Abstract
Chromosome axis-associated HORMA domain proteins (HORMADs), e.g. ASY1 in Arabidopsis, are crucial for meiotic recombination. ASY1, as other HORMADs, is assembled on the axis at early meiosis and depleted when homologous chromosomes synapse. Puzzlingly, both processes are catalyzed by AAA+ ATPase PCH2 together with its cofactor COMET. Here, we show that the ASY1 remodeling complex is temporally and spatially differently assembled. While PCH2 and COMET appear to directly interact in the cytoplasm in early meiosis, PCH2 is recruited by the transverse filament protein ZYP1 and brought to the ASY1-bound COMET assuring the timely removal of ASY1 during chromosome synapsis. Since we found that the PCH2 homolog TRIP13 also binds to the ZYP1 homolog SYCP1 in mouse, we postulate that this mechanism is conserved among eukaryotes. Deleting the PCH2 binding site of ZYP1 led to a failure of ASY1 removal. Interestingly, the placement of one obligatory crossover per homologous chromosome pair, compromised by ZYP1 depletion, is largely restored in this separation-of-function zyp1 allele suggesting that crossover assurance is promoted by synapsis. In contrast, this zyp1 allele, similar to the zyp1 null mutant, showed elevated type I crossover numbers indicating that PCH2-mediated eviction of ASY1 from the axis restricts crossover formation.
INTRODUCTION
Sexual reproduction relies on the generation of gametes that contain only half of the genetic material of the parental cells; this reduction in chromosome number is achieved through meiosis. Besides its role in maintaining genome size over generations, meiosis generates genetic diversity through a random selection of either the maternal or the paternal homologous chromosome (homolog) and by an exchange of DNA segments between homologs by meiotic recombination.
Meiotic recombination is initiated by the formation of programmed double-strand breaks (DSBs) catalyzed by a complex containing the topoisomerase-like protein SPO11 and other accessory proteins (1–4). DSBs are resected by the MRN/MRX complexes, leading to single-strand DNA ends, which are bound by the recombinases RAD51 and DMC1. These recombinases promote the search for and initial invading steps into the sister chromatid of either the same chromosome or the homolog, resulting in either inter-sister or inter-homolog intermediates. The inter-homolog intermediates become associated with the ZMM proteins (including MSH4, MSH5, SHOC1(ZIP2), HEI10 (ZIP3), ZIP4 and MER3 in Arabidopsis) to form the presumptive double-Holliday junctions (dHJs). dHJs can be subsequently resolved either into class I (interference-sensitive) crossovers (COs) through the ZMM pathway together with MutLγ endonuclease, the MLH1-MLH3 heterodimer, or processed into non-crossovers (NCOs) by other mechanisms. In addition, a small fraction of inter-homolog intermediates is processed by another, less-understood non-ZMM pathway (including the endonuclease MUS81) to form class II (interference-insensitive) COs (1,2,5). As a consequence of crossing-over, physical linkages (chiasmata) between homologs are formed at late prophase I that ensure the equal segregation of homologs during the first meiotic division (meiosis I) (1,5).
Recombination is critically dependent on the chromosome axis, a conserved meiosis-specific proteinaceous structure assembled along the entire length of chromosomes at early meiotic prophase I (1,5–9). The chromosome axis anchors and organizes the sister chromatids to form linear loop-arrays, enabling efficient DSB formation and inter-homolog recombination as observed in diverse taxa including, e.g. yeast, worms, mice and rice (5,10–14).
The chromosome axis is composed of multiple proteins. In Arabidopsis, several axial components have been identified, including cohesin complexes, which build the basic scaffold of the axis; the HORMA domain-containing protein (HORMAD) ASY1 (homolog of Hop1 in yeast, HORMAD1/2 in mammals); and two coiled-coil proteins known as ‘axis core’, i.e. ASY3 (homolog of Red1 in yeast, SYCP2 in mammals) and ASY4 (homolog of SYCP3 in mammals) (15–21). Plants deficient in any of these components show strong defects in CO formation, highlighting the importance of the chromosome axis in recombination that has also been found to be the case in several other organisms (15,16,21–24). As chromosomes synapse, the chromosome axes of the homologs (now referred to as lateral elements) become connected by transversal filaments (TFs), which form together with the central element the synaptonemal complex (SC) (1,2,9).
HORMADs are dynamically present on chromosomes in a stage-dependent manner, namely being assembled on chromosomes before synapsis and getting depleted from the axis when homologous chromosomes synapse (25–28). Recently, it has been shown in Arabidopsis that PCH2 orchestrates both the chromosomal recruitment and dissociation of ASY1/Hop1 by mediating its conformational change (29,30). This mechanism is likely also present in mice and budding yeast (28,31–34). In Arabidopsis, PCH2 requires the cofactor COMET. Whether the necessity for a co-factor is conserved is currently not clear since no COMET homolog has so far been identified in budding yeast and a role of COMET in meiosis in mice has not been described so far (32,35).
While recently light has been shed on the role of the PCH2-COMET complex in ASY1 recruitment at early prophase I (29,30), the question of how this complex specifically dissociates ASY1 from the synapsed axes remains unclear. In particular, it is not understood how the two functions of PCH2 could be temporally separated, i.e. the assistance in ASY1’s recruitment to and the facilitation of the dissociation of ASY1 from the chromosome axis (26,29,30).
Here, we demonstrate that recruitment of PCH2 to the SC is neither dependent on the chromosome axis components ASY1 and ASY3 nor on the PCH2 cofactor COMET but relies on the installation of the TF protein ZYP1. We further show that PCH2 directly binds to the C-terminal region of ZYP1, which is supposed to be proximal to the axis (36). Deletion of the PCH2-binding sequence in ZYP1 still allows to a large extent synapsis with no PCH2 recruitment to the SC and results in a prolonged presence of ASY1 on the chromosome axis. Interestingly, this separation-of-function zyp1 allele largely restores bivalent formation that is reduced in zyp1 null mutants, suggesting that synapsis per se likely promotes CO assurance. However, the number of type I COs in this separation-of-function zyp1 allele is still elevated resembling the situation in zyp1 null mutants. Therefore, our results provide evidence for the hypothesized necessity of ASY1 removal from the axis for the complete polymerization of the SC and prevention of excess class I CO formation. Since we found that TRIP13, the ortholog of PCH2 in mouse, also directly binds to the C-terminus of the mouse ZYP1 homolog SYCP1, the here-identified mechanism controlling ASY1 dynamics might also apply to other organisms, such as mammals.
MATERIALS AND METHODS
Plant materials
The Arabidopsis thaliana accession Columbia (Col-0) was used as the wild-type reference throughout this research. The T-DNA insertion lines SALK_046272 (asy1-4) (25), SAIL_423H01 (asy3-1) (21), SALK_040213 (zyp1a) (37), SALK_050581 (zyp1b) (37), mlh1-3 (SK25975) and SALK_031449 (pch2-2) (26) were obtained from the Salk Institute Genomics Analysis Laboratory (SIGnAL, http://signal.salk.edu/cgi-bin/tdnaexpress) via NASC (http://arabidopsis.info/). The PROPCH2:PCH2:GFP, PROCOMET:COMET:GFP, PROASY3:ASY3:RFP and PROMLH1:MLH1:GFP reporter constructs were generated and described previously (25,30). All plants were grown in growth chambers with a 16 h light/21°C and 8 h dark/18°C at the humidity of 60%.
Plasmid construction
To generate the PROZYP1B:ZYP1BΔ661–728 line, the coding sequence for the amino acids 661–725 in ZYP1B was deleted via PCR using the entry clone PROZYP1:ZYP1B/pDONR221 previously generated as a template. The PCR fragments were ligated and subsequently integrated into the destination vector pGWB501 by gateway LR reaction. For creating the yeast two-hybrid construct of ZYP1B-BD, the full length CDS of ZYP1B was amplified via PCR using primers containing a NcoI (forward primer) and a BamHI (reverse primer) restriction sites (Supplemental Table S1). The PCR fragments were inserted into the pGBKT7 vector via restriction-mediated cloning. To generated the ZYP1B truncations (ZYP1B1-300-BD, ZYP1B301-600-BD, ZYP1B601-856-BD), the coding sequence of relevant fragments was amplified via PCR using primers flanked with attB1 and attB2 sites followed by the gateway BP reaction with the pDONR223 vector (Supplemental Table S1). The other ZYP1B truncations (ZYP1B601-725-BD, ZYP1B726-834-BD, ZYP1B601-660-BD, and ZYP1B661-725-BD) were constructed by deleting the additional coding sequences of ZYP1B using the entry clones of ZYP1B601-856-BD, ZYP1B726-856-BD, or ZYP1B601-725-BD as the PCR template (Supplemental Table S1). Next, the resulting entry clones were integrated into the pGBKT7-GW vector by a gateway LR reaction. The PCH2-related AD (activation domain) constructs were described previously (30).
Generation of zyp1 null mutants
For generating zyp1a/b double null mutants, a CRISPR-Cas9-based gene editing approach was applied. Two single guide RNA sequences were designed to target the N-terminal and central regions of ZYP1B, respectively (target 1: GCCAGTCTCATTGAGAAGAA; target 2: GAGTGAGAGCCATCAGCTGC). The ZYP1B gene editing constructs were transformed into the zyp1a single null mutant (SALK_040213). T1 transformants were analysed by sequencing after PCR amplification using two primer pairs, which specifically bind to ZYP1B (ZYP1B-target1-F/R and ZYP1B-target2-F/R) (Supplemental Table S1). In the T2 generation, CRISPR-Cas9 construct free plants were selected via PCR. The exact mutations of zyp1b were analyzed by integrating the PCR amplicons surrounding the targeting sites into T-vector via TA cloning followed by sequencing. The loss of ZYP1 proteins in zyp1a/b mutants was verified by the absence of a signal in immunodetection using ZYP1 antibody on male meiocytes.
Yeast two-hybrid assay
To perform the yeast two-hybrid interaction test, the relevant AD and BD constructs were co-transformed into the auxotrophic yeast strain AH109 using the polyethylene glycol/lithium acetate method according to the manufacturer's manual (Clontech). Yeast cells haboring the relevant combinations of constructs were dotted on plates with double (-Leu-Trp), triple (-Leu-Trp-His) and quadruple (-Leu-Trp-His-Ade) synthetic dropout medium to assay growth.
Cytological analysis
The imaging of reporters in male meiocytes was performed according to (38). In brief, Arabidopsis anthers harboring the fluorescent reporters at appropriate meiotic stage were dissected and immediately imaged using a Leica TCS SP8 inverted confocal microscope. The meiotic stages were determined by the criteria including chromosome morphology, nucleolus position, and cell shape (38).
Meiotic chromosome spread analysis was performed according to (29). In brief, fresh flower buds were fixed in the ethanol: acetic acid (3:1) fixative for 48 h at 4°C followed by two times of washing with 75% ethanol, and then stored in 75% ethanol at 4°C. For chromosome spreading, flower buds at appropriate stage were initially digested in the enzyme solution (10 mM citrate buffer containing 1.5% cellulose, 1.5% pectolyase and 1.5% cytohelicase) for 3 h at 37°C. Next, single flowers were dissected and smashed with a bended needle on the microscopy slide (note: avoid drying the samples). The spreading step was performed on a 46°C hotplate with 10 μl of 45% acetic acid and then the slide was rinsed with ice-cold Carnoy's fixative. After drying at 37°C for 12h, the slides were mounted with anti-fade DAPI solution (Vector Laboratories).
Immunostaining experiment was performed as described previously (29). Briefly, fresh flower buds at appropriate meiotic stage were dissected and macerated in 10 μl digestion enzyme mixture (0.4% cytohelicase, 1% polyvinylpyrrolidone, and 1.5% sucrose) on the poly-lysine coated slide for 7 mins in a moisture chamber at 37°C followed by a squashing. Next, the samples were incubated further for 8 mins after adding another 10 μl enzyme solution. Subsequently, the samples were smashed thoroughly in 20 μl 1% Lipsol solution. Next, the spreading was performed after adding 35 μl fixative (4% (w/v) paraformaldehyde, pH 8.0) and the slides were dried at room temperature for 2–3 h. For the immunostaining, the slides were washed three times in PBST buffer and blocked in PBST containing 1% BSA for 1 h at room temperature in a moisture chamber. Next, the slides were incubated with relevant antibodies (anti-GFP: gb2AF488 from Chromotek (1:300 dilution), anti-ASY1 (1:500 dilution), anti-ZYP1 (1:500 dilution) at 4°C for 48 h. After three times of washing (10 mins each) in PBST, the slides were incubated with fluorescein-conjugated secondary antibodies for 24 h at 4°C. Next, the DNA was counterstained with anti-fade DAPI solution (Vector Laboratories) following three times of washing. Images were acquired using the Leica TCS SP8 inverted confocal microscopy.
RESULTS
Nucleation of PCH2 on the SC is independent of ASY1, ASY3 and COMET
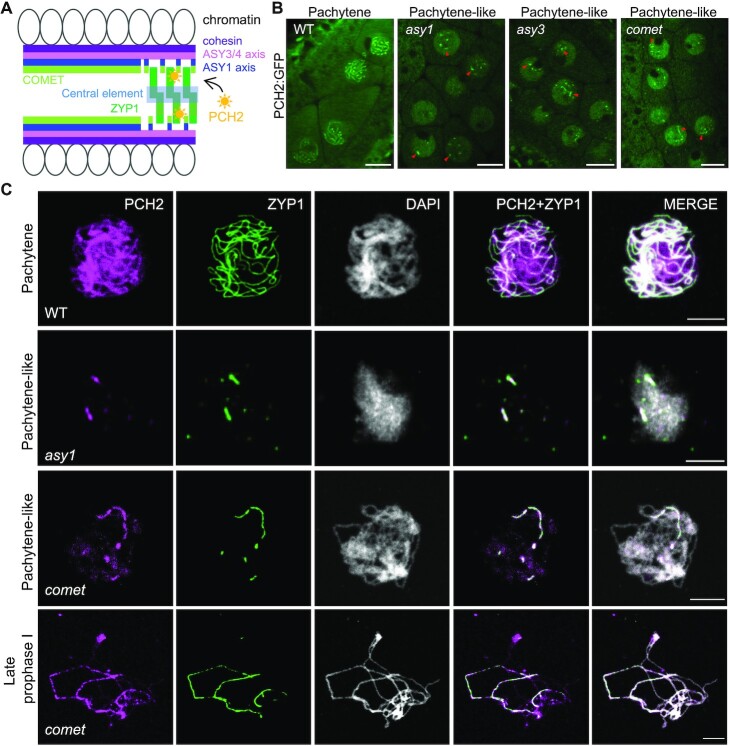
In the wildtype, PCH2 is recruited to the SC coinciding with chromosome synapsis at zygotene and stays along synapsed chromosomes throughout pachytene (Figure 1A, B) (25,26). We therefore asked whether this localization pattern depends on the chromosome axis protein ASY1 and/or ASY3 as parts of the lateral element (LE) of the SC. To address this question, we introduced a previously generated functional PCH2 reporter line, PCH2:GFP (25), into asy1 and asy3 mutants and followed the localization of PCH2 in male meiocytes by laser scanning confocal microscopy. In asy1 and asy3 mutants, chromosome synapsis and formation of the SC are largely, yet not completely, impaired (21,39). Since we could detect short stretches of PCH2:GFP (Figure 1B), we conclude that PCH2 can still be recruited to the remnants of the SC present in these mutants, consistent with a previous report using immuno-detection techniques (Figure 1C) (26). Thus, ASY1 and ASY3 are not required for the localization of PCH2 to the SC.
Figure 1.
Recruitment of PCH2 to the synaptonemal complex is independent of the chromosome axis and its co-factor COMET. (A) A schematic representation of the structure of synapsing chromosomes and the key proteins involved. (B) Localization of PCH2:GFP in the male meiocytes of wild-type (WT), asy1, asy3, and comet mutant plants at pachytene or pachytene-like stages using confocal laser scanning microscope. Red arrowheads indicate the short stretches of PCH2 signal localizing at synapsed chromosomal regions. Bars: 10 μm. (C) Co-immunolocalization of PCH2 and ZYP1 in WT, asy1 and comet mutants at pachytene or pachytene-like stages and at late prophase I. Bars: 5 μm.
Previously, we found that COMET, in its function as an adaptor for PCH2, is associated with chromosomes by binding likely directly to ASY1 in early prophase (30). Therefore, we wondered if the recruitment of PCH2 to the SC relies on COMET. In comet mutants, chromosome synapsis is also largely compromised, but occasional stretches of SC can be formed (30). Similar to the situation in asy1 and asy3 mutants, we found that PCH2 could still assemble into short thread-like structures in male meiocytes of comet mutants (Figure 1B), resembling the situation in mutants for CMT-1, the COMET homolog in C. elegans (40). This finding was confirmed by co-immunolocalization of PCH2 and ZYP1 in comet mutants (Figure 1C).
Taken together, these results suggest that PCH2’s localization on the SC is not dependent on the axis-associated proteins ASY1, ASY3 and COMET, yet tightly correlates with chromosome synapsis and SC formation.
ZYP1 is required for the recruitment of PCH2 to paired chromosomes
Next, we asked whether PCH2’s nucleation on the SC might rely on ZYP1, which is up to now the only component identified in plants belonging to the central region of the SC. However, ZYP1 function in Arabidopsis is distributed to two redundantly-acting and in tandem arranged genes, ZYP1A and ZYP1B, which are positioned tail to tail and separated by <2 kb. Since no zyp1a zyp1b double mutant existed when we started this work, we mutated ZYP1B via CRISPR-Cas9 in the background of a zyp1a T-DNA insertion line (SALK_040213), leading to four different double mutant combinations: zyp1a/b-1, zyp1a/b-2, zyp1a/b-3 and zyp1a/b-4, harboring a C insertion, a 17 bp deletion, an A insertion, and a TG deletion in ZYP1B, respectively (Supplemental Figure S1A). All these mutations in ZYP1B lead to shifts in the reading frame and the formation of premature stop codons, thus likely representing null mutants (Supplemental Figure S1A). Consistent with a complete loss of ZYP1 function in these alleles, we did not detect a ZYP1 signal in any of the four zyp1a/b double mutant combinations by immunodetection of ZYP1 using an antibody that recognizes both paralogs (Supplemental Figure S1B).
Next, we performed a detailed phenotypic analysis of the zyp1a/b mutants. However, while this work was in progress, two other reports presented the analysis of zyp1a/b double mutants (36,41). Our results of the fertility and chromosome behavior in zyp1a/b mutants shown in Supplemental Figures S1C-G and S2A are largely consistent with these two reports except for reduced pollen viability, which we detected in our assays. Briefly, we found that, compared to the wildtype, there were no obvious growth defects in zyp1a/b double mutants. Additionally, no obvious difference in the silique length of the double mutants in comparison with the wildtype could be seen, suggesting that zyp11a/b mutants are largely fertile (Supplemental Figure S1C). However, zyp1a/b mutants showed a slight reduction in pollen viability, i.e. ∼2–3% decrease in comparison to the wildtype and zyp1a single mutants (one-way ANOVA with Dunnett's correction for multiple comparison, P < 0.001) (Supplemental Figure S1D and E). In addition, we observed a significant though mild reduction in seed set (Supplemental Figure S1F and G) (P < 0.001), suggesting very likely slight defects in female meiosis and/or abortion of early embryos.
Consistent with the previous reports (36,41), chromosome spread analyses revealed that chromosome pairing and coalignment can function largely independently of ZYP1 in Arabidopsis (Supplemental Figure S2A [a-l]). In contrast, ZYP1 is required for tight homolog interaction and CO assurance as judged by the presence of univalents in zyp1a/b mutants (17.6% in zyp1a/b-1, n = 74 meiocytes; 10.3% in zyp1a/b-2, n = 78 meiocytes; 12.5% in zyp1a/b-3, n = 64 meiocytes; 14.3% in zyp1a/b-4, n = 21 meiocytes) (Supplemental Figure S2A [m-s] and B).
The finding that loss of ZYP1 does not affect the pairing/coalignment of chromosomes allowed us to assess whether pairing as opposed to the presence of ZYP1 is required for the recruitment of PCH2 to chromosomes. To this end, we introduced the PCH2:GFP reporter, along with an ASY3:RFP construct to stage meiotic nuclei (42), into the zyp1a/b-1 double mutants. While PCH2 nucleated on synapsed chromosomes at pachytene in the wildtype, we observed only a diffuse GFP signal in the zyp1a/b-1 mutant. ASY3:RFP, on the other hand, showed a wild-type-like localization pattern on paired chromosomes (Figure 2A, B, Supplemental Figure S5A).
Figure 2.
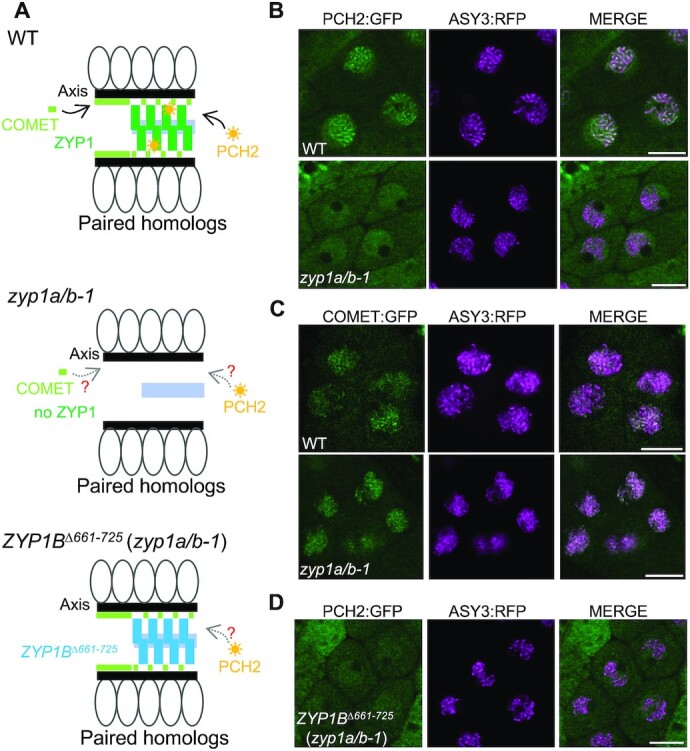
Recruitment of PCH2 to the synaptonemal complex is dependent on ZYP1. (A) Schematic depiction of the structure of synapsing/co-aligning chromosomes in WT, zyp1a/b-1, and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants. The question marks represent the questions raised in the relevant mutants. (B) Localization patterns of PCH2:GFP together with ASY3:RFP in male meiocytes of WT and zyp1a/b-1 mutant plants at pachytene or pachytene-like stages. Bars: 10 μm. (C) Localization patterns of COMET:GFP together with ASY3:RFP in male meiocytes of WT and zyp1a/b-1 mutant plants at early prophase. Bars: 10 μm. (D) Localization pattern of PCH2:GFP together with ASY3:RFP in male meiocytes of WT and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants at pachytene-like stage. Bars: 10 μm.
Since COMET functions as the cofactor of PCH2 in Arabidopsis (30), we also asked whether the absence of ZYP1 would influence the chromosome association of COMET, too. To address this, a previously generated functional COMET reporter, COMET:GFP (30), was introduced into the zyp1a/b-1 mutants together with ASY3:RFP. Following the localization of COMET in the male meiocytes of wild-type and zyp1a/b-1 mutant plants revealed that COMET’s association with chromosomes was not affected in zyp1a/b-1 mutants (Figure 2C).
Overall, these data show that the two components of the ASY1 dissociation machinery are brought together via two different pathways: COMET’s localization is independent of the TF protein ZYP1 but relies on the LE and ASY1 itself (30); conversely, PCH2 depends on ZYP1 for SC localization but does neither require ASY1 nor ASY3 to be present on the axis.
PCH2 directly interacts with the C-terminus of ZYP1
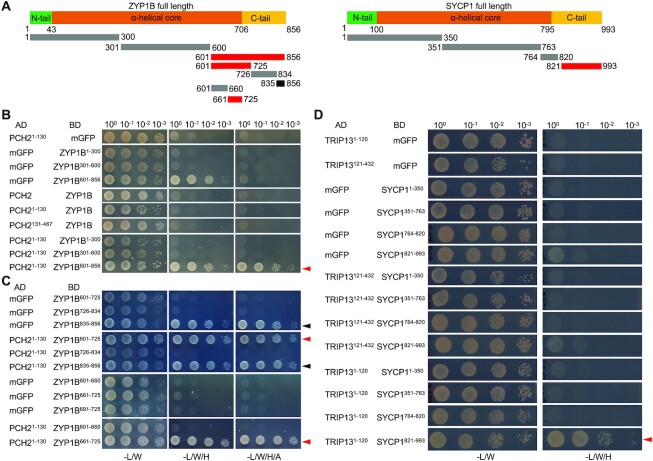
The findings above raised the question whether PCH2 would directly bind to ZYP1. To answer this question, we performed yeast two-hybrid assays. An initial assay between the full length PCH2 protein and the entire ZYP1B protein, as a representative of the two ZYP1 proteins, did not reveal an interaction (Figure 3A, B). However, the binding of two truly interacting proteins in vivo can be masked when tested in vitro or in a heterologous system due to incorrect folding, improper conformation, or lack of secondary modifications as, for instance, seen for the interaction between ASY1 and ASY3 (25). Therefore, we first tested the binding of ZYP1B to two earlier generated PCH2 truncations: PCH21–130, which we have previously found to interact with COMET, and PCH2131-467, which does not bind to COMET (30). Nevertheless, again no interaction could be detected (Figure 3B). However, when we next divided ZYP1B into three parts (1–300 aa, 301–600 aa and 601–856 aa), we found that the C-terminal fragment (ZYP1B601–856) interacted with PCH21–130 (Figure 3B).
Figure 3.
PCH2/TRIP13 interacts with ZYP1/SYCP1. (A) Schematic graph of Arabidopsis ZYP1B and mouse SYCP1 showing the central α-helical core domain flanked by the unstructured N- and C-terminal tails. The lines below indicate the truncations used in the yeast two-hybrid interaction assay. Red lines indicate the positive interaction and black line indicates the autoactivation. (B–D) Yeast two-hybrid assay of PCH2 and ZYP1 (B and C) or TRIP13 and SYCP1 (D) with different truncations. Red and black arrowheads indicate positive interaction and autoactivation, respectively.
To narrow down the binding domain of ZYP1B with PCH2, we further divided the C-terminal part of ZYP1B into three segments: 601–725 aa, 725–834 aa and 835–856 aa. ZYP1B835–856 showed a strong autoactivation in our assay and hence, could not be evaluated. While PCH21–130 did not bind to ZYP1B726–834, a strong interaction of ZYP1B601-725 with PCH21–130 was observed (Figure 3C). This interaction was corroborated using a split-luciferase assay (Supplemental Figure S3A).
It is worth mentioning that ZYP1B661–725 is almost identical to ZYP1A in the region between amino acids 661 and 725 (Supplemental Figure S4), with only one differing amino acid. Thus, we conclude that PCH2 binds with its N-terminal region (1–130 aa) to the C-terminal parts of ZYP1B (661–725 aa) and likely ZYP1A (661–725 aa).
Since the recruitment of PCH2/TRIP3 to the SC was found to depend on the installation of ZYP1 (and its orthologs) in other organisms, including rice, yeast, mouse, and C. elegans (17,43–45), we asked whether PCH2 directly interacts with other TF proteins. In support of a broadly conserved regulation of PCH2-type proteins, we found that mouse TRIP13 binds in Y2H and BiFC assays through its N-terminal region (1–120 aa) to the C-terminal part of SYCP1 (homolog of ZYP1) (821–993 aa) (Figure 3D, Supplemental Figure S3B and C).
A separation-of-function mutant of ZYP1B reveals the necessity of the SC-localized PCH2 for ASY1 depletion
Seeing the elaborated system that assures the tightly temporally controlled presence of ASY1 at the chromosome axis, we next asked what the consequences of an alteration of its residence time are. The failure of bringing the ASY1 dissociation complex to the paired/coaligned chromosomes explains the previous findings that loss of ZYP1 results in a prolonged/continuous presence of ASY1 on the axis (36,41). The normal localization of ASY1 in zyp1a/b mutants at early prophase suggests that the SC-localized PCH2 is dispensable for the chromosome assembly of ASY1 (Supplemental Figure S6A and B). At pachytene-like stage (visualized by the DAPI-stained thick thread-like chromosomes), we observed no obvious removal of ASY1 from the paired axes in zyp1a/b double mutants (Figure 4A-C and Supplemental Figure S5B), in accordance with previous reports (36,41). This observation was confirmed by imaging the live male meiocytes of zyp1a/b-1 double mutants harboring a previously generated ASY1:GFP reporter together with the above-used ASY3:RFP (25) (Supplemental Figure S6A and B). This gave rise to the hypothesis that the recombination defects in mutants without ZYP1 function are at least partially due to the prolonged presence of ASY1 on the axis.
Figure 4.
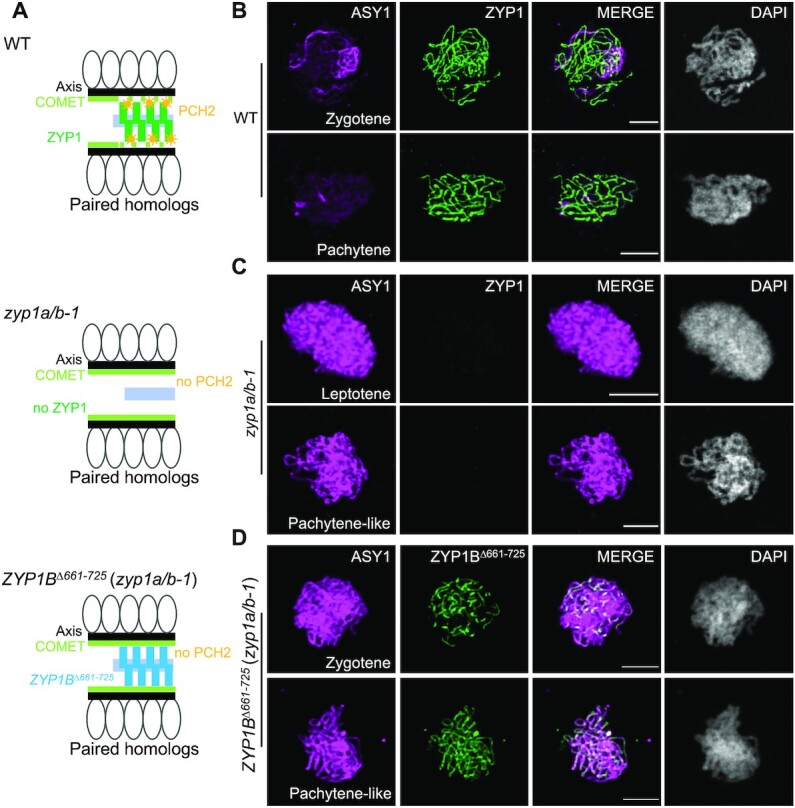
Absence of ZYP1 results in the deficient removal of ASY1 from the chromosome axis. (A) Schematic depiction of the structure of paired chromosomes in WT, zyp1a/b-1, and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants. (B–D) Co-immunolocalization of ASY1 with ZYP1 or ZYP1BΔ661–725 in WT (B), zyp1a/b-1 (C) and ZYP1BΔ661–725 (zyp1a/b-1) (D) mutant plants at different prophase I stages. Bars: 5 μm.
However, the absence of ZYP1 and, with that, the lack of synapsis might have further and possibly indirect and not yet understood consequences, resulting in the observed increase in CO formation and independently of an altered ASY1 pattern (36,41). To tackle this problem, we generated a separation-of-function version of ZYP1B which keeps the ability to polymerize along the paired axes but cannot bind to PCH2. To this end, we used a previously generated ZYP1B genomic construct (42) and deleted the PCH2 binding domain in ZYP1B located between amino acids 661–725 (PROZYP1B:ZYP1bΔ661–725, called ZYP1BΔ661–725), as mapped above by the yeast two-hybrid assays (Figure 3C).
We first introduced the ZYP1BΔ661–725 construct into zyp1a/b-1 mutants, called ZYP1BΔ661–725 (zyp1a/b-1) (Figure 2A), and found that the deletion of 661–725 aa still allowed the polymerization of ZYP1B on the SC as revealed by immunodetection of ZYP1 (Supplemental Figure S7A). Despite the formation of an SC, PCH2 was not recruited to the chromosomes in ZYP1BΔ661–725 (zyp1a/b-1) at pachytene-like stages as determined by the chromosome morphology and cell shape (Figure 2D). Instead, PCH2:GFP showed a diffuse localization pattern in the nucleus in contrast to the wildtype (Figure 2B and D), corroborating that the domain between 661–725 in ZYP1 binds to PCH2 in planta. Importantly, we found by immunodetection that ASY1 could not be removed from the synapsed axes of ZYP1BΔ661–725 (zyp1a/b-1) plants, resembling the situation in zyp1a/b-1 double mutants (Figure 4A and D). This result was confirmed by the imaging of an ASY1:GFP reporter in male meiocytes of ZYP1BΔ661–725 (zyp1a/b-1) plants (Supplemental Figure S7B). Notably, the retention of ASY1 at synapsed chromosomes is reminiscent of the yeast zip1-4LA mutation that uncouples the SC assembly and PCH2 recruitment/stabilization, also leading to an extended occupancy of Hop1 on paired/synapsed chromosomes (46,47). These data suggest that the ZYP1-dependent nucleation of PCH2 on the SC is crucial for ASY1 removal from the synapsing axes. Furthermore, we asked whether the prolonged presence of ASY1 would affect the chromosomal residence of COMET. In the wildtype, the chromosome-localized fraction of COMET was largely reduced at pachytene and only a few bright dots were observed, corresponding likely to the rDNA-containing NOR regions (Supplemental Figure S6C). However, in ZYP1BΔ661–725 (zyp1a/b-1) plants, COMET was prolonged present on paired/synapsed chromosomes (Supplemental Figure S6C), consistent with its localization dependency on ASY1 (30).
A detailed analysis of the SC in male meiocytes of ZYP1BΔ661–725 (zyp1a/b-1) plants by ZYP1 immunolocalization revealed that ZYP1 polymerization was nearly completed in only 3 out of 75 meiocytes with only very few and short chromosome regions having no ZYP1 signals. In the remaining 72 meiocytes, ZYP1 did not accumulate along the entire length of chromosome axes (Figure 5A). In contrast, the SC appeared to be completely assembled along with the regular depletion of ASY1 in wild-type plants expressing the ZYP1BΔ661–725 construct (n = 20 meiocytes) at pachytene stage (Figure 5B). These results suggest that the assembly of ZYP1BΔ661–725per se on paired chromosomes has probably no dominant impact on the SC extension. However, we currently cannot exclude whether and, if so, to what extent the wild-type ZYP1B outcompetes the ZYP1bΔ661–725 for SC assembly.
Figure 5.
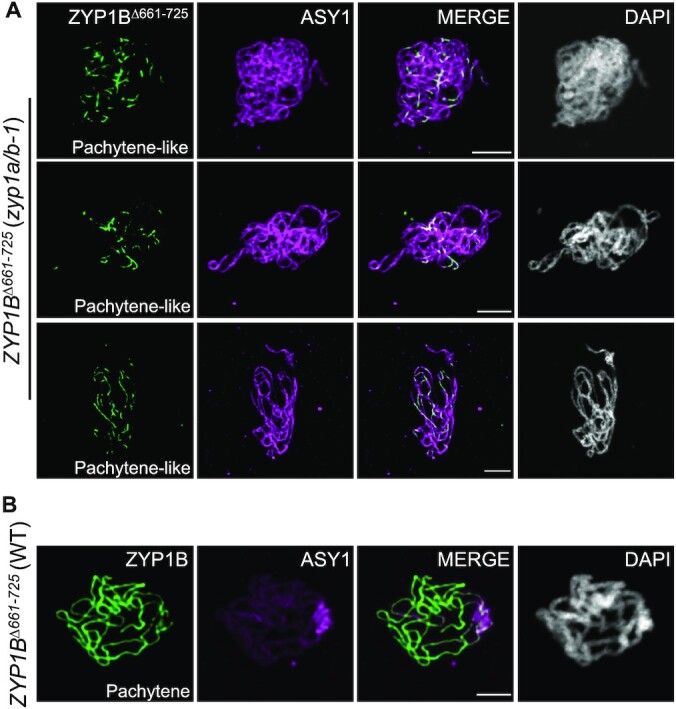
ZYP1△661–725 does not completely polymerize in zyp1 mutants. (A) Co-immunolocalization of ZYP1BΔ661–725 and ASY1 in ZYP1BΔ661–725 (zyp1a/b-1) mutant plants at pachytene-like stage. Bars: 5 μm. (B) Co-immunolocalization of ZYP1BΔ661–725 and ASY1 in WT plants at pachytene. Bars: 5 μm.
ZYP1bΔ661-725 partially restores bivalent formation, yet still leads to an increase of type I COs
This ZYP1bΔ661–725 allele allowed us then to evaluate the effects triggered by the complete loss of ZYP1 versus the prolonged axial presence of ASY1 in plants that form an SC. First, we compared the number of chiasmata in zyp1a/b mutants versus the ZYP1BΔ661–725 (zyp1a/b-1) plants according to the cytological configuration of the bivalents at metaphase I in male meiocytes as previously described in (48). In the wildtype, the estimated minimum number of chiasmata was 8.30 ± 0.89 (n = 50 meiocytes) per meiosis. This number was slightly, yet significantly (one-way ANOVA with Tukey's correction for multiple comparison test) reduced to 7.76 ± 1.03 (n = 51 meiocytes, P = 0.0319), 7.45 ± 0.92 (n = 38 meiocytes, P = 0.0004) and 7.52 ± 1.02 (n = 64 meiocytes, P = 0.0002) in zyp1a/b-1, zyp1a/b-2 and zyp1a/b-3 mutants, respectively (Figure 6A). These data are in accordance with previously published results (36,41). In comparison, we found that the estimated minimum number of chiasmata in ZYP1BΔ661–725 (zyp1a/b-1) plants (8.45 ± 0.97, n = 95 meiocytes) was slightly increased and reached wild-type levels. Interestingly, the number of meiocytes containing univalents significantly decreased from over 10% in zyp1 double mutants (see above) to 4.2% (n = 120 meiocytes, P = 0.047, chi-square test) in zyp1 mutants expressing ZYP1bΔ661–725 while no univalent was observed in the wildtype (n = 85 meiocytes), indicating that either ZYP1 or synapsis per se are able to promote the formation of the obligatory CO without ASY1 depletion.
Figure 6.
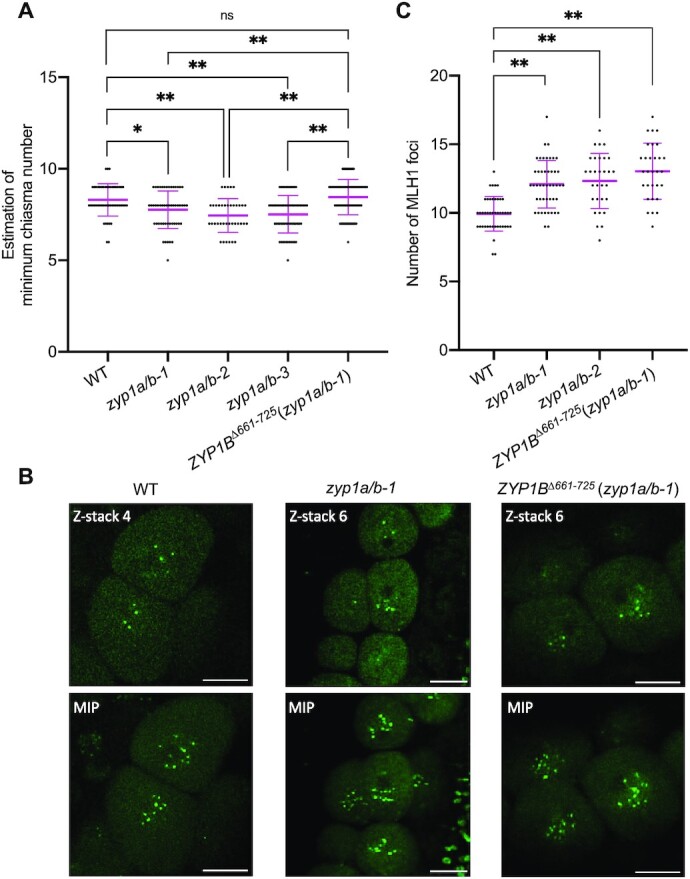
ZYP1 and ZYP1-mediated ASY1 removal regulate the formation of wild-type level of COs. (A) Scatter dot plot of the estimation of minimum chiasma number in WT, zyp1a/b-1, zyp1a/b-2, zyp1a/b-3 and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants. The statistical analysis was performed by one-way ANOVA with Tukey's correction for multiple comparison test. (B) Z-stack imaging of MLH1:GFP in WT, zyp1a/b-1 and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants at late prophase of male meiocytes. The upper panel shows one representative z-stack image for each genotype and lower panel depicts the maximum intensity projection (MIP) of z-stack images displaying all MLH1 foci from each meiocyte. Bars: 10 μm. (C) Scatter dot plot of the number of MLH1 foci in WT, zyp1a/b-1, zyp1a/b-2, and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants. The statistical analysis was performed by one-way ANOVA with Dunnett's correction for multiple comparison test. * and ** indicate significant differences at P < 0.05 and P < 0.01, respectively.
Previous reports showed that the number of type I COs increases in the absence of ZYP1, despite a reduced number of distinguishable chiasmata. This discrepancy can be explained by more closely located COs, as evidenced by sequencing, that cannot be resolved cytologically, for instance in asy1 mutants (36,41). To understand whether the elevation of type I COs in zyp1 mutants is due to the absence of ZYP1 per se or related to the defective ASY1 removal, we analyzed whether the ZYP1BΔ661–725-mediated formation of the SC, which fails to recruit PCH2 for ASY1 removal, would affect the number of type I COs in zyp1 mutants. To answer this, we decided to count the number of MLH1 foci present in late pachytene cells. MLH1 is one of the MutLγ endonucleases crucial for the resolution of CO intermediates and has been widely used in different organisms, including Arabidopsis, to estimate the number of type I COs (2). To achieve this, we employed a previously generated functional MLH1 reporter line (49) and introgressed it into zyp1a/b, and ZYP1BΔ661–725 (zyp1a/b-1) mutant plants for MLH1 foci counting. To detect all of the MLH1 foci and properly count them, z-stack images were acquired with 0.8 μm interval (Supplemental Movie 1 and 2). We found 9.93 ± 1.26 (n = 44 meiocytes) MLH1 foci per meiocyte in the wildtype (Figure 6B and C, Supplemental Movie 1), consistent with previous reports using an immunodetection method (21,26,50). Similar to the findings in (36,41), this number is slightly, yet significantly increased to 12.02 ± 1.79 (n = 46 meiocytes) and 12.33 ± 2.01 (n = 30 meiocytes) (one-way ANOVA with Dunnett's correction for multiple comparison, both P < 0.001) in zyp1a/b-1 and zyp1a/b-2 mutants, respectively (Figure 6B and C, Supplemental Movie 2). These data suggest that ZYP1 has a dual function in CO formation. On the one hand, ZYP1 assures the formation of at least one CO for each pair of homologs as indicated by the reduced number of chiasmata and enhanced occurrence of univalents in zyp1 a/b mutants. On the other hand, it limits the total number of type I COs as indicated by the elevated number of MLH1 foci in zyp1a/b.
To understand whether the ZYP1BΔ661–725-mediated formation of the SC, which fails to recruit PCH2 for ASY1 removal, would affect the number of type I COs in zyp1 mutants, we counted the MLH1 foci in ZYP1BΔ661–725 (zyp1a/b-1) plants using our MLH1:GFP reporter system. Interestingly, compared to the wildtype (9.93 ± 1.26, n = 44 meiocytes), we found that the number of MLH1 foci in ZYP1BΔ661–725 (zyp1a/b-1) plants also significantly increased to 12.93 ± 2.05 (n = 25 meiocytes, one-way ANOVA with Dunnett's correction for multiple comparison, P < 0.001), and displayed no significant difference from the zyp1a/b double mutants (Figure 6B and C, Supplemental Movie 3). While we cannot fully exclude the possibility that the precise number of type I COs is strongly dependent on the complete polymerization of ZYP1, this result suggests that the increased formation of type I COs in zyp1 mutants is likely not (at least not solely) attributed to the absence of ZYP1 per se. Instead, we speculate that the prolonged occupancy of ASY1 and/or the absence of PCH2 on paired chromosomes are probably decisive for the increase in recombination activity seen in zyp1 and ZYP1BΔ661–725 plants.
DISCUSSION
Developmental processes usually follow a defined order in space and time. Often these processes are unidirectional, preventing cells and tissues from being kept in futile loops. Meiosis is a paradigm for this biological organization principle, which avoids, for instance, that meiocytes are held in repeated cycles of DSB formation and their repair through COs. Important for the formation and placement of COs is the dynamic localization of the meiotic HORMADs, first present on the chromosome axis at early meiosis and subsequently removed as prophase progresses. The fact that this dynamic behavior is well conserved across many sexually reproducing organisms including yeast, mammals, and plants (17,26,27,51,52), suggests that it is key to meiosis. Here, we have revealed an elegant mechanism of how ASY1 assembly and disassembly can be temporally separated. With this, our work has also shed light on the question of what the biological relevance of this dynamic behavior of HORMADs is.
Previous studies have shown that the loading of meiotic HORMADs on the axis depends on their interacting partners, i.e. the coiled-coil axial core proteins such as ASY3/Red1 in Arabidopsis and yeast, respectively (21,53,54). These axial core proteins are proposed to bind via their closure motif to the HORMA domain of the meiotic HORMADs, thereby recruiting the HORMADs to the chromosome axis (25,55). However, HORMADs also have a closure motif that is hypothesized to binds to their own HORMA domain, thus leading to a so-called closed conformation and precluding the recruitment of HORMADs to the axis (25,51,56). The AAA+ ATPase PCH2 catalyzes the transition of HORMADs from a closed to an open conformation (56,57). The PCH2-mediated conformational change is thought to enable the nuclear targeting and chromosomal assembly of ASY1 in Arabidopsis resembling a similar functional interplay between PCH2 and Hop1 in yeast (25,29–31).
Puzzlingly, PCH2 also catalyzes the removal of ASY1 from the axis and we have shown here that the ASY1-dismantling activity of PCH2 is brought about by the installation of the SC, which directly recruits PCH2 via binding to the TF protein ZYP1. In contrast, COMET, the adaptor protein for PCH2, does not require ZYP1 for its localization at the axis and probably only ASY1 itself is responsible for the loading of COMET to the axis (Figure 2C) (30). Thus, despite a direct interaction between PCH2 and COMET, it seems that they do not mutually recruit each other to the SC. Instead, a bipartite loading process is at work that brings the two parts of the ASY1-dismantling complex together. By this mechanism, progression of meiosis is linked with the establishment of a new protein complex providing a means to the unidirectionality of meiosis. Other examples of this biological organization principle are the formation of the DNA replication complexes and the assembly of the spliceosome, for which the next step can only be executed if the preceding protein is correctly loaded and, at the same time, the binding of the following protein often causes the release of the previously bound factor (58,59).
It is tempting to speculate that ASY1 bound to the axis is sterically protected from an attack by freely diffusing PCH2 and only the incoming SC causes a conformational change of ASY1 and/or stabilizes the interaction between COMET and PCH2 to allow the efficient removal of ASY1 from the axis. An alternative, yet not necessarily mutually exclusive scenario is that an increased concentration of PCH2 on the SC, promoted by the installation of ZYP1, might trigger the removal of ASY1 by PCH2.
Orthologs of PCH2 in different organisms exhibit a similar SC-dependent localization pattern (32,33,43,44,46,60). In C. elegans, SYP1 (the ortholog of ZYP1) is required for the localization of PCH2 on paired chromosomes (43). Similarly, the accumulation of CRC1 (the ortholog of PCH2) on chromosomes is abolished in rice zep1 (the ortholog of ZYP1) mutants (44). Interestingly, a special allele of Zip1 in budding yeast, called zip1-4LA, in which four leucine residues (amino acids of 643, 650, 657 and 664) in the coiled-coil region close to the C-terminal end of Zip1 have been replaced by alanines, allows wild-type like synaptonemal complex formation, yet cells arrest prior to the first meiotic division in a Pch2-dependent manner (47). Subsequent work showed that Pch2 is not loaded to the SC in the zip1-4LA mutant (46), leading to the persistent presence of Hop1 on paired/synapsed chromosomes as seen here in the Arabidopsis ZYP1BΔ661–725 (zyp1a/b-1) plants. Thus, although a direct interaction between Pch2 and Zip1 has so far not been identified, it is tempting to speculate that this leucine-rich region in Zip1 is the docking platform for Pch2 in budding yeast.
Notably, the zip1-4LA mutation in budding yeast Zip1 is more N-terminally located than the here-identified binding site of PCH2 in ZYP1 of Arabidopsis. However, assuming an overall similar structure of the yeast and Arabidopsis transversal elements with the human SYCP1, for which a global structural model has been obtained (61), both PCH2 binding sites in ZYP1/Zip1 fall into a region where the C-terminal ends of two transversal filament dimers build an antiparallel tetrahelical bundle running along the chromosome axis (Figure 7A). Postulating an upright binding of two hexameric PCH2 wheels in the tetrameric bundle, which would be possible considering the dimensions based on the known structure of the PCH2 hexamer and the SYCP1 tetrahelical bundle, PCH2 in yeast and Arabidopsis may bind to the same topological position, assuming that the more N-terminal part of one transverse filament dimer interacts with the more C-terminal part of the other dimer in the antiparallel tetrahelical region (Figure 7B and C). We want to emphasize that only for humans, a structural understanding of SYCP1 has so far been developed and, although they show great similarity in function, TF proteins are largely not conserved at the sequence level. Thus, the scheme presented in Figure 7 is still highly speculative and needs further experimental support.
Figure 7.
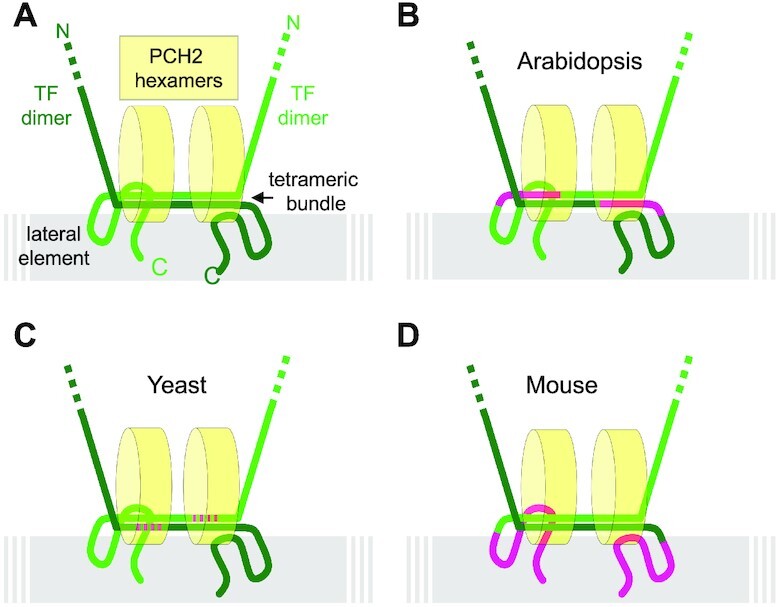
Model for a conserved recruitment mechanism of PCH2 to the SC. (A) Hypothetical generic model of PCH2 binding to the transversal filament that combines information from different organisms and assumes the formation of a similar sized tetrahelical bundle formed by the antiparallel oriented C-terminal ends of two transverse filament dimers, as published for human SYCP1 (57), also occurs in Arabidopsis, mouse and yeast. Two hexameric PCH2 wheels might bind through probably their N-termini to the region of the tetrahelical bundle parallel to the chromosome axis. Alternatively, one hexameric PCH2 wheel might lay flat on top of the tetrahelical bundle. (B–D) The localization of PCH2 at the SC is possibly conserved between diverse organisms although the domains in the transverse filament proteins making the main contact with PCH2 have slightly shifted in evolution. The domains found to be relevant for correct PCH2 binding are indicated in magenta for Arabidopsis (this study), mouse (this study) and Yeast (37).
None-the-less, we have shown here that TRIP13, the ortholog of PCH2 in mouse, also directly binds to the C-terminus of the murine ZYP1 homolog SYCP1 (Figure 3D, Supplemental Figure S3B). However, the binding site of TRIP13 in SYCP1, i.e. 821–993 aa, does not seem to be in the presumptive tetrahelical bundle region but to the C-terminal tail of the protein (61). Given the similarity of the anchoring of PCH2 via a transverse filament protein in Arabidopsis and yeast, we postulate that the C-terminal tail of SYCP1 in mouse might fold towards the tetrahelical bundle resulting in a conserved positioning of TRIP13 in mouse (Figure 7D). In summary, the existing data from yeast, mouse and Arabidopsis are compatible with a similar localization of PCH2/TRIP13 at the SC through binding to the C-terminal regions of TF proteins while in each case a different region in the C-terminus of the TF protein has likely evolved to facilitate binding.
Deleting the binding site of PCH2 in ZYP1 (ZYP1BΔ661–725) allowed us then to shed light on the question of which aspects of zyp1 mutants are related to a malfunctioning SC as opposed to secondary consequences, such as the failure to remove ASY1 and subsequent consequences. The appearance of univalents in zyp1 mutants, despite an increase in total number of COs, suggests that CO assurance is affected. The reason for the CO assurance defects of zyp1 is not clear yet but possibly could go back to the finding that, although chromosomes co-align in zyp1 mutants, the homologs are not as closely juxtaposed as in the wildtype (36,41), presumably destabilizing and/or reducing homolog interaction. The here-observed substantial restoration of the formation of the obligatory CO in ZYP1BΔ661–725 plants implies that defective CO assurance in zyp1 mutants is likely attribute to a failure of synapsis.
However, synapsis is not completely restored in ZYP1BΔ661–725 plants, a finding that could be accounted for by multiple, not mutually exclusive reasons: First, compared to the wild-type ZYP1, the depletion of 661–725 aa in ZYP1B could possibly result in a compromised and/or unstable SC structure and further studies are needed to resolve whether the deletion of the PCH2 binding site in ZYP1BΔ661–725 has additional (structural) consequences. Second, the compromised removal of ASY1 might provide a structural hindrance on the axes and hence, could interfere with the completion of SC formation. Previously, we found that ASY1 without its C-terminal closure motif, locking it in a constantly open state, could assemble normally on the axis but cannot be released from the axis at late prophase although PCH2 is very likely recruited to the SC (29). Notably, the presence of ASY1 without closure motif in the wildtype also results in a partially incomplete synapsis (29). Third, PCH2 might have a role in regulating synapsis, e.g. work in C.elegans has shown that PCH2 together with its co-factor CMT-1 (the ortholog of COMET) plays a role in proofreading homolog interactions to ensure synapsis fidelity (40).
An important aspect of zyp1 mutants in Arabidopsis is the increase in type I COs and the concomitant reduction of CO interference (36,41). However, in contrast to the large restoration of bivalent formation, expression of ZYP1BΔ661–725 does not reverse the elevation of type I COs in zyp1 mutants, suggesting that the ZYP1BΔ661–725 allele possibly has no or reduced activity in restricting CO formation. This shows that the CO assurance function of ZYP1 can be genetically separated from its role in CO interference.
Interestingly, the replacement of Zip1 in budding yeast with its ortholog from Kluyveromyces lactis resulted in a separation-of-function of Zip1 allele. This Zip1 allele was shown to be non-functional for SC formation while exhibiting high activity for promoting Mlh3-dependent CO formation independently of Msh4/Msh5. This finding suggested that the assembled SC acts to constrain MutLγ- and promote MutSγ-dependent CO formation during normal meiosis in S. cerevisiae (62). In addition, Pch2 in yeast was proposed to modulate CO interference by regulating the chromosome axis morphogenesis (60,63) Therefore, another possibility to explain the increase in COs in zyp1 and ZYP1BΔ661–725 plants could be the loss of PCH2 localization on SC that would be needed for remodelling of the chromosome axis and establishment of CO interference.
Yet another alternative is that removal of ASY1 represses the activity of DSB-forming complexes preventing excess DSBs and/or destabilizes the recombination machinery (3,23). Support for such a function comes from the recent finding that ASY1 directly interacts with several DSB proteins such as MTOPVIB, PRD2, and PRD3 (3), and possibly anchors these proteins to the axis. Moreover, loss of ASY1 shortens the residence time of the recombinase DMC1 on chromosomes at early meiosis (23), suggesting that ASY1 is likely involved in the stabilization and/or recruitment of (at least some of) the components of the recombination machinery. Therefore, the removal of ASY1 and possibly its homologs in other species represents another means of introducing unidirectionality by terminating meiotic recombination. It is then crucial that the re-association of ASY1 with the axis is prevented possibly by the sequestration of PCH2 to ZYP1 and a fast self-closing of ASY1 present in the nucleoplasm (29).
Notably, the finding that zyp1 mutants and ZYP1BΔ661–725 plants do not resemble the meiotic defects seen in pch2 mutants indicates that PCH2 has additional roles beyond HORMAD removal (26,28,64,65). Indeed, loss of Trip13 in mice has no effect on synapsis but reduces DSB repair efficiency (17,66). PCH2 in C. elegans promotes CO assurance independent of HORMAD removal (43). Pch2 in budding yeast is required for the meiotic recombination checkpoint independent of its accumulation on the SC and Hop1 release (31,33,46,47,67). Pch2 in budding yeast also regulates proper CO levels and distribution (63), but the connection to Hop1 dynamics remains to be explored.
Besides the necessity for synapsis and recombination (26), PCH2 might have also further roles in meiosis in Arabidopsis, such as the involvement in a meiotic checkpoint by monitoring synapsis and meiotic recombination. Indeed, it was recently shown that Arabidopsis and likely other plants also have a pachytene checkpoint (68). However, whether this checkpoint relies on PCH2 needs to be tested in future studies.
Taken together, PCH2/TRIP13 has acquired many, often species-specific roles in and outside of meiosis during evolution. Thus, a comparative approach studying PCH2’s function in many different species will be informative to reveal its ancient function and address principles of its action, e.g. to what extent the defects in meiotic recombination in pch2/trip13 mutants are attributed to a compromised release of meiotic HORMADs.
DATA AVAILABILITY
The data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Attila Toth (Technical University Dresden, Germany), Ihsan Dereli (Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany) and Mariana Motta as well as Jason Sims (both University of Hamburg, Germany) for the critical reading of the manuscript. We are grateful to Attila Toth for providing murine TRIP13 and SYCP1 cDNAs. C.Y. was supported by funds from the National Natural Science Foundation of China (32170354 to C.Y.) and Huazhong Agricultural University (101-11042110006 to C.Y.). C.Y. received a postdoc-first grant of the Department of Biology of the University of Hamburg. This work was further supported by funds from the University of Hamburg to A.S.
Author contributions: C.Y. conceived the research and designed the experiments. C.Y. performed most of the experiments. K.S., B.H. and M.B. carried out chromosome spreads, Y.H. performed the BiFC assays and additional microscopy work, H.T.E. conducted the Y2H analysis of murine proteins, L.C. did the split luciferase assay, D.S. performed the immunolocalization of PCH2 and ZYP1 in asy1 mutants. A.S. provided reagents and chemicals, C.Y., H.M. and A.S. analyzed the data. C.Y. and A.S. wrote the manuscript.
Notes
Present address: Kostika Sofroni, Department of Meiosis, Max Planck Institute for Multidisciplinary Sciences, 37077 Göttingen, Germany.
Contributor Information
Chao Yang, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Kostika Sofroni, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Yuki Hamamura, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Bingyan Hu, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Hasibe Tunçay Elbasi, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Martina Balboni, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Lei Chu, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China.
Dagmar Stang, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Maren Heese, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
Arp Schnittger, Department of Developmental Biology, Institute of Plant Science and Microbiology, University of Hamburg, Hamburg 22609, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [32170354 to C.Y.]; Huazhong Agricultural University [101-11042110006 to C.Y.]; Universität Hamburg [Departmental budget, Postdoc-first award to C.Y.]. Funding for open access charge: Departmental budget.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang Y., Copenhaver G.P.. Meiotic recombination: mixing it up in plants. Annu. Rev. Plant Biol. 2018; 69:577–609. [DOI] [PubMed] [Google Scholar]
- 2. Mercier R., Mezard C., Jenczewski E., Macaisne N., Grelon M.. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015; 66:297–327. [DOI] [PubMed] [Google Scholar]
- 3. Vrielynck N., Schneider K., Rodriguez M., Sims J., Chambon A., Hurel A., De Muyt A., Ronceret A., Krsicka O., Mézard C.et al.. Conservation and divergence of meiotic DNA double strand break forming mechanisms in Arabidopsis thaliana. Nucleic Acids Res. 2021; 49:9821–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001; 52:1–53. [DOI] [PubMed] [Google Scholar]
- 5. Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015; 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blat Y., Protacio R.U., Hunter N., Kleckner N.. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002; 111:791–802. [DOI] [PubMed] [Google Scholar]
- 7. Panizza S., Mendoza M.A., Berlinger M., Huang L., Nicolas A., Shirahige K., Klein F.. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011; 146:372–383. [DOI] [PubMed] [Google Scholar]
- 8. Zickler D., Kleckner N.. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999; 33:603–754. [DOI] [PubMed] [Google Scholar]
- 9. Zickler D., Kleckner N.. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015; 7:a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xue Z., Liu C., Shi W., Miao Y., Shen Y., Tang D., Li Y., You A., Xu Y., Chong K.et al.. OsMTOPVIB is required for meiotic bipolar spindle assembly. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:15967–15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hollingsworth N.M., Ponte L.. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997; 147:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M.. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005; 16:5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodyer W., Kaitna S., Couteau F., Ward J.D., Boulton S.J., Zetka M.. HTP-3 Links DSB Formation with Homolog Pairing and Crossing Over during C. elegans Meiosis. Dev. Cell. 2008; 14:263–274. [DOI] [PubMed] [Google Scholar]
- 14. Carballo J.A., Johnson A.L., Sedgwick S.G., Cha R.S.. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008; 132:758–770. [DOI] [PubMed] [Google Scholar]
- 15. Chambon A., West A., Vezon D., Horlow C., Muyt A.D., Chelysheva L., Ronceret A., Darbyshire A.R., Osman K., Heckmann S.et al.. Identification of ASYNAPTIC4, a component of the meiotic chromosome axis. Plant Physiol. 2018; 178:233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Armstrong S.J. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002; 115:3645–3655. [DOI] [PubMed] [Google Scholar]
- 17. Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C.R., Cooke H.J., Jasin M., Keeney S.et al.. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009; 5:e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hollingsworth N.M., Johnson A.D.. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. Genetics. 1993; 133:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukuda T., Daniel K., Wojtasz L., Toth A., Höög C.. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp. Cell Res. 2010; 316:158–171. [DOI] [PubMed] [Google Scholar]
- 20. Lammers J.H., Offenberg H.H., Aalderen M.van, Vink A.C., Dietrich A.J., Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell Biol. 1994; 14:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferdous M., Higgins J.D., Osman K., Lambing C., Roitinger E., Mechtler K., Armstrong S.J., Perry R., Pradillo M., Cunado N.et al.. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 2012; 8:e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lambing C., Tock A.J., Topp S.D., Choi K., Kuo P.C., Zhao X., Osman K., Higgins J.D., Franklin F.C.H., Henderson I.R.. Interacting genomic landscapes of REC8-cohesin, chromatin, and meiotic recombination in Arabidopsis. Plant Cell. 2020; 32:1218–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchez-Moran E., Santos J.L., Jones G.H., Franklin F.C.H.. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007; 21:2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daniel K., Lange J., Hached K., Fu J., Anastassiadis K., Roig I., Cooke H.J., Stewart A.F., Wassmann K., Jasin M.et al.. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat. Cell Biol. 2011; 13:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang C., Sofroni K., Wijnker E., Hamamura Y., Carstens L., Harashima H., Stolze S.C., Vezon D., Chelysheva L., Orban-Nemeth Z.et al.. The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. EMBO J. 2020; 39:e101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambing C., Osman K., Nuntasoontorn K., West A., Higgins J.D., Copenhaver G.P., Yang J., Armstrong S.J., Mechtler K., Roitinger E.et al.. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 2015; 11:e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen C., Jomaa A., Ortega J., Alani E.E.. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E44–E53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roig I., Dowdle J.A., Toth A., Rooij D.G.de, Jasin M., Keeney S. Mouse TRIP13/PCH2 Is Required for Recombination and Normal Higher-Order Chromosome Structure during Meiosis. PLoS Genet. 2010; 6:e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang C., Hu B., Portheine S.M., Chuenban P., Schnittger A.. State changes of the HORMA protein ASY1 are mediated by an interplay between its closure motif and PCH2. Nucleic Acids Res. 2020; 48:11521–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balboni M., Yang C., Komaki S., Brun J., Schnittger A.. COMET functions as a PCH2 cofactor in regulating the HORMA domain protein ASY1. Curr. Biol. 2020; 30:4113–4127. [DOI] [PubMed] [Google Scholar]
- 31. Herruzo E., Lago-Maciel A., Baztán S., Santos B., Carballo J.A., San-Segundo P.A.. Pch2 orchestrates the meiotic recombination checkpoint from the cytoplasm. PloS Genet. 2021; 17:e1009560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silva R.C.da, Vader G. Getting there: understanding the chromosomal recruitment of the AAA+ ATPase Pch2/TRIP13 during meiosis. Curr. Genet. 2021; 67:553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raina V.B., Vader G.. Homeostatic control of meiotic prophase checkpoint function by Pch2 and Hop1. Curr. Biol. 2020; 30:4413–4424.e5. [DOI] [PubMed] [Google Scholar]
- 34. Ma H.T., Poon R.Y.C.. TRIP13 functions in the establishment of the spindle assembly checkpoint by replenishing O-MAD2. Cell Rep. 2018; 22:1439–1450. [DOI] [PubMed] [Google Scholar]
- 35. Corbett K.D. Molecular Mechanisms of Spindle Assembly Checkpoint Activation and Silencing. Prog. Mol. Subcell. Biol. 2017; 56:429–455. [DOI] [PubMed] [Google Scholar]
- 36. Capilla-Pérez L., Durand S., Hurel A., Lian Q., Chambon A., Taochy C., Solier V., Grelon M., Mercier R.. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2023613118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C.H.. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005; 19:2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prusicki M.A., Keizer E.M., Rosmalen R.P.van, Komaki S., Seifert F., Muller K., Wijnker E., Fleck C., Schnittger A. Live cell imaging of meiosis in Arabidopsis thaliana. Elife. 2019; 8:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lambing C., Kuo P.C., Tock A.J., Topp S.D., Henderson I.R.. ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:13647–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giacopazzi S., Vong D., Devigne A., Bhalla N.. PCH-2 collaborates with CMT-1 to proofread meiotic homolog interactions. PloS Genet. 2020; 16:e1008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. France M.G., Enderle J., Röhrig S., Puchta H., Franklin F.C.H., Higgins J.D.. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2021671118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang C., Hamamura Y., Sofroni K., Bower F., Stolze S.C., Nakagami H., Schnittger A.. SWITCH 1/DYAD is a WINGS APART-LIKE antagonist that maintains sister chromatid cohesion in meiosis. Nat. Commun. 2019; 10:1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deshong A.J., Ye A.L., Lamelza P., Bhalla N.. A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 2014; 10:e1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miao C., Tang D., Zhang H., Wang M., Li Y., Tang S., Yu H., Gu M., Cheng Z.. CENTRAL REGION COMPONENT1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell. 2013; 25:2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Borner G.V., Barot A., Kleckner N.. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Subramanian V.V., MacQueen A.J., Vader G., Shinohara M., Sanchez A., Borde V., Shinohara A., Hochwagen A.. Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol. 2016; 14:e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mitra N., Roeder G.S.. A novel nonnull ZIP1 allele triggers meiotic arrest with synapsed chromosomes in Saccharomyces cerevisiae. Genetics. 2007; 176:773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Santos J.L., Alfaro D., Sanchez-Moran E., Armstrong S.J., Franklin F.C.H., Jones G.H.. Partial diploidization of meiosis in autotetraploid Arabidopsis thaliana. Genetics. 2003; 165:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pochon G., Henry I.M., Yang C., Lory N., Jiménez N.F.-, Böwer F., Hu B., Carstens L., Tsai H.T., Pradillo M.et al.. The Arabidopsis Hop1 homolog ASY1 mediates cross-over assurance and interference. 2022; bioRxiv doi:17 March 2022, preprint: not peer reviewed 10.1101/2022.03.17.484635. [DOI] [PMC free article] [PubMed]
- 50. Chelysheva L., Grandont L., Vrielynck N., Guin S.le, Mercier R., Grelon M.. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 2010; 129:143–153. [DOI] [PubMed] [Google Scholar]
- 51. Rosenberg S.C., Corbett K.D.. The multifaceted roles of the HORMA domain in cellular signaling. J. Cell Biol. 2015; 211:745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nonomura K.-I., Nakano M., Eiguchi M., Suzuki T., Kurata N.. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 2006; 119:217–225. [DOI] [PubMed] [Google Scholar]
- 53. Smith A.V., Roeder G.S.. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997; 136:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang F., Fuente R.D.L., Leu N.A., Baumann C., McLaughlin K.J., Wang P.J.. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 2006; 173:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. West A.M., Rosenberg S.C., Ur S.N., Lehmer M.K., Ye Q., Hagemann G., Caballero I., Uson I., MacQueen A.J., Herzog F.et al.. A conserved filamentous assembly underlies the structure of the meiotic chromosome axis. Elife. 2019; 8:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. West A.M.V., Komives E.A., Corbett K.D.. Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Res. 2018; 46:279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ye Q., Kim D.H., Dereli I., Rosenberg S.C., Hagemann G., Herzog F., Toth A., Cleveland D.W., Corbett K.D.. The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 2017; 36:2419–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siddiqui K., On K.F., Diffley J.F.X.. Regulating DNA Replication in Eukarya. Cold Spring Harb. Perspect. Biol. 2013; 5:a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joshi N., Barot A., Jamison C., Borner G.V.. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009; 5:e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dunce J.M., Dunne O.M., Ratcliff M., Millán C., Madgwick S., Usón I., Davies O.R.. Structural basis of meiotic chromosome synapsis through SYCP1 self-assembly. Nat. Struct. Mol. Biol. 2018; 25:557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Voelkel-Meiman K., Johnston C., Thappeta Y., Subramanian V.V., Hochwagen A., MacQueen A.J.. Separable crossover-promoting and crossover-constraining aspects of Zip1 activity during budding yeast meiosis. PLoS Genet. 2015; 11:e1005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zanders S., Alani E.. The pch2Δ mutation in Baker's yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 2009; 5:e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cuacos M., Lambing C., Pachon-Penalba M., Osman K., Armstrong S.J., Henderson I.R., Sanchez-Moran E., Franklin F.C.H., Heckmann S.. Meiotic chromosome axis remodelling is critical for meiotic recombination in Brassica rapa. J. Exp. Bot. 2021; 72:3012–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. San-Segundo P.A., Roeder G.S.. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999; 97:313–324. [DOI] [PubMed] [Google Scholar]
- 66. Li X.C., Li X., Schimenti J.C.. Mouse pachytene checkpoint 2 (Trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007; 3:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. San-Segundo P.A., Roeder G.S.. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999; 97:313–324. [DOI] [PubMed] [Google Scholar]
- 68. Jaeger-Braet J.D., Krause L., Buchholz A., Schnittger A.. Heat stress reveals a specialized variant of the pachytene checkpoint in meiosis of Arabidopsis thaliana. Plant Cell. 2021; 34:433–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.