamide adenine dinucleotide (NAD) is an indispensable reduction-oxidation (redox) co-enzyme that shuttles electrons in the form of a hydride ion (H-) by interconverting between its oxidized (NAD+) and reduced (NADH) forms. It is also a substrate for signaling enzymes including sirtuins, poly-ADP ribose polymerases (PARPs), CD38 and SARM1. NAD-consuming enzymes degrade the NAD molecule and release nicotinamide (NAM), thus necessitating a balance between synthesis and consumption to maintain NAD homeostasis. NAD levels have been reported to decline in a spectrum of pathological conditions including aging, obesity, inflammatory, cardiovascular, and neurodegenerative diseases. In many cases, the decline in cellular NAD content correlates with mitochondrial and metabolic dysfunction. NAD boosting ameliorates metabolic perturbations in rodent models of disease 1–6 and has shown promise in some human clinical trials 7,8, albeit with modest or no effects in others 9–11.
Animals have coevolved with commensal microorganisms, including the gut microbiome. The microbes that dwell in the gut lumen are unique in that they can potentially access both ingested food and host-derived circulating nutrients to support their metabolic activity. That said, nutrients that are absorbed in the proximal parts of the small intestine are largely inaccessible to the microorganisms residing in the distal gut lumen. Moreover, it is unclear which, if any, circulating host nutrients permeate the epithelial barrier sufficiently to drive microbiome metabolism. Two established nutrient sources of the colonic microbiome are poorly digestible complex carbohydrates and host mucus 12–18.
As with host cells, NAD is essential to microorganisms. Tremendous effort has been undertaken over the past century to understand NAD metabolism in mammalian tissues and free-living microbes. In contrast, NAD metabolism in the gut microbiome is less extensively studied. A recent study revealed a profound decrease in the NADH/NAD+ redox ratio (but not absolute NAD levels) in colonocytes of germ-free mice 19. Compared to conventional mice, germ free mice fed a western diet had higher liver NAD 20. Intriguingly, gut microbiota are required for the full NAD-boosting effect of orally delivered precursors including both NAM and the common nutraceutical nicotinamide riboside (NR) 21.
In mammals, three major routes of NAD biosynthesis are (1) de novo synthesis from tryptophan, (2) salvage synthesis from NAM via nicotinamide phosphoribosyltransferase (NAMPT) or NR via nicotinamide riboside kinases 22, and (3) Preiss-Handler synthesis from nicotinic acid (NA, also called “niacin”) by nicotinic acid phosphoribosyltransferase (NAPRT). De novo synthesis of NAD from tryptophan is quantitatively important mainly in the liver, but the kidney and some immune cells also utilize this pathway to make NAD 23–25. Turnover of NAD by NAD-consuming enzymes generates NAM, which is exchanged with the systemic circulation. This provides a common pool of precursor that can be used to generate NAD through the salvage pathway. NA is obtained from the diet or microbiome, circulates at lower concentrations, and makes a quantitatively smaller contribution to NAD synthesis in most tissues 23. NAD synthesis can be increased by exogenous supplementation with intermediates such as NR or nicotinamide mononucleotide (NMN) 1,6,26–28. The degree to which this reflects direct incorporation of the parent molecule vs. breakdown and resynthesis is an active area of investigation 23,29.
Most microbial species encode one or more enzymes required to synthesize NAD either de novo from amino acids or through salvage of “vitamin B3” (a collective term for NA, NAM and related derivates such as NR and nicotinic acid riboside (NAR)) (Figure 1A) 30,31. In unicellular microorganisms, de novo synthesis can occur either from tryptophan (as in mammals) or more commonly from aspartate by L-aspartate oxidase (nadB) 32. Importantly, many bacteria (but not mammals) encode the enzyme to convert NAM to NA: nicotinamide deamidase (pncA) 21,33,34.
Figure 1. The majority of microbial NAD synthesis is not accounted for by dietary precursors.
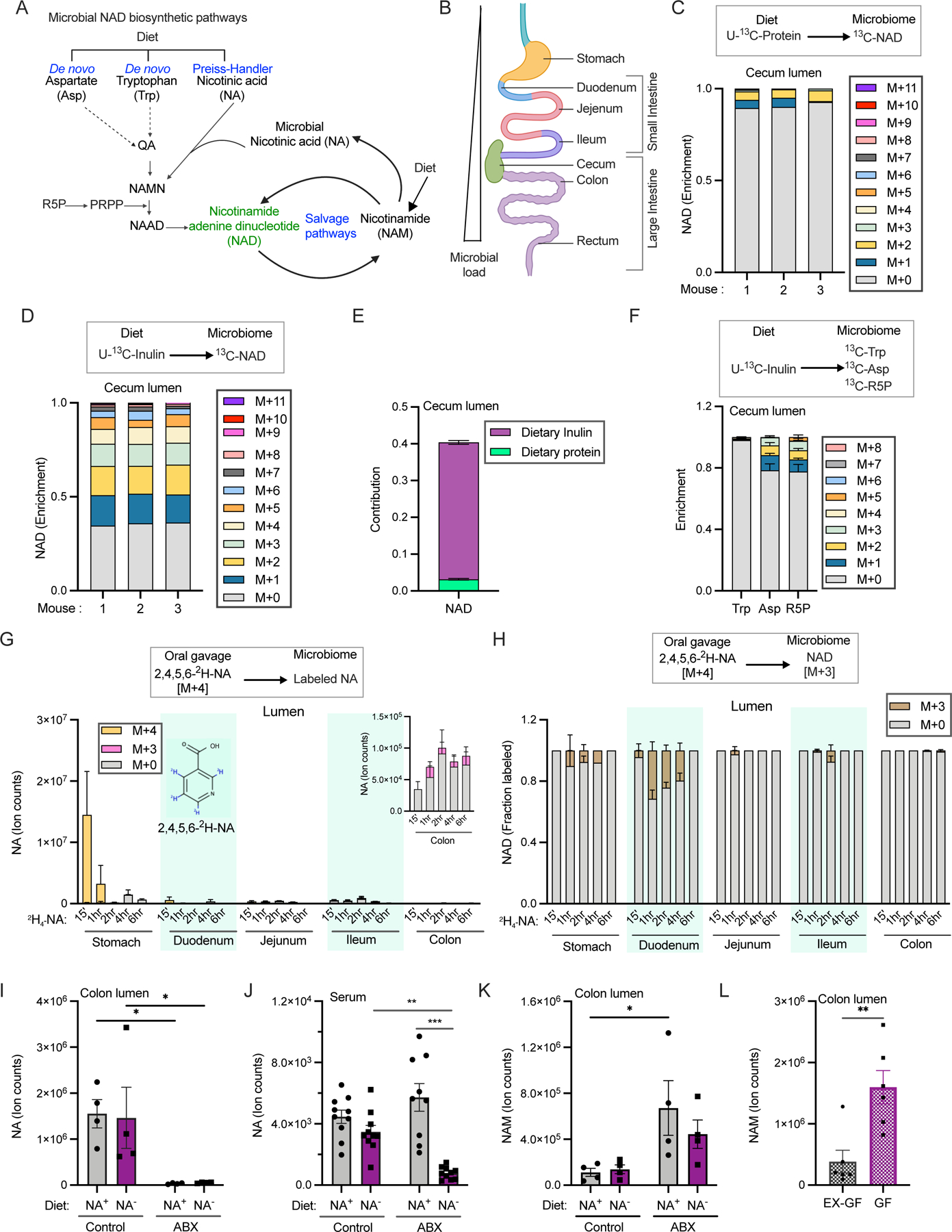
A. Pathways for NAD biosynthesis in microbes. (abbreviations for pathway intermediates QA-quinolinic acid; NAMN – nicotinic acid mononucleotide; NAAD - nicotinic acid adenine dinucleotide).
B. Schematic illustration of different parts of the mouse gastrointestinal tract.
C. Enrichment of carbon labeled NAD in the cecal lumen of mice fed with U-13C-protein for 24h. n=3 mice.
D. Enrichment of carbon labeled NAD in the cecal lumen of mice fed with U-13C-inulin for 24h. n=3 mice.
E. Contribution of U-13C-protein and U-13C-inulin to NAD synthesis in the cecal lumen of mice treated as in C and D. n=3 mice per group.
F. Enrichment of carbon labeled tryptophan (trp), aspartate (asp), and ribose phosphate (R5P) in cecal lumen of mice fed with U-13C-inulin as in D. n=3 mice.
G. Labeled and unlabeled NA content in different parts of the gut lumen after oral gavage of 1.96 μmoles of [2,4,5,6-2H]-NA, equivalent to one-third daily dietary nicotinic acid intake. Inset shows ion counts for NA in colon lumen. n=2–3 mice per time point.
H. Fraction of labeled NAD in different parts of the gut lumen of mice treated as in G. n=2–3 mice per time point.
I. NA content in the colonic lumen of conventional mice fed diet with (NA+) or without nicotinic acid (NA−) that were on drinking water or antibiotics cocktail (ABX). (n=4 per group; Holm-Sidak test following two-way ANOVA, * = p < 0.05).
J. NA ion counts in the serum of mice treated as in I. (n=9–10 mice per group; Holm-Sidak test following two-way ANOVA, ** = p < 0.01, *** = p < 0.001).
K. NAM content in the colonic lumen of mice treated as in I. (n=4 per group; Holm-Sidak test following two-way ANOVA, * = p < 0.05).
L. Abundance of NAM in the colonic lumen of germ-free (GF) and germ-free mice colonized with microbiota from specific pathogen free mice (Ex-GF). (n=4 per group; two-tailed unpaired t-test, ** = p < 0.01).
In this study, we use isotopic tracers to determine how the microbial NAD pool is established. We find that dietary protein and vitamin B3 are not primary precursors for NAD biosynthesis in gut microbiota. Instead, NAM in the host circulation enters the gut lumen to feed NAD biosynthesis, with fiber catabolism also feeding into NAD in the colonic microbiome. Beyond consuming host-derived NAM, the microbiome converts NAM into NA, which provides an alternative NAD precursor to host tissues, restoring NAD levels in intestinal cells when salvage synthesis of NAD from NAM is blocked. The gut microbiome also converts orally delivered NR into NA, which circulates at high levels post oral NR and is critical to NR’s capacity to boost host NAD. In short, we identify a NAM-NA metabolic cycle that allows sharing of NAD precursors between host and microbiome.
RESULTS
Soluble fiber contributes to microbiome NAD synthesis in the large intestine
We explored whether dietary fiber or protein provide precursors for de novo NAD biosynthesis by the gut microbiome, which is present along the length of the gastrointestinal (GI) tract (Figure 1B). Grain-based chow contains around 15–20% insoluble fiber (generally not metabolized) and 3–5% soluble fiber (fermented by colon bacteria) 35–37. Mice were fed a refined diet mixed with 5% unlabeled inulin w/w for 14 days and then switched to diet with U-13C-inulin for 24h before sacrifice. Alternatively, they were fed standard chow containing U-13C-labeled protein for 24h. After accounting for the natural abundance of 13C, we calculated that ~65% of NAD molecules detected in the cecum lumen contained one or more labeled carbon atoms derived from inulin, versus less than 10% from protein (Figures 1C and 1D). Quantitative analysis accounting for precursor (e.g., amino acids) labeling in the cecum lumen and all carbon atoms in NAD revealed that fiber accounts for ~37% of the total carbon contribution to NAD synthesis, versus only ~3% for protein (Figures 1E, and Supplementary Figure S1A). The contribution of inulin to microbial NAD synthesis was lower in other parts of the gut lumen and intestinal tissues (Supplementary Figure S1B). Dietary fiber contributes to microbial NAD in the large intestinal lumen primarily via aspartate and ribose 5-phosphate (Figure 1F). Consistent with aspartate-dependent de novo synthesis of NAD, we were able to detect labeling in the NAM moiety of NAD by MS/MS (Supplementary Figures S1C-S1E).
Simple sugars such as fructose are normally absorbed in the proximal part of the GI tract but may reach distal parts of the gut lumen under conditions of excessive nutrient intake 38. We found that an oral bolus of U-13C-fructose labeled NAD in the GI lumen and host tissues (Supplementary Figures S1F and S1G). The mass shift of NAD in the large intestinal lumen (up to M+19) indicates that all moieties of NAD can be labeled, including the nicotinamide ring. Consistently, we detected labeling of intermediates in NAD synthesis such as aspartate, quinolinate and ribose 5-phosphate, and were able to discern labeling of the nicotinamide ring by MS/MS fragmentation of NAD in the colon lumen (Supplementary Figures S1H-S1K). On the other hand, utilization of ribose phosphate from the pentose phosphate pathway can account for the observed NAD labeling in the lumen of the small intestine and host tissues. Thus, carbohydrates that reach the colon will feed into de novo NAD synthesis, but a large portion of NAD synthesis remains unaccounted for after considering dietary protein and fiber.
Dietary nicotinic acid is not available to microorganisms residing in the distal gut
Standard mouse chow contains NA, which could provide a precursor for NAD synthesis by gut microbes. To examine this possibility, we gavaged mice with labeled NA (2,4,5,6-2H-NA) at a dose equivalent to 40% of daily intake. High levels of fully labeled NA (M+4 NA) were detected in the stomach lumen at the earliest time point assayed (Figure 1G). Labeled NA in the serum peaked at 15 min and gradually declined (Supplementary Figure S1L). Incorporation of this tracer into NAD results in loss of label from the redox-active (4) position to generate M+3 metabolites. A small amount of unmetabolized M+4 NA was found in the lumen of the duodenum and jejunum, whereas only M+3 NA was detected in the distal small intestine and colon. While oral NA made a significant contribution to microbial NAD synthesis in the duodenal lumen (up to ~32% at one-hour post-bolus), the contribution was negligible in distal small intestine and large intestine (Figure 1H). These data are consistent with oral NA being absorbed or metabolized before reaching the lumen of the ileum and colon.
To establish the relative contribution of microbes to NA in the gut lumen and circulation, we next fed mice a refined diet with or without NA. Mice on each of the diets were either given normal drinking water or water treated with a cocktail of antibiotics to deplete microbiota. While antibiotics alone were sufficient to deplete NA in the colon lumen, a combination of NA-deficient diet and antibiotics were required to decrease circulating NA (Figures 1I and 1J).
Circulating nicotinamide is a major precursor for luminal NAD and nicotinic acid
Strikingly, while antibiotics depleted NA in the gut lumen, they augmented luminal NAM (Figure 1K). A similar decrease in luminal NA and increase in NAM were observed in the intestinal lumens of germ-free mice (Figure 1L, Supplementary Figures S1M and S1N). These data raise the possibility that there is a microbiome-independent source of gut lumen NAM, and that microbes convert this NAM into NA.
What is the source of gut lumen NAM? One possibility is the entry of circulating NAM originating in the host 39. To explore this possibility, we intravenously injected mice with 2,4,5,6-2H-NAM (Figure 2A). We detected fully labeled NAM in the lumen all along the GI tract (Figure 2B). In addition, M+4 labeled NA was found in the luminal contents along the length of the GI tract (Figure 2C). Low NAM but high NA levels in the lumen of the large intestine indicate rapid microbial deamidation of NAM to NA and directly correlate with dense microbial load. NA generated from infused NAM reenters the circulation as evidenced by labeled NA in serum (Figure 2D). The absence of NAD in the large intestinal lumens of antibiotics treated and germ-free mice suggests that luminal NAD is predominantly microbiota-derived (Supplementary Figures S1O and S1P). Labeled NAD molecules were detected in the luminal contents of both the small and large intestines, indicating incorporation into the microbial pools (Supplementary Figure S2A). Similarly, intravenously delivered NAM tracer accumulated in the serum and GI lumens of germ-free mice (Figure 2E and Supplementary Figure S2B) but was not converted into NA and NAD (Figure 2F, Supplementary Figures S2C and S2D).
Figure 2. Circulating NAM enters the gut lumen and supports microbial NAD biosynthesis.
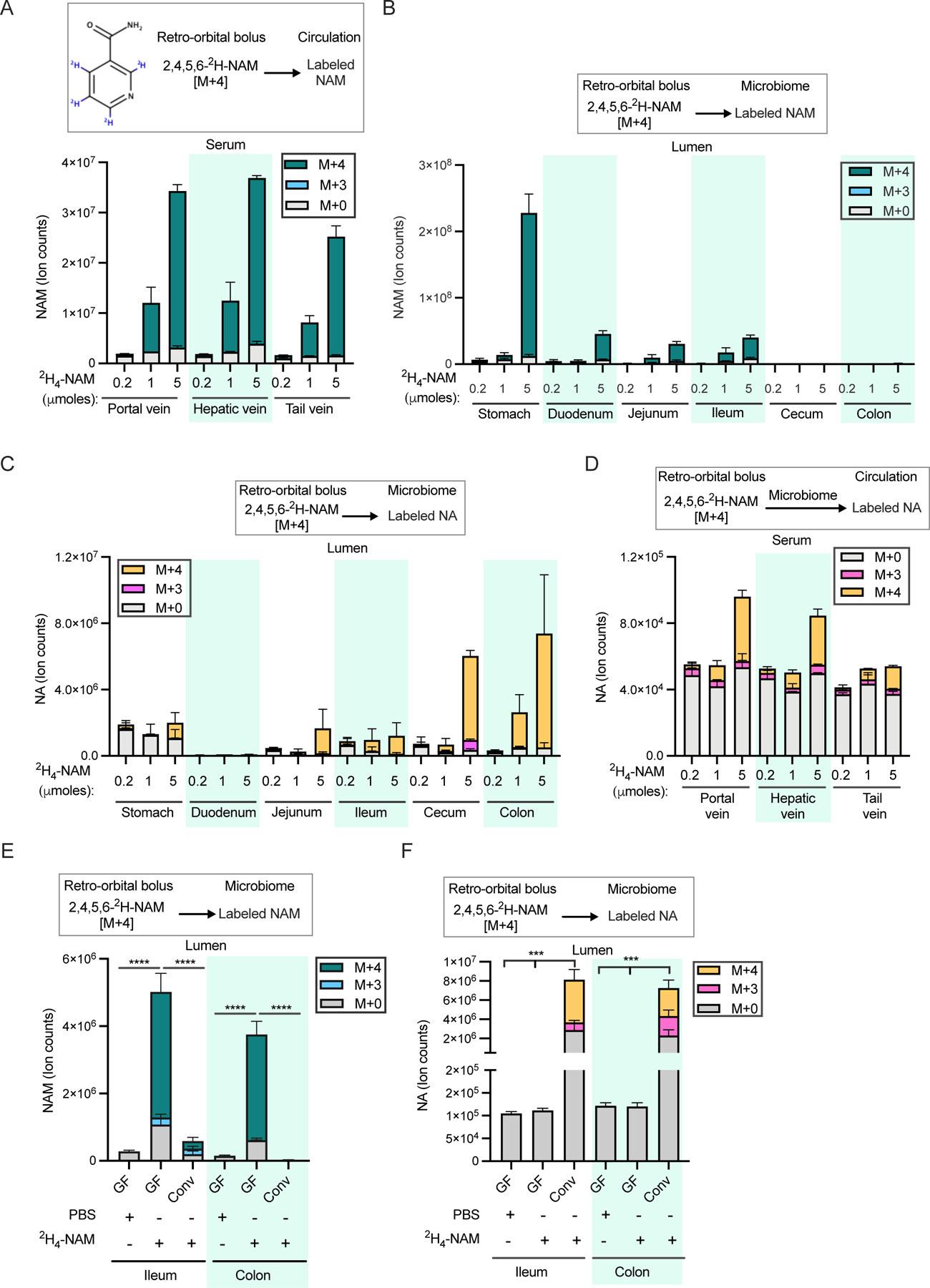
A. NAM levels in the serum samples collected from portal, hepatic and tail vein 15 min after retro-orbital injection of different doses of [2,4,5,6-2H]-NAM. n=2–3 mice per group.
B. Labeled NAM detected in the gut lumen of mice treated as in A.
C. Labeled and unlabeled NA detected in the luminal samples collected from different parts of the gastrointestinal tract of mice treated as in A.
D. Labeled and unlabeled NA detected in the serum samples collected from mice treated as in A.
E. Labeled and unlabeled NAM in luminal samples collected from germ-free (GF) and conventional (Conv) mice retro-orbitally injected with 5 μmoles of [2,4,5,6-2H]-NAM and sacrificed after 2h. n=4 mice per group. (n=4 per group; Tukey multiple comparison test following two-way ANOVA, **** = p < 0.0001).
F. Labeled and unlabeled NA in the luminal samples collected from mice treated as in E. (n=4 per group; Tukey test following two-way ANOVA, **** = p < 0.0001).
To rule out that the above results are due to large transient perturbations in circulating NAM levels following intravenous injection and to enable more quantitative downstream analyses, we next carried out experiments involving continuous intravenous infusion of 2,4,5,6-2H-NAM at a minimally perturbative rate (0.2 nmol/g/min). NAM infusion resulted in a rapid increase in serum M+4 NAM labeling and slower accumulation of circulating M+3 NAM derived from cycle(s) of NAD synthesis and breakdown in tissues (Figure 3A) 23,40. Both infused (M+4) and recycled (M+3) NAM were detected in the gut lumen, along with luminal and serum M+4 and M+3 NA (Supplementary Figures S3A-S3C). NAD was labeled throughout the small intestine and colon, reaching steady-state in the jejunum and ileum lumen within 12h and colon at 18h (Figure 3B, Supplementary Figures S3D and S3E). Despite higher uptake of NAM, labeling of NAD was slow in the stomach lumen. By normalizing to fractional NAM labeling in serum, we estimated the quantitative contribution of circulating NAM to microbiome NAD synthesis in different parts of the gastrointestinal tract (Figure 3C). Circulating NAM accounts for 80–90% of microbial NAD synthesis in the ileum and jejunum, and 45% in the colon (consistent with the colon microbiome synthesizing about half of its NAD de novo from fiber). Thus, circulating host NAM enters the gut lumen and is used by the microbiome for both NAD and NA synthesis.
Figure 3. Vitamin B3 cycles between host and gut microbiome.
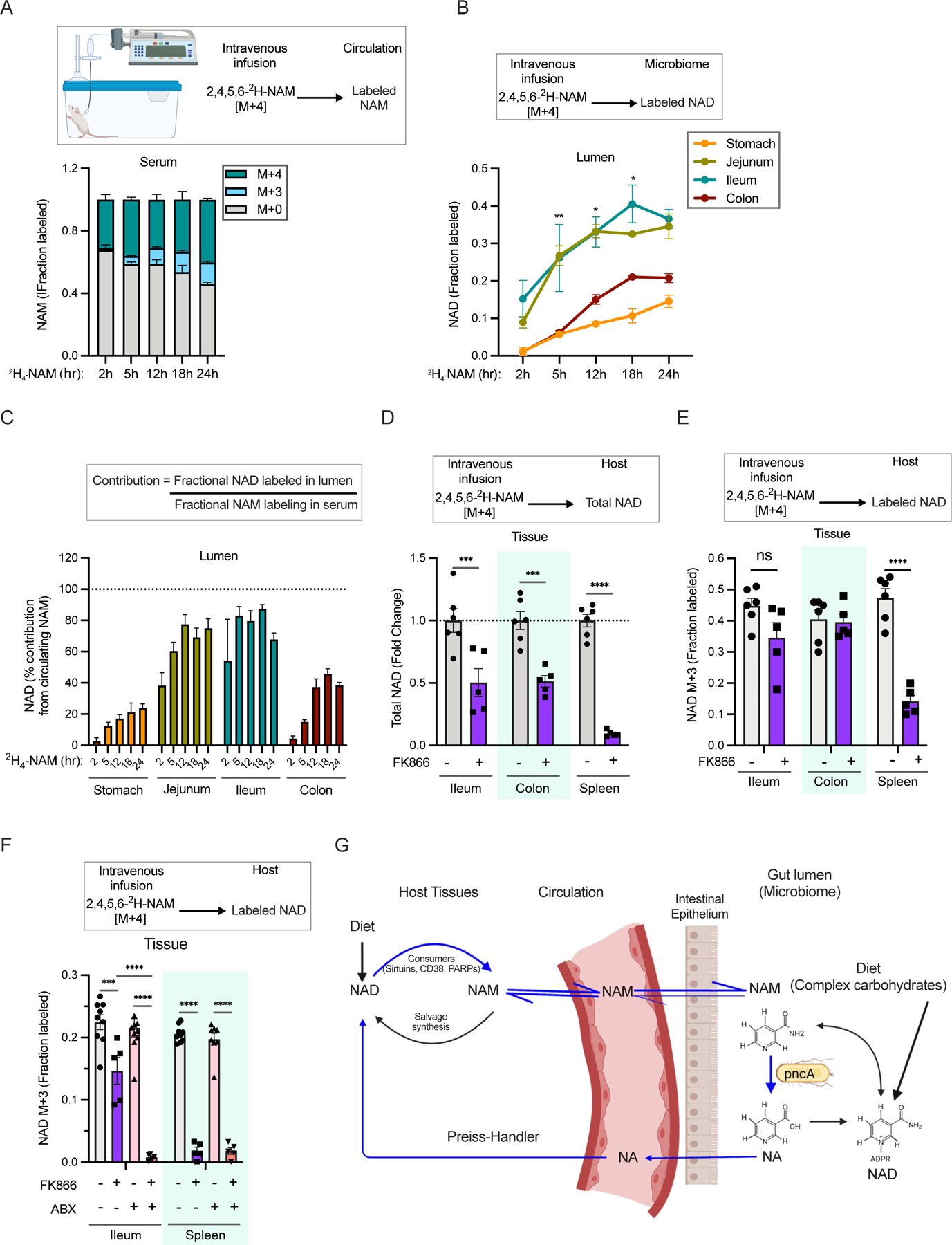
A. Fraction labeling of nicotinamide in the serum of mice infused with 4mM [2,4,5,6-2H]-NAM for different time points.
B. Fraction labeled NAD in the luminal content of mice infused as in A. (n=2–4 per group; Tukey’s multiple comparison test of fractional labeling in the ileum lumen and colon lumen following two-way ANOVA, * = p < 0.05, ** = p < 0.01).
C. Percent contribution of circulating NAM to NAD synthesis in different parts of the gastrointestinal tract estimated from mice infused as in A.
D. Relative levels of total NAD in mice intraperitoneally injected with vehicle or FK866 and infused with [2,4,5,6-2H]-NAM for 23h. (n=5–6 mice per treatment group; Sidak’s multiple comparison test following two-way ANOVA, *** = p < 0.001, **** = p < 0.0001).
E. Fraction labeled NAD in mice treated as in D. (n=5–6 mice per treatment group; Sidak’s multiple comparison test following two-way ANOVA, **** = p < 0.0001). Data for spleen in D and E was previously reported in 40.
F. Fraction labeled NAD in tissues collected from control and antibiotics (ABX) treated mice intraperitoneally injected with vehicle or FK866 and infused with [2,4,5,6-2H]-NAM for 5h. (n=5–10 mice per treatment group; Tukey’s multiple comparison test following two-way ANOVA, *** = p < 0.001, **** = p < 0.0001).
G. Model showing a vitamin B3 cycle between host and microbes. Host-derived NAM in the circulation enters the gut lumen and is deamidated to NA by the microbiome. In turn, host tissues use microbiome-produced NA for synthesis of NAD, which is turned over to release NAM.
Microbial deamidation bypasses salvage synthesis in host intestine and liver
Most tissues rely on the salvage pathway for NAD synthesis 23. Inhibiting NAMPT, the rate-limiting enzyme in the salvage pathway, generally depletes NAD levels in tissues in proportion to their NAD consumption rates 40. Intestine is an exception to this trend. For example, intestine and spleen have similar rapid NAD turnover rates, but treatment with the NAMPT inhibitor FK866 depletes >90% of NAD in spleen 40, but only about ~50% in intestine (Figure 3D). Using 2,4,5,6-2H-NAM tracer, we observed persistent labeling of intestinal (but not spleen) NAD from the infused NAM even in the presence of NAMPT inhibitor (Figure 3E). We hypothesized that this persistent synthesis reflects intestinal incorporation of NA made by microbial deamidation of NAM, and thus would be blocked by depletion of gut microbiome. Indeed, FK866 treatment dramatically decreased NAD labeling from NAM in antibiotics treated mice (Figure 3F). Nevertheless, NAD pool size is maintained in the intestines of mice treated with antibiotics alone, potentially via an increase in the expression of Nampt to facilitate greater flux through NAM-dependent synthesis (Supplementary Figures S3F and S3G). In the liver, treatment of mice with either FK866 or antibiotics decreased labeling of NAD from infused NAM, and combined treatment completely prevented intravenous NAM from contributing to hepatic NAD (Supplementary Figure S3H and S3I). Thus, gut microbiota provide an alternative route from NAM to NAD, bypassing the salvage synthesis pathway, in intestine and liver (Figure 3G).
Nicotinamide riboside boosts tissue NAD pools via breakdown and deamidation to nicotinic acid
Nicotinamide riboside (NR) is widely promoted and consumed as an NAD-boosting nutraceutical supplement 5. Intact NR can be taken up by cells and converted to NAD without the need for the energetically expensive endogenous metabolite phosphoribosyl pyrophosphate or the key salvage enzyme NAMPT. However, the majority of orally delivered NR has been shown to be rapidly cleaved to NAM before entering circulation and peripheral tissues 23,29. Moreover, oral NR supplementation has recently been shown to be much less effective at boosting liver and intestinal tissue NAD levels in the absence of gut microbiota, consistent with its being incorporated via a deamidated route 21. We measured circulating levels of vitamin B3 species in mice following NR oral gavage (500 mg/kg). Circulating NA increased >100-fold, peaking at >50 µM (Figure 4A). NAM showed a similar trend, with a more extended duration and modestly higher peak concentration (Figure 4B). The peak levels of NA observed after NR gavage are comparable to those in humans receiving therapeutic NA supplementation to lower plasma lipids and lipoproteins 41–43 and in excess of the EC50 for activation of GPR109A (the “niacin receptor”) 44–46.
Figure 4. Oral NR supports NAD synthesis in mammalian tissues mainly via conversion to NA.
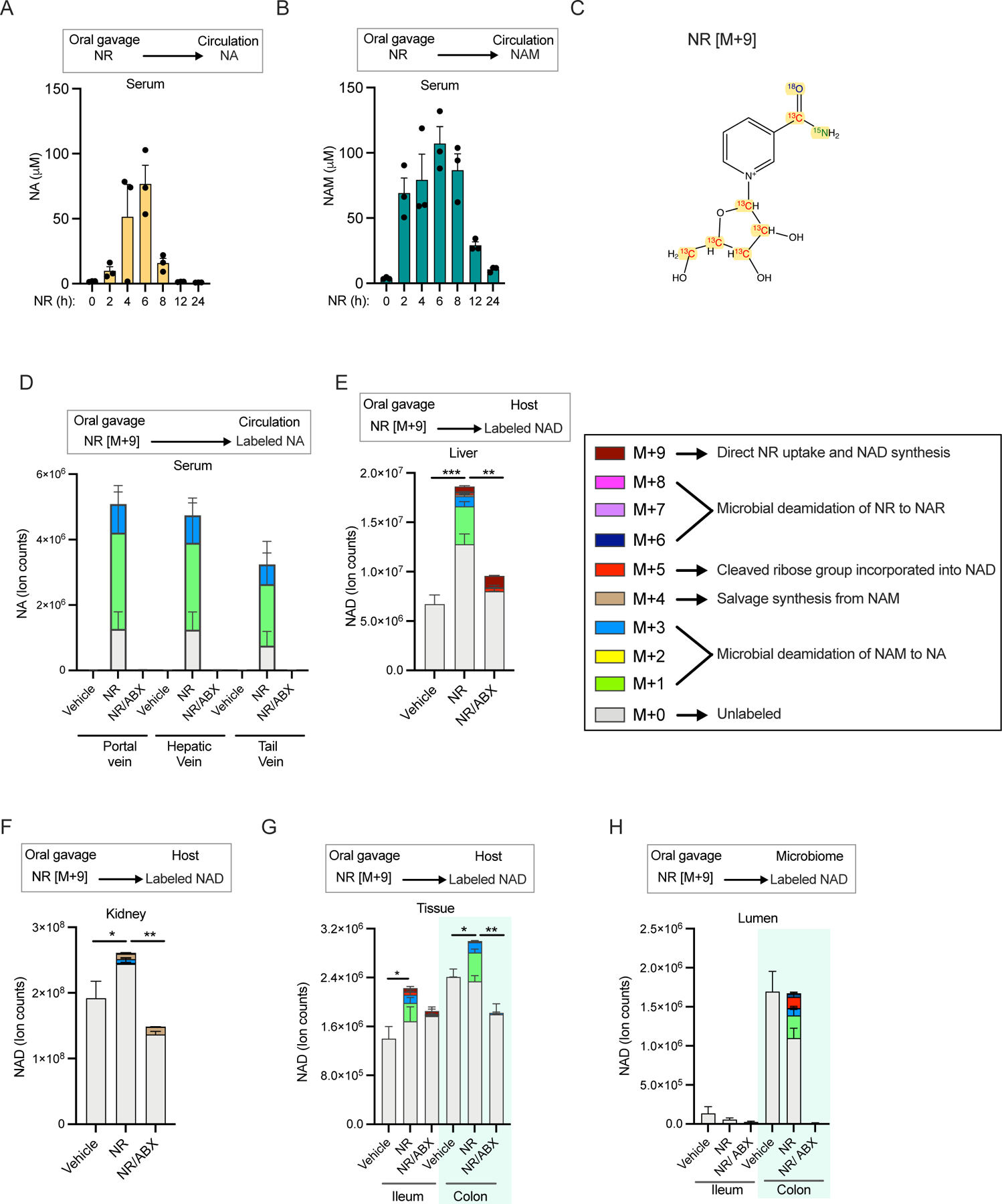
A. Serum concentration of NA in mice orally gavaged with NR at a dose of 500mg/kg body weight n=3 per group.
B. Serum NAM concentration in mice treated as in A.
C. Schematic of M+9 NR isotopically labeled with 15N, 13C and 18O on the nicotinamide moiety and 13C carbon on ribose ring to allow tracing of the molecular fate of NR in host and microbes.
D. Relative level of NA in the serum of control and antibiotics (ABX) treated mice orally gavaged with PBS (vehicle) or NR at a dose of 600 mg/kg body weight. Serum samples were collected from portal, hepatic and tail veins after 3h of treatment (n=3–5 per treatment group).
E. NAD isotopologues detected in the liver of mice treated as in D. (n=4–5 per treatment group; Tukey’s multiple comparison test following one-way ANOVA, ** = p < 0.01, *** = p < 0.001).
F. NAD isomers in the kidney of mice treated as in D. (n=4–5 per treatment group; Tukey’s multiple comparison test following one-way ANOVA, * = p < 0.05, ** = p < 0.01).
G. Isotopologues of NAD in the intestinal tissues of mice treated as in D. (n=4–5 per treatment group; Tukey’s multiple comparison test following two-way ANOVA, * = p < 0.05, ** = p < 0.01).
H. Labeling pattern of NAD in the luminal samples collected from mice treated as in D. (n=4–5 per treatment group).
Production of NAD and NA from NR can occur multiple ways, e.g., (i) NR to NAD to NAM to NA; (ii) NR to NAM to NA to NAD; (iii) NR to NAR to NAD and/or NA. Given that NR has been reported to be more effective than equivalent doses of NAM or NA 47 and that metabolism of oral NMN was recently suggested to proceed via the deamidated mononucleotide 48, we were particularly intrigued by possibility (iii). To distinguish these routes, we synthesized an M+9 isotopologue, on which 15N, 13C and 18O atoms have been introduced on the amide group in the NAM moiety and the five carbon atoms of the ribose ring are 13C (Figure 4C). Deamidation always removes the labeled nitrogen from the amide group and reamidation has a 50% chance of removing the labeled oxygen as well 49. Orally delivered NR was found in the GI tract but not in other peripheral tissues, indicating rapid metabolism of NR before it enters the circulation (Supplementary Figure S4A). Only a small fraction of NAD in the liver and small intestine were M+9, indicating minimal direct assimilation of NR (Supplementary Figure S4B). Thus, route (i) is a minor contributor.
In both routes (ii) and (iii), the nicotinamide moiety of NAD is expected to be labeled as a mixture of M+1 and M+3, indicating synthesis via a deamidated species: NA or NAR. The routes can be distinguished because (iii) retains the original labeled ribose moiety, which would result in production of M+6 and M+8 NAD. Only M+1 and M+3 labeled NAD molecules were detected in the host tissues and gut microbiome (Supplementary Figure S4B), indicating separation of the NAM/NA moiety from the labeled ribose, i.e., assimilation via route (ii). Consistent with this, we detected M+1 and M+3 labeled nicotinic acid adenine dinucleotide (NAAD) in tissues (Supplementary Figure S4C). As expected for NR assimilation via a deaminated species, depletion of gut microbiota with antibiotics completely eliminated NR-derived NA, despite increased NAM levels in serum (Figure 4D and Supplementary Figure S4D). Similarly, depletion of microbiota led to loss of NA and NAAD while increasing NAM in the gut lumen and host tissues (Supplementary Figures S4E-G). NR supplementation increased NAD levels in multiple host tissues including liver, kidney and intestine and this effect was dramatically abrogated in antibiotics-treated mice (Figures 4E-G). While direct incorporation of NR into NAD was higher in antibiotics-treated mice, NAD synthesis from this route was not sufficient to significantly increase the total NAD pool in the absence of gut microbiota. Interestingly, large intestinal microbiota use both the NA and ribose phosphate generated from NR for NAD synthesis (Figure 4H). Thus, NR is catabolized to NA by gut microbiota, and the resulting NA supports NAD synthesis in host tissues.
DISCUSSION
Hosts and their gut microbiomes have coevolved to have complex symbiotic relationships. While it is widely perceived that microbes extend digestive capabilities and produce metabolites such as short-chain fatty acids, flux of metabolites from host to microbiome remains limited to a few examples. These include the feeding of microbes on host-derived mucus 12–14,17,18, incorporation of urea nitrogen into microbial protein 50–52, and bidirectional transport of lactate 53. In addition, host-derived bile acids are metabolized by microbes and have a suppressive effect to limit microbiome growth 54–56. In each of these cases, metabolism of host molecules by the microbiome may be considered opportunistic, even if it is ultimately mutually beneficial. Here we describe a unique bidirectional interaction between the host and gut microbiome in sharing precursors for NAD biosynthesis.
Our results also shed light on the metabolism of dietary NAD precursors. The primary dietary precursors for NAD synthesis, amino acids derived from dietary proteins and NA, are readily absorbed in the upper GI tract, and thus not available to microbes residing in the distal parts of the small intestine and large intestine. Even with a diet free of both NAM and NA, microbial NA production remains high enough to maintain circulating levels in the host. Soluble fiber (traced with 13C-inulin) accounts for just under half of NAD synthesis in the large intestine. We find that host-derived NAM almost completely supports microbiome NAD biosynthesis in the small intestine and makes up the other approximately half of NAD synthesis in the large intestine. Microbial NAD generated by any of these routes is turned over to produce NAM and NA that can be taken up by host intestinal tissues and used to regenerate NAD via the Preiss-Handler pathway.
In this study we established that the large intestinal microbiome uses inulin as a dietary source for NAD synthesis. Dietary supplementation with inulin favors the growth of lactic acid bacteria such as Bifidobacteria and Lactobacilli while depleting proinflammatory bacterial families and genera 57,58. Therefore, we cannot rule out an effect of microbiome composition on NAD labeling in inulin-fed mice. Furthermore, diverse microbiota-accessible polysaccharides including pectin, xylan, mannan and hemicellulose are present in rodent and human diets. Thus, further work is needed to determine whether the type and source of dietary fiber impacts the pathways utilized by intestinal microbiota for NAD synthesis.
The relative importance of NA-dependent NAD synthesis (Preiss-Handler pathway) in mammals has been difficult to ascertain. Compared to NAM, NA circulates at a concentration that is nearly an order of magnitude lower and has a much lower turnover flux 23,45,59. The rate limiting enzyme in the Preiss-Handler pathway, NAPRT, is expressed in multiple host tissues but the highest enzymatic activity levels are found in the liver and kidney 60, suggesting that NA might primarily contribute to NAD synthesis in those organs. However, exogenous NA is readily incorporated into NAD in many human cell lines 60,61 and some tumors amplify the NAPRT gene and depend primarily on the Preiss-Handler pathway for NAD 62–64.
Under conditions of increased NA availability, such as direct supplementation or provision of alternate precursors that can be converted to NA by the microbiome, the Preiss-Handler pathway may become more quantitatively important and other NA-dependent mechanisms may also be engaged. An important feature of NAPRT, as compared to NAMPT (NAM-dependent synthesis) is that NAPRT is not subjected to feedback inhibition 60. Incubation of the purified enzymes with NAD results in inhibition of NAMPT 65, but not NAPRT 66, and preincubation of cells with NA prevents NAM-dependent NAD synthesis, but not vice versa 67. Thus, NAD levels can theoretically be increased more by NA than by NAM. In addition, NA at pharmacological doses has lipid-lowering properties and can engage GPR109A, which is responsible for a common side effect – flushing 45,68. The mechanism of lipid lowering remains somewhat controversial but may involve inhibition of the diacylglycerol synthesis enzyme DGAT2 69–71. We find that high dose NR can lead to peak circulating NA concentrations over 50 µM in mice. This concentration is comparable to NA levels in plasma following oral NA intake in humans 41–43 and thus may be sufficient to transiently activate GPR109A or inhibit DGAT2, mechanisms of action that have not previously been considered relevant for NR. Importantly, if NA-dependent mechanisms play a role in the beneficial effects of NR in mice, it could help explain the lack of translation in some clinical trials - the lower doses used for human studies would be unlikely to support comparable elevation of circulating NA.
We also clarified the metabolic route from NR to NAD. Like Shats et al., 21 we find that NAD boosting effect of NR depends on microbial deamidation. Our use of NR labeled at multiple different atom types additionally distinguishes whether NAD synthesis from NR occurs via NAR or NA. The NAR-dependent route is attractive and would explain how NR could be more effective than either NA or NAM for boosting NAD 47. Our data, however, conclusively demonstrate that NR contributes to NAD mainly via NA.
NAAD is an effective biomarker for NAD supplementation using NR 9–11,47. Although it was originally suggested to result from deamidation of NAD 47, the discovery that microbes deamidate precursors led to the suggestion that it originates from NA 21. The labeling patterns in our study strongly support the latter hypothesis, as labeling of NAAD matches that of NA and not NAD.
Our findings suggest some intriguing hypotheses for future investigation. The dependence of microbes on host NAM raises the possibility that the host might modulate microbiome composition or function by limiting luminal NAM availability. In addition, it is unclear whether luminal transport of NAM or microbiome NA production are influenced by stresses such as metabolic syndrome 72–74. As the colonic microbiome additionally depends on complex carbohydrates that are fermented in the large intestine for NAD synthesis, increased intake of heavily processed food rich in sugar and low in dietary fiber may have a deleterious effect on NAD homeostasis. Such diets are already known to disturb microbiome composition and metabolic activity and are associated with wide range of metabolic diseases in the host 17,75,76. High doses of fructose can also support NAD synthesis in the colon, consistent with recent work showing that fructose can fuel production of microbial metabolites that drive hepatic lipogenesis 38,77. It will be important to determine how the cycling of NAD precursors between host and microbiome interacts with host physiology under conditions of metabolic stress.
In conclusion, we have determined the primary precursors used by small and large intestinal microbiota for NAD synthesis, leading to the identification of a vitamin B3 cycle that effectively shares NAD precursors between the host and microbes. Our findings imply that perturbation of NAD metabolism either in the host or microbes has the potential to disrupt NAD homeostasis and impact physiology in the other.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Lead author, Joseph Baur (baur@pennmedicine.upenn.edu) should be contacted for further information and requests for resources and reagents.
Materials availability
NR+9 (15N,13C6,18O-NR) was synthesized in the laboratory of Marie Migaud (mmigaud@southalabama.edu) in limited quantity for this study.
Data and code availability
Source data for the graphs are included in Data S1. All metabolomics data reported have been deposited at Mass Spectrometry Interactive Virtual Environment (MassIVE) and are publicly available as of the date of publication. Accession numbers for metabolomics data, software and code used for data analysis and carbon correction are listed in the Key Resources Table.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Reagents | ||
| [2,4,5,6-2H]NAM | Cambridge Isotope Laboratories | Catalog# DLM-6883 |
| [2,4,5,6-2H]NA | Cambridge Isotope Laboratories | Catalog# DLM-4578 |
| U-13C6-Fructose | Cambridge Isotope Laboratories | Catalog# CLM-1553 |
| U-13C-Inulin (from chicory) | Cambridge Isotope Laboratories Sigma Aldrich |
Catalog# CLM-9181 Catalog# 900388 |
| Algal crude protein extract-13C | Sigma-Aldrich | Catalog# 642878 |
| 15N,13C6,18O-NR | This paper | N/A |
| NR | Elysium | N/A |
| FK866 | Selleck Chemicals | Catalog# S2799 |
| Ampicillin | Sigma Aldrich | Catalog# A9518 |
| Neomycin | Sigma Aldrich | Catalog# N1876 |
| Metronidazole | Sigma Aldrich | Catalog# M3761 |
| Ciprofloxacin | Sigma Aldrich | Catalog# 17850 |
| Vancomycin | Mylan Institutional LLC | NDC Code 67457-340-01 |
| Imipenem | Fresenius Kabi USA, LLC | NDC Code 63323-322-25 |
| Rodent Diets | ||
| Laboratory rodent diet 5010 | Animal Specialties and Provisions | LabDiet Catalog# 0006524 |
| Niacin+ and Niacin− diets | Research Diets | Modified D11112202 |
| Diet premix for inulin and protein labeling experiments | Research Diets | Modified D22020201 diets. Labeled inulin experiment: replaced 50g inulin per Kg D22020201 diet with a 50g mixture of labeled + unlabeled inulin (1:3 13C:12C). Labeled protein experiment: Replaced 25% casein in D22020201 with 13C-Algal protein. |
| Experimental Models: Organisms/ Strains | ||
| C57BL/6J | Jackson laboratory | Stock No: 000664 |
| C57BL/6J.Nia | National Institute on Aging Rodent Colony | N/A |
| C57BL/6 Germ free mice | University of Pennsylvania Gnotobiotic Mouse Facility | N/A |
| Software and Algorithms | ||
| El-MAVEN software | Elucidata | https://www.elucidata.io/el-maven |
| Accucor | GitHub | https://github.com/XiaoyangSu/AccuCor |
| Deposited data | ||
| Metabolomics and isotope tracing raw dataset | This paper | https://massive.ucsd.edu/ProteoSAFe/QueryMSV?id=MSV000090481 |
| Source data for Figures1C-L, 2, 3A-F, 4, Supplementary Figures S1, S2, S3 and S4 | This paper | Data S1 - Source Data |
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL DETAILS
Animal studies were conducted according to protocols approved by the University of Pennsylvania and Princeton University Institutional Animal Care and Use Committees. 10–12-week-old C57BL/6J male mice purchased from The Jackson Laboratory (Stock No: 000664). Three month old male C57BL/6J.Nia mice obtained from the National Institute on Aging rodent colony were used for infusion studies with FK866 treatment. 8–12-week-old germ-free C57BL/6 male mice were obtained from University of Pennsylvania Gnotobiotic Mouse Facility. Mice in this study are conventional mice under specific pathogen free (SPF) conditions except when explicitly indicated as germ-free or re-colonized germ-free mice. All mice were acclimatized for at least two weeks at the animal housing facility before use in experiments with ad libitum access to chow diet (LabDiet, Laboratory Rodent Diet 5010) and water and maintained on 12h light: dark cycle (7AM-7PM) at ~21°C. Sentinel mice housed in the animal facility are checked routinely for pathogen infections.
METHOD DETAILS
In vivo tracing of dietary protein and inulin
Dietary protein and inulin tracing experiments were conducted at Princeton University. 13C labeled diets were prepared by adding 13C-Crude Protein extract from algae (Sigma-Aldrich, 642878) or 13C-Inulin (Sigma-Aldrich, 900388) to a premix (modified from normal diet with reduced protein, inulin and starch content, Research Diets Inc., D22020201). The final enrichment for each labeled macronutrient was 25%, and both labeled diets share the same macronutrient composition (63% carbohydrate, 20% protein, 7% fat, 5% Inulin, 2.5 % cellulose). 8-week-old male C57BL/6NCrl mice (strain 027; Charles River Laboratories) were first adapted to a composition-matched non-labeled diet for 2 weeks to ensure minimal microbiome perturbation upon diet switch. The non-labeled diet was replaced with a diet that contained a mixture of labeled and unlabeled protein or inulin (1:3) at 9am in the morning, 24 hours before sacrifice, during which mice were given free access to water. In diet labeling experiments, we first calculated NAD enrichment by normalizing ion counts of each of the isotopologues to total ion counts for each metabolite. Then the carbon contribution of protein or inulin to NAD was calculated by determining the fraction of labeled carbons in NAD and normalizing to the precursor labeling in the diet (25%).
In vivo tracing of fructose
Random fed 12–14-week-old C57BL/6J male mice were orally gavaged with 2g/ kg body weight U-13C-Fructose (Cambridge Isotope Laboratories, Tewksbury, MA) dissolved in normal saline. Mice were sacrificed after 2h to collect tissues and luminal contents from the GI tract.
In vivo tracing of orally delivered NA
10 to 12-week-old C57BL/6J male mice were orally gavaged with 1.96 µmoles of 2,4,5,6-2H-NA (Cambridge Isotope Laboratories, Tewksbury, MA). Oral dose was calculated based on NA content in chow diet (120ppm; 0.98 mM) and average intake of 5g chow/ day by a mouse, this is equivalent to 40% of diet. Blood samples were collected before gavage, 30 min after NA delivery and at the time of sacrifice. Luminal contents and intestinal tissues were collected from the indicated parts of the GI tract.
Antibiotic treatment
To deplete gut microbiota mice were given a combination of ampicillin (1 g/L), vancomycin (0.5 g/L), neomycin (1 g/L), metronidazole (0.5 g/L), ciprofloxacin (0.2 g/L), and primoxin (0.5 g/L) in drinking water for 3–5 weeks before use in experiments.
NA diet feeding experiments
Custom diets with or without NA were purchased from Research Diets Inc. Cellulose (BW200) and inulin were used as fiber source, and macronutrient and fiber levels were matched to 5010 chow diet. NA diets used to feed germ-free mice were made with 1.5X vitamins and mineral acid casein and subjected to two rounds of gamma-irradiation. 14-week-old C57BL/6J male mice housed in SPF conditions were fed the diets with or without NA and given normal drinking water or drinking water with antibiotics cocktail for five weeks. Germ-free and conventional mice were fed with double-irradiated diets for two weeks.
Colonization of germ-free mice
For colonization of germ-free mice (Ex-GF), two adult germ-free mice were co-housed with one special pathogen free (SPF) mouse in each cage for 18 days before tissue collection.
In vivo tracing of isotopically labeled NAM
2,4,5,6-2H-NAM was purchased from Cambridge Isotope Laboratories (Tewksbury, MA). NAM tracer was either dissolved in PBS (retro-orbital injection) or normal saline (intravenous infusions). C57BL/6J male mice were retro-orbitally injected with 0.2–5 µmoles of 2,4,5,6-2H-NAM in 100µl volume, blood samples were collected from the tail vein either 15min or 2h after injection as indicated in the figure legend and mice were anesthetized to collect blood samples from the portal vein and hepatic vein. Mice were immediately euthanized by cervical dislocation and tissues were collected and clamped in liquid nitrogen.
For intravenous infusions, mice were surgically implanted with a catheter in the right jugular vein and the infusion was performed within a week. NAM tracer infusion was performed as described previously 23,40. Briefly, 4mM 2,4,5,6-2H-NAM tracer was infused via the jugular catheter at a constant rate of 0.2 nmol/g body weight/min for different period as indicated in the figure legends. Blood samples (~20µl) were collected from the tail vein using microvette blood collection tubes (Sarstedt, Cat. # 16.440.100). At the end of the infusion period, mice were euthanized by cervical dislocation and tissues were quickly dissected and clamped in liquid nitrogen.
FK866 treatment
C57BL6/J male mice were given normal drinking water or pre-treated with antibiotics cocktail for four weeks to deplete gut microbiota. C57BL6/J or C57BL/6J.Nia mice were injected with vehicle (45% Propylene glycol, 5% Tween 80, 50% water) or 50 mg/kg FK866 (Selleck Chemicals, Houston, Tx) once (5h NAM infusion) or three times at 8hr intervals (23h NAM infusion). NAM infusion was started 1h after the first FK866 treatment and mice were sacrificed after 5h or 23h.
Synthesis of M+9 NR
[A] Synthesis of 13C, 18O and 15N labelled M+4 nicotinamide (3, Scheme 1) : Compound (3) was prepared according to reported procedure 21.
Scheme 1:
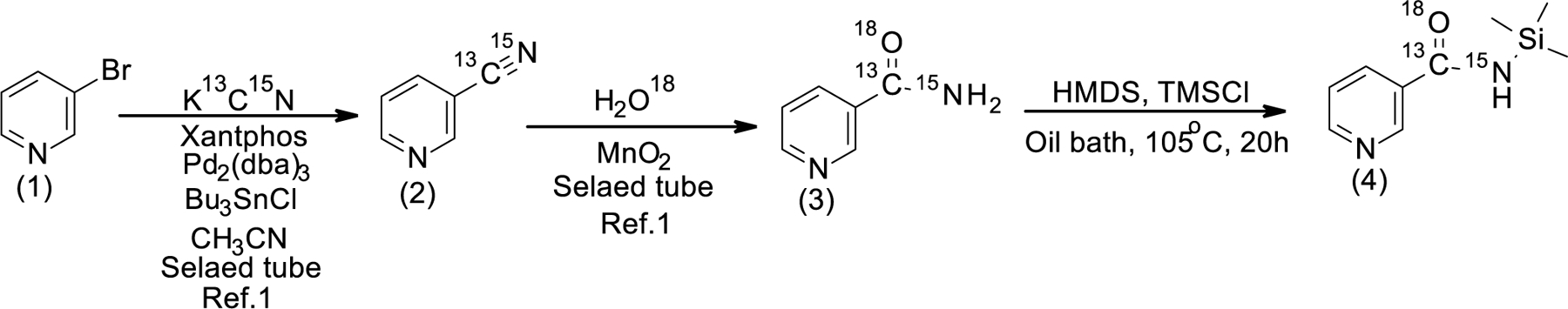
Synthesis of silylated M+4 NAM
[B] Synthesis of Silylated M+4 NAM (4, Scheme 1): A mixture of 13C, 18O and 15N labelled M+4 NAM (0.40 g; 0.032 mol), HMDS (9.00 mL) and TMSCl (0.81 mL, 0.70 g, 0.0064 mol) was heated in an oil bath at 105°C (oil bath temperature) for 20 hours. Upon completion, the reaction mixture was allowed to cool down to room temperature and clear colorless solution was transferred into a 25 mL round-bottom single-neck flask under argon through a cannula and evaporated to dryness on a rotary evaporator followed by drying under high vacuum to give a white crystalline product (0.623 g, 100%).
[C] Synthetic of 1,2,3,5-Tetra-O-acetyl-α/β-D-[13C5]ribofuranoside ([13C5]RTA, 6, Scheme 2): Compound (6) was prepared according to reported procedure 78.
Scheme 2:
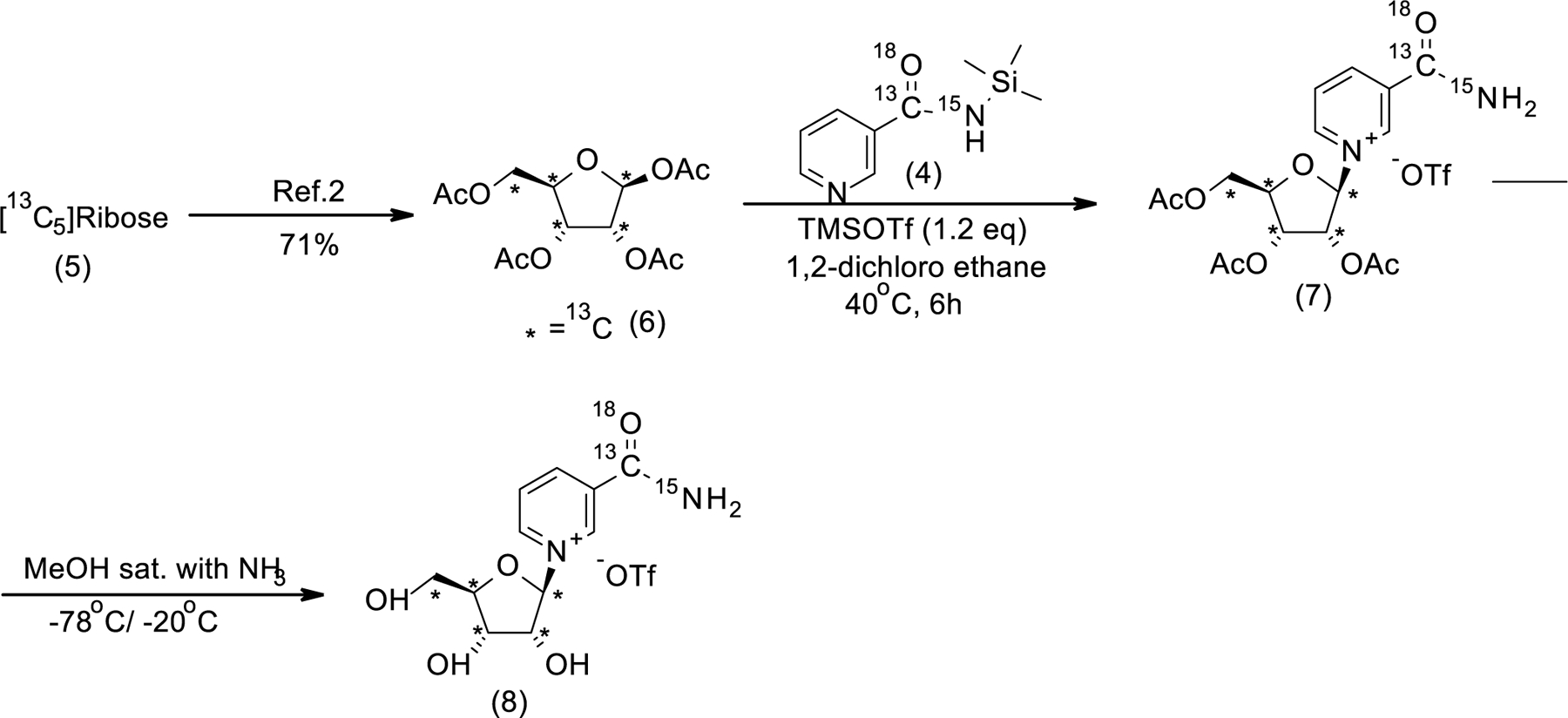
Synthesis of silylated M+9 Nicotinamide riboside
[D] General procedure for the synthesis of M+9 NR Synthesis of M+9 NR 2,3,5-triacetate (Vorbruggen glycosylation, compound 7, Step-1): In a flame dried flask under an argon atmosphere, M+4 silylated NAM (0.37g, 0.00187 mol, 1eqv) and M+5 ,13C labelled -d-ribose tetraacetate (0.664g, 0.00206 mol, 1.1 eqv) were added to dry 1,2-dichloroethane (12mL). After 5 minutes stirring at rt, TMSOTf (0.40mL, 0.00224 mol, 1.2eqv) was added to the stirred 1,2-dichloroethane solution at the same temperature. Now, the resulting mixture was stirred at 40 °C until complete disappearance of the starting materials. The reaction progress was monitored by 1H NMR analysis of the crude mixture. Upon completion, the resulting solution was concentrated, and the yellow oily crude product was used directly for the next step without purification. Synthesis of M+9 NR (de-esterification, compound 8, step-4,): In a pressure tube closed with a septum, evacuated and filled with argon, [13C,18O,15N]-NRTA -OTf (1.14g, 0.00187 mol, 1eqv) was dissolved in anhydrous methanol (30 mL). The solution was cooled down to −78°C at stirring, and ammonia gas (passed through a tube filled with NaOH) was bubbled into the solution through a long metal needle for ca. 5 min. The reaction solution was additionally stirred at − 78°C for 10 min, and, subsequently, the septum was removed, and the tube was immediately closed with a threaded PTFE cap and transferred into a freezer (−20°C) and kept at −20°C for 6 days. The pressure tube was transferred into an ice bath, and, by using a cannula, the content of the tube was transferred into a recovery flask, cooled down in the same ice bath to 0°C. The recovery flask was attached to a rotary evaporator, and ammonia gas was evaporated without any external heating and immersion in a water bath which resulted in a continuous maintaining of the solution temperature below 0°C. After removal of ammonia, residual methanol was removed at ca. 25°C and an oily residue was kept under high vacuum to give a viscous yellow liquid (1.0g). According to the 1H NMR data, the product was contained with the mixture of acetamide, methanol and 2.9 % of M+4 Nam. 1H NMR (400 MHz, D2O, δ, ppm): 9.53 (s, 1H, Ar-H), 9.20 (s, 1H, Ar-H), 8.93–8.91 (m, 1H, Ar-H), 8.23–8.19 (m, 1H, Ar-H), 6.18 (d, J = 177Hz, 1H, H-1), 4.66–3.99 (m, 3H, H-2, H-4&H-3), 3.82–3.79 (m, 1H, H-5a), 3.28–3.27 (m, 1H, H-5b); 13C NMR (100 MHz; D2O, δ, ppm): 170.60 (d, J=17.5Hz, CO), 165.69 (d, J=18.7Hz, CH3CONH2), 145.62 (d, J= 1.18Hz, C-6NAM), 142.60 (C-2NAM), 140.34 (C-4NAM), 133.40 (d, J= 56.8Hz, C-3NAM). 128.33 (d, J= 3.29Hz, C-5NAM), 99.91 (dd, J= 40.02 Hz &3.08Hz, C-1ribose), 87.67 (t, J= 39.9 Hz, C-4ribose), 77.42 (t, J=38.32 Hz, C-2ribose), 69.75 (t, J= 36.19 Hz, C-3ribose), 60.17 (d, J= 41.06 Hz, C-5ribose); 19F NMR (377 MHz, D2O) δF: −78.82 (s); HRMS calcd for 13C6 12C5H15 15N14N18O16O4 [M]+ 264.1189found 264.1194.
NR oral gavage
Mice were orally gavaged with mixture of unlabeled NR and custom-synthesized M+9 NR at a ratio of 1:2.6 in PBS (600 mg/kg body weight). Blood samples were collected at different time points. Three hours after oral dosing, mice were anaesthetized to collect blood samples from hepatic and tail veins and sacrificed by cervical dislocation to collect tissues. In cases where the tracer was diluted with unlabeled molecules, the ratio was verified by mass spectrometry and used to correct for the total contribution of the administered molecule to a given metabolite of interest. This method is expected to be accurate for following any single molecule, but could underestimate the frequency of two labeled molecules coming together by chance, such as regeneration of labeled NR or NAR in the intestinal lumen from labeled ribose and NAM or NA.
Metabolite extraction from serum and tissue samples
Serum samples were thawed on wet ice before metabolite extraction. 50ul of 100% methanol was added to 5ul serum, vortexed and incubated on dry ice for 10 minutes, and centrifuged at 16,000 g for 20 minutes, and 10 ul of water was added to 40ul of serum extracts and use for LC-MS analysis. Frozen tissues were weighed and ground with liquid nitrogen in a mortar and pestle or cryomill (Retsch). Cryomilled tissues were extracted with 40:40:20 acetonitrile:methanol:water (25 mg tissue / ml extraction buffer), vortexed and incubated on ice for 10 min. Tissue samples were then centrifuged at 16,000 g for 30 minutes. The tissue supernatants were transferred to new Eppendorf tubes and then centrifuged again at 16,000 g for 30 minutes to remove residual debris before LC-MS analysis.
Metabolite Measurements
Extracts were analyzed within 36 hours by liquid chromatography coupled to a mass spectrometer (LC-MS). The LC–MS method involved hydrophilic interaction chromatography (HILIC) coupled to the Q Exactive PLUS mass spectrometer (Thermo Scientific) as reported previously 79. The LC separation was performed on a XBridge BEH Amide column (150 mm 3 2.1 mm, 2.5 mm particle size, Waters, Milford, MA). Solvent A is 95%: 5% H2O: acetonitrile with 20 mM ammonium bicarbonate, and solvent B is acetonitrile. The gradient was 0 min, 90% B; 2 min, 90% B; 3 min, 75%; 7 min, 75% B; 8 min, 70%, 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 14 min, 25% B; 16 min, 0% B, 20.5 min, 0% B; 21 min, 90% B; 25 min, 90% B. Other LC parameters are: flow rate 150 µl/min, column temperature 25°C, injection volume 10 µL and autosampler temperature was 5°C. The mass spectrometer was operated in both negative and positive ion mode for the detection of metabolites. Other MS parameters are: resolution of 140,000 at m/z 200, automatic gain control (AGC) target at 3e6, maximum injection time of 200 ms and scan range of m/z 75–1000.
Nicotinic acid detection:
Nicotinate was detected with a 13-min method using the same LC buffers and column but a different gradient at a flow rate of 300 µl/min: 0 min, 90% B; 2 min, 90% B; 5 min, 0 %B; 8 min, 0 % B; 9 min, 90% B; 13 min, 90% B. A SIM scan was used covering m/z 120 – 130 in positive mode.
MS/MS of unlabeled and labeled NAD:
Targeted MS/MS was performed using the PRM function with the parent ions at m/z 664.1, 669.1, 674.1 and NCE 20%. Other parameters are, resolution 17500, AGC target 5e5, maximum injection time 200 ms, isolation window 1.5 m/z. LC condition was the same as the above 25-min method for water soluble metabolites.
Raw LC/MS data were converted to mzXML format using the command line “msconvert” utility 80. Data were analyzed via MAVEN software using an in-house metabolite library established from authentic standards. Isotope labeling patterns were corrected for natural 13C abundance using AccuCor 81. Standard curves for NA, NAM, and NAD in extraction buffer were run to verify that ion count varied linearly with concentration over the ranges being studied and to calculate absolute concentrations of metabolites.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analysis was performed using Graphpad Prism 9 for macOS version 9.4.1 (Graphpad Software, San Diego, California USA). Unless otherwise noted, data are presented as mean ± SEM. Statistical significance for each individual figure is described in the figure legend. One-way or two-way ANOVA was used with Tukey’s post-hoc test for comparisons of 3 or more groups. Studenťs t-test was used for 2 group comparisons or to determine nominal significance. Asterisks displayed in the figure denote statistical significance (p < 0.05; **, p < 0.01; ***, p<0.001). No statistical approach was employed to test for normal distribution of the data presented in the manuscript.
Supplementary Material
Highlights.
Nicotinamide from the host circulation enters the gut lumen
Circulating nicotinamide and dietary fiber support microbial NAD synthesis
Microbiome-generated nicotinic acid bypasses the NAD salvage pathway in host tissues
Oral nicotinamide riboside feeds NAD synthesis mainly via microbial nicotinic acid
Acknowledgments:
This work was funded and supported by a Crohn’s and Colitis Career Development Award to KC; the Howard Hughes Medical Institute and Burroughs Wellcome Fund via the PDEP and Hanna H. Gray Fellow Programs awarded to MRM; NIH grants CA211437 to WL, DP1DK113643 to JDR, R01DK098656 and R01AG043483 to JAB. We acknowledge support from the Regional Metabolomics and Fluxomics Core and the Rodent Metabolic Phenotyping Core of the Penn Diabetes Research Center P30-DK19525 and S10-OD025098, as well as the CINJ Cancer Center Support Grant, Rutgers Cancer Institute of New Jersey Metabolomics Shared Resource, supported, in part, with funding from NCI-CCSG P30CA072720–5923.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Inclusion and Diversity Statement:
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research.
Competing Interests Statement:
J.D.R. is a consultant to Pfizer and an advisor and stock owner in Colorado Research Partners, L.E.A.F. Pharmaceuticals, Rafael Pharmaceuticals, Raze Therapeutics, Kadmon Pharmaceuticals, and Agios Pharmaceuticals. J.D.R. is co-inventor of SHIN2 and related SHMT inhibitors, which have been patented by Princeton University, and that he is a co-founder of Toran Therapeutics. J.A.B. is consultant to Pfizer and Cytokinetics, an inventor on a patent for using NAD precursors in liver injury and has received research funding and materials from Elysium Health and Metro International Biotech, both of which have an interest in NAD precursors. The remaining authors have nothing to declare.
References
- 1.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. (2012). The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab 15, 838–847. 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardell SJ, Hopf M, Khan A, Dispagna M, Hampton Sessions E, Falter R, Kapoor N, Brooks J, Culver J, Petucci C, et al. (2019). Boosting NAD(+) with a small molecule that activates NAMPT. Nat Commun 10, 3241. 10.1038/s41467-019-11078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay P, Horvath B, Rajesh M, Varga ZV, Gariani K, Ryu D, Cao Z, Holovac E, Park O, Zhou Z, et al. (2017). PARP inhibition protects against alcoholic and non-alcoholic steatohepatitis. J Hepatol 66, 589–600. 10.1016/j.jhep.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Tarrago MG, Chini CCS, Kanamori KS, Warner GM, Caride A, de Oliveira GC, Rud M, Samani A, Hein KZ, Huang R, et al. (2018). A Potent and Specific CD38 Inhibitor Ameliorates Age-Related Metabolic Dysfunction by Reversing Tissue NAD(+) Decline. Cell Metab 27, 1081–1095 e1010. 10.1016/j.cmet.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshino J, Baur JA, and Imai SI (2018). NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab 27, 513–528. 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshino J, Mills KF, Yoon MJ, and Imai S (2011). Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 14, 528–536. 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirinen E, Auranen M, Khan NA, Brilhante V, Urho N, Pessia A, Hakkarainen A, Ulla Heinonen JK, Schmidt MS, Haimilahti K, et al. (2020). Niacin Cures Systemic NAD(+) Deficiency and Improves Muscle Performance in Adult-Onset Mitochondrial Myopathy. Cell Metab 32, 144. 10.1016/j.cmet.2020.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Yoshino M, Yoshino J, Kayser BD, Patti GJ, Franczyk MP, Mills KF, Sindelar M, Pietka T, Patterson BW, Imai SI, and Klein S (2021). Nicotinamide mononucleotide increases muscle insulin sensitivity in prediabetic women. Science 372, 1224–1229. 10.1126/science.abe9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dollerup OL, Christensen B, Svart M, Schmidt MS, Sulek K, Ringgaard S, Stodkilde-Jorgensen H, Moller N, Brenner C, Treebak JT, and Jessen N (2018). A randomized placebo-controlled clinical trial of nicotinamide riboside in obese men: safety, insulin-sensitivity, and lipid-mobilizing effects. Am J Clin Nutr 108, 343–353. 10.1093/ajcn/nqy132. [DOI] [PubMed] [Google Scholar]
- 10.Elhassan YS, Kluckova K, Fletcher RS, Schmidt MS, Garten A, Doig CL, Cartwright DM, Oakey L, Burley CV, Jenkinson N, et al. (2019). Nicotinamide Riboside Augments the Aged Human Skeletal Muscle NAD(+) Metabolome and Induces Transcriptomic and Anti-inflammatory Signatures. Cell Rep 28, 1717–1728 e1716. 10.1016/j.celrep.2019.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remie CME, Roumans KHM, Moonen MPB, Connell NJ, Havekes B, Mevenkamp J, Lindeboom L, de Wit VHW, van de Weijer T, Aarts S, et al. (2020). Nicotinamide riboside supplementation alters body composition and skeletal muscle acetylcarnitine concentrations in healthy obese humans. Am J Clin Nutr 112, 413–426. 10.1093/ajcn/nqaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens EC, Chiang HC, and Gordon JI (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, and Chervonsky AV (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salyers AA, Vercellotti JR, West SE, and Wilkins TD (1977). Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol 33, 319–322. 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommer F, and Backhed F (2013). The gut microbiota--masters of host development and physiology. Nat Rev Microbiol 11, 227–238. 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 17.Sonnenburg ED, and Sonnenburg JL (2014). Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab 20, 779–786. 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, and Gordon JI (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 19.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, and Bultman SJ (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 13, 517–526. 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backhed F, Manchester JK, Semenkovich CF, and Gordon JI (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 104, 979–984. 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shats I, Williams JG, Liu J, Makarov MV, Wu X, Lih FB, Deterding LJ, Lim C, Xu X, Randall TA, et al. (2020). Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metab 31, 564–579 e567. 10.1016/j.cmet.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bieganowski P, and Brenner C (2004). Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117, 495–502. 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Su X, Quinn WJ 3rd, Hui S, Krukenberg K, Frederick DW, Redpath P, Zhan L, Chellappa K, White E, et al. (2018). Quantitative Analysis of NAD Synthesis-Breakdown Fluxes. Cell Metab 27, 1067–1080 e1065. 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minhas PS, Liu L, Moon PK, Joshi AU, Dove C, Mhatre S, Contrepois K, Wang Q, Lee BA, Coronado M, et al. (2019). Macrophage de novo NAD(+) synthesis specifies immune function in aging and inflammation. Nat Immunol 20, 50–63. 10.1038/s41590-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Tao J, Ling Y, Li F, Zhu X, Xu L, Wang M, Zhang S, McCall CE, and Liu TF (2019). Switch of NAD Salvage to de novo Biosynthesis Sustains SIRT1-RelB-Dependent Inflammatory Tolerance. Front Immunol 10, 2358. 10.3389/fimmu.2019.02358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giroud-Gerbetant J, Joffraud M, Giner MP, Cercillieux A, Bartova S, Makarov MV, Zapata-Perez R, Sanchez-Garcia JL, Houtkooper RH, Migaud ME, et al. (2019). A reduced form of nicotinamide riboside defines a new path for NAD(+) biosynthesis and acts as an orally bioavailable NAD(+) precursor. Mol Metab 30, 192–202. 10.1016/j.molmet.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Zhang N, Zhang G, and Sauve AA (2020). NRH salvage and conversion to NAD(+) requires NRH kinase activity by adenosine kinase. Nat Metab 2, 364–379. 10.1038/s42255-020-0194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zapata-Perez R, Tammaro A, Schomakers BV, Scantlebery AML, Denis S, Elfrink HL, Giroud-Gerbetant J, Canto C, Lopez-Leonardo C, McIntyre RL, et al. (2021). Reduced nicotinamide mononucleotide is a new and potent NAD(+) precursor in mammalian cells and mice. FASEB J 35, e21456. 10.1096/fj.202001826R. [DOI] [PubMed] [Google Scholar]
- 29.Frederick DW, Loro E, Liu L, Davila A Jr., Chellappa K, Silverman IM, Quinn WJ 3rd, Gosai SJ, Tichy ED, Davis JG, et al. (2016). Loss of NAD Homeostasis Leads to Progressive and Reversible Degeneration of Skeletal Muscle. Cell Metab 24, 269–282. 10.1016/j.cmet.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, and Brenner C (2007). Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484. 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 31.Magnusdottir S, Ravcheev D, de Crecy-Lagard V, and Thiele I (2015). Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet 6, 148. 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurnasov O, Goral V, Colabroy K, Gerdes S, Anantha S, Osterman A, and Begley TP (2003). NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem Biol 10, 1195–1204. 10.1016/j.chembiol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Bockwoldt M, Houry D, Niere M, Gossmann TI, Reinartz I, Schug A, Ziegler M, and Heiland I (2019). Identification of evolutionary and kinetic drivers of NAD-dependent signaling. Proc Natl Acad Sci U S A 116, 15957–15966. 10.1073/pnas.1902346116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, and Brenner C (2009). Microbial NAD metabolism: lessons from comparative genomics. Microbiol Mol Biol Rev 73, 529–541, Table of Contents. 10.1128/MMBR.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaur A, Rose DJ, Rumpagaporn P, Patterson JA, and Hamaker BR (2011). In vitro batch fecal fermentation comparison of gas and short-chain fatty acid production using "slowly fermentable" dietary fibers. J Food Sci 76, H137–142. 10.1111/j.1750-3841.2011.02172.x. [DOI] [PubMed] [Google Scholar]
- 36.Pellizzon MA, and Ricci MR (2018). The common use of improper control diets in diet-induced metabolic disease research confounds data interpretation: the fiber factor. Nutr Metab (Lond) 15, 3. 10.1186/s12986-018-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Hee B, and Wells JM (2021). Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol 29, 700–712. 10.1016/j.tim.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Jang C, Hui S, Lu W, Cowan AJ, Morscher RJ, Lee G, Liu W, Tesz GJ, Birnbaum MJ, and Rabinowitz JD (2018). The Small Intestine Converts Dietary Fructose into Glucose and Organic Acids. Cell Metab 27, 351–361 e353. 10.1016/j.cmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson LM, and Gross CJ (1979). Metabolism of niacin and niacinamide in perfused rat intestine. J Nutr 109, 654–662. 10.1093/jn/109.4.654. [DOI] [PubMed] [Google Scholar]
- 40.McReynolds MR, Chellappa K, Chiles E, Jankowski C, Shen Y, Chen L, Descamps HC, Mukherjee S, Bhat YR, Lingala SR, et al. (2021). NAD(+) flux is maintained in aged mice despite lower tissue concentrations. Cell Syst 10.1016/j.cels.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlson LA, Oro L, and Ostman J (1968). Effect of a single dose of nicotinic acid on plasma lipids in patients with hyperlipoproteinemia. Acta Med Scand 183, 457–465. 10.1111/j.0954-6820.1968.tb10508.x. [DOI] [PubMed] [Google Scholar]
- 42.Menon R, Tolbert D, and Cefali E (2007). The comparative bioavailability of an extended-release niacin and lovastatin fixed dose combination tablet versus extended-release niacin tablet, lovastatin tablet and a combination of extended-release niacin tablet and lovastatin tablet. Biopharm Drug Dispos 28, 297–306. 10.1002/bdd.555. [DOI] [PubMed] [Google Scholar]
- 43.Reiche I, Westphal S, Martens-Lobenhoffer J, Troger U, Luley C, and Bode-Boger SM (2011). Pharmacokinetics and dose recommendations of Niaspan(R) in chronic kidney disease and dialysis patients. Nephrol Dial Transplant 26, 276–282. 10.1093/ndt/gfq344. [DOI] [PubMed] [Google Scholar]
- 44.Soga T, Kamohara M, Takasaki J, Matsumoto S, Saito T, Ohishi T, Hiyama H, Matsuo A, Matsushime H, and Furuichi K (2003). Molecular identification of nicotinic acid receptor. Biochem Biophys Res Commun 303, 364–369. 10.1016/s0006-291x(03)00342-5. [DOI] [PubMed] [Google Scholar]
- 45.Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, and Offermanns S (2003). PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med 9, 352–355. 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- 46.Wise A, Foord SM, Fraser NJ, Barnes AA, Elshourbagy N, Eilert M, Ignar DM, Murdock PR, Steplewski K, Green A, et al. (2003). Molecular identification of high and low affinity receptors for nicotinic acid. J Biol Chem 278, 9869–9874. 10.1074/jbc.M210695200. [DOI] [PubMed] [Google Scholar]
- 47.Trammell SA, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW, Li Z, Abel ED, Migaud ME, and Brenner C (2016). Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun 7, 12948. 10.1038/ncomms12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim L-J, Chalmers TJ, Smith GC, Das A, Poon EWK, Wang J, Tucker SP, Sinclair DA, Quek L-E, and Wu LE (2020). Nicotinamide mononucleotide (NMN) deamidation by the gut microbiome and evidence for indirect upregulation of the NAD+ metabolome. bioRxiv, 2020.2009.2010.289561. 10.1101/2020.09.10.289561. [DOI] [Google Scholar]
- 49.French JB, Cen Y, Vrablik TL, Xu P, Allen E, Hanna-Rose W, and Sauve AA (2010). Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry 49, 10421–10439. 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuller MF, and Reeds PJ (1998). Nitrogen cycling in the gut. Annu Rev Nutr 18, 385–411. 10.1146/annurev.nutr.18.1.385. [DOI] [PubMed] [Google Scholar]
- 51.Ni J, Shen TD, Chen EZ, Bittinger K, Bailey A, Roggiani M, Sirota-Madi A, Friedman ES, Chau L, Lin A, et al. (2017). A role for bacterial urease in gut dysbiosis and Crohn's disease. Sci Transl Med 9. 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart GS, and Smith CP (2005). Urea nitrogen salvage mechanisms and their relevance to ruminants, non-ruminants and man. Nutr Res Rev 18, 49–62. 10.1079/NRR200498. [DOI] [PubMed] [Google Scholar]
- 53.Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, Wibowo MC, Wurth RC, Punthambaker S, Tierney BT, et al. (2019). Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med 25, 1104–1109. 10.1038/s41591-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borgstrom B, Dahlqvist A, Lundh G, and Sjovall J (1957). Studies of intestinal digestion and absorption in the human. J Clin Invest 36, 1521–1536. 10.1172/JCI103549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding JW, Andersson R, Soltesz V, Willen R, and Bengmark S (1993). The role of bile and bile acids in bacterial translocation in obstructive jaundice in rats. Eur Surg Res 25, 11–19. 10.1159/000129252. [DOI] [PubMed] [Google Scholar]
- 56.Lorenzo-Zuniga V, Bartoli R, Planas R, Hofmann AF, Vinado B, Hagey LR, Hernandez JM, Mane J, Alvarez MA, Ausina V, and Gassull MA (2003). Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology 37, 551–557. 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 57.Gibson GR, Beatty ER, Wang X, and Cummings JH (1995). Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108, 975–982. 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 58.Muthyala SDV, Shankar S, Klemashevich C, Blazier JC, Hillhouse A, and Wu CS (2022). Differential effects of the soluble fiber inulin in reducing adiposity and altering gut microbiome in aging mice. J Nutr Biochem 105, 108999. 10.1016/j.jnutbio.2022.108999. [DOI] [PubMed] [Google Scholar]
- 59.Jacobson EL, Dame AJ, Pyrek JS, and Jacobson MK (1995). Evaluating the role of niacin in human carcinogenesis. Biochimie 77, 394–398. 10.1016/0300-9084(96)88152-1. [DOI] [PubMed] [Google Scholar]
- 60.Hara N, Yamada K, Shibata T, Osago H, Hashimoto T, and Tsuchiya M (2007). Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J Biol Chem 282, 24574–24582. 10.1074/jbc.M610357200. [DOI] [PubMed] [Google Scholar]
- 61.Zamporlini F, Ruggieri S, Mazzola F, Amici A, Orsomando G, and Raffaelli N (2014). Novel assay for simultaneous measurement of pyridine mononucleotides synthesizing activities allows dissection of the NAD(+) biosynthetic machinery in mammalian cells. FEBS J 281, 5104–5119. 10.1111/febs.13050. [DOI] [PubMed] [Google Scholar]
- 62.Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, Liu F, Turner K, Yang H, Brunk E, et al. (2019). NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569, 570–575. 10.1038/s41586-019-1150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li XQ, Lei J, Mao LH, Wang QL, Xu F, Ran T, Zhou ZH, and He S (2019). NAMPT and NAPRT, Key Enzymes in NAD Salvage Synthesis Pathway, Are of Negative Prognostic Value in Colorectal Cancer. Front Oncol 9, 736. 10.3389/fonc.2019.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piacente F, Caffa I, Ravera S, Sociali G, Passalacqua M, Vellone VG, Becherini P, Reverberi D, Monacelli F, Ballestrero A, et al. (2017). Nicotinic Acid Phosphoribosyltransferase Regulates Cancer Cell Metabolism, Susceptibility to NAMPT Inhibitors, and DNA Repair. Cancer Res 77, 3857–3869. 10.1158/0008-5472.CAN-16-3079. [DOI] [PubMed] [Google Scholar]
- 65.Elliott GC, and Rechsteiner MC (1982). Evidence for a physiologically active nicotinamide phosphoribosyltransferase in cultured human fibroblasts. Biochem Biophys Res Commun 104, 996–1002. 10.1016/0006-291x(82)91348-1. [DOI] [PubMed] [Google Scholar]
- 66.Gaut ZN, and Solomon HM (1971). Inhibition of nicotinate phosphoribosyltransferase in human platelet lysate by nicotinic acid analogs. Biochem Pharmacol 20, 2903–2906. 10.1016/0006-2952(71)90202-4. [DOI] [PubMed] [Google Scholar]
- 67.Hillyard D, Rechsteiner MC, and Olivera BM (1973). Pyridine nucleotide metabolism in mammalian cells in culture. J Cell Physiol 82, 165–179. 10.1002/jcp.1040820205. [DOI] [PubMed] [Google Scholar]
- 68.Benyo Z, Gille A, Kero J, Csiky M, Suchankova MC, Nusing RM, Moers A, Pfeffer K, and Offermanns S (2005). GPR109A (PUMA-G/HM74A) mediates nicotinic acid-induced flushing. J Clin Invest 115, 3634–3640. 10.1172/JCI23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganji SH, Tavintharan S, Zhu D, Xing Y, Kamanna VS, and Kashyap ML (2004). Niacin noncompetitively inhibits DGAT2 but not DGAT1 activity in HepG2 cells. J Lipid Res 45, 1835–1845. 10.1194/jlr.M300403-JLR200. [DOI] [PubMed] [Google Scholar]
- 70.Hu M, Chu WC, Yamashita S, Yeung DK, Shi L, Wang D, Masuda D, Yang Y, and Tomlinson B (2012). Liver fat reduction with niacin is influenced by DGAT-2 polymorphisms in hypertriglyceridemic patients. J Lipid Res 53, 802–809. 10.1194/jlr.P023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romani M, Hofer DC, Katsyuba E, and Auwerx J (2019). Niacin: an old lipid drug in a new NAD(+) dress. J Lipid Res 60, 741–746. 10.1194/jlr.S092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ibrahim GR, Shah I, Gariballa S, Yasin J, Barker J, and Salman Ashraf S (2020). Significantly Elevated Levels of Plasma Nicotinamide, Pyridoxal, and Pyridoxamine Phosphate Levels in Obese Emirati Population: A Cross-Sectional Study. Molecules 25. 10.3390/molecules25173932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kouassi Nzoughet J, Chao de la Barca JM, Guehlouz K, Leruez S, Coulbault L, Allouche S, Bocca C, Muller J, Amati-Bonneau P, Gohier P, et al. (2019). Nicotinamide Deficiency in Primary Open-Angle Glaucoma. Invest Ophthalmol Vis Sci 60, 2509–2514. 10.1167/iovs.19-27099. [DOI] [PubMed] [Google Scholar]
- 74.Odum EP, and Wakwe VC (2012). Plasma concentrations of water-soluble vitamins in metabolic syndrome subjects. Niger J Clin Pract 15, 442–447. 10.4103/1119-3077.104522. [DOI] [PubMed] [Google Scholar]
- 75.Daien CI, Pinget GV, Tan JK, and Macia L (2017). Detrimental Impact of Microbiota-Accessible Carbohydrate-Deprived Diet on Gut and Immune Homeostasis: An Overview. Front Immunol 8, 548. 10.3389/fimmu.2017.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, and Macia L (2014). The role of short-chain fatty acids in health and disease. Adv Immunol 121, 91–119. 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 77.Zhao S, Jang C, Liu J, Uehara K, Gilbert M, Izzo L, Zeng X, Trefely S, Fernandez S, Carrer A, et al. (2020). Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate. Nature 579, 586–591. 10.1038/s41586-020-2101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Makarov MV, Harris NW, Rodrigues M, and Migaud ME (2019). Scalable syntheses of traceable ribosylated NAD(+) precursors. Org Biomol Chem 17, 8716–8720. 10.1039/c9ob01981b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Xing X, Chen L, Yang L, Su X, Rabitz H, Lu W, and Rabinowitz JD (2019). Peak Annotation and Verification Engine for Untargeted LC-MS Metabolomics. Anal Chem 91, 1838–1846. 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adusumilli R, and Mallick P (2017). Data Conversion with ProteoWizard msConvert. Methods Mol Biol 1550, 339–368. 10.1007/978-1-4939-6747-6_23. [DOI] [PubMed] [Google Scholar]
- 81.Su X, Lu W, and Rabinowitz JD (2017). Metabolite Spectral Accuracy on Orbitraps. Anal Chem 89, 5940–5948. 10.1021/acs.analchem.7b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the graphs are included in Data S1. All metabolomics data reported have been deposited at Mass Spectrometry Interactive Virtual Environment (MassIVE) and are publicly available as of the date of publication. Accession numbers for metabolomics data, software and code used for data analysis and carbon correction are listed in the Key Resources Table.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Reagents | ||
| [2,4,5,6-2H]NAM | Cambridge Isotope Laboratories | Catalog# DLM-6883 |
| [2,4,5,6-2H]NA | Cambridge Isotope Laboratories | Catalog# DLM-4578 |
| U-13C6-Fructose | Cambridge Isotope Laboratories | Catalog# CLM-1553 |
| U-13C-Inulin (from chicory) | Cambridge Isotope Laboratories Sigma Aldrich |
Catalog# CLM-9181 Catalog# 900388 |
| Algal crude protein extract-13C | Sigma-Aldrich | Catalog# 642878 |
| 15N,13C6,18O-NR | This paper | N/A |
| NR | Elysium | N/A |
| FK866 | Selleck Chemicals | Catalog# S2799 |
| Ampicillin | Sigma Aldrich | Catalog# A9518 |
| Neomycin | Sigma Aldrich | Catalog# N1876 |
| Metronidazole | Sigma Aldrich | Catalog# M3761 |
| Ciprofloxacin | Sigma Aldrich | Catalog# 17850 |
| Vancomycin | Mylan Institutional LLC | NDC Code 67457-340-01 |
| Imipenem | Fresenius Kabi USA, LLC | NDC Code 63323-322-25 |
| Rodent Diets | ||
| Laboratory rodent diet 5010 | Animal Specialties and Provisions | LabDiet Catalog# 0006524 |
| Niacin+ and Niacin− diets | Research Diets | Modified D11112202 |
| Diet premix for inulin and protein labeling experiments | Research Diets | Modified D22020201 diets. Labeled inulin experiment: replaced 50g inulin per Kg D22020201 diet with a 50g mixture of labeled + unlabeled inulin (1:3 13C:12C). Labeled protein experiment: Replaced 25% casein in D22020201 with 13C-Algal protein. |
| Experimental Models: Organisms/ Strains | ||
| C57BL/6J | Jackson laboratory | Stock No: 000664 |
| C57BL/6J.Nia | National Institute on Aging Rodent Colony | N/A |
| C57BL/6 Germ free mice | University of Pennsylvania Gnotobiotic Mouse Facility | N/A |
| Software and Algorithms | ||
| El-MAVEN software | Elucidata | https://www.elucidata.io/el-maven |
| Accucor | GitHub | https://github.com/XiaoyangSu/AccuCor |
| Deposited data | ||
| Metabolomics and isotope tracing raw dataset | This paper | https://massive.ucsd.edu/ProteoSAFe/QueryMSV?id=MSV000090481 |
| Source data for Figures1C-L, 2, 3A-F, 4, Supplementary Figures S1, S2, S3 and S4 | This paper | Data S1 - Source Data |
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


