Abstract
The glucocorticoid receptor (GR) is a ubiquitously expressed transcription factor that controls metabolic and homeostatic processes essential for life. Although numerous crystal structures of the GR ligand-binding domain (GR-LBD) have been reported, the functional oligomeric state of the full-length receptor, which is essential for its transcriptional activity, remains disputed. Here we present five new crystal structures of agonist-bound GR-LBD, along with a thorough analysis of previous structural work. We identify four distinct homodimerization interfaces on the GR-LBD surface, which can associate into 20 topologically different homodimers. Biologically relevant homodimers were identified by studying a battery of GR point mutants including crosslinking assays in solution, quantitative fluorescence microscopy in living cells, and transcriptomic analyses. Our results highlight the relevance of non-canonical dimerization modes for GR, especially of contacts made by loop L1–3 residues such as Tyr545. Our work illustrates the unique flexibility of GR’s LBD and suggests different dimeric conformations within cells. In addition, we unveil pathophysiologically relevant quaternary assemblies of the receptor with important implications for glucocorticoid action and drug design.
INTRODUCTION
Nuclear receptors (NRs) are transcription factors that control central physiological processes ranging from reproduction and development to metabolism, homeostasis, and ultradian rhythms (1,2). Steroid receptors are a NR subclass that includes the glucocorticoid receptor (GR/NR3C1) (Figure 1A), the androgen receptor (AR/NR3C4), the progesterone receptor (PR/NR3C3), the mineralocorticoid receptor (MR/NR3C2), as well as estrogen receptors α and β (ERα/NR3A1 and ERβ/NR3A2) (3–5). NRs display a modular architecture with an unstructured N-terminal domain (NTD) followed by a ‘core’ formed by a highly conserved DNA-binding domain (DBD), an interdomain linker or hinge, and a C-terminal ligand-binding domain (LBD) (Figures 1A and Supplementary Figure S1) (6–8). GR binds glucocorticoids (GCs; either endogenous such as cortisol, or synthetic, e.g. dexamethasone (DEX)) in an internal cavity of the LBD. This ligand-binding pocket (LBP) is allosterically coupled to activation function 2 (AF-2), a pocket that binds coregulators (9,10). A nearby cavity, topologically equivalent to AR binding function-3 (BF-3) interacts with cochaperones (11,12). Finally, the LBDs of GR and the related oxosteroid receptors (AR, PR and MR) feature a unique C-terminal extension termed F-domain (5,13) (Figure 1B).
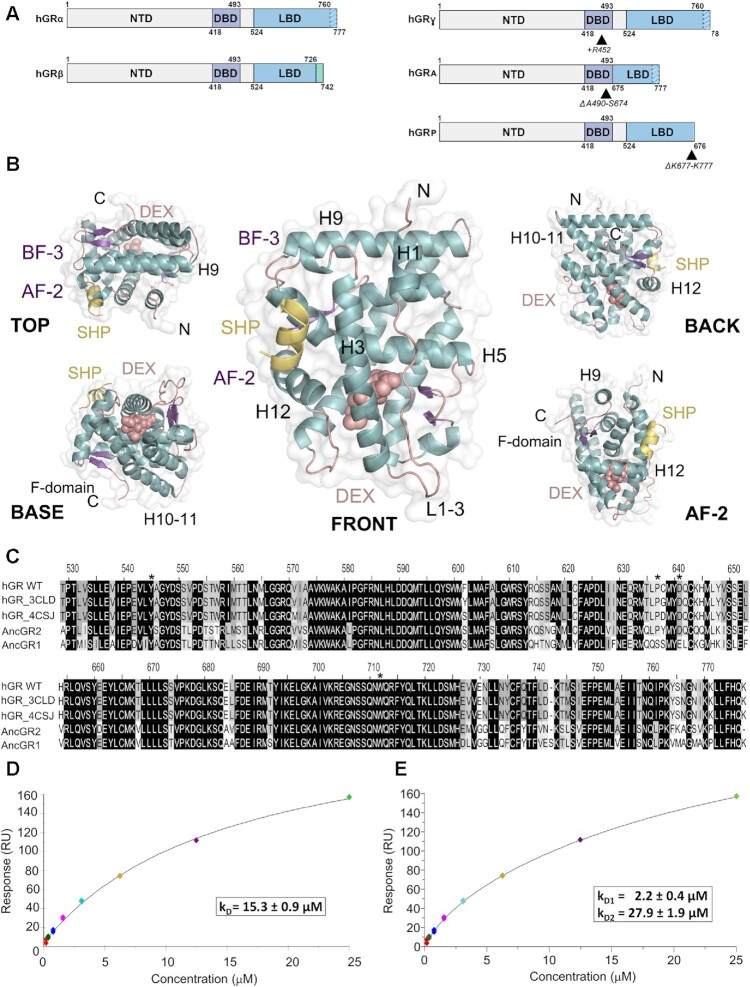
Figure 1.
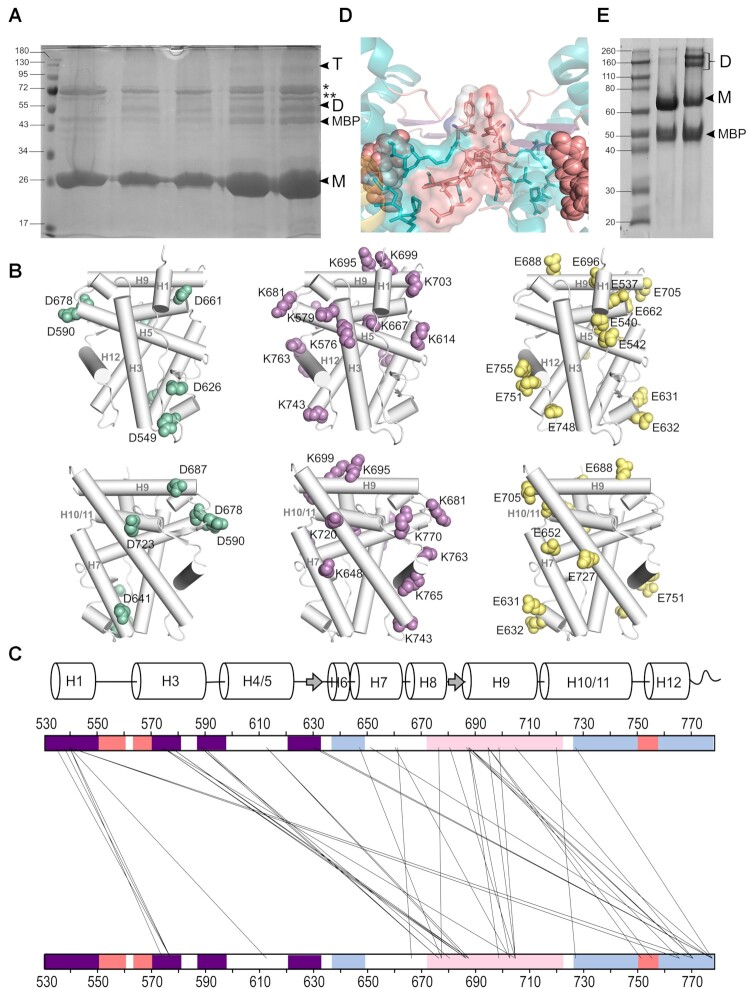
The ligand-binding domain of GR self-associates in solution. (A) Schematic representation of GR domain organization. GRα and GRβ are identical up to residue 727 (H10/11), but the last 50 (in GRα) and 15 residues (in GRβ, green box) are unrelated. Other GR isoforms are shown (right). (B) Overall structure of the GR-LBD monomer. The module is shown in standard orientation in the middle of the panel (i.e. with H1 and H3 displayed facing the viewer and the AF-2 pocket on the left). Four additional views are shown to present other LBD areas. Models are depicted as cartoons with helices (blue), loops (pink) and beta-sheets (purple). DEX (salmon spheres) and SHP peptide (yellow cartoon) are shown. The BF-3 pocket is also labeled. (C) Sequence alignment of LBDs between WT human GR, two engineered variants used in several structures (PDBs 3CLD and 4CSJ), and the ancGR1 and 2 forms. Strictly conserved residues are white with black shading; other conservatively replaced residues are shaded gray. Residues mutated in the current study are marked with asterisks. (D, E) SPR analysis of GR-LBD self-association according to (D) 1:1 or (E) multisite models. The results of experiments conducted in duplicate are shown along with the calculated affinity constants (kD). Data were fitted to the 1:1 and multisite models with χ2 values of 4.52 and 1.25, respectively.
NR3C1 is constitutively expressed in nearly all vertebrate cells. Upon ligand binding, GR translocates to the nucleus (14) where it binds to chromatin (15) and integrates signals ranging from available ligands to chromatin remodeling complexes (16) to control cognate target genes (up to 17% of the human transcriptome (17)) to regulate inflammation, cell proliferation, and differentiation in a tissue-specific manner (18–20). GR also antagonizes the activity of other transcription factors such as activator protein 1 (AP-1) and nuclear factor kappaB (NF-κB) (21). Even though monomeric GR is believed to play a DNA-independent role in modulating inflammation (22), more recent work suggests the relevance also of both direct DNA binding of GR dimers/tetramers (23–28). In line with these manifold activities, alterations in GR signaling pathways due to mutations in NR3C1 lead to impaired sensitivity to GCs, which may manifest as either GC resistance (Chrousos syndrome) or hypersensitivity (29). GR is thus an important pharmacological target to treat inflammatory diseases, but chronic GC use results in drug resistance and side effects (16).
Structures of the DBD dimer, both free and DNA-bound have been solved (30–33), and the LBD has been extensively studied in complex with agonists or antagonists (3,15,34–37). To date, however, no structure of full-length (FL) GR has been solved. Thus, several key issues regarding GR tertiary and quaternary structure remain unresolved: what is the conformation adopted by dimeric GR and DNA-bound tetramers in vivo, and how do LBDs associate in these multimers? Are topologically distinct GR conformations possible, and are they linked to specific biological functions (e.g. activation vs. repression of transcription)? Knowledge of the oligomeric conformations of oxosteroid receptors would aid in the design of novel selective, potent GR modulators that may lessen side effects of current drugs.
Here we present a systematic structure-and-function investigation of GR multimerization using X-ray crystallography, state-of-the-art bioinformatics tools, surface plasmon resonance (SPR) and crosslinking experiments in solution, quantitative fluorescence microscopy in live cells, and RNA-seq analysis of relevant GR variants. We report five new structures of DEX-bound GR-LBD (GR-LBD·DEX) and integrate this information into the wealth of previous structural data to generate the most thorough catalog to date of possible GR homodimers. Four distinct interfaces have been observed to participate in 20 topologically different GR-LBD homodimers. We have identified the most favored GR homodimers and suggest how they can combine into pathophysiologically relevant oligomeric assemblies in cells.
MATERIALS AND METHODS
Peptides and proteins
A peptide corresponding to residues Gln18-Lys27 of the small heterodimer partner (SHP/NR0B2, box 1 motif; NH2-Q18GAASRPAILYALLSSSLK27-OH) was custom-synthesized at Pepmic. Recombinant ancGR2-LBD (corresponding to residues 529–777 of the human receptor) cloned into a pMALCH10T vector was expressed as fusion protein with an N-terminal maltose-binding protein (MBP) and a hexahistidine tag and purified to homogeneity using standard chromatographic procedures (38).
Crystallization and structure determination
Purified, concentrated DEX-bound ancGR2-LBD was combined with a 3-fold molar excess of SHP peptide and incubated for one hour at RT. Drops of the ancGR2-LBD-SHP mixture were equilibrated against 0.1 M Tris–HCl, pH 8.0, 0.2 M sodium chloride, 2.0 M ammonium sulfate (P31, I4122 and I4132 crystals); 0.1 M PIPES, pH 7.0, 0.1 M ammonium acetate, 2.5 M sodium formate (P61 crystals); or 85 mM sodium cacodylate trihydrate, pH 6.5, 0.17 M sodium acetate trihydrate, 25.5% (w/v) PEG8000, 15% (v/v) glycerol (C2 crystals) using the sitting drop vapor-diffusion method. Diffraction data were collected at 100 K at the ALBA-CELLS synchrotron and processed using MOSFLM (http://www.mrc-lmb.cam.ac.uk/harry/mosflm/) and CCP4 (http://www.ccp4.ac.uk/). The crystal structures were solved and refined using MOLREP, REFMAC5 and COOT from the CCP4 package. Crystal packing was analyzed using PISA (http://www.ebi.ac.uk/), model quality was assessed with MolProbity (http://molprobity.biochem.duke.edu/), and figures were prepared with PyMOL (http://www.pymol.org).
Surface plasmon resonance (SPR) analyses
SPR analyses were performed at 25°C in a BIAcore T200 instrument (GE Healthcare). Highly purified, DEX-bound recombinant WT ancGR2-LBD and its Y545C and Y545A mutants were diluted in 10 mM sodium acetate, pH 5.0, and directly immobilized on CM5 chips (GE Healthcare) by amine coupling at densities between 300 and 400 resonance units (RU). As a reference, one of the channels was also amine-activated and blocked in the absence of protein. The running buffer was 50 mM HEPES, pH 7.2, 50 mM Li2SO4, 5% glycerol, 5 μM DEX, supplemented with various concentrations of dithiothreitol (DTT). Sensorgrams were analyzed with the BIAcore T200 Evaluation software 3.0 and fitted according to the Langmuir 1:1 and multisite models.
Crosslinking experiments
Purified recombinant ancGR2-LBD (33 μM) was incubated with 100-fold molar excess of crosslinkers 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, Pierce) or disuccinimidyl dibutyric acid (DSBU) for 1–2 h at 37°C following the manufacturer's instructions. In some experiments the Y545C variant was incubated after affinity purification for 30 min at room temperature without further treatment. Samples of the reaction mixtures were boiled in the presence of Laemmli sample buffer, either reducing (EDC- and DSBU-crosslinked proteins) or non-reducing (in the case of the Y545C variant) and resolved by SDS-PAGE.
Nano-LC–MS/MS mass spectrometry
CBB-stained bands corresponding to monomeric, dimeric and tetrameric GR-LBD after crosslinking with EDC or DSBU were excised from the gels and subjected to in-gel digestion following standard protocols. Briefly, excised bands were reduced (10 mM DTT) in 50 mM bicarbonate buffer, pH 8.0, for 45 min at 56°C, alkylated (50 mM iodoacetamide in 50 mM ammonium bicarbonate buffer for 30 min at 25°C) and digested with either trypsin alone or followed by GluC treatment, or with chymotrypsin overnight at 37°C in 100 mM ammonium acetate buffer, pH 8.0 (sequencing-grade endoproteases were from Promega). In the case of the Y545C mutant, proteins in the excised bands were directly alkylated without previous DTT treatment to prevent reduction of the Cys545–Cys545’ disulfide bridge.
Tryptic peptides were diluted in 1% formic acid and loaded onto a 180 μm × 20 mm C18 Symmetry trap column (Waters) at a flow rate of 15 μl/min using a nanoAcquity Ultra Performance LCTM chromatographic system (Waters). Peptides were separated using a C18 analytical column (BEH130 C18, 75 mm × 25 cm, 1.7 μm; Waters) with a 120-min run, comprising three consecutive linear gradients: from 1 to 35% B in 100 min, from 35 to 50% B in 10 min, and from 50 to 85% B in 10 min (A = 0.1% formic acid in water, B = 0.1% formic acid in CH3CN). The column outlet was directly connected to an Advion TriVersa NanoMate (Advion) fitted on an LTQ-FT Ultra mass spectrometer (Thermo), which was operated in positive mode using the data-dependent acquisition mode. Survey MS scans were acquired in the FT-ICR cell with the resolution (defined at 400 m/z) set to 100,000. Up to six of the most intense ions per scan were fragmented and detected in the linear ion trap. The ion count target value was 1,000,000 for the survey scan and 50,000 for the MS/MS scan. Target ions already selected for MS/MS were dynamically excluded for 30 s. Spray voltage in the NanoMate source was set to 1.70 kV. Capillary voltage and tube lens on the LTQ-FT were tuned to 40 and 120 V, respectively. The minimum signal required to trigger MS to MS/MS switch was set to 1,000 and activation Q value was set at 0.25. Singly charged precursors were rejected for fragmentation.
Differential scanning fluorimetry
Thermofluor experiments were performed in an iQ5 Multicolor Real Time PCR Detection System (BIO-RAD) using 96-well plates (Hard-Shell® High-Profile Semi-Skirted PCR Plate, BIO-RAD) and a 25 μl total volume for each reaction. Melting curves were acquired from eight replicates to determine the average melting temperature (Tm). GR-LBD samples (0.5 mg/mL) were prepared in 20 mM HEPES, pH 8.0, 200 mM NaCl, 10% glycerol, 50 mM imidazole, 1 mM DTT, 50 μM DEX, and centrifuged 5 min. at 14,000 rpm immediately before measurements. SYPRO Orange dye (Sigma-Aldrich) was firstly prepared at 80× in the same buffer, starting from a 5,000× commercial dilution. The final concentration of the dye in each well was 5×. The plates were sealed with optical-quality sealing film (Microseal® B Seals, BIO-RAD) and centrifuged at 2,000 × g for 30 s. Samples were equilibrated for 60 s and analyzed using a linear gradient from 16 to 95°C with increments of 1°C/min, recording the SYPRO orange fluorescence throughout the gradient using the iQ5 Optical System Software 2.0. Values were fitted using the online tool JTSA with the four-parameter logistic equation, and the calculated fluorescence midpoints were compared with an unpaired t-test for equal variances using GraphPad Prism 8.
Cell line generation and culture
Mammary adenocarcinoma 3617-derived GRKO cells for number and brightness assays (N&B) were transiently transfected with the various forms of mouse GR (WT, W718A, Y551A, Y551A-Dim, P643A, D647V, P643A-Tetra, D647V-Tetra) using jetOPTIMUS™ reagent (PolyPlus) according to the manufacturer's instructions. The cell lines used for total RNA-sequencing (RNA-Seq) experiments were developed in the same GRKO cell line (25,39). Briefly, GFP-tagged versions of mouse GR (WT, Y551A, Y551A-Dim, D647V, D647V-Tetra) were stably integrated into the GT-Rosa26 locus via Crispr Cas9/homology directed repair with puromycin selection and FACS sorting. Cells for the RNA-Seq and N&B assays were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 5 μg/ml tetracycline (Sigma), 10% fetal bovine serum (Sigma), sodium pyruvate, non-essential amino acids, and 2 mM l-glutamine maintained in a humidifier at 37°C. The FBS-supplemented media was replaced with media supplemented with charcoal/dextran-treated FBS (CSS) to remove glucocorticoids from cells for 24 h prior to hormone treatment (100 nM DEX). All variants of GFP-GR for RNA-Seq and N&B were generated with the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene).
Number and brightness (N&B) analysis
Images were taken at the CCR, LRBGE Optical Microscopy Core facility in a LSM 780 laser scanning microscope (Carl Zeiss, Inc.) equipped with an environmental chamber. Cells were imaged from 20 min up to a maximum of 2 h after DEX addition. We used a 63× oil immersion objective (NA = 1.4). The excitation source was a multi-line Ar laser tuned at 488 nm. Fluorescence was detected with a GaAsP detector in photon-counting mode.
N&B measurements were done as previously described (24). For each studied cell, a single-plane stack of 150 images (256 × 256 pixels) were taken with a pixel size of 80 nm and the pixel dwell time of 6.3 μs. In all stacks, we discarded the first 10 images to reduce the effect of image bleaching on the results. The frame time under these conditions is 0.97 s, which guarantees independent sampling of molecules according to previously reported FCS measurements (40). Each stack was further analyzed using the N&B routine of the SIMFCS 2.0 software (Global Dynamics). In this routine, the average fluorescence intensity (<I>) and its variance (σ2) at each pixel of an image are determined from the intensity values obtained at the given pixel along the image stack. The apparent brightness (B) is calculated as the ratio of σ2 to <I> while the apparent number of moving particles (N) corresponds to the ratio of <I> to B (41). We used SIMFCS 2.0 software to classify pixels as occurring in the nucleus or the MMTV array (42) according to their intensity values. Selection of cells for analysis followed these criteria. (i) In the case of stimulated cells, an accumulation of signal at the MMTV array must be visible. (ii) The average apparent number of molecules (N) in the nuclear compartment must have a range of 3–30 units in all cases, (iii) no saturation of the detector at any pixel (N < 60), and (iv) bleaching cannot exceed 5–10%. In a previous work it has been demonstrated that B is equal to the real brightness ϵ of the particles plus one (41). Therefore, ϵ at every pixel of images can be easily extracted from B measurements. Importantly, this analysis only provides information regarding the moving or fluctuating fluorescent molecules since fixed molecules (relative to our frame time) will give B values equal to 1. The experiments were independently repeated two times for each treatment/condition.
Total RNA collection and sequencing
RNA isolations were performed using the PureLink RNA kit (Thermo Fisher Scientific 12183018A) as per the manufacturer's instructions. We collected three biological replicates of each condition and sample quality was assessed using the Agilent Bioanalyzer. Strand-specific sequencing libraries were generated from rRNA-depleted (Illumina RS-122-2301) total-RNA samples, using Illumina stranded total RNA (Illumina20020596) according to the manufacturer's instructions. Raw reads were demultiplexed into Fastq format allowing up to one mismatch using Illumina Bcl2fastq v2.17. Reads of the samples were trimmed for adapters and low-quality bases using Cutadapt1.18. RNA-Seq alignment to the mouse mm10 genome was performed by STAR (43). DESEQ2 (44) was used to normalize the data by read depth, identify differentially expressed genes for each form of GR (DEX/vehicle) and calculate log2 fold changes and false discovery rates (FDR) for each gene. RNA-Seq analyses was performed in two batches that were sequenced and analyzed separately: (i) GFP-GR WT (batch1), GFP-GR-Y551A, GFP-GRdim-Y551A; (ii) GFP-GR WT (batch2), GFP-GR-D647V, GFP-GRtetra-D647V. The two sets of GR WT RNA-Seq data are biological replicates collected and sequenced separately with each batch.
Impact of GR point mutations on protein folding/stability
We estimated the impact of the generated mutations on the overall protein stability with the FoldX empirical force (http://foldxsuite.crg.eu/), which has an estimated error of ∼0.7 kcal/mol. Ten iterations were conducted for each mutation, and later averaged. Free energy differences between mutant and WT proteins (ΔΔG) < 1 kcal/mol were considered not significant, those between 1 and 2, 2 and 4, and >4 kcal/mol as slightly, mildly, and strongly destabilizing, respectively.
Sequence conservation analyses
The sequence of human GR-LBD was used as query against the whole NR database, from which we selected ∼880 sequences and included the ancestral GR (38). We also downloaded the sequences corresponding to the LBD regions from a representative fraction of proteomes at PFAM rp55 (PF00104_rp55). (Note that the sequences included in the PFAM alignment are truncated, as they lack for instance the non-conserved F-domain). We followed three different approaches to ensure sequence and alignment diversity and thus stability of the analyses. First, we aligned the ∼880 sequences to a structure-based profile from entries 5UFS (GR-LBD) and 5JJM (AR-LBD). The resulting alignment, 880_aln, was used to run pySCA in addition to the original SCA5 method. Secondly, we aligned our ∼880 sequences to a profile generated from the PF00104_rp55 removing fragments to generate the 840_aln. Finally, we used the PFAM alignment as retrieved from the PFAM database, which contains ∼13,000 sequences (PF00104_rp55).
Multiple correspondence analysis (MCA): identifying specificity-determining positions
We have performed both supervised and unsupervised runs of the S3DET method (http://csbg.cnb.csic.es/JDet/) on 840_aln using default parameters to maintain a large sequence diversity.
Statistical coupling analysis (SCA)
We have used the SCA5 8/2011 version with the three different alignment versions given above, and the updated version pySCA with 880_aln. Each alignment produced unique sets of residues termed Sca5_880, Sca5_840 and Sca5_onPFAMrp55, respectively. Next, we labeled residues in the three different sectors that emerged as outputs of the program with letters A, B and C, following their order of appearance. The stability of the identified sectors was assessed with a statistical test based on hypergeometric calculations of the groups of residues belonging to given sectors between pairs of alignments. P-values were adjusted using FDR. Next, specific residues from the significant sectors were extracted and selected according to their rank. For instance, if a particular residue appears only in one sector on a low-ranking pair of alignments (e.g. rank 19, with a borderline P-value) this residue will not be selected as part of a sector. On the contrary, if a residue appears in high-ranking pairs, it will be retained. Residues termed as ‘A’ and ‘B’ appeared to be equivalent in different pairs of alignments, so they were assigned to the class sector 2, while residues belonging to the ‘C’ group were stable, and thus assigned to class sector 1.
Clustering of interaction surfaces in GR-LBD dimers
We grouped the interaction surfaces observed in the 20 topologically different GR-LBD homodimers by a hierarchical clustering analysis, using ad-hoc R scripts. For each dimer, the interaction surface was defined as the set of solvent-exposed residues in the monomers (i.e. residues with >25% relative accessible surface area, ASA) that became buried (<25% relative ASA) in the corresponding dimer. Relative ASA were calculated using ICM (Molsoft LLC). Then, we calculated the center of coordinates of the residues forming each interaction interface. To compare two pairs of homodimers, we computed the Euclidean distances between the centers of their interaction surfaces after superimposition on a common monomer. The 40 × 40 distance matrix representing the distances between all pairs of interaction surfaces was used to perform hierarchical clustering with Ward's method. Finally, the dendrogram generated from this analysis was sorted in order of increasing distance.
Docking experiments and analysis
Homodimeric models of GR-LBD were built using pyDock docking and scoring method (45). First, protein models were prepared by removing all cofactors and heteroatoms, and missing side chains were modeled with SCWRL 3.0 (46). Then, the Fast Fourier Transform (FFT)-based docking programs FTDock (47) (with electrostatics and 0.7-Å grid resolution) and ZDOCK 2.1 (48) were used to generate 10,000 and 2,000 rigid-body docking poses, respectively. These were merged in a single pool for subsequent pyDock scoring, based on energy terms previously optimized for rigid-body docking. The pyDock binding energy is basically composed of ASA-based desolvation, Coulombic electrostatics, and Van der Waals (VdW) energy terms. Electrostatics and VdW contributions were limited to –1.0/+1.0 and 1.0 kcal/mol for each inter-atomic energy value, respectively, to avoid excessive penalization from possible clashes derived from the rigid-body approach.
Predicted dimer interfaces
Optimal docking areas (ODA) per surface-exposed protein residues were obtained by computing surface patches with optimal desolvation energy based on the selection of low-energy docking regions generated from each surface residue (49). ODA hot spots (residues with low ODA values, usually less than –10.0 kcal/mol) indicate regions with favorable desolvation energy upon interaction with a partner protein.
From the resulting docking poses, normalized interface propensity (NIP) values were obtained for each residue with the built-in patch module of pyDock, implementing the pyDockNIP algorithm (50). A NIP value of 1 indicates that the corresponding residue is involved in all predicted interfaces of the 100 lowest energy docking solutions, while a value of 0 means that it appears as expected by random chance. Finally, a negative NIP value implies that the residue appears at the low-energy docking interfaces less often than expected by random chance. Usually, residues with NIP ≥ 0.2 are considered as hot-spot residues when using FTDock.
Energetic characterization of GR dimers
The binding energy of the different crystal dimers was computed with the pyDock bindEy module, using the same scoring function as in docking.
RESULTS
GR-LBD self-associates in solution
To characterize GR-LBD behavior in solution, we performed SPR experiments with the ancient variant of the human GR (ancGR2; Figure 1C). This construct has been repeatedly used in GR structure-function studies because of its higher solubility and stability in vitro (7). For simplicity, we will refer to all variants of the receptor as GR-LBD, unless otherwise specified. Briefly, purified GR-LBD·DEX was expressed and purified, immobilized on CM5 chips using amine coupling and increasing concentrations of the same agonist-bound protein (between 0.2 and 25 μM) were run over as analyte. Although GR-LBD immobilization to the CM5 chip might occlude some interaction surfaces, SPR experiments clearly demonstrate interactions between soluble and immobilized GR. Several affinity models were used to interpret SPR data (Figure 1D, E and Supplementary Figure S2A, B). GR-LBD self-association behavior could be fitted to a non-covalent, 1:1 Langmuir model with an affinity constant (kD) of 15.3 ± 0.9 μM (Figure 1D). Interestingly, we obtained a statistically significant (F-test, P-value = 0.001) better fit of the data by using a model with non-covalent multisite interactions, with two independent binding sites (kD1 = 2.2 ± 0.4 μM and kD2 = 27.9 ± 1.9 μM, respectively; Figure 1E). These results are consistent with GR-LBD tetramer formation, as previously reported for FL-GR in living cells (24).
Novel crystal structures of agonist-bound GR-LBD highlight its versatility for self-association
Next, we performed crystallization trials with GR-LBD·DEX in the presence of the AF-2 targeting peptide Gln12-Lys30 from SHP. Coregulator peptides are usually added to induce the active conformation of H12 forming the AF-2 pocket (38). We conducted solubility screens using all commercially available kits (over 4,800 conditions) to identify several unreported crystallization conditions. Diffraction data from crystals that belong to five different space groups (C2, P31, P61, I4122 and I4132, from lower to higher symmetry) were collected using synchrotron radiation. The major features of the inter-monomer contacts observed in these new GR-LBD structures are shown in Figure 2 and Supplementary Table S1.
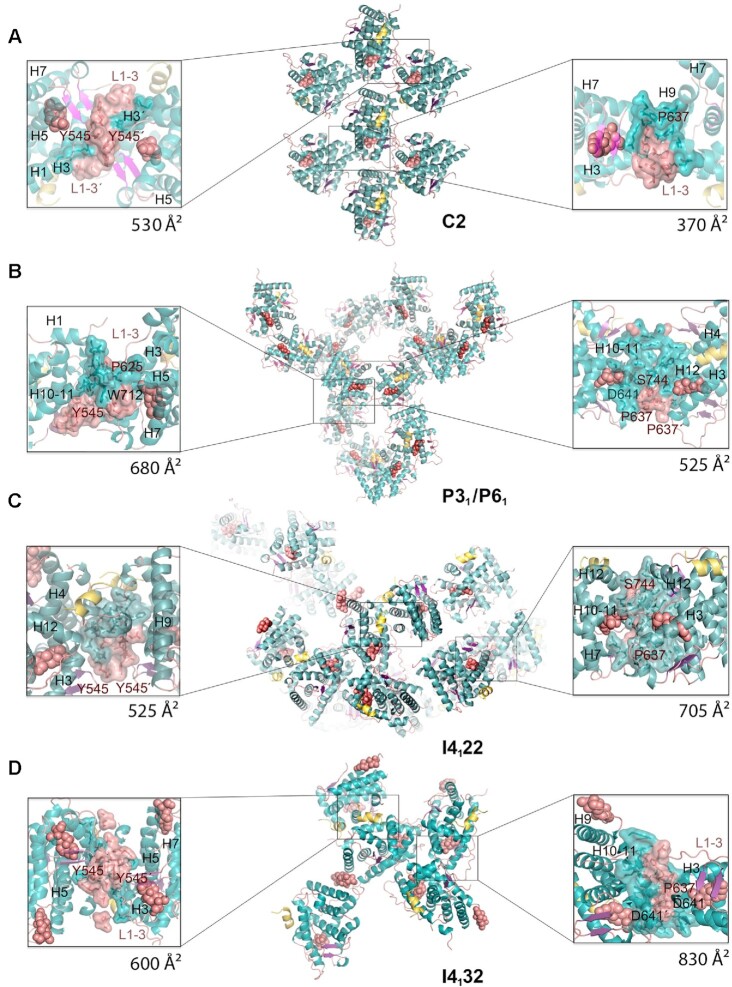
Figure 2.
New crystal structures of GR-LBD·DEX reveal a variety of quaternary assemblies. For all structures, the overall crystal packing is shown in the central panels. Monomers are depicted as cartoons with helices colored blue and loops colored salmon. DEX molecules are represented as salmon spheres and SHP peptides as yellow ribbons. Details of intermonomer interfaces are given in the lateral panels, in which the side chains of interacting residues are shown as sticks. (A) C2 crystals. Note that major contacts are centered on L1–3 with stacked Tyr545 phenol rings from two neighboring molecules. A monoclinic structure of ancGR2-LBD bound to another synthetic GC, triamcinolone acetonide, and complexed to a shorter SHP peptide had been previously reported (5UFS; (38)). Interestingly, 5UFS features a Tyr545-centered parallel dimer almost identical to the topologically equivalent arrangement in our current C2 crystals. (B) Related P31 and P61 crystals. Tyr545 engages in heterologous contacts with a neighboring molecule in these crystals (see the position of the Trp712’ side chain). Structures of ancGR2-LBD bound to either DEX or a different GC (mometasone furoate) and complexed to a TIF-2 peptide had been previously reported in a similar hexagonal crystal form (3GN8 and 4E2J; a and b axes are ∼5% longer in our crystals, while the c axis is ∼4% shorter). These relatively small differences in the cell constants compared to the current P61 structure result in a markedly different small intermonomer interface, however, which is asymmetric in 3GN8/4E2J. (C, D) Common packing of I4122 and I4132 crystals. Note that the phenol rings of two Tyr545 residues stack as in the C2 crystals, although the two interacting monomers are fully differently oriented relative to each other. Note also that the largest interface in these crystals features abutting Asp641 side chains from three monomers organized around a local (I4122) or exact 3-fold axis (I4132; right side panel in D).
C2 crystals contain a single molecule of GR-LBD·DEX complexed with the SHP peptide, which is well defined by electron density. Two different inter-monomer contacts were identified: the larger, symmetric interaction surface is centered on the L1–3 loops of both monomers and is stabilized by aromatic π-stacking interactions between opposite Tyr545/Tyr545’ residues (Figure 2A; residues from the second monomer are primed). Further stability is provided by a network of hydrogen bonds (H-bonds), involving most notably Asp549 (L1–3), Arg569 (H3), and Asp626 (β-strand S1). A significantly smaller, asymmetric interface features Glu688 (H9), whose carboxylate engages in strong H-bonds with the main chain N atom and the hydroxyl of Ser556’ (L1–3). Additional interactions involve H1 (Leu532) and H9 residues (Lys695, Lys699) from one LBD molecule facing H6 (Glu632’) from the neighbor. Residue Pro637’ (L5–6) is part of this interface, making strong VdW interactions with Glu688.
Two additional, related structures were solved in the enantiomorphic trigonal and hexagonal space groups, P31 and P61 (Figure 2B and Supplementary Table S1). In the GR-LBD·DEX homodimer with the larger interface, the cleft between H9 and H10–11 is filled with side chains from neighboring L1–3’ and L5–6’ loops. In particular, the aromatic side chains of Trp712 and Phe715 dock into a shallow groove formed by residues at the C-terminal end of H5 and the following loop. This arrangement is thus topologically unrelated to the canonical dimerization mode, in which H10–11 helices from two monomers run parallel to each other, resulting in much larger interaction areas of ∼1,000 Å2. This interface is strengthened by salt bridges between residues Arg690 and Asp549’ and by several H-bonds (e.g. between the carbonyl oxygen of Phe774 and the hydroxyl of Ser550’). Noteworthy, the side chain of Tyr545’ engages also in important contacts at this interface, docking on H9 from a neighboring monomer. A second, symmetric homodimer is centered on the aromatic Tyr638 (L6–7) and Phe735/Tyr738 (C-terminal end of H11) facing each other. However, since positions 638 and 738 are occupied by smaller polar residues in WT GR (Cys and Gln, respectively; Figure 1C), this arrangement is unlikely to be significant in vivo.
Finally, two related, medium-resolution structures of GR-LBD·DEX·SHP were obtained in the tetragonal and cubic space groups (I4122 and I4132, respectively; Figure 2C, D). Also in this case, we observe a symmetric homodimer in which the Tyr545/Tyr545’ aromatic rings are stacked, although the overall arrangement of LBD modules differs strongly from the Tyr545-directed dimer found in C2 crystals. Additional H1-H3’ contacts result in a more compact conformation, which is stabilized by H-bonds between both main- and side-chain atoms of the two monomers, including a Glu542–Arg569’ salt bridge. The largest interaction interface in these forms features a trimeric arrangement in which loops L1–3’/H3’, S2-L6’ and L11–12’ dock perpendicularly onto H10–11. The large, buried surface area in this trimer appears to compensate the electrostatic repulsion of abutting Asp641 carboxylates from the three monomers around a pseudo (in the tetragonal form) or exact 3-fold axis (in the cubic cell).
A thorough catalog of homodimeric arrangements illustrates the multivalent potential of the GR-LBD
The fact that even minor changes in protein complexes and crystallization conditions result in different GR-LBD arrangements, as demonstrated by the variety of crystal contacts described above (Figure 2A–D), prompted us to systematically analyze protein-protein contacts in all crystal structures of the domain previously deposited in the PDB (Figure 3A, B; Supplementary Tables S2 and S3). GR-LBD residues involved in homodimer formation cluster in four areas on the surface, the ‘front’, ‘back’, ‘top’ and ‘base’ of the domain (Figure 3B–D), in the standard view of NRs (Figure 1B).
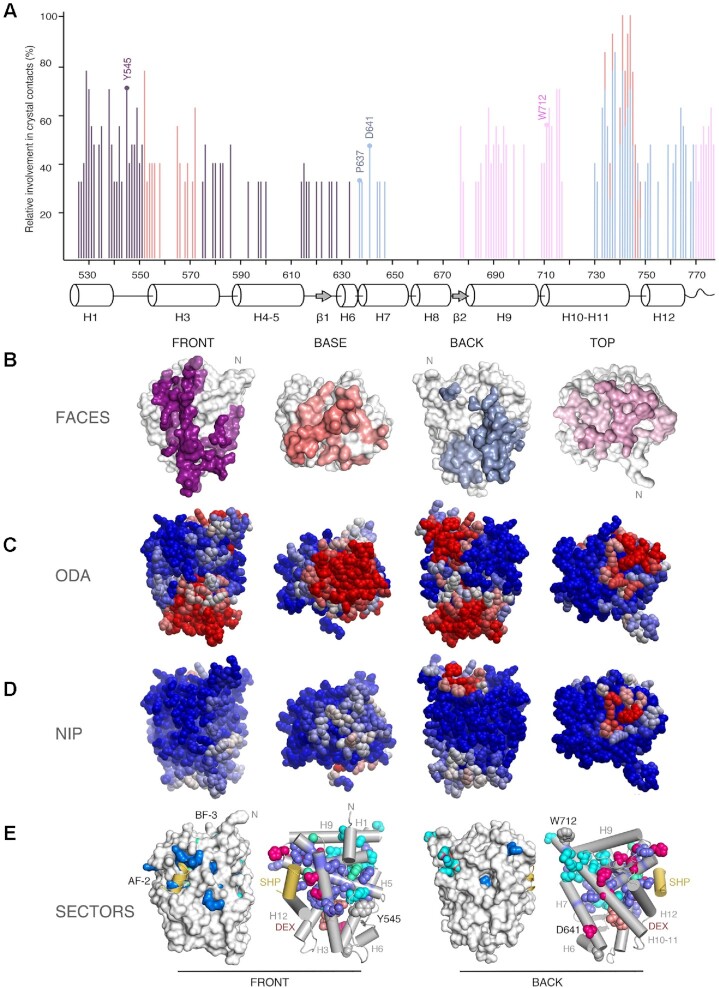
Figure 3.
Experimental structures and bioinformatics analyses unveil four major homodimerization surfaces on GR-LBD. (A) Relative frequencies of residue involvement in GR-LBD homodimer formation. Bar height indicates how often a given residue engages in crystal contacts in all available structures of GR-LBD, normalized to the residue most frequently found in homodimer interfaces, Leu741. Secondary structure elements given below the plot correspond to the crystal structure of human GR-LBD resolved at the highest resolution, 6NWL. (B) Residues involved in GR-LBD homodimerization cluster in continuous patches on its front (colored purple), base (coral), back (blue) and top (pink) faces. The association of these four faces yields the catalog of GR-LBD dimers represented in Figure 4. Models are shown in the same orientation and at the same magnification in panels (C)–(E). (C) Predicted protein-protein interaction ODA. ODA ‘hotspots’ (residues with favorable docking energy; ODA < –10.0 kcal/mol) are colored red, residues with ODA > 0 kcal/mol are shown in blue, and intermediate values are scaled accordingly. ODA hotspots form continuous surface patches that essentially overlap with the four protein-protein interaction interfaces shown in panel (B). (D) Hotspot interface residues predicted from docking experiments. Surface residues are colored according to their NIP. Residues with NIP > 0.4 and < 0 are colored red and blue, respectively; intermediate values are scaled accordingly. (E) SCA identifies two sectors of clustered, physically connected residues in GR-LBD. The front and back orientations of GR-LBD are depicted, and in both cases the module is represented as a solid surface and as a cartoon, with helices shown as rods and labeled. Residues belonging to sectors I and II are shown with their side chains atoms as spheres, colored cyan and dark blue, respectively. Residues belonging to both sectors are colored pink. SHP peptide is colored yellow, and DEX is shown as salmon spheres.
Next, we analyzed which combinations of these four homodimerization interfaces have been encountered in crystal structures. This analysis revealed 20 topologically distinct homodimers (see Supplementary Table S3 for interacting residues in all monomer pairs) numbered #1–20 throughout the manuscript. These homodimeric arrangements appear to cover the whole GR-LBD self-association landscape. Along with 11 symmetric (isologous) dimeric arrangements (i.e. between the same secondary structure elements / residues, such as in the Tyr545-mediated dimers described above), asymmetric or heterotypic homodimers (i.e. where the contacting GR-LBDs engage in interactions using different elements) are also common (nine arrangements).
We further explored the homodimerization potential of GR-LBD with a state-of-the-art protein-protein docking procedure and a total of 12,000 generated docking dimers. This analysis revealed the existence of at least one docking orientation close to 16 of the 20 representative ‘crystal homodimers’ (with a root-mean-square deviation (RMSD) ≤ 10 Å) (Supplementary Figure S3). Interestingly, the 2nd best-scoring docking orientation was close (7.3 Å RMSD) to one of the dimers (#20). In two further cases (#8, #10), there were docking orientations within 5 Å from the crystal structures, although with no optimal docking scoring.
For an unbiased estimate of the similarity between different homodimers, we first considered the overlapping of shared contact residues. To this end, we mapped these sets of residues into a multiple sequence alignment and calculated distances between the resulting vectors using different metrics (Figure 4A). This Jaccard analysis confirmed e.g. the topological similarity between two front-to-front homodimers: the first described non-canonical conformation (#1, 1M2Z (3)) and #2, an arrangement observed in 4P6W (51) (Supplementary Tables S2 and S3). On the other hand, front-to-front homodimer pairs #6 and #11, although sharing interface residues, differ strongly in that the two monomers are arranged parallel and antiparallel to each other, respectively. Similarly, homodimers #9 and #10 share the important residue Ile628 at the center of their intermonomer interfaces, but the two modules are quite differently oriented relative to each other. The way the four distinct areas on the GR-LBD surface assemble to generate the 20 topologically different homodimers is schematically presented in Figure 4B.
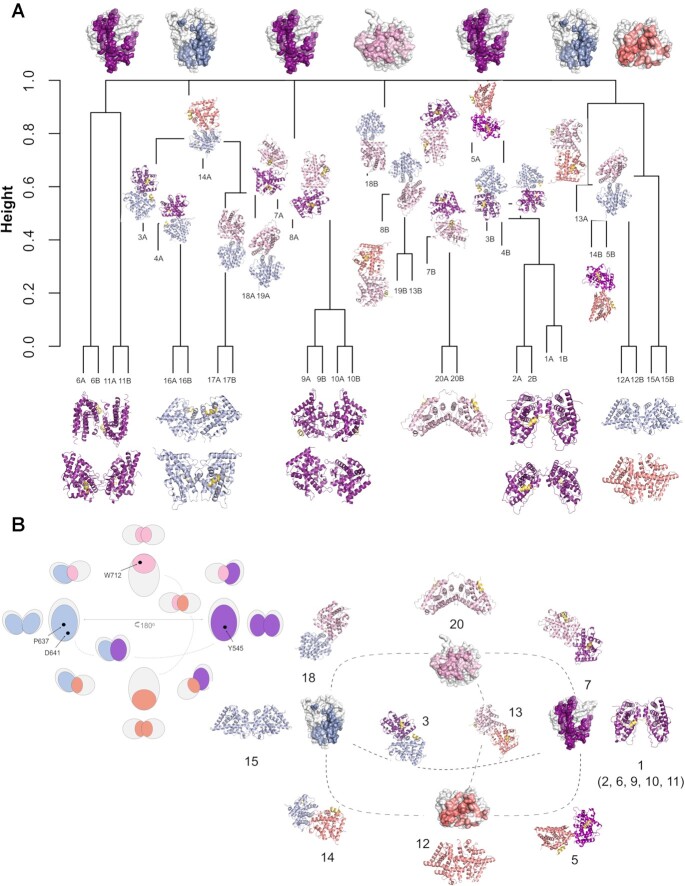
Figure 4.
An integrated catalog of GR-LBD homodimers. The four distinct GR-LBD protein-protein interfaces associate to generate 20 topologically different homodimers. (A) A dendrogram based on a hierarchical analysis of protein-protein contacts using Jaccard's index groups the 20 unique GR-LBD assemblies into six different clusters. (B) Relationships between the different GR-LBD homodimers. For orientation, monomers highlighting the four interacting surfaces are placed at the cardinal points in this panel (top, nord; front, east; base, south; and back, west), colored-coded as in Figure 3. Monomers in 10 representative homodimers are depicted as cartoons; each monomer is colored according to the face used to associate with its partner. Dimers are placed closest to the generating monomers. An equivalent schematic representation of GR-LBD homodimerization potential is shown at the upper left corner, with the position of major interacting residues indicated.
Alternatively, GR-LBD homodimers were superimposed on a common origin and classified by mapping the centers of coordinates of their interaction surfaces (Supplementary Figure S3A). Supplementary Figure S3B shows the orientations of these surfaces, represented by vectors between the common center of coordinates and each interaction surface. A hierarchical clustering analysis based on the Euclidean distance between these vectors grouped all homodimers into six clusters (Supplementary Figure S3C). From the spherical coordinates of the vectors representing the orientations of the interaction surfaces (Supplementary Figure S3E), we found that these clusters can be associated with combinations of the previously defined surfaces: cluster 1 (top-front), cluster 2 (top-back), cluster 3 (base-back), cluster 4 (base-front), cluster 5 (front), and cluster 6 (base). While each cluster may contain surfaces with different binding energy values (Supplementary Figure S3D), arrangements corresponding to top and back interaction surfaces have in general more favorable binding energy. The distribution of interfaces in the top 1,000 docking models also shows significant clustering around regions with favorable binding energy (Supplementary Figure S3F). However, docking solutions also clustered in other regions with less optimal energy, such as front and base surfaces.
Two sectors define the internal circuits linking key GR-LBD interaction sites
Positions differentially conserved within protein subfamilies (termed ‘specificity determining positions’, SDPs) are related to functional specificity (e.g. binding of different cofactors). Recent bioinformatics methods based on MCA have allowed identification of the subtle patterns of conservation of SDPs within large protein families (52). Our initial MCA run did not replicate the previously reported family classification (38), and only identified three residues that divide the NR superfamily into two clusters. This prompted us to use more sophisticated statistical tools to search for evolutionary conserved units in GR-LBD. To analyze if self-association surfaces may be allosterically coupled to other functional regions, we performed a SCA, which entirely relies on correlated amino acid variations across the domain without considering its 3D structure (53,54). Indeed, this analysis identified 40 residues that decompose the GR-LBD sequence into two quasi-independent groups of correlated residues or ‘sectors’ (Figure 3E). Sector 1 comprises 17 residues in and around H1 and H10, most notably LBP residues Met601 and Arg611 along with the nearby Phe606, whereas sector 2 features 20 residues mostly from H3 and H5 (e.g. Met604 in the LBP, Lys579, Phe584, Gln597 of AF-2 and Trp577 in an internal path connecting AF-2 to the LBP). Finally, Gly583 (BF-3), Leu596 (AF-2) and the internal Tyr663 belong to both sectors. Interestingly, all residues cluster in the upper half of the LBD, where both sectors are physically interconnected (Figure 3E). Sector 1 residues cluster around the N-terminus of the LBD and are thus likely candidates to interact with the hinge and DBD. Perhaps more relevantly, sector 2 comprising residues profusely innervate the LBP and AF-2 regions while Arg611 from sector 1 is essential to position the hormone. The three residues that belong to both sectors are strategically located to link the LBP with AF-2 and BF-3. Taken together, our results suggest that both sectors couple functionally relevant regions (e.g. ligand and coregulator binding or chaperone docking/release).
In vitro crosslinking experiments corroborate non-canonical GR-LBD dimerization
The results presented above suggest that many surface-exposed residues of GR-LBD engage in a variety of crystal contacts. To clarify their role in homodimer formation in solution, we took advantage of intermolecular H-bonds between Glu/Asp and Lys at some of the crystal interfaces, which can be ‘frozen’ upon incubation with the zero-length crosslinker, EDC.
Indeed, we observed rapid formation of a covalent dimer upon incubation of GR-LBD with EDC, as well as a fainter band corresponding to a tetrameric arrangement(s) (Figure 5A). To identify crosslinked Asp/Glu-Lys pairs, gel bands corresponding to oligomeric GR-LBD were excised, enzymatically digested with either trypsin or chymotrypsin, and analyzed by MS (Supplementary Figure S4A, Supplementary Tables S4 and S5). Most notably, we found that elements essential for top-to-top (#20) and front-to-front (#1, #2, #6, #9, #10 and #11), non-canonical dimerization are overrepresented among EDC-linked peptides, with the most common contacts involving (i) residues of H1 and L1–3, on the one side, and from H3, on the other, which would correspond to front-to-front interactions, as well as (ii) H9 and the L9-10 linker from two monomers, which is compatible with homodimer #20 (Figure 5B, C and Supplementary Figure S4B, Supplementary Table S4). Similar results were obtained with the MS-cleavable, urea-based crosslinker, DSBU (Supplementary Table S6).
Figure 5.
Several GR-LBD homodimers are populated in solution. (A) SDS-PAGE analysis of GR-LBD after incubation with EDC. Notice bands with relative molecular masses corresponding to GR-LBD dimers (D) and tetramers (T) in all the lanes except control lane 1 (no EDC added). Lanes 2 and 3, protein incubated at about 0.37 mg/ml; lanes 4 and 5, protein incubated at about 1.5 mg/ml. Samples in lanes 2 and 4 were treated at RT; those in lanes 3 and 5 at 30°C. Bands corresponding to co-purified DnaK and GroEL E. coli chaperones are marked with (*) and (**), respectively. (B) Schematic representation of GR-LBD structure, with side side chains of residues identified using EDC shown as spheres (Asp in green, Lys in lilac, and Glu in yellow). (C) Crosslink map of EDC-treated GR-LBD showing all intermonomer crosslinks identified by MS analysis. Regions corresponding to the top, front, base, and back surfaces are colored as in Figures 3 and 4. A secondary structure plot is shown above the map. (D) Closeup of the major homodimerization interface in C2 crystals (front-to-front homodimer #11), dominated by stacked Tyr545/Tyr545’ residues. (E) Non-reducing SDS-PAGE analysis of purified GR-LBD(Y545C) (lane 3) shows spontaneous dimerization in solution. Note that the WT protein does not form dimers when incubated at the same concentration (lane 2).
Mutant Y545C exhibits GR-LBD non-canonical homodimerization in solution
Inspection of homodimer interfaces in GR-LBD revealed several symmetric arrangements in which the side chain of a solvent-exposed residue from one monomer is located within VdW distance of the same residue from a crystal neighbor (Figure 2A and Figure 5D). To verify whether some of these conformations are present in solution, we have generated several Cys mutants of the GR-LBD. All studied mutants were properly folded, as indicated by only minor decreases in melting temperatures in DSF analysis, in line with the results of a systematic bioinformatics analysis of mutant stability performed using Fold-X (not shown). Incubation of purified Y545C in low-reducing conditions resulted in the rapid formation of covalent dimers (Figure 5E). By contrast, neither the WT protein nor other Cys mutants tested (e.g. D641C, S744C) dimerized under the same conditions (Figure 5E). These findings strongly suggest that the side chains of Tyr545 from two monomers are close enough in solution, at least in a subset of GR-LBD molecules.
To directly confirm that residues Cys545/Cys545’ are responsible for disulfide bridge-mediated dimerization in solution, bands corresponding to the dimer were excised from the gel, treated with iodoacetamide to block free Cys residues, and subjected to enzymatic digestion with trypsin and GluC. MS analysis of these digests allowed indeed the identification of peaks corresponding to peptide V543LCSGYD549 crosslinked to either V543’LCSGYD549’ or V543’LCSGYDSTLPDTSTR558’, thus confirming Cys545-mediated covalent bond formation (Supplementary Figure S4C and Supplementary Table S7).
To further assess the contribution of Tyr545 to GR homodimerization in solution, we performed SPR assays with its Y545C and Y545A mutants. Indeed, experiments conducted with the Cys mutant revealed significant increases in affinity. The increase was the highest when the mutant was used as both ligand (i.e. chip-immobilized) and analyte, as indicated by dramatic increases in affinity (kD = 0.85 vs. 13.5 μM for the WT-WT association at 100 μM DTT; Supplementary Figure S2C, D) as well as in complex lifetime, with a 4-fold increase at 10 μM DTT (t1/2 = 1,130 vs. 270 s). By contrast, truncation to Ala at position 545 led to a slightly less tight association (Supplementary Figure S2E). We also crystallized and solved the structure of the Y545A variant, thus confirming proper folding of the generated point mutants. Noteworthy, Y545A crystallized in the P61 space group, which does not involve symmetric contacts between the side chains of residues at position 545 (Figure 2B). Altogether, our results confirm that residue Tyr545 plays an important role in GR-LBD homodimerization in solution.
Residues Tyr545 and Asp641 modulate multimerization of FL-GR
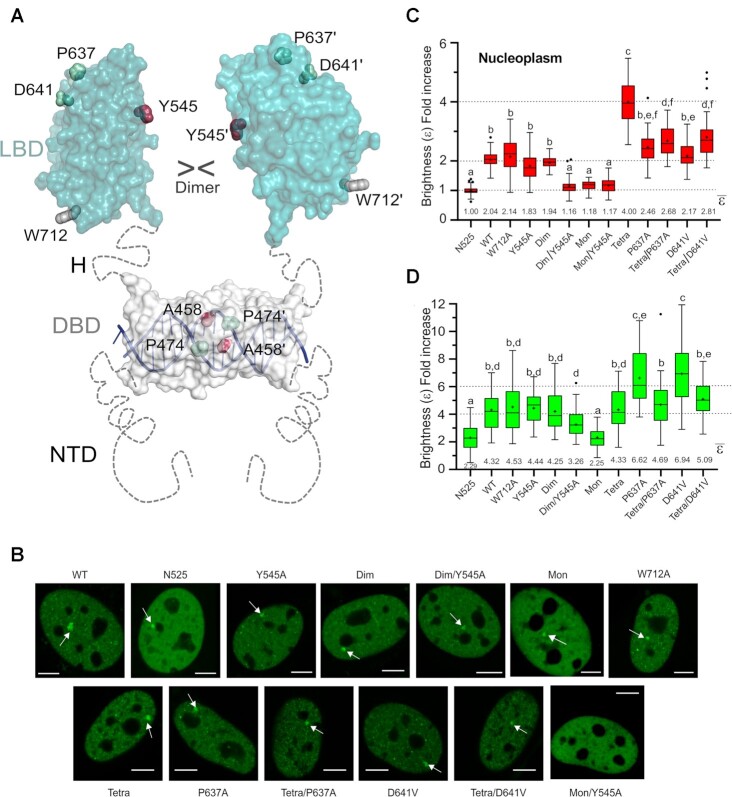
Quantitative fluorescence microscopy in living cells (the number and brightness method, N&B) allows to estimate the average oligomeric state of a fluorescent protein from its molecular brightness (ϵ; (41)). For N&B experiments we use mouse adenocarcinoma cells, which possess a tandem array of DNA binding sites for GR, the MMTV array (42). We have knocked out expressed GR protein in these cells, thus designated GRKO (25). These cells are transiently transfected with GFP-tagged full length mouse GR (GFP-mGR) or mutants thereof, and the oligomeric state of fluorescently tagged GR molecules is quantified by comparing to a constitutively monomeric GR variant (N525*) (24). The mutated residues of the mouse GR variants correspond to the human versions discussed above and are noted in the figure legends. Further, presence of the MMTV array in the GRKO cells allows us to differentially assess GR oligomerization not only in the entire nucleoplasm but also in a region highly enriched in GR binding sites that is visible under the microscope as a bright spot (Figure 6B, arrowheads).
Figure 6.
Mutation of LBD–LBD interface residues profoundly affects the multimerization behavior of FL-GR. (A) Schematic 3D model of FL-GR. The NTD is highly disordered and is followed by the globular DBD (DNA-bound conformation is shown) and the LBD. Note that the length and flexibility of the hinge (H) allows for the formation of various homodimers. The side chains of all residues mutated to assess the multimerization behavior of GR are shown as spheres. (B) Subcellular localization of WT GFP-mGR and indicated mutants in 3617-GRKO cells, as assessed by fluorescence microscopy. Variant N525* lacks the entire LBD and remains monomeric, irrespective of DEX treatment (23). White arrowheads point to the MMTV arrays. Scale bar: 5 μm. Data for WT_GR, GRdim, GRmon and GRtetra were taken from (24) and are shown for comparison purposes. All N&B experiments have been performed with mouse GR as in (23). However, residue numbers correspond to the human protein to facilitate comparisons. (C) GR oligomerization in the nucleus, as determined in N&B assays. The fold increase in molecular brightness (ϵ) relative to the N525* monomeric control is shown (N = 428 total cells with 21 < N < 37 between treatments). (D) Quaternary structure of DNA-bound GR. The results of N&B assays at the MMTV arrays are represented as in panel C (N = 338 total cells with 17 < N < 35 between treatments). Note that simultaneous disruption of intermonomer interactions mediated by the DBD (Ala458Thr) and the LBD (Tyr545Ala) in the GRdim/Y545A double mutant results in a variant that is monomeric in the nucleus, while at the array it formed mostly trimers. In panels (C) and (D), centered lines show the medians and crosses represent sample means (average numbers below each box-plot). Box limits indicate the 25th and 75th percentiles; whiskers extend 1.5-fold the interquartile range from the 25th and 75th percentiles, with outliers represented by dots. Boxes with different superscript letters are significantly different from each other (P < 0.05; one-way ANOVA followed by Tukey's multiple comparison test).
To test the relevance for the full-length receptor (FL-GR) of key surface-exposed residues identified in GR-LBD homodimer interfaces in vitro, we generated Ala mutants of mouse GR at positions topologically equivalent to human residues Tyr545, Pro637, Asp641 and Trp712, all of which are conserved in ancGR2 (Figure 1C). Further, all residues but Asp641 are strictly conserved from fish to humans, and Asp641 is conservatively replaced by a glutamate in non-mammals (Supplementary Figure S1). To study the impact of the Tyr545→Ala exchange in the background of two other variants previously shown to be important for GR homodimerization (23), we also generated the double mutant (Ala458Thr, Tyr545Ala) (in following termed GRdim/Y545A) and the triple mutant (Ala458Thr, Tyr545Ala, Ile628Ala), or GRmon/Y545A. Finally, we also generated double mutants in which a GR variant that tetramerizes both in the nucleus and at the array (Pro474Arg, (24)) was combined with either P637A or D641V (These variants, (Pro474Arg, Pro637Ala) and (Pro474Arg, Asp641Val), are in following termed GRtetra/P637A and GRtetra/D641V, respectively; Figure 6A).
Next, we transiently transfected GFP-mGR and the generated mutants into GRKO cells and performed N&B experiments as previously described (23,24,55). All mutants translocate to the nucleus upon hormone stimulation (Figure 6B), indicating the mutants can bind ligand at the tested hormone concentration of 100 nM DEX. Further, all variants except those carrying the GRmon double mutant (Ala458Thr, Ile628Ala) were visible at the MMTV array, suggesting that DNA binding was possible (Figure 6B, arrowheads). Of note, reduced genome-wide chromatin binding for GRmon has recently been shown (39), and only a very small percentage of GRmon cells form visible arrays (23,24). Interestingly, we did not detect any arrays in cells with GRmon/Y545A (Figure 6B), suggesting an even more drastic phenotype for this triple mutant.
While W712A oligomerizes as the WT receptor both in the nucleoplasm and at the array (Figure 6C, D), Tyr545Ala substitution appears to slightly decrease dimerization in the nucleoplasm (ϵ = 1.83). By contrast, Pro637Ala produces a slight increase in oligomerization (ϵ = 2.46), even though neither difference achieves statistical significance. Since FL-GR dimerizes at least through both DBD and LBD (Figure 6A), we tested the effect of the Tyr545Ala mutation in the GRdim background, which has impaired DBD-DBD contacts yet mostly dimerizes in the nucleoplasm (Figure 6C, (23,24)). Indeed, the GRdim/Y545A double mutant shows significant tendency to remain monomeric in the nucleus (ϵ = 1.16), like the previously characterized GRmon (Figure 6C, (23,24,56)). Taken together, these results confirm the important role of Tyr545 in the dimerization of GR in live cells.
On the other hand, the Asp641Val mutation, linked to a GC resistance phenotype known as Chrousos syndrome (57), and Pro637Ala promoted higher-order oligomerization at the array (Figure 6D), possibly hexamers or a mixture of tetra- and octamers. These findings prompted us to analyze the impact of these two mutants when combined with a DBD mutation that enforces GR tetramerization, Pro474Arg. Unexpectedly, instead of a synergistic effect, the combined mutations reversed to GRtetra oligomerization in the nucleus (Figure 6C). By contrast, the GRtetra mutation abrogated the ability of P637A and D641V variants to form higher-order oligomers at the array level (Figure 6D). These observations highlight a complex relationship between the different structural domains of the NR.
Role of residues Tyr545 and Asp641 on GR transcriptional activity
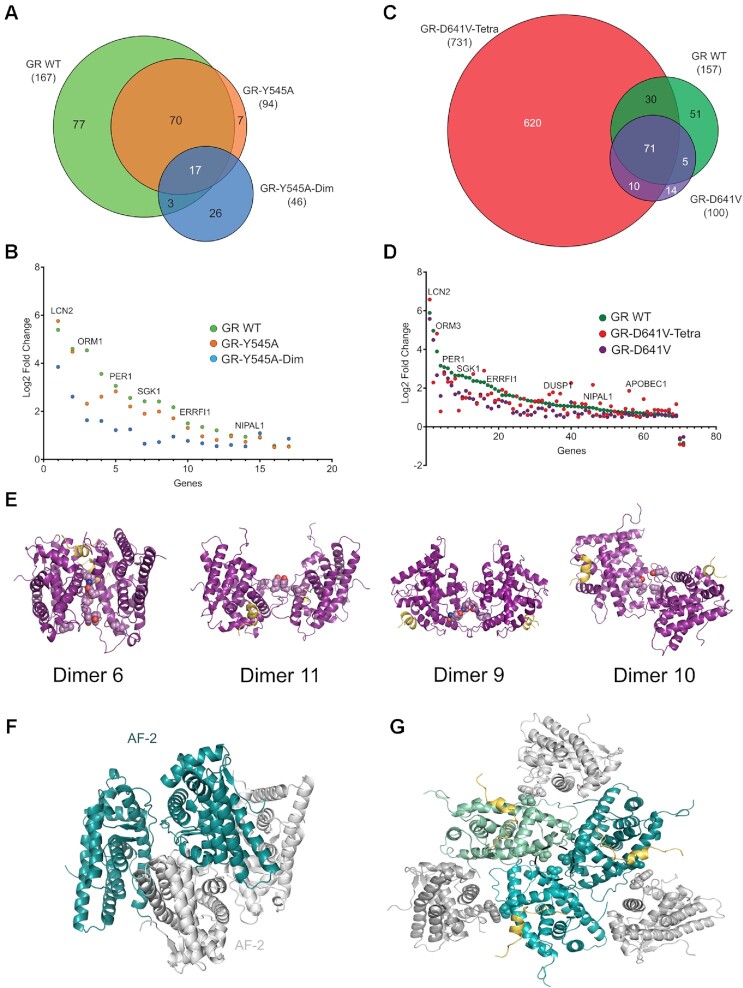
Having demonstrated that residues Tyr545 and Asp641 play essential roles in FL-GR oligomerization, both in solution and in live cells, we wondered if they would modulate the activity of the receptor. To this end, we generated stable cell lines expressing GFP-tagged mouse versions of the GRY545A and GRdim/Y545A variants (see methods), and compared their transcriptional programs to that of mouse GR WT by RNA-seq, after 2h of 100 nM Dex stimulation (Supplementary Table S8 and Supplementary Figure S5A–F). As summarized in Figure 7A, GRY545A upregulates only 56% of the genes compared to WT GR, and only three out of the 21 genes downregulated by the WT receptor. In line with the more significant impact of the GRdim/Y545A double mutant on receptor homodimerization, its transcriptional activity was more compromised than GRY545A single mutant, with only 28% of the number of genes being modulated respect to WT GR (Figure 7A). Of the 17 common genes between the three GR types, all but 2 have only modestly lower fold change with Dex between GR WT and GRY545A, while most of the same common genes exhibit a more severe impact upon hormone treatment in the GRdim/Y545A cells (Figure 7B).
Figure 7.
Residues Tyr545 and Asp641 have major roles in GR quaternary structures and transcriptional activity. Stable cell lines expressing GFP-tagged variants of the indicated mouse GR mutants were treated with 100 nM DEX for 2 h (note than numbers correspond to hGR, as in Figure 6). (A, C) Venn diagrams of differentially expressed genes from RNA-Seq (|log2 Fold change| > 0.5, False-discovery-rate < 0.01). The two WT GR sets (b1, b2) are biological replicates collected and sequenced separately for each batch. (B, D) Scatter plots of shared hormone-regulated genes (17 and 71 common genes, respectively) between GR mutants, compared and plotted against their respective WT GR control. (E) Models of human GR-LBD homodimers based on the observed assemblies #10, #6, #9 and #11. The critical homodimerization residues, Tyr545 and Ile628, are shown as color-coded spheres in all cases. (F) Model of GR-LBD tetramer generated by docking dimer #11 onto itself. The model is compatible with the results obtained with the Y545C mutant and EDC-crosslinking. (G) Putative GR-LBD hexamer favored by the Asp641Val mutation. The central trimer corresponds to an arrangement observed in tetragonal and cubic crystal forms (#14; Figures 2C, D and 4), while the peripheral monomers dock according to conformation #4. Asp641 residues from the central trimer are encircled. This generates a closed hexamer by additional interactions between the N-terminal end of H10 and H12’ at the third interface. In addition to D641V, other GR mutants associated with Chrousos disease might favor multimeric forms that are incompatible with active GR tetramers on DNA. For instance, replacement of Thr556 by an aliphatic Ile would stabilize this arrangement through contacts with e.g. residues Met560 and Pro637 from its ‘own’ monomer and/or His645’/Asn731’ from a neighboring molecule. Similar considerations apply to mutations such as Arg714Gln, Phe737Leu, Ile747Met and Leu773Pro as well as to variant Pro637Ala, which also forms higher-order oligomers at the array (Figure 6D).
RNA-Seq analysis of cells expressing the higher oligomeric variant, D641V, revealed that the mutant modulated only two thirds of the hormone-responsive genes compared to WT GR (Figure 7C), consistent with the in vivo phenotype described as Chrousos syndrome (GC resistance) (57). The double mutant GRtetra/D641V shows a much larger number of hormone responsive genes with respect to GR WT, similar to the GRtetra single mutation reported elsewhere (25). This suggests that the constitutively tetrameric conformation exhibited by this DBD mutant can exert a strong effect on gene response even in tandem with the D641V variant of the LBD. However, the tetramer mutation cannot completely rescue the relative fold change of many common DEX-responsive genes, especially genes with the highest response (Figure 7D). These observations reinforce the importance of both DBD and LBD modules of the GR in transcription regulation, and underscore the functional relevance of two surface-exposed LBD residues, Tyr545 and Asp641.
DISCUSSION
Although many structures of GR-LBD have been reported (Supplementary Table S2), the physiologically relevant conformation(s) of GR and other oxosteroid NRs remain controversial. The new structures presented here highlight the ability of different GR-LBD surfaces to engage in various quaternary arrangements depending on the bound agonists/antagonists, cofactors, and other biochemical parameters. These observations prompted us to dissect the oligomerization capability of GR-LBD. Since the ancient LBD structure may not accurately mimic the multimerization behavior of modern, wild-type GR, our structural predictions were experimentally verified by characterizing several key mutations of exposed residues in the background of mouse FL-GR in live cells. The results of these experiments demonstrate that the multimerization landscape of ancGR2 reproduces that of modern GR, in the context of the full-length receptor. Careful inspection of intermonomer contacts in all available 3D structures, irrespective of the variant used for the structural analysis, allowed us to identify 20 topologically different homodimeric architectures (Figure 4, Supplementary Table S3). These experimental GR conformations could be grouped into six different clusters considering relationships between the interacting residues (Figure 4A, Supplementary Figures S3C and S3E), which correspond to three topologically distinct front-to-front homodimers (#1, #2, #6, #9, #10 and #11), along with base-to-base (#12), top-to-top (#20), and back-to-back arrangements (#15, #16 and #17). SPR and XL-MS experiments in solution coupled with quantitative fluorescence microscopy of FL-GR WT and several mutants in live cells along with RNA-seq analyses of most relevant variants allow us to postulate the more likely quaternary arrangements of GR-LBD associated with its different genetic variants (Supplementary Table S9).
It has long been accepted that the ‘canonical’, H10-mediated homodimeric conformation adopted by ERα-LBD (Supplementary Figure S6B) is hindered by the F-domain in oxosteroid receptors (5,13). Several observations suggested that the non-canonical dimer reported for AR (Supplementary Figure S6A), (5,13,58) could be adopted by other NRs. However, none of the 20 GR-LBD homodimers can be considered as topologically equivalent to the one observed in the crystal structure of AR-LBD, illustrating a more complex multimerization behavior than previously anticipated. Replacement of AR interface residues such as Thr656 by positively charged Lys/Arg residues and of the following Asn657 by bulkier Gln/His residues in all other members of the subfamily might preclude the formation of AR-like homodimers in GR/MR/PR. This highlights the difficulty to extrapolate the quaternary structure of one NR to other, even closely related NRs, and the need for experimental evidence to identify physiologically relevant conformations (13). A related arrangement had been previously observed in some structures of agonist- or antagonist-bound GR-LBD (3), but its biological relevance has been repeatedly questioned (59,60). Here we identify and discuss several unrelated multimeric assemblies of GR-LBD that might illuminate different structural arrangements of the chromatin-bound full-length protein.
GR can adopt several topologically distinct dimeric conformations
Our data indicates that ‘non-canonical’ GR homodimers centered on the Tyr545 phenol ring are critical for GR homodimerization. In live cells, variant Y545A is mostly monomeric when combined with an exchange that also disrupts DBD-DBD contacts (Ala458Thr, GRdim). Consistently, the transcriptional program of the Y545A variant is significantly affected, and the double mutant GRdim/Y545A is practically inactive (Figure 7A). Further, an important fraction of GR-LBD Y545C molecules rapidly and spontaneously forms covalent homodimers in vitro mediated by the Cys545–Cys545’ disulfide bond (Figures 5C, D). Finally, Ala replacement of Try545 led to a reduced monomer-monomer affinity in SPR experiments (Supplementary Figure S2E).
Taken together, we propose four agonist-bound plausible GR dimers (Figure 7E). Formation of the Cys545–Cys545’ disulfide bridge in the Y545C variant is compatible with two topologically different, roughly parallel and antiparallel symmetric homodimers (#6 and #11; Figure 7E). Incomplete dimerization of GR-LBD(Y545C) suggests that other dimeric states are also populated in solution. Indeed, in a third, also antiparallel GR-LBD homodimer the Tyr545 side chains are located at the borders of the protein-protein interface (#9, Figure 7E). This arrangement is centered on residues Ile628/Ile628’, which are important for receptor dimerization (23). Indeed, the I628A mutant decreased GR multimerization of FL-GR in living cells to a larger extent than the Y545A variant, both in the nucleoplasm and DNA-bound (24). The most likely explanation for these differences is that Tyr545 and Ile628 affect two topologically distinct although interconnected dimerization interfaces of GR-LBD, in line with our structural analysis (Figures 3 and4). In this regard, it is noteworthy that this side chain is less exposed than the Tyr545 phenolic ring, engaging in close VdW contacts with Leu620, Cys622 and Pro625. Thus, the larger impact of the I628A mutant might simply reflect disruption of an extended protein-protein interface area centered on the Ile628 side chain. Finally, a second Ile628-centered, parallel conformation is found in crystals of ancGR1 (homodimer #10; Figure 7E, (56)). Therefore, both parallel and antiparallel arrangements of LBD modules with different involvement of the Tyr545 and Ile628 valences are compatible with current data and are equally possible. Some of these quaternary arrangements in solution could be verified in our XL-MS experiments (Figure 5A, Supplementary Table S6). GR-LBD features several strictly conserved Tyr/Trp residues, two of which, Tyr545 and Trp712, are repeatedly found at monomer-monomer interfaces (Figure 2 and Supplementary Table S3). Against expectations, elimination of the bulky indole ring at position 712 had no effect on the oligomerization properties of FL-GR in live cells, as the W712A variant behaved as WT both in the nucleoplasm and DNA-bound (Figures 6C and D). These findings strongly suggest that Trp712 is not important for GR multimerization in vivo. This would exclude, among others, the symmetric homodimer #20, which had been proposed as the most likely conformation of dimeric GR-LBD (61). Nevertheless, the fact that Trp712 participates in six topologically different homodimeric arrangements suggests an enhanced propensity for protein-protein interactions. Indeed, the recently reported cryo-EM structure of GR-LBD bound to the ‘client-maturation complex’ Hsp90-p23 has revealed that Trp712 occupies the central position in a major binding epitope for the p23 co-chaperone (62).
Interestingly, antagonist binding appears to result in completely unrelated GR-LBD assemblies: the symmetric back-to-back (#15) and base-to-base homodimers (#12), which have much larger interface areas and much lower energies than all other GR-LBD conformations (Supplementary Table S3). These dimers are indeed only observed in GR-LBD bound to the antagonist, mifepristone/RU-486 (1NHZ and 5UC3, respectively, (59)), and share several important features. First, the bound antagonist displaces H12, which partially disrupts the LBP. More importantly, in both cases the AF-2 of one monomer is partially covered by either H12 (#15) or H3 (#12) from a neighboring molecule, thus interfering with coregulator binding (Supplementary Figures S5G, H). Further, essentially the same arrangements are found in crystals of the dominant negative GRβ, which is known to bind only antagonists (63). Even though tetrameric assemblies in 1NHZ/5UC3 and in a third structure of mifepristone-bound GR (3H52; (64)) have been reported, these are not compatible with the results of our XL experiments with EDC (Figure 5A and Supplementary Figure S4B) or DSBU (Supplementary Tables S4 and S6). Inactive RXRα also adopts a disc-like tetrameric conformation, in which H12 from one molecule protrudes away and docks on the AF-2 of an adjacent monomer (Supplementary Figure S5I; (65)). Although the overall arrangement differs from the ones adopted by agonist-bound GR-LBD, the fact that AF-2 is occluded in both cases points to a general feature of self-repressed or inactive conformations in NRs. Altogether, these findings strongly suggest that the most stable, back-to-back (#15) and base-to-base conformations (#12) of GR-LBD are associated with inactive, self-repressed states, which are induced or stabilized by antagonist binding. Previous experiments had demonstrated that FL-GR bound to mifepristone is fully dimeric in the nucleus, but its ability to form tetramers on DNA is compromised (24). Thus, impaired tetramerization of RU486-bound GR might account for its transcriptional deficiency.
Towards a model for FL-GR quaternary arrangements and their role in physiology and disease
Our earlier live-cell imaging studies have revealed that dimeric GR might be an intermediate state towards transcriptionally active tetramers bound to target DNA sequences (24,25,39) also in line with the current SPR results (Figure 1D, E). Although more speculative, it is possible to generate tentative models of DNA-bound GR tetramers that satisfy the constraints derived from current structure-function information (Figure 7F). First, we reasoned that since tetramers but not higher order multimers are detected on chromatin, none of the known intermonomer ‘anchors’, Tyr545 and Ile628, would be available for protein-protein interactions in active tetramers. In other words, all GR-LBD valences should be satisfied in bona fide GR tetramers. Since the Ile628 side chain is exposed in both conformations with opposing Tyr545 rings (#6 and #11), different combinations with homodimers #9 and #10 are conceivable (Supplementary Figures S6E-J). These hypothetical multimeric arrangements reconcile the role of residues Tyr545 and Ile628 for GR multimerization. The additional interactions predicted in these multimeric arrangements, in addition to DBD-DBD interactions (23) and condensation provided by the NTD (27) would overcome the energy loss due to Tyr545→Ala or Ile628→Ala exchanges, explaining why variants Y545A and I628A are still tetrameric on chromatin (Figure 6D). However, the triple mutant (A458T, Y545A, I628A) did not bind the array, indicating that simultaneous elimination of the Tyr545 and Ile628 valences would generate a well folded, but fully inactive variant.
On the other hand, our results suggest that formation of non-physiological multimers might underlie the deleterious effect of glucocorticoid resistance mutants. Unexpectedly, we observed that a GR mutant linked to Chrousos syndrome, D641V (57), preferentially formed higher-order oligomers when bound to DNA (Figure 6D), which was accompanied by a significant reduction in its transcriptional activity (Figure 7C). A three-fold reduced affinity of the Val641 mutant for DEX was previously proposed as the underlying molecular defect in the D641V variant. However, since Asp641 is exposed on the GR surface and is located 8 Å away from the closest DEX atom, it is unlikely to play any relevant role in hormone recognition. This non-conservative mutation has a particularly strong stabilizing impact of 3.4 kcal/mol for each dimer pair on the GR-LBD trimer observed in current tetragonal and cubic crystals (#14; see Figure 2D, Supplementary Table S3). Therefore, these structures provide a plausible model for a trimeric arrangement in which the three Asp641 side chains face each other around a three-fold axis. The positive effect of the mutation results from the relief of strong electrostatic repulsion between the three abutting negatively charged Asp641 residues, coupled with favorable VdW contacts made by the aliphatic Val641 side chains with each other and with Cys638 from a neighboring molecule. Notably, conformation #4 is fully compatible with this trimer and generates a closed hexamer (Figure 7G). This conformation would occlude the AF-2 pockets of the ‘external’ monomers and is thus incompatible with coregulator binding, impeding GR transcription. Several additional residues linked to Chrousos disease are located at or close to one of the three monomer-monomer interfaces, and their mutations might promote local conformations that also stabilize this hexamer (Figure 7G). This is, to the best of our knowledge, the first time in which non-productive multimers of a NR are postulated as the molecular basis of a human disease.
In conclusion, our results suggest that GR multimerization needs to be reconsidered from the perspective of multiple, energetically roughly equivalent homodimers. It is therefore conceivable that several different GR dimers coexist within a cell, all of which, in principle, recruit different cofactors and thus activate different transcription pathways. ‘Selection’ of a given conformation might be determined not only by the availability of GR-specific agonist/antagonist but also by many other parameters, including cell state. Current structure-function information suggests that at least four different GR-LBD homodimers have roughly equal probabilities to be formed in vivo, which would in turn generate different tetrameric arrangements on DNA. We are tempted to speculate that these individual conformations of tetrameric GR are associated with specific transcriptional programs. Future investigations should verify the validity of this hypothesis and establish whether specific GR conformations result in unique expression patterns. The high plasticity of GR-LBD for self-association and its potential modulation by different agonists and/or antagonists opens new venues for the potential treatment of GR-related diseases. The ability to promote or stabilize these specific GR multimeric states might be an essential step towards the development of novel GCs with reduced side effects.
DATA AVAILABILITY
The atomic coordinates and structure factors have been deposited in the PDB with accession codes 7YXC (C2), 7YXD (P31), 7YXN (P61), 7YXO (I4122), 7YXP (I4132) and 7YXR (P61, Y545A). The PDB data has been designed ‘for immediate release on publication’. MS data have been deposited to the ProteomeXchange Consortium (PRIDE partner repository: dataset identifier PXD028039). Bioinformatics data and code for the clustering and SCA are available at https://github.com/ibn90/GRPROJECT2021. The Gene Expression profiles are deposited in the Gene Expression Omnibus (GEO) database and the code assigned is GSE212973. All relevant data are available upon request.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession codes 7YXC (C2), 7YXD (P31), 7YXN (P61), 7YXO (I4122), 7YXP (I4132) and 7YXR (P61, Y545A). The Gene Expression profiles are deposited in the Gene Expression Omnibus (GEO) database and the code assigned is GSE212973.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Erick Ortlund (Emory University) for providing the plasmid for ancGR2-LBD, Arnold T. Hagler for his comments on the manuscript, and Ildefonso Cases (CABD) for his help with code. We thank ALBA-Cells synchrotron XALOC team for beamline support.
Author contributions: P.F.-P. and E.E.-P. designed and supervised the project, obtained financial support, and share overall responsibility. A.J.P., A.A.M., M.A.M. and R.A. obtained recombinant proteins. M.A.M., A.A.M. and A.J.P. performed crystallization trials. A.A.M, J.F.D. and R.A. generated mutants and analyzed proteins. I.N.B. and A.R.M. performed and interpreted SCA analyses. J.F.R. performed and interpreted docking, NIP, and ODA calculations. T.T., R.L.S. and T.A.J. generated FL-GR mutants. G.F. and D.M.P. performed and analyzed N&B experiments. A.J.P., A.A.M., R.A., P.F.-P. and E.E.-P. performed and interpreted MS SPR. A.J.P., P.F.-P. and E.E.-P. collected and interpreted X-ray diffraction data, P.F.-P. solved and refined structures. T.T., R.L.S., T.A.J., A.V.F., C.C. and P.P. contributed tools. T.T., T.A.J. and SK performed and analyzed the genomics data. D.M.P., and G.L.H. supervised cell experiments. A.J.P., P.F.-P. and E.E.-P. drafted the article. A.R.M, T.A.J., J.F.R., D.M.P., G.L.H., P.F.-P. and E.E.-P. critically reviewed the manuscript. All authors approved the manuscript.
Contributor Information
Alba Jiménez-Panizo, Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, University of Barcelona (UB), 08028 Barcelona, Spain; Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain; National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
Andrea Alegre-Martí, Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, University of Barcelona (UB), 08028 Barcelona, Spain; Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain.
Theophilus T Tettey, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
Gregory Fettweis, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
Montserrat Abella, Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, University of Barcelona (UB), 08028 Barcelona, Spain; Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain.
Rosa Antón, Biomedical Research Institute Sant Pau (IIB Sant Pau), 08041 Barcelona, Spain.
Thomas A Johnson, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
Sohyoung Kim, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
R Louis Schiltz, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
Israel Núñez-Barrios, Andalusian Center for Developmental Biology (CABD-CSIC). Campus Universitario Pablo de Olavide, 41013 Sevilla, Spain.
Joan Font-Díaz, Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain; Department of Cell Biology, Physiology and Immunology, Faculty of Biology, University of Barcelona, 08028 Barcelona, Spain.
Carme Caelles, Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain; Department of Biochemistry and Physiology, Faculty of Pharmacy and Food Sciences, University of Barcelona, Barcelona 08028, Spain.
Annabel F Valledor, Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain; Department of Cell Biology, Physiology and Immunology, Faculty of Biology, University of Barcelona, 08028 Barcelona, Spain.
Paloma Pérez, Instituto de Biomedicina de Valencia (IBV)-CSIC, 46010, Valencia, Spain.
Ana M Rojas, Andalusian Center for Developmental Biology (CABD-CSIC). Campus Universitario Pablo de Olavide, 41013 Sevilla, Spain.
Juan Fernández-Recio, Instituto de Ciencias de la Vid y del Vino (ICVV), CSIC - Universidad de La Rioja - Gobierno de La Rioja, 26007 Logroño, Spain.
Diego M Presman, IFIBYNE, UBA-CONICET, Universidad de Buenos Aires, Facultad de Ciencias Exactas y Naturales, Buenos Aires C1428EGA, Argentina.
Gordon L Hager, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892-5055, USA.
Pablo Fuentes-Prior, Biomedical Research Institute Sant Pau (IIB Sant Pau), 08041 Barcelona, Spain.
Eva Estébanez-Perpiñá, Department of Biochemistry and Molecular Biomedicine, Faculty of Biology, University of Barcelona (UB), 08028 Barcelona, Spain; Institute of Biomedicine of the University of Barcelona (IBUB), University of Barcelona (UB), 08028 Barcelona, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
E.E.-P. thanks the generosity of the Gemma E. Carretero Fund; MINECO [BFU2017-86906-R, SAF2017-71878-REDT, SAF2015-71878-REDT to E.E.-P., RTI2018-101500-B-I00 to P.F.-P., RTI2018-096735-B-100 to A.R.M., PID2019-110167RB-I00 to J.F.-R., SAF2017-89510-R to A.V.F. and C.C.]; G.L.H thanks the NIH Intramural Research Program; D.M.P was supported by CONICET. Funding for open access charge: Spanish Ministry of Science (MINECO).
Conflict of interest statement. None declared.
REFERENCES
- 1. Conway-Campbell B.L., Pooley J.R., Hager G.L., Lightman S.L.. Molecular dynamics of ultradian glucocorticoid receptor action. Mol. Cell Endocrinol. 2011; 348:383–393. [DOI] [PubMed] [Google Scholar]
- 2. Busada J.T., Cidlowski J.A.. Mechanisms of glucocorticoid action during development. Curr. Top Dev. Biol. 2017; 125:147–170. [DOI] [PubMed] [Google Scholar]
- 3. Bledsoe R.K., Montana V.G., Stanley T.B., Delves C.J., Apolito C.J., McKee D.D., Consler T.G., Parks D.J., Stewart E.L., Willson T.M.et al.. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002; 110:93–105. [DOI] [PubMed] [Google Scholar]
- 4. Evans R.M., Mangelsdorf D.J.. Nuclear receptors, RXR, and the big bang. Cell. 2014; 157:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jimenez-Panizo A., Perez P., Rojas A., Fuentes-Prior P., Estebanez-Perpina E.. Non-canonical dimerization of the androgen receptor and other nuclear receptors: implications for human disease. Endocr. Relat. Cancer. 2019; 26:R479–R497. [DOI] [PubMed] [Google Scholar]
- 6. Housley P.R., Sanchez E.R., Danielsen M., Ringold G.M., Pratt W.B.. Evidence that the conserved region in the steroid binding domain of the glucocorticoid receptor is required for both optimal binding of hsp90 and protection from proteolytic cleavage. A two-site model for hsp90 binding to the steroid binding domain. J. Biol. Chem. 1990; 265:12778–12781. [PubMed] [Google Scholar]
- 7. Ortlund E.A., Bridgham J.T., Redinbo M.R., Thornton J.W.. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007; 317:1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R.. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009; 324:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rogatsky I., Wang J.C., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R.. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13845–13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfaff S.J., Fletterick R.J.. Hormone binding and co-regulator binding to the glucocorticoid receptor are allosterically coupled. J. Biol. Chem. 2010; 285:15256–15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Estebanez-Perpina E., Arnold L.A., Nguyen P., Rodrigues E.D., Mar E., Bateman R., Pallai P., Shokat K.M., Baxter J.D., Guy R.K.et al.. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:16074–16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jehle K., Cato L., Neeb A., Muhle-Goll C., Jung N., Smith E.W., Buzon V., Carbo L.R., Estebanez-Perpina E., Schmitz K.et al.. Coregulator control of androgen receptor action by a novel nuclear receptor-binding motif. J. Biol. Chem. 2014; 289:8839–8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuentes-Prior P., Rojas A., Hagler A.T., Estebanez-Perpina E.. Diversity of quaternary structures regulates nuclear receptor activities. Trends Biochem. Sci. 2019; 44:2–6. [DOI] [PubMed] [Google Scholar]
- 14. Vandevyver S., Dejager L., Libert C.. On the trail of the glucocorticoid receptor: into the nucleus and back. Traffic. 2012; 13:364–374. [DOI] [PubMed] [Google Scholar]
- 15. Weikum E.R., Knuesel M.T., Ortlund E.A., Yamamoto K.R.. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell. Biol. 2017; 18:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clark A.R., Belvisi M.G.. Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol Ther. 2012; 134:54–67. [DOI] [PubMed] [Google Scholar]
- 17. Franco L.M., Gadkari M., Howe K.N., Sun J., Kardava L., Kumar P., Kumari S., Hu Z., Fraser I.D.C., Moir S.et al.. Immune regulation by glucocorticoids can be linked to cell type-dependent transcriptional responses. J. Exp. Med. 2019; 216:384–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sevilla L.M., Bayo P., Latorre V., Sanchis A., Perez P.. Glucocorticoid receptor regulates overlapping and differential gene subsets in developing and adult skin. Mol. Endocrinol. 2010; 24:2166–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cain D.W., Cidlowski J.A.. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 2017; 17:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oh K.S., Patel H., Gottschalk R.A., Lee W.S., Baek S., Fraser I.D.C., Hager G.L., Sung M.H.. Anti-inflammatory chromatinscape suggests alternative mechanisms of glucocorticoid receptor action. Immunity. 2017; 47:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Bosscher K., Desmet S.J., Clarisse D., Estebanez-Perpina E., Brunsveld L.. Nuclear receptor crosstalk - defining the mechanisms for therapeutic innovation. Nat. Rev. Endocrinol. 2020; 16:363–377. [DOI] [PubMed] [Google Scholar]
- 22. Louw A. GR dimerization and the impact of GR dimerization on GR protein stability and half-life. Front. Immunol. 2019; 10:1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Presman D.M., Ogara M.F., Stortz M., Alvarez L.D., Pooley J.R., Schiltz R.L., Grontved L., Johnson T.A., Mittelstadt P.R., Ashwell J.D.et al.. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 2014; 12:e1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Presman D.M., Ganguly S., Schiltz R.L., Johnson T.A., Karpova T.S., Hager G.L.. DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8236–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paakinaho V., Johnson T.A., Presman D.M., Hager G.L.. Glucocorticoid receptor quaternary structure drives chromatin occupancy and transcriptional outcome. Genome Res. 2019; 29:1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Escoter-Torres L., Greulich F., Quagliarini F., Wierer M., Uhlenhaut N.H.. Anti-inflammatory functions of the glucocorticoid receptor require DNA binding. Nucleic Acids Res. 2020; 48:8393–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia D.A., Johnson T.A., Presman D.M., Fettweis G., Wagh K., Rinaldi L., Stavreva D.A., Paakinaho V., Jensen R.A.M., Mandrup S.et al.. An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol. Cell. 2021; 81:1484–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weikum E.R., de Vera I.M.S., Nwachukwu J.C., Hudson W.H., Nettles K.W., Kojetin D.J., Ortlund E.A.. Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes. Nucleic Acids Res. 2017; 45:8596–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chrousos G.P., Loriaux D.L., Tomita M., Brandon D.D., Renquist D., Albertson B., Lipsett M.B.. The new world primates as animal models of glucocorticoid resistance. Adv. Exp. Med. Biol. 1986; 196:129–144. [DOI] [PubMed] [Google Scholar]
- 30. Luisi B.F., Xu W.X., Otwinowski Z., Freedman L.P., Yamamoto K.R., Sigler P.B.. Crystallographic Analysis of the Interaction of the Glucocorticoid Receptor with DNA. Nature. 1991; 352:497–505. [DOI] [PubMed] [Google Scholar]
- 31. Hudson W.H., Youn C., Ortlund E.A.. The structural basis of direct glucocorticoid-mediated transrepression. Nat. Struct. Mol. Biol. 2013; 20:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frank F., Okafor C.D., Ortlund E.A.. The first crystal structure of a DNA-free nuclear receptor DNA binding domain sheds light on DNA-driven allostery in the glucocorticoid receptor. Sci. Rep. 2018; 8:13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hard T., Kellenbach E., Boelens R., Maler B.A., Dahlman K., Freedman L.P., Carlstedt-Duke J., Yamamoto K.R., Gustafsson J.A., Kaptein R.. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990; 249:157–160. [DOI] [PubMed] [Google Scholar]
- 34. Schacke H., Berger M., Rehwinkel H., Asadullah K.. Selective glucocorticoid receptor agonists (SEGRAs): novel ligands with an improved therapeutic index. Mol. Cell Endocrinol. 2007; 275:109–117. [DOI] [PubMed] [Google Scholar]
- 35. Biggadike K., Bledsoe R.K., Coe D.M., Cooper T.W., House D., Iannone M.A., Macdonald S.J., Madauss K.P., McLay I.M., Shipley T.J.et al.. Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:18114–18119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hurt D.E., Suzuki S., Mayama T., Charmandari E., Kino T.. Structural analysis on the pathologic mutant glucocorticoid receptor ligand-binding domains. Mol. Endocrinol. 2016; 30:173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X., Wang Y., Ortlund E.A.. First high-resolution crystal structures of the glucocorticoid receptor ligand-binding domain-peroxisome proliferator-activated gamma coactivator 1-alpha complex with endogenous and synthetic glucocorticoids. Mol. Pharmacol. 2019; 96:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weikum E.R., Okafor C.D., D’Agostino E.H., Colucci J.K., Ortlund E.A. Structural analysis of the glucocorticoid receptor ligand-binding domain in complex with triamcinolone acetonide and a fragment of the atypical coregulator, small heterodimer partner. Mol. Pharmacol. 2017; 92:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson T.A., Paakinaho V., Kim S., Hager G.L., Presman D.M.. Genome-wide binding potential and regulatory activity of the glucocorticoid receptor's monomeric and dimeric forms. Nat. Commun. 2021; 12:1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mikuni S., Tamura M., Kinjo M.. Analysis of intranuclear binding process of glucocorticoid receptor using fluorescence correlation spectroscopy. FEBS Lett. 2007; 581:389–393. [DOI] [PubMed] [Google Scholar]
- 41. Digman M.A., Dalal R., Horwitz A.F., Gratton E.. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008; 94:2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McNally J.G., Mueller W.G., Walker D., Wolford R.G., Hager G.L.. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000; 287:1262–1265. [DOI] [PubMed] [Google Scholar]
- 43. Dobin A., Gingeras T.R.. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinformatics. 2015; 51:11.14.1–11.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cheng T.M., Blundell T.L., Fernandez-Recio J.. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein-protein docking. Proteins. 2007; 68:503–515. [DOI] [PubMed] [Google Scholar]
- 46. Canutescu A.A., Shelenkov A.A., Dunbrack R.L. Jr. A graph-theory algorithm for rapid protein side-chain prediction. Protein. Sci. 2003; 12:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gabb H.A., Jackson R.M., Sternberg M.J.. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 1997; 272:106–120. [DOI] [PubMed] [Google Scholar]
- 48. Chen R., Li L., Weng Z.. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003; 52:80–87. [DOI] [PubMed] [Google Scholar]
- 49. Fernandez-Recio J., Totrov M., Skorodumov C., Abagyan R.. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins. 2005; 58:134–143. [DOI] [PubMed] [Google Scholar]
- 50. Grosdidier S., Fernandez-Recio J.. Identification of hot-spot residues in protein-protein interactions by computational docking. BMC Bioinformatics. 2008; 9:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He Y., Yi W., Suino-Powell K., Zhou X.E., Tolbert W.D., Tang X., Yang J., Yang H., Shi J., Hou L.et al.. Structures and mechanism for the design of highly potent glucocorticoids. Cell Res. 2014; 24:713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chagoyen M., Garcia-Martin J.A., Pazos F.. Practical analysis of specificity-determining residues in protein families. Brief Bioinform. 2016; 17:255–261. [DOI] [PubMed] [Google Scholar]
- 53. Halabi N., Rivoire O., Leibler S., Ranganathan R.. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009; 138:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lockless S.W., Ranganathan R.. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999; 286:295–299. [DOI] [PubMed] [Google Scholar]
- 55. Presman D.M., Ball D.A., Paakinaho V., Grimm J.B., Lavis L.D., Karpova T.A., Hager G.L.. Quantifying transcription factor dynamics at the single-molecule level in live cells. Methods. 2017; 123:76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu X., Wang Y., Gutierrez J.S., Damsker J.M., Nagaraju K., Hoffman E.P., Ortlund E.A.. Disruption of a key ligand-H-bond network drives dissociative properties in vamorolone for duchenne muscular dystrophy treatment. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:24285–24293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hurley D.M., Accili D., Stratakis C.A., Karl M., Vamvakopoulos N., Rorer E., Constantine K., Taylor S.I., Chrousos G.P.. Point mutation causing a single amino acid substitution in the hormone binding domain of the glucocorticoid receptor in familial glucocorticoid resistance. J. Clin. Invest. 1991; 87:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nadal M., Prekovic S., Gallastegui N., Helsen C., Abella M., Zielinska K., Gay M., Vilaseca M., Taules M., Houtsmuller A.B.et al.. Structure of the homodimeric androgen receptor ligand-binding domain. Nat. Commun. 2017; 8:14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kauppi B., Jakob C., Farnegardh M., Yang J., Ahola H., Alarcon M., Calles K., Engstrom O., Harlan J., Muchmore S.et al.. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J. Biol. Chem. 2003; 278:22748–22754. [DOI] [PubMed] [Google Scholar]
- 60. Helsen C., Claessens F.. Looking at nuclear receptors from a new angle. Mol. Cell Endocrinol. 2014; 382:97–106. [DOI] [PubMed] [Google Scholar]
- 61. Bianchetti L., Wassmer B., Defosset A., Smertina A., Tiberti M.L., Stote R.H., Dejaegere A.. Alternative dimerization interfaces in the glucocorticoid receptor-alpha ligand binding domain. Biochim. Biophys. Acta. Gen. Subj. 2018; 1862:1810–1825. [DOI] [PubMed] [Google Scholar]
- 62. Noddings C.M., Wang R.Y., Johnson J.L., Agard D.A.. Structure of Hsp90-p23-GR reveals the hsp90 client-remodelling mechanism. Nature. 2022; 601:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Min J., Perera L., Krahn J.M., Jewell C.M., Moon A.F., Cidlowski J.A., Pedersen L.C.. Probing dominant negative behavior of glucocorticoid receptor beta through a hybrid structural and biochemical approach. Mol. Cell Biol. 2018; 38:e00453-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schoch G.A., D’Arcy B., Stihle M., Burger D., Bar D., Benz J., Thoma R., Ruf A.. Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations. J. Mol. Biol. 2010; 395:568–577. [DOI] [PubMed] [Google Scholar]
- 65. Gampe R.T. Jr, Montana V.G., Lambert M.H., Wisely G.B., Milburn M.V., Xu H.E. Structural basis for autorepression of retinoid x receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000; 14:2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the PDB with accession codes 7YXC (C2), 7YXD (P31), 7YXN (P61), 7YXO (I4122), 7YXP (I4132) and 7YXR (P61, Y545A). The PDB data has been designed ‘for immediate release on publication’. MS data have been deposited to the ProteomeXchange Consortium (PRIDE partner repository: dataset identifier PXD028039). Bioinformatics data and code for the clustering and SCA are available at https://github.com/ibn90/GRPROJECT2021. The Gene Expression profiles are deposited in the Gene Expression Omnibus (GEO) database and the code assigned is GSE212973. All relevant data are available upon request.