Abstract
Efforts have been made to reduce the genomes of living cells, but phage genome reduction remains challenging. It is of great interest to investigate whether genome reduction can make phages obtain new infectious properties. We developed a CRISPR/Cas9-based iterative phage genome reduction (CiPGr) approach and applied this to four distinct phages, thereby obtaining heterogeneous genome-reduced mutants. We isolated and sequenced 200 mutants with loss of up to 8–23% (3.3–35 kbp) of the original sequences. This allowed the identification of non-essential genes for phage propagation, although loss of these genes is mostly detrimental to phage fitness to various degrees. Notwithstanding this, mutants with higher infectious efficiency than their parental strains were characterized, indicating a trade-off between genome reduction and infectious fitness for phages. In conclusion, this study provides a foundation for future work to leverage the information generated by CiPGr in phage synthetic biology research.
INTRODUCTION
Bacteriophages (phages) are the most abundant (1) and genetically diverse (2–4) biological entities on earth. For more than a century, phage research has been critical to numerous fundamental biological discoveries (5) and has provided important biotechnical tools in molecular genetics (6). Recent advances in viral metagenomic sequencing have characterized a large number of phage sequences in the human gut (2,3) and various other environments (4). The majority of these sequences (>75%) are novel (4,7), and >95% belong to tailed phages with double-stranded (ds) DNA genomes (8,9). The novel tailed phages limit our understanding of phage biology and the role of phages in the ecosystem (4). In addition, phages have been recognized as natural antimicrobial agents for treating bacterial infections (10,11). However, some major concerns, e.g. extremely high host specificity for a small set of bacterial strains and the fact that some phages harbor virulence genes, remain regarding the use of wild-type (WT) phages in phage therapy (12). Therefore, many efforts have been made to genetically engineer phages to overcome these limitations (6,13).
Phage synthetic biology can integrate functional genes or gene circuits into phage genomes, bolstering their antibacterial activity and potential in various bioengineering applications (6,13). For example, phages have been integrated with additional function genes to reduce bacterial biofilms (14), detect hosts rapidly (15) and remove antibiotic resistance or virulence genes specifically from the microbiome (16,17). Such efforts for tailed phages were mostly limited to the model phages, such as T7 and T4, and the phages closely related to these models (14,18,19). This is due to the highly divergent nature of phages and the lack of knowledge of the importance of each gene of the novel phages. Moreover, the limited DNA encapsulation capability of phage particles does not allow large gene circuit integration into phage genomes (20,21). The accessory genes of phages may help phages better adapt to a wide range of ecological niches but may be non-essential for phage development under certain conditions (22). A reduction of these non-essential genes can create some space in phage genomes. This raises questions regarding the extent to which a phage genome can be reduced, especially in the case of novel phages that possess relatively large numbers of unknown functional genes (4,7). The removal of redundant genes from the phage genome can make it possible to design a phage genome that possesses more functions (23,24), paving the way for phage therapy.
Tailed phages often have relatively large (14–735 kbp) and extraordinarily diverse genomes (25). It remains challenging to reduce tailed phage genomes on a large scale. One challenge is the identification of the non-essential genes of a given phage (26). A summary of the non-essential gene set of phage T7 (25 genes, 41.7%) was obtained from accumulated knowledge derived from numerous studies conducted in recent decades (27,28). These genes are interspersed throughout the genome, hindering efforts at genome reduction in phage T7. Moreover, for some phages that package using pac or cos sites, such as T7 and lambda (28), large-scale deletion of the genome could lead to phage genome packaging failure (29,30). Approaches that are widely used in genome reduction in living cells, i.e. Escherichia coli (31), yeast (32), Bacillus subtilis (33) and Mycoplasma mycoides (26), may not be suitable for application in tailed phages due to the phages’ exclusive characteristic of self-propagation within specific hosts. Moreover, the bioinformatic method of comparing complete bacterial genomes to identify the essential gene set for cellular viability in a given bacterial species (34) often fails for phage genomes because of their highly divergent nature (4).
Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) system has been used in phage genome editing (35–39). Nevertheless, the stepwise deletion of many genomic regions is laborious and time-consuming, especially if simultaneous screening of mutant phages that are capable of maintaining robust growth is required. J. Craig Venter and colleagues divided M. mycoides JCVI-syn1.0 genes into three categories, namely essential, quasi-essential and non-essential genes, based on transposon mutagenesis. Quasi-essential genes are not critical for viability but are nevertheless required for robust growth (26). It is likely that phages also possess quasi-essential genes, and this represents another challenge for screening genome-reduced phages with robust growth from among numerous heterogeneous mutants (40).
In this work, to address these challenges, we show a top-down genome reduction approach termed CRISPR/Cas9-based iterative phage genome reduction (CiPGr). In the CiPGr process, host bacterial cells harboring a dual-plasmid system consisting of pTarget with heterogeneous spacers (41) and spCas9 are infected by a specific phage. During phage infection and injection of phage DNA into the cell, the single-guide RNA (sgRNA) encoded by pTarget causes the Cas9 nuclease (encoded by spCas9) to cleave the gene at the designed site, and the resulting break is subsequently repaired via homologous recombination (HR) induced by the HR sequence (pTarget), causing gene deletion or disruption (35). If the disrupted or deleted gene is not essential for phage propagation, the mutant phage progeny can multiply. The mutant phage population is continuously transferred to fresh cells harboring the dual-plasmid system, and gene deletions or disruptions accumulate in the phage genome. The generated genome-reduced phages that possess growth advantages will be predominant in the population and are finally selected. We applied CiPGr to four diverse tailed phages (T7, T4, seszw and selz) and characterized the resulting mutant phages that possessed growth advantages compared with their WT strains. The results demonstrate the high value of CiPGr in phage synthetic biology.
MATERIALS AND METHODS
Bacteria, phages, plasmids and media
Escherichia coli strain MG1655, phage T4 and phage T7 are model strains. Salmonella enterica strain was isolated from the Shenzhen University General Hospital (Shenzhen). Genome sequencing showed a high similarity (average nucleotide identity = 99.8%) to that of Typhimurium strain ST56 (CP050739.1). Two WT phages, namely seszw and selz, were isolated in-house from water samples collected in Shenzhen and Lanzhou, China, respectively, using S. enterica (ST56) as the host bacterium. T7 (39.9 kbp) and T4 (168.9 kbp) are model phages that specifically infect E. coli strains and belong to the families Autographiviridae and Straboviridae, respectively. Phages seszw (45.8 kbp) and selz (154.8 kbp) were newly isolated from sewage water samples using S. enterica (ST56) as the host (Supplementary Figure S1A). Phage selz was classified as a member of the family Ackermannviridae (42), and seszw was classified as a member of the genus Skatevirus (Supplementary Figure S2). To our knowledge, T7 uses pac sites to produce specific ends by endonuclease at the terminal repeat (TR); T4 uses a headful packaging strategy (43). The packaging mechanism of the newly isolated phage selz is probably similar to that of T4, because the large subunit terminase (TerL) of phage selz is highly similar to that of T4. The packaging mechanism of phage seszw is unknown.
We adapted the type II CRISPR/Cas9 system from Streptococcus pyogenes for phage genome editing (35,44). The plasmids pTarget and spCas9 (45) were obtained from the laboratory of Chenli Liu, Shenzhen Institute of Synthetic Biology. The backbone of the spCas9 plasmid was amplified from the pCas plasmid (Addgene plasmid id: 62225). The plasmid of spCas9 consists of Cas9, lambda-red, an antibiotic-resistant gene and the SacB gene. The temperature-sensitive replicon of spCas9 has two point mutations (9697 and 10 395, A to G) compared with that of pCas. The backbone of the pTarget plasmid was amplified from the plasmid (Addgene plasmid id: 62226) with the modifications of an antibiotic-resistant gene and spacer (Supplementary Figure S1B). The spCas9 plasmid was transformed into E. coli MG1655 and S. enterica (ST56) grown in Luria–Bertani (LB) broth supplemented with 50 μg/ml kanamycin, to generate the E. coli MG1655 (spCas9) and S. enterica ST56 (spCas9) strains. LB containing 0.7% agar was used as a solid medium.
Design, construction and delivery of the CiPGr plasmid library
We designed a 200 bp CiPGr cassette to meet the requirement for on-chip DNA synthesis. The sequences of the barcode (20 bp), the HR template (50 × 2 bp), the promoter (36 bp), the spacer (20 bp) and the primer (24 bp) were arranged sequentially on the cassette (Figure 1A). For each gene, cassette-100 and cassette-gene used the same spacer while we designed different homologous arms for each of the cassette-100 spacers and the same homologous arm for all cassette-gene spacers. All gene sequences of the four phages were predicted and annotated (Supplementary Table S1). The phage genomes were examined using sgRNAcas9 V3.0 (46) to identify all possible protospacer sequences, and the most suitable protospacers were selected. Known essential genes that have been confirmed in other studies or by a homology search in NCBI were excluded (Supplementary Table S2). We synthesized the designed cassette oligonucleotides on a 6000-format chip (Genewiz). The cassette-100 and cassette-gene cassettes for each phage were separated by polymerase chain reaction (PCR; Supplementary Table S3; Phusion DNA polymerase, New England BioLabs, M0530). The cycling conditions were as follows: 98°C for 2 min; 24 cycles of 98°C for 10 s, 58°C for 20 s and 72°C for 6 s; 72°C for 10 min; hold at 4°C.
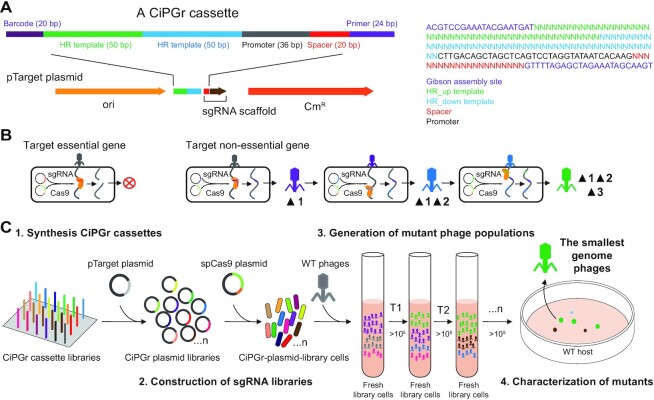
Figure 1.
Schematic diagram illustrating the CiPGr approach. (A) Design of the CiPGr cassette. Heterogeneous barcodes (20 bp) were designed and used to separate the cassettes into eight libraries by PCR using primer (24 bp) sequences; the primer was part of the sgRNA sequence and was designed as the Gibson assembly site for the cassettes and the pTarget plasmid. CmR, chloramphenicol resistance gene; HR, homologous recombination; ori, origin of replication. (B) Continuous genome editing of the phages using the dual-plasmid system. Essential genes were resistant to editing; non-essential genes were deleted or disrupted, and mutations accumulated in the phage genome. (C) Workflow of the CiPGr approach. Step 1: the CiPGr cassettes were synthesized on a DNA chip. Step 2: the CiPGr cassettes were inserted into pTarget and delivered to MG1655 or Salmonella (ST56) host strains containing the spCas9 plasmid, resulting in eight CiPGr plasmid libraries. Step 3: the iterative CiPGr process was initiated by adding WT phages (T7, seszw, T4 and selz) to cultured cells carrying the libraries, followed by repeated transfer of the mutant phage populations to fresh cultures. Step 4: the mutant phages were isolated, characterized and sequenced, and those with the smallest genomes were identified.
The plasmid pTarget backbone was amplified using PCR. Both the plasmid backbone and the pooled cassette sequences were purified by gel electrophoresis. Each cassette pool and the plasmid backbone were assembled in four parallel Gibson assembly reactions (New England BioLabs, E2611 L), and the resulting DNA was pooled and purified using the GeneClean Turbo Kit (MP Biomedicals, 111102400). The purified Gibson assembly products were transformed into DH5α electrocompetent cells. A total of 2–10 parallel transformation assays were performed. The cells were pooled and plated on 15 cm2 Petri dishes with 20 μg/ml chloramphenicol and 50 μg/ml kanamycin. Colonies were allowed to grow overnight at 37°C. The total number of colony-forming units was estimated to be 105–106 (Supplementary Table S4).
The colonies were then scraped off the plates, and the plasmids were extracted from the colonies using the TIANprep Mini Plasmid Kit (Tiangen, DP103). The eight plasmid libraries (one cassette-100 and one cassette-gene library for each phage) were electrotransformed into E. coli MG1655 (spCas9) and S. enterica ST56 (spCas9) cells, yielding >105 colonies on LB plates containing 20 μg/ml chloramphenicol and 50 μg/ml kanamycin. This transformation generated a dual-plasmid system [spCas9 and pTarget (cassette-100 or cassette-gene)] in each bacterial cell. To evaluate the coverage of the designed cassette sequences in these plasmid libraries, Phusion PCR (New England BioLabs, M0530) targeting the cassette fragments was conducted, and the PCR products were sequenced on a HiSeq 2500 sequencer (Illumina). The raw sequencing data from the adaptor region were removed to produce clean data. We mapped the sequencing data to the designed cassette sequences and determined the ratio of each cassette sequence with 100% identity in the plasmid libraries.
Generation and metagenomic sequencing of mutant phage populations
The bacterial cells harboring the plasmid libraries were grown from 300 μl aliquots frozen at −80°C in 15 ml of LB medium with antibiotics and l-arabinose overnight at 37°C. Each phage (109) was co-cultured for 2–12 h with cells (109) containing the corresponding library at 37°C, as shown in Supplementary Table S5 (T1), to generate heterogeneous mutant phages; then, 1 ml of the supernatant containing the mutant phages was transferred to a fresh library cell culture. These transfers were conducted hundreds of times. The remainder of the supernatant generated at each transfer was stored at 4°C for further analysis. The titer of the mutant phages was measured at appropriate time points by a spotting assay. If the number of mutant phages was <105 plaque-forming units (PFU)/ml, the original host cells were used to cultivate the mutant phage population to expand the population. PCR was used to target some genes to monitor the occurrence of phage gene editing. A 1 μl aliquot of the culture supernatant was used as the template for PCR using Ex Taq DNA polymerase (Takara, RR01AM). Amplification was performed using appropriate primers flanking the genes of interest. The obtained products were verified by agarose gel electrophoresis and Sanger sequencing.
To monitor the efficiency of gene deletion, total DNA was extracted from the mutant phage populations according to the instructions provided with the Phage DNA Isolation Kit (NORGEN, 46850) and subjected to metagenomic sequencing using an Illumina HiSeq1500 sequencer. We sequenced the mutant phage populations present at the 20th, 30th, 40th and 50th transfers of phages T7 and T4 and sequenced all of the mutant populations every 50–100 transfers.
Screening, characterization and genome sequencing of the isolated mutant phages
To obtain isolates of the mutant phages, up to 103 PFU of mutant phages, 300 μl of the original host cells and 3 ml of 0.7% top LB agar were mixed in a tube and poured onto LB plates. The plates were incubated at 37°C overnight. The eight largest and eight smallest plaques were picked and purified. To determine the bacterial killing curves of the isolated mutant phages, the host cells were grown overnight, 10 μl of the culture was added to 96-well plates with 200 μl of fresh LB medium and the single plaques were purified and transferred to the wells. The OD600 was monitored on a microplate reader at 37°C with shaking for 12 h. Each experiment was repeated three times.
The mutant phages were selected according to their infection dynamics and further purified by the plate streaking method. Briefly, 300 μl of suspensions of the original E. coli or Salmonella cells (∼108 cells/ml) were mixed with 3 ml of 0.7% top agar and poured onto LB plates. The mutant phages were streaked on the plate and cultured overnight. Single plaques were then picked using a sterile pipette tip and transferred to a 1.5 ml Eppendorf tube containing 200 μl of LB medium. Streaking of the samples was performed three times to isolate single plaques. Genome sequencing of the isolated mutant phages was performed as described above.
The one-step growth experiment was conducted as previously described (47,48). Briefly, 10 ml of exponential-phase host cells (∼2 × 108 cells/ml) were centrifuged (7000 g, 3 min) and resuspended in 1 ml of fresh LB medium. The selected mutant phages (100 μl, ∼2 × 108 phages/ml) were added to obtain a multiplicity of infection (MOI) of 0.01, and phages were allowed to adsorb for 5 min at 37°C. After this, 9 ml of fresh LB medium was added and incubated at 37°C with agitation (220 rpm). One sample (10 μl) was immediately taken before incubation. Samples were withdrawn from the mixture every 3 or 10 min until 60 min of growth. The samples (10 μl) were serially diluted and plated to count the PFUs. The burst size was the ratio of the final number of PFUs during the latent period to the initial number of PFUs during the plateau period (48). Each experiment was repeated three times.
Screening, characterization and genome sequencing of mutants with increased infectious efficiency
To screen the mutants with high infectious efficiency, for each of the four phages, the mutant populations (10 μl of the culture supernatant) generated by the library of cassette-100 or the library of cassette-gene were collected every 10 transfers and the mutant populations generated by the same library were mixed together (30–60 populations for each library). Each mixture of mutant phages (105) was co-cultured with cells of the corresponding host strain (logarithmic growth, 109) for 2 h (T1), and 10 μl of the resulting culture supernatant was then transferred to a fresh host cell culture. These transfers were conducted up to eight times (T8, 16–20 generations). To obtain isolates of the mutant phages with the highest infectious efficiency, up to 103 PFU of T8 mutant phages, 300 μl of the original host cells and 3 ml of 0.7% top agar were mixed in a tube and poured onto LB plates. The plates were incubated at 37°C overnight, and three plaques were then picked and purified. To characterize and evaluate the infectious efficiency, we conducted an assay to determine the bacterial killing curves for each of the isolated mutants following a previously described protocol (49,50). Briefly, the host cells were incubated in fresh LB medium at 37°C until reaching an OD600 of 0.3–0.4 (∼108 cells/ml), 200 μl of the bacterial culture was added to 96-well plates and the mutant phages were transferred to the wells at an MOI of 0.1. The OD600 was monitored every 5 min for 12 h on a microplate reader at 37°C with shaking. Each experiment was repeated three times.
Phage competition assays
WT phage (105) and corresponding optimal mutant (105) were mixed equally, and the mixture was co-cultured with cells of the corresponding host strain (original MG1655 or ST56) (logarithmic growth, 109) for 2 h (T1), and 10 μl of the resulting culture supernatant was then transferred to a fresh host cell culture. These transfers were conducted up to four times (T4, 8–10 generations). The relative abundance of WT phage and optimal mutant phage in the culture supernatant were determined by SYBR Green QPCR (51,52). A 10 μl PCR mixture was prepared with 5 μl of 2 x SYBR Green Master Mix, 0.2 μl 10 μM primers, 3.6 μl of UltraPure dH2O and 1 μl of denatured phages. The cycling conditions were as follows: 95°C for 5 min; 40 cycles of 95°C for 10 s, 60°C for 30 s; followed by the melt curve setting of one cycle of 95°C for 15 s, 60°C for 60 s and 95°C for 15 s. For each run, duplicates of five 10-fold dilutions of phages, samples and no template controls were simultaneously subjected to analysis.
Fitness of WT phages and optimal mutants
Phage fitness refers to the maximum doubling rate and is expressed as doublings per hour of the phage population under conditions of unlimited hosts (53–55). The fitness measure to determine the phage growth rate is directly comparable among phages, regardless of differences in phage generation time, burst size or other fitness components. The fitness experiment was conducted as previously described (40). Briefly, the fitness of the phages was determined by adding the phages (105 PFU/ml) and exponentially grown original MG1655 or Salmonella (ST56) cells (108 PFU/ml) to 3 ml of LB broth at an MOI of 0.001. The mixture was incubated at 37°C for 30 min. Then, 10 μl of the mixture was transferred to a new tube with fresh original WT host cells and broth every 30 min, and the phage titer was determined with double layer plaque assays. Low phage densities (<107 PFU/ml) were maintained throughout the assay to ensure the condition of unlimited hosts.
Integration of foreign DNA into the mutant phage genome
Integration of the DNA fragments into the genome of T7 300L (behind gp19) was performed using the CRISPR/Cas9-assisted method (35). Three DNA fragments with no predicted open reading frames (ORFs) were obtained from Saccharomyces cerevisiae chromosome I (I: 485 bp, 6546–7030; II: 1000 bp, 359–6358; III: 999 bp, 4001–4999) by PCR. We integrated fragment I into the genome of T7 300L to obtain the 300L-0.5k, fragment II into 300L-0.5k to obtain 300L-1.5k, and fragment III into 300L-1.5k to obtain 300L-2.5k. The spCas9 (45) plasmid was first transformed into MG1655. The 1 kbp HR templates, the DNA fragments (I, II and III) and the sgRNA scaffold were assembled into the pTarget plasmid backbone using Gibson assembly reactions (New England BioLabs, E2611 L). The resulting products were amplified in E. coli DH5α and transformed into MG1655. Subsequently, the dual-plasmid-containing MG1655 cells (fragment I) were infected with the T7 300L mutant phage, and the 300L-0.5k mutant was purified by the plate streaking method as described above. The 300L-1.5k mutant was obtained by using 300L-0.5k to infect dual-plasmid-containing MG1655 cells (fragment II) and the 300L-2.5k mutant was obtained by using 300L-1.5k to infect dual-plasmid-containing MG1655 cells (fragment III). The mutants (300L-0.5k, 300L-1.5k and 300L-2.5k) were verified by PCR and Sanger sequencing.
Data analysis
The frequency of gene deletion or disruption is shown as the percentage of edited genes within one population. We used the open-source computational pipeline breseq (version 0.28.0) to predict both point mutations and gene deletions or disruptions based on mapping of the reads in each population to the reference genome sequences of the original phages (56,57). The lists of gene deletions or disruptions were manually resolved from the unassigned new junction and missing coverage evidence in the breseq output. The frequency of occurrence of gene deletion or disruption was manually confirmed by counting the number of mutant reads and WT reads in the metagenomic sequencing data of the populations. The pheatmap R package (version 1.0.12) (https://cran.r-project.org/web/packages/pheatmap/) was used to generate heatmaps of the frequency of gene deletion or disruption in the populations. Pearson correlation coefficients and P-values (unpaired t-test) (50) were obtained using SPSS software (version 17). Regression analyses of the number of deleted or disrupted genes and the genome size as a function of the number of transfers were performed using SPSS software. Statistical analysis of quantitative PCR (QPCR) was carried out using triplicate cycle threshold (CT) values as determined by automated threshold analysis with Jena qPCRsoft 4.1. CT data were normalized by log10 transformation using SPSS and CT-values which were converted to QPCR-derived phage titer (PFU/ml) using a calibration curve (58). The fitness was calculated as ω = log2 (P2/P1), where P2 and P1 are the numbers of phages at time points of 2 h and 1 h, respectively (54,59–61). The sequence reads of the mutant phages were de novo assembled using SOAP-denove (V2.04-r241) (62). To calculate the ratio of non-synonymous (Ka) to synonymous (Ks) substitutions, TBtools software (63) was used. The CRISPR/Cas9 system might accelerate phage evolution in the spacer and protospacer adjacent motif (PAM) regions (64). To eliminate bias, all the genes for which we designed spacers were uniformly ignored in the downstream analyses of Ka/Ks.
RESULTS
Development of the CiPGr approach
The CiPGr approach was performed according to the scheme presented in Figure 1. First, to delete or disrupt the phage gene of interest, the CRISPR spacer and HR templates were designed as parts of a single oligonucleotide (CiPGr cassette). The 200 bp CiPGr cassette harbored a 20 bp spacer sequence, a 36 bp promoter sequence and a 100 bp HR template sequence with 50 bp arms (Figure 1A). We added barcode (20 bp) and primer (24 bp) sequences to the ends of the cassette. This design ensured that the cassette sequences could be synthesized on a DNA chip. Second, the synthesized CiPGr cassettes for different phages were separated using the primer and the barcodes, and integrated into the plasmid pTarget (Figure 1B). The CiPGr pTarget plasmids were pooled as different libraries and used to transform E. coli MG1655 (spCas9) or S. enterica ST56 (spCas9) cells, thereby generating CiPGr plasmid library cells. Third, the phage genome reduction process was initiated by adding the WT phages to cultures of the corresponding CiPGr plasmid library cells (Figure 1C). Due to the heterogeneous pTarget plasmids that made up each library, various mutant phages were generated. These mutant phage populations were continuously transferred to fresh cultures of CiPGr plasmid library cells, and gene deletions or disruptions accumulated in the mutant progenies as the phage populations were amplified. Because the mutant phages in the amplified populations were expected to exhibit varying levels of growth fitness, mutants that had a growth advantage were expected to predominate in the population after a series of transfers.
We applied CiPGr to four different tailed phages: T7, T4, seszw and selz (Supplementary Figure S1A). We designed two types of CiPGr cassettes for each gene; one cassette (cassette-100) was designed to delete a 100 bp fragment, and the other (cassette-gene) was designed to delete the entire gene. Our initial reason for using cassette-100 was to disrupt phage genes while not greatly reducing the size of the genome. We selected eight spacers for each gene due to the likely variation in the on-target activities of different sgRNAs; these spacers spanned the entire gene, ensuring successful gene editing. For some genes, fewer spacers were selected because the gene sequences were short or contained few predicted spacers. We did not design CiPGr cassettes that target annotated structural genes, previously reported essential genes (13,27) or likely regulatory elements.
In total, we designed and synthesized 5828 unique CiPGr cassettes targeting 926 genes of the four phages with an average of 5–6 cassettes per gene. Cloning and delivery of the eight pooled CiPGr plasmid libraries into E. coli MG1655 (spCas9) or S. enterica ST56 (spCas9) yielded 2 × 105 to 2 × 106 colonies and achieved 176–5865 colonies per individual plasmid in the libraries (Supplementary Table S3). We assessed the CiPGr plasmid libraries by next-generation sequencing (NGS) and observed that 42.38% (±2.98%) of the cloned CiPGr cassette sequences in the libraries shared 100% identity with the designed sequences. These correct sequences showed that each of the eight libraries captured 97.34% (±3.36%) of the designed cassette sequences, covering 99.33% (±0.98%) of the genes of interest, and indicated the completeness of our genome-wide engineering approach (Supplementary Table S5; Supplementary Figure S3). Some of the designed CiPGr cassette sequences (218, 3.74%) were not detected in the libraries, probably due to the low sequencing depth or to high error rates in DNA synthesis. Next, the four WT phages were used to infect CiPGr plasmid library cells (Figure 1C). Serial transfer of the mutant populations of the four phages in the eight libraries was conducted 300–600 times, corresponding to >2000 generations of the phage life cycle (Supplementary Table S6). Taking the genes gp4.7 and gp5.3 of phage T7 as examples, we detected gene deletions by PCR in the mutant phage populations derived by serial transfer and found that the mutant with disrupted gp5.3 and the mutant with disrupted gp4.7 gradually increased and the mutant with disrupted gp5.3 became dominant in the populations (Supplementary Figure S4A), confirming the feasibility of the approach.
Identification of the removable gene sets of the phages
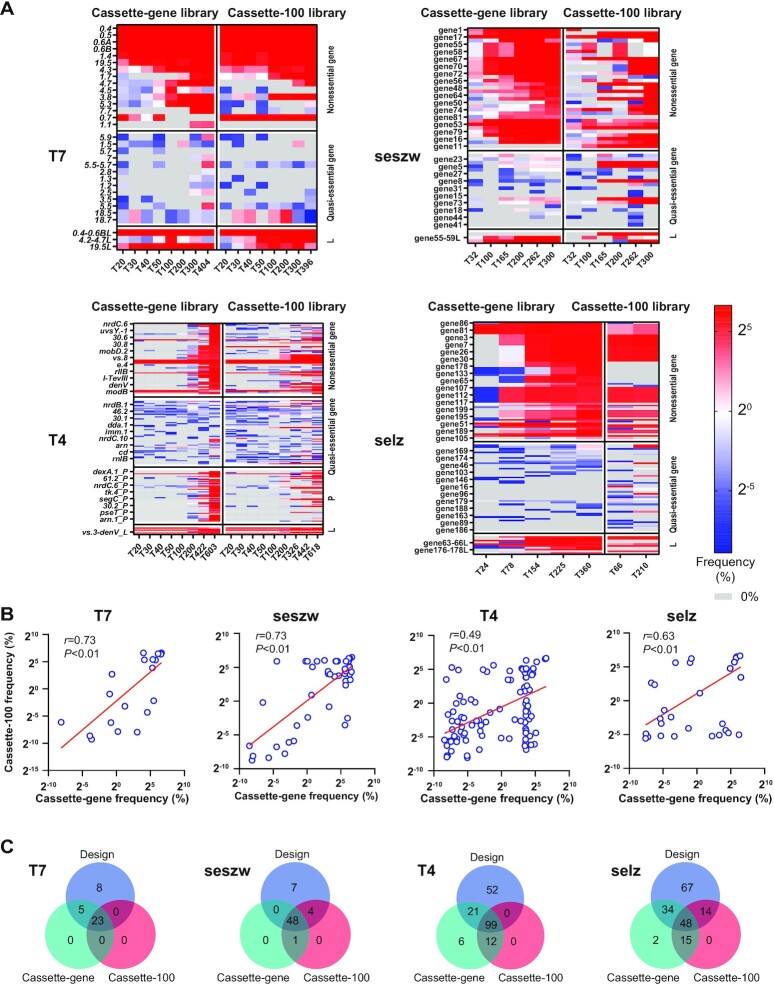
We initially assumed that the removal of some genes from the phage genomes would not impair the phage life cycle but that it might have various levels of impact on their growth fitness. To detect the removable gene sets of the phages, we performed metagenomic sequencing of the mutant phage populations generated during serial transfer. The metagenomic sequencing results obtained using the two types of plasmid libraries (cassette-gene and cassette-100) demonstrated that the numbers of disrupted and deleted genes that we designed were 28 in T7 (46.7% of 60 genes, 39 937 bp), 53 in seszw (65.4% of 81 genes, 45 881 bp), 119 in T4 (42.9% of 277 genes, 168 903 bp) and 95 in selz (47.8% of 201 genes, 154 811 bp). Loss of large fragments from the genomes of the mutants of the four phages was also observed; this caused the number of deleted or disrupted genes to increase by 0 in T7, by 1 in seszw, by 7 in T4 and by 18 in selz (Figure 2A). We detected three different large fragments (gp0.3–0.6, gp4.3–4.7 and gp19.5) in phage T7, three (genes 3–10, 55–59 and 69–71) in seszw, seven (modA–B, nrdG–D, nrdC.1–6, nrdC.11–mobD.3, vs.3–denV, ipIII and e.2–e.8) in T4 and 10 (genes 2–7, 23–31, 51–52, 63–66, 78–83, 107–117, 130–133, 176–178, 189–195 and 200) in phage selz (Figure 2A; Supplementary Table S1). Moreover, 1322 one- or two-base deletions were detected in the metagenomic sequencing reads in the mutant populations of T4 only; these resulted in the disruption of an additional 12 genes. These losses were not part of the experimental design, and some of them can probably be explained by the occurrence of CRISPR-escape mutations (CEMs) (65).
Figure 2.
Removable gene sets of the phages. (A) The frequencies of mutation (deletion and disruption) of genes in the removable gene sets of the phages T7, seszw, T4 and selz were measured in the mutant phage populations obtained at the nth transfer (T, transfer; n, number of transfers) generated by the two types of plasmid libraries (cassette-gene and cassette-100). The frequency represents the percentage (log2) of the corresponding reads of the gene (deleted or disrupted) in the population; accordingly, heatmaps were generated by the pheatmap R package. ‘L’ denotes the deletion of a large fragment that we did not design; ‘P’ denotes the premature termination codons in the coding regions that cause gene disruption by one- or two-base deletions. (B) Scatter plots of the average frequencies of the mutated genes in the two types of plasmid libraries. The Pearson correlation coefficients (r) of the frequencies for the two types of libraries and the P-values (p) were calculated using SPSS software. Each blue circular ring represents a gene in the removable gene set. The red lines are y–x regression lines for the frequencies of mutated genes. (C) Venn diagrams showing the overlap of the removable gene sets detected by the two types of libraries with the gene set that we designed for mutation.
For the model phage T7, 25 non-essential genes have been identified in previous studies (28). In this study, we detected 80% (20) of these non-essential genes (five genes, gp0.3, gp1.6, gp1.8, gp6.3 and gp6.5, were not detected), and an additional eight genes were deleted or disrupted (gp1.3, gp1.7, gp2.5, gp3.5, gp5.5–5.7, gp7.7, gp18.5 and gp18.7). For another model phage, T4, 16 non-essential genes and 60 auxiliary genes were reported in a previous study (66). Here, CiPGr deleted or disrupted 138 genes, including 13 (81.3%) non-essential genes, 32 (53.3%) auxiliary genes, 86 (67.7%) hypothetical genes and 7 other functional genes (Figure 2A; Supplementary Table S1). Overall, the numbers of genes in the removable gene sets were 28 in T7 (46.7%), 138 in T4 (50%), 53 in seszw (65.4%) and 113 in selz (56.2%) (Supplementary Table S7). Among the detected removable genes, 57.1% of those in T7, 61.9% of those in T4, 75.5% of those in seszw and 75.2% of those in selz were annotated as hypothetical genes (Supplementary Table S7), indicating that the four phages have distinct tolerances to gene deletion.
The removable gene sets of these phages detected by the two types of libraries (cassette-100 and cassette-gene) were for the most part consistent (Figure 2B; Supplementary Table S1). The cassette-100 plasmid library was designed to disrupt genes by deleting 100 bp, and the cassette-gene plasmid library was designed to delete entire genes. Due to this design, the cassette-100 plasmid library resulted in less genome reduction than did the cassette-gene plasmid library. The consistent results obtained using the two types of libraries suggest that the amount of genome reduction did not affect the identification of the removable gene sets in our study. For both T7 and seszw, the removable gene sets exhibited high correlations (r = 0.73, T7; r = 0.73, seszw) in the frequency of gene deletion or disruption in the populations derived using the two types of CiPGr plasmid libraries (Figure 2B); for T4 and selz, the correlations were lower (r = 0.49, T4; r = 0.63, selz) (Figure 2B). More importantly, we observed no correlation between the frequency of gene deletion or disruption and the abundance of the corresponding CiPGr cassette in the plasmid library (Supplementary Figure S5A, B), implying that the frequency of gene deletion or disruption did not depend on the abundance of the corresponding CiPGr cassette in the plasmid library.
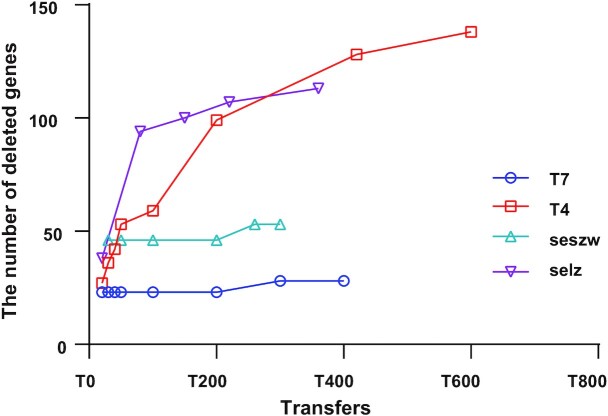
In examining the occurrence of gene deletion and disruption after serial transfers, we observed that the removable gene sets detected by CiPGr were becoming saturated. Between the 20th transfer and the last, the number of removable genes increased by 18.5% for T7, by 8.16% for seszw, by 57% for selz and by 78% for T4. However, in the last ∼100 transfers, the number of removable genes increased by 0% for T7 and by only 6.1% for seszw, 7.6% for selz and 10.8% for T4 (Figure 3). When we took only the genes that we designed for deletion and disruption into account, we obtained a similar result (Supplementary Figure S4B). The majority of the removable genes of T7 and seszw (81.5% of T7 and 91.84% of seszw) were removed in the early transfers (by the 20th transfer in T7 and by the 32th transfer in seszw), while the number of removable genes of T4 and selz gradually increased as the number of transfers increased. This was probably caused by the poor cleavage efficiency of the CRISPR system due to DNA modification (64,67,68). Nevertheless, our results demonstrate the versatility of CiPGr in detecting removable gene sets in a variety of tailed phages.
Figure 3.
Numbers of removable genes detected in the mutant phage populations. The y-axis shows the number of deleted or disrupted genes in the mutant phage populations, and the x-axis shows the number of transfers.
Phenotypes and genotypes of the mutant phages
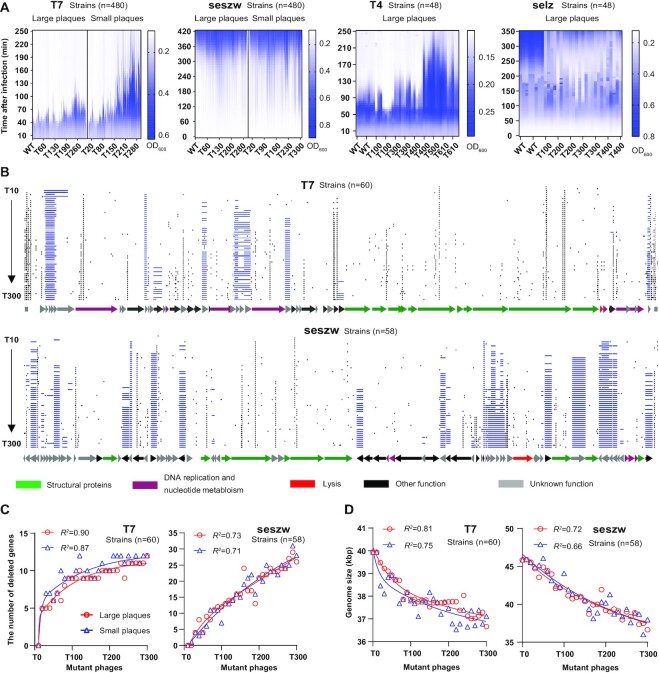
To determine the impact of gene deletion on phage phenotype, we first determined the dynamics of infection of bacterial cells by these mutants on a large scale as a way of evaluating their ability to kill bacterial cells [E. coli strain MG1655 and S. enterica (ST56) that had not been transfected with the dual plasmid system]. To study the effects of removing the genes in the removable gene sets, we mainly focused on the mutant phages of T7 and seszw that were generated in the cassette-gene plasmid libraries. Eight large and eight small plaques (Supplementary Figure S6A) were picked, and the phages obtained from these plaques were purified every 10 transfers, resulting in 480 mutants for each phage from the mutant populations over a total of 300 transfers. The infection dynamics indicate that deletion of genes decreases the ability of phages to kill their hosts to various degrees. We observed that the performance of most of the large-plaque mutants in killing their hosts (killing time of T7 mutants: 66.5 ± 10 min and of seszw mutants: 270 ± 76 min) was better than that of the small-plaque mutants (killing time of T7 mutants: 108 ± 36 min and of seszw mutants: 240 ± 76 min) (Figure 4A). Moreover, the killing abilities of both large- and small-plaque mutants of T7 and seszw became weaker as the number of transfers increased [killing time of T7 mutants: 40–250 min (WT, 50 min); killing time of seszw mutants: 340–90 min (WT, 320 min)] (Figure 4A). Only a small number of the mutants acquired increased ability to kill their hosts (17.9% in T7, 1.5% in seszw) compared with the parental phages.
Figure 4.
Phenotypes and genotypes of the isolated mutant phages. (A) Concentrations of the cells co-cultured with the isolated mutant phages shown as OD600 values. L, large plaques; S, small plaques. Tn, the nth transfer. The vertical axis displays the culture time, and the horizontal axis represents the isolated mutant phages sampled at the nth transfer. We use killing time to evaluate the ability of phages to kill their hosts. For the phages seszw and selz, phage-resistant bacteria emerged rapidly and caused an increase in OD600. The longer time required for the OD600 of S. enterica to sharply increase (seszw, OD600<0.2: selz, OD600<0.4) indicates the higher killing abilities of the phages. (B) Genome maps of the T7 and seszw mutant phages. Blue lines show gene deletion regions. Black lines show point mutations. Arrows represent genes; the directions in which the arrows point correspond to the directions of transcription and translation. (C) Increase in the number of genes that were absent from the genomes of the isolated mutant phages as a function of the number of transfers. The red and blue lines show the regression coefficients between the number of transfers of the phages and the number of mutated genes accumulated in the genomes of their large- and small-plaque mutants, respectively. (D) The red and blue lines show the regression coefficients between the number of transfers and the sizes of the genomes of the isolated mutant phages.
For T4 and selz, we picked and purified eight plaques every 100 transfers (T4, from the 100th transfer to the 600th, n = 48; selz, from the 100th transfer to the 400th, n = 48) and evaluated their infection dynamics. The T4 mutants showed a trend similar to that of the T7 mutants (Figure 4A), in that the ability of the phages to kill the host cells became weaker (the killing time increased from 80 min to 250 min) as the number of transfers increased; however, a high percentage (25%) of the mutants showed an increased ability to kill cells (killing time <115 min) compared with WT T4 (Figure 4A). Interestingly, gene deletion in the phage selz increased the killing time (OD600 <0.4) from 235 min to 345 min (Figure 4A). These results demonstrate that the impact of phage genome reduction on phage growth was heterogeneous but was mostly detrimental to phage propagation.
To link the genotypes of the mutant phages to their phenotypes, we next sequenced 60 mutant phages selected from among the 480 sampled plaques of T7 and seszw. For each sampling point, we selected the mutants with the strongest and weakest host-killing abilities based on their killing curves (Supplementary Figure S6B). When we compared these 60 mutant phage genomes of T7 (Figure 4B; Supplementary Figure S7) and seszw (Figure 4B; Supplementary Figure S8), we observed that gene deletions accumulated in the genomes of the T7 and seszw mutants as the number of transfers increased, as expected. In the genomes of the T7 mutants, nine genes (2 kbp) were deleted in transfers 1–100, one gene (0.1 kbp) was deleted in transfers 100–200 and two genes (1.1 kbp) were deleted in transfers 200–300; in the genomes of the seszw mutants, 13 genes (4.7 kbp) were deleted in transfers 1–100, eight genes (1.9 kbp) were deleted in transfers 100–200 and five genes (2.5 kbp) were deleted in transfers 200–300. Plotting the number of serial transfers versus the number of deleted genes (T7: R2 >0.87, seszw: R2 >0.71) and versus the sizes of the mutant phage genomes (T7: R2 >0.75, seszw: R2 >0.66) yielded rarefaction curves (Figure 4C, D). The T7 mutants with the strongest and weakest host-killing abilities showed highly similar rates of gene deletion from their genomes as the number of transfers increased. Thus, we were able to extrapolate from these observations (Figure 4C, D) that the number of deleted genes approached a maximum in the T7 genome. In the case of seszw, the number of deleted genes continued to increase, implying that more genes could have been deleted if more rounds of CiPGr had been conducted.
Screening and characterization of mutant phages with increased infectious efficiency
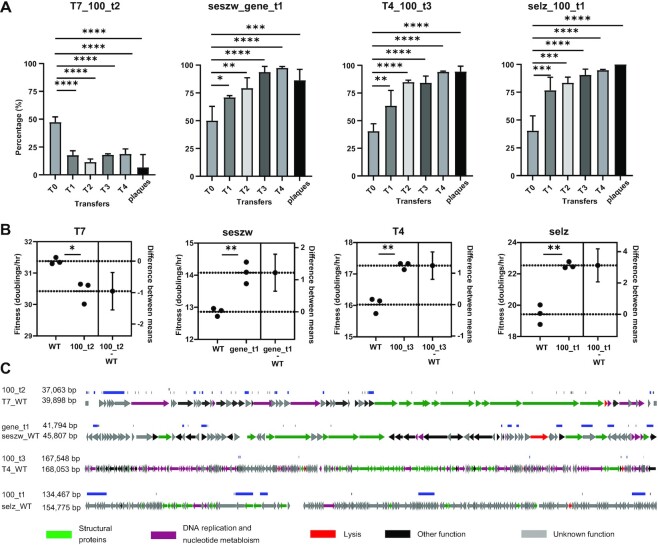
Next, to screen for mutant phages that possessed higher infectious efficiency among the heterogeneous mutants generated by CiPGr, the mutant populations obtained for each of the four phages were mixed and co-cultured with the original host cells that had not been transfected with the dual plasmid system. After eight serial transfers, the mutants with higher infectious efficiency killed their hosts faster and became predominant in the populations. We eventually isolated 24 mutants from the populations in the final transfer (six for each phage). To compare the infectious efficiency, we determined the killing curves for each of these isolated mutants and their WT strains (69). We observed that the killing times of the optimal mutants in infecting their bacterial hosts decreased by 10% for T7_100_t2 (P = 0.05), by 64.8% for T4_100_t3 (P < 0.01), by 53.1% for seszw_gene_t1 (P <0.01) and by 63.1% for selz_100_t1 (P <0.01) compared with the killing times of their parental phages (Supplementary Figure S9). WT T7 was nearly optimal in killing its host. The observation regarding T7 is consistent with a previous result obtained by simulation (55).
To verify rigorously the results by the killing curves, we thus selected the optimal mutant and its WT strain for each of the four phages to perform the phage competition assay and the fitness assay. For the phage competition assay (70), the mixtures of the optimal mutant and the corresponding WT phage for each of the four phages were made equally, and serial transfers with their original hosts were conducted. The mutant or WT phage that had a high infection efficiency was expected to become dominant in the population after serial transfer. To detect the abundance of the optimal mutant and the WT phage in the population, eight primer pairs were designed for further QPCR investigation (Supplementary Figure S10). We observed that the optimal mutants for seszw, T4 and selz had an increased abundance with the serial transfer compared with the WT strains during the competition assay. After four transfers, the relative abundances of the optimal mutants for seszw (97.45 ± 1.23%), T4 (94.22 ± 0.69%) and selz (94.86 ± 0.58%) were >90%, while that of the optimal mutant of T7 (18.75 ± 4.55%) was <20% (Figure 5A). The result of the last transfer was verified by double layer plaque. Thirty-six plaques were picked randomly from the last transfer products for each phage and identified by PCR. The observation was consistent with the result obtained by QPCR (Figure 5A). The fitness of the optimal T7 mutant (T7_100_t2) decreased by 0.9 ± 0.2 (P = 0.01) doublings per hour compared with that of the WT strain (Figure 5B), but that of the optimal seszw mutant (seszw_gene_t1) increased by 1.2 ± 0.2 (P <0.005) doublings per hour, that of the optimal T4 mutant (T4_100_t3) increased by 1.2 ± 0.17 (P <0.005) doublings per hour and that of the optimal selz mutant (selz_100_t1) increased by 3.1 ± 0.38 (P <0.005) doublings per hour compared with their WT strains (Figure 5B). This indicated that the infection efficiency of the optimal mutants for seszw, T4 and selz was indeed higher than those of the corresponding WT phages, allowing these mutants to become dominant in the phage competition assay.
Figure 5.
Characterization of genome-reduced mutant phages with high infectious efficiency. (A) Infection competition between the WT phages and the corresponding optimal mutants against their hosts. Tn, the nth transfer, The vertical axis represents the abundance of the mutant phages in the population. Plaques: the plaques were picked randomly from the last transfer products (double layer agar plate) and identified by PCR. The significance was determined using unpaired t-test (50). *P <0.02; **P <0.005; ***P <0.001; ****P <0.0001. (B) Fitness of the WT phages and the corresponding optimal mutants. The data are shown as the mean ± standard deviation from three repeated experiments. The significance was determined using unpaired t-test (50). *P = 0.01; **P <0.005. Difference between means: (mutant–WT) ± standard error of the mean. (C) Genome maps of the mutant phages that displayed high infectious efficiency. Blue lines show the gene deletion regions, and black lines show the point mutations. Arrows represent genes; the directions in which the arrows point correspond to the directions of transcription and translation.
Next, we sequenced the genomes of these optimal mutants and observed that the genome was reduced by 7.1% in the T7 mutant (T7_100_t2), by 0.3% in the T4 mutant (T4_100_t3), by 8.76% in the seszw mutant (seszw_gene_t1) and by 13.1% in the selz mutant (selz_100_t1) (Figure 5C; Supplementary Figure S11). These results illustrated the feasibility of obtaining optimal mutants by genome reduction, although some WT phages (e.g. T7, in this study) have been evolved nearly optimally.
Characterization of mutant phages with the smallest genomes generated by CiPGr
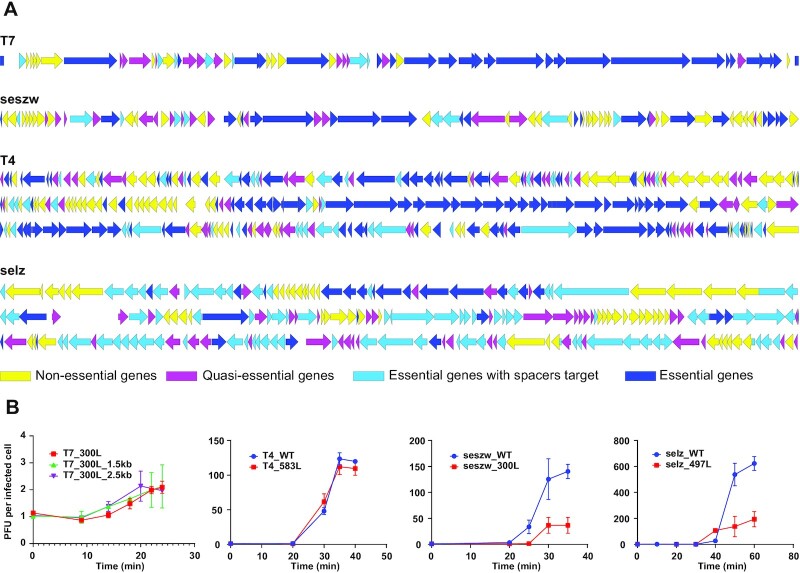
Examination of the sequenced genomes of the isolated mutant phages (mutants: T7, n = 91; seszw, n = 69; T4, n = 32; selz, n = 8) showed that the number of deleted or disrupted genes in these mutant phage genomes was approximately half the number of genes in the removable gene sets (14/28 in T7, 34/53 in seszw, 79/138 in T4 and 64/113 in selz) (Supplementary Tables S1 and S7). The majority of gene deletions and disruptions (100% in T7, 92.4% in T4, 94.1% in seszw and 98.4% in selz) that were detected in the isolated mutant phage genomes occurred at relatively high frequencies (>5%) across the mutant phage populations in the serial transfers (Figure 2B; Supplementary Table S1). This suggests that the genes that were deleted or disrupted in the isolated mutant phage genomes are less important for phage propagation than other genes in the removable gene sets. Thus, we tentatively divided the phage genes into three groups (26): (i) genes that were absent in the isolated mutant phage genomes were classified as non-essential; (ii) other genes in the removable gene sets (frequency <5% in the mutant phage populations) were classified as quasi-essential because the loss of these genes seriously impacts phage propagation, leading to rapid recession of the corresponding mutants in the populations during the serial transfers; and (iii) genes that were not detected in the removable gene sets were classified as essential for these phages (Figure 6A; Supplementary Table S1).
Figure 6.
Characterization of the genome-reduced mutant phages with the smallest genomes. (A) Detailed maps showing the genes that were deleted from the phage genomes. Arrows represent genes. Non-essential phage genes are indicated by yellow arrows, quasi-essential genes are represented by magenta arrows, essential genes with spacer target are displayed in light blue and essential genes are shown in dark blue. (B) One-step growth curves of the T7, seszw, T4 and selz mutants. The data shown represent the mean ± standard deviation of the values obtained in three experiments. The phage titers were determined by PFU counting. The letter L denotes mutant phages that formed large plaques.
Among the heterogeneous mutant phages generated by CiPGr whose genomes were sequenced, the smallest genomes were found in the mutant T7_300L (36 937 bp, genome reduced by 8.4%), seszw_300L (36 655 bp, genome reduced by 20.1%), T4_583L (151 790 bp, genome reduced by 10.1%) and selz_497L (119 098 bp, genome reduced by 23%) (Supplementary Figure S11; Supplementary Table S8). Comparison of the protein sequences encoded by the genomes of these four WT phages shows that a small number of homologous genes (identity ≥30% and e-value ≤1e-10) shared low identities (<46%) in two of the tested phages; 33 such genes were found in a comparison between selz and T4, one was found in a comparison between seszw and selz, and one was found in a comparison between T7 and seszw. In examining the smallest genomes that were obtained for each of these four phages, we found that most of the genes (n = 29; 10 related to DNA replication, recombination and repair, three related to nucleotide metabolism, 10 related to phage structure, one related to packaging and five encoding conserved hypothetical proteins) that were shared between selz and T4 remained in the smallest genomes, suggesting that these genes are highly important for phages in the families Straboviridae and Ackermannviridae (Supplementary Table S9) and highlighting the potential value of our approach in the study of phage biology.
Compared with those of their WT phages, the burst size of T7_300L decreased from 143 (±39) to 2.1 (±0.2), that of seszw_300L decreased from 140 (±13) to 37 (±15), that of T4_583L decreased from 120 (±2) to 109 (±9) and that of selz_497L decreased from 622 (±53) to 194 (±58) (Figure 6B). We performed 12 transfers (80–100 generations) of these mutant phages onto their original hosts and sequenced the genomes of the progeny phages. The results show that these mutants are genetically stable; only a few point mutations (T7_T12, gp17, Asn158 to Asp; T4_583L_T12, gene 249, Thr277 to Ala; T4_583L_T12, gene 252, Ala955 to Thr) were observed (Supplementary Figure S11). Phage particles of the parental T7 and seszw phages and their mutants (T7_300L and seszw_300L) were observed using transmission electron microscopy (TEM), and no significant (P >0.05) changes in particle size caused by genome reduction were apparent for either phage (Supplementary Figure S12). Next, we integrated 1.5 and 2.5 kbp yeast genome fragments that do not encode known genes into the genome of T7_300L and found that its burst size remained unchanged after incorporation of these fragments (Figure 6B), implying that gene loss rather than alteration in the genome size per se was likely to be the most important factor affecting phage fitness in this study. Nevertheless, we have to accept the trade-off between phage genome reduction and infectious efficiency when considering the application of this method in phage genome design and engineering.
DISCUSSION
The results of numerous studies of the minimal genomes of free-living cells (26,31,32,34,71) have expanded the understanding of fundamental biological principles and generated great interest in reduction of the genomes of tailed phages. Using CiPGr, we demonstrated the feasibility of compacting the genomes of different tailed phages to generate viable genome-reduced phages. The discrepancy between the removable gene sets detected by metagenomic sequencing of the mutant phage populations and the gene sets that were absent from the isolated mutant phage genomes suggests that these genes are of different importance for phage fitness. Although genome reduction is in most cases detrimental to phage propagation, we screened for mutants that displayed improved infectious efficiency compared with their parental phages.
Studies indicate that multiple factors, including spacer selection and extensive base modification (36,64), impact the efficiency of phage genome editing by the CRISPR/Cas9-assisted HR method (35–38). We designed multiple spacer sequences for each gene of interest to avoid editing failure (72). Our results reveal that the majority of the genes of interest were disrupted or deleted from the genomes of the phages T7 (28/36, 77.8%), seszw (52/60, 86.7%), T4 (120/188, 63.8%) and selz (96/179, 53.6%) during the iterative CiPGr process (Figure 2C), showing the feasibility of our design. Nevertheless, editing of some genes was not detected. We think there may be three reasons for this. First, the corresponding designed spacers could be invalid. We calculated the number of designed spacers and the cassette abundance in the libraries for the genes that were not edited (Supplementary Figure S5C). The results showed that these genes had an average of 6, 5, 6 and 6.5 spacers per gene in T7, T4, seszw and selz, respectively, confirming the occurrence of gene editing (Supplementary Table S1). Second, base modifications could be present within the designed sites. It has been shown that the Cas9 nuclease acts on the modified T4 genome, although with lower efficiency than in the case of the unmodified genome (36). We examined the genomes of the other three phages but failed to find any known genes akin to the T4 genes that encode cytosine hydroxymethylase (g42) and dCTPase (g56). Consistent with previous results (36), the efficiency of gene deletion and gene disruption was much lower in T4 than in T7 (Figure 3). Nevertheless, the percentage of removable genes in T4 (50%) did not differ greatly from the percentages in the other three tested phages (46.7% in T7, 65.4% in seszw and 56.2% in selz). This suggests that the effect of genome modification on determining the removable gene sets of phages during the CiPGr process was negligible. Third, and most probably, the genes that were not edited could be essential to these phages, although we were not able to completely exclude the other two reasons. For example, in this study, we failed to detect five genes (gp0.3, gp1.6, gp1.8, gp6.3 and gp6.5) in the removable gene set of T7, despite the fact that these genes were previously considered non-essential (28). This was apparently not caused by the absence of the corresponding cassettes from the plasmid libraries (the abundance of the cassettes, log2 >5, Supplementary Figure S5C). A likely explanation for this is that these genes are important for T7 growth in MG1655 [e.g. gp0.3 acts to overcome the DNA restriction system of the host (73)] and that their loss severely impacts the growth fitness of T7 within its host, leading to rapid recession of the corresponding mutants in the populations in our study. Thus, we can conclude that the hypothetical genes that were resistant to deletion in our study (Supplementary Table S1) must play a critical role in phage propagation.
The iterative process ensures that mutants that possess a large growth advantage will predominate in the mutant population and will thus be preferentially selected for the next round of CiPGr from among the numerous heterogeneous mutants. Therefore, the order in which the genes were lost from the mutant phage genomes is likely to suggest that there are differences in the importance of these genes in phage propagation, i.e. the earlier a gene was deleted or disrupted, the less important the gene is for phage propagation. This was evident from the metagenomic sequencing data of the mutant populations (Figure 2A) and the genomes of the isolated mutant phages (Supplementary Figures S7 and S8), as indicated by the increased frequency (64.86 ± 2.92%) of the removable genes of the four phages across the populations as the number of transfers increased (Figure 2A; Supplementary Table S1).
CiPGr resulted in the production of thousands of generations of mutant phage progeny (Supplementary Table S6). A previous study indicated that Cas9 can cause rapid evolution of phage T4 mutants in a short time (64). Unexpectedly, a number of point mutations were observed in the genomes of the isolated phage mutants. The number of point mutations in the mutant T7 and seszw phages was counted and was found to be 60 and 58, respectively, and there was a linear relationship between the number of transfers and the cumulative number of point mutations in the mutant phage genomes (Supplementary Figure S13A). These mutations probably cannot be attributed to the CEM only, as 23.5 ± 8.3% (T7) and 12 ± 7.7% (seszw) of them were located in the spacer and PAM regions, and most of them were spontaneous mutations (74). Moreover, 90 ± 6% (T7) and 86.7 ± 6.9% (seszw) of these point mutations were missense mutations, leading to the corresponding amino acid changes. Considering the positive selection exerted by CiPGr on mutants with increased fitness, these mutations probably benefit the propagation of the mutant phages to some extent, as we showed that the ratio of non-synonymous to synonymous substitutions (Ka/Ks, including only the genes without designed spacers) of most of the mutant phages (94.4% of T7 and 76.9% of seszw) was >1 (2.89 ± 1.66 of T7, 1.82 ± 0.85 of seszw) (Supplementary Figure S13B).
In conclusion, our study demonstrated that genome redundancy is common in tailed phages, and the impact of reducing phage genomes on phage fitness is heterogeneous. Considering that the phage propagation is highly dependent on the hosts, the results may vary for different hosts. Nevertheless, within the specific host strains, we used CiPGr to reduce the sizes of four different phage genomes and thereby generated genetic maps for essential, non-essential and quasi-essential genes within the phage genomes under the given conditions in this study. This information is of great interest for the redesign of phage genomes and in the study of phage biology.
DATA AVAILABILITY
The complete genome sequences of the phage and the raw sequencing data used in this study have been deposited in the GenBank database with the accession numbers shown in Supplementary Table S10. All data, plasmids and strains used for this study are available from the authors upon request.
Supplementary Material
Contributor Information
Shengjian Yuan, Shenzhen Key Laboratory of Synthetic Genomics, Guangdong Provincial Key Laboratory of Synthetic Genomics, CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China; University of Chinese Academy of Sciences, Beijing, 100049, China.
Juan Shi, Shenzhen Key Laboratory of Synthetic Genomics, Guangdong Provincial Key Laboratory of Synthetic Genomics, CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China.
Jianrong Jiang, Shenzhen Key Laboratory of Synthetic Genomics, Guangdong Provincial Key Laboratory of Synthetic Genomics, CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China.
Yingfei Ma, Shenzhen Key Laboratory of Synthetic Genomics, Guangdong Provincial Key Laboratory of Synthetic Genomics, CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work received support from the Ministry of Science and Technology of China, National Key R&D Program of China [http://www.most.gov.cn;2018YFA0903100]; the Strategic Priority Research Program of the Chinese Academy of Sciences [XDB29050500]; Guangdong Provincial Key Laboratory of Synthetic Genomics [2019B030301006]; Shenzhen Key Laboratory of Synthetic Genomics [ZDSYS201802061806209]; and Shenzhen Institute of Synthetic Biology Scientific Research Program [JCHZ20200001].
Conflict of interest statement. None declared.
REFERENCES
- 1. Potera C. Phage renaissance: new hope against antibiotic resistance. Environ. Health Perspect. 2013; 121:a48–a53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma Y., You X., Mai G., Tokuyasu T., Liu C.. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome. 2018; 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camarillo-Guerrero L.F., Almeida A., Rangel-Pineros G., Finn R.D., Lawley T.D.. Massive expansion of human gut bacteriophage diversity. Cell. 2021; 184:1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C.. Uncovering Earth's virome. Nature. 2016; 536:425–430. [DOI] [PubMed] [Google Scholar]
- 5. Salmond G.P.C., Fineran P.C.. A century of the phage: past, present and future. Nat. Rev. Microbiol. 2015; 13:777–786. [DOI] [PubMed] [Google Scholar]
- 6. Lemire S., Yehl K.M., Lu T.K.. Phage-based applications in synthetic biology. Annu. Rev. Virol. 2018; 5:453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gregory A.C., Zayed A.A., Conceição-Neto N., Temperton B., Bolduc B., Alberti A., Ardyna M., Arkhipova K., Carmichael M., Cruaud C.et al.. Marine DNA viral macro- and microdiversity from pole to pole. Cell. 2019; 177:1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brussow H., Hendrix R.W.. Phage genomics: small is beautiful. Cell. 2002; 108:13–16. [DOI] [PubMed] [Google Scholar]
- 9. Roux S., Páez-Espino D., Chen I.M.A., Palaniappan K., Ratner A., Chu K., Reddy T.B.K., Nayfach S., Schulz F., Call L.et al.. IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021; 49:D764–D775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kupferschmidt K. Resistance fighters. Science. 2016; 352:758–761. [DOI] [PubMed] [Google Scholar]
- 11. Reardon S. Phage therapy gets revitalized. Nature. 2014; 510:15–16. [DOI] [PubMed] [Google Scholar]
- 12. Lu T.K., Koeris M.S.. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 2011; 14:524–531. [DOI] [PubMed] [Google Scholar]
- 13. Lenneman B.R., Fernbach J., Loessner M.J., Lu T.K., Kilcher S.. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol. 2021; 68:151–159. [DOI] [PubMed] [Google Scholar]
- 14. Lu T.K., Collins J.J.. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl Acad. Sci. USA. 2007; 104:11197–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meile S., Sarbach A., Du J., Schuppler M., Saez C., Loessner M.J.. Engineered reporter phages for rapid bioluminescence-based detection and differentiation of viable Listeria cells. Appl. Environ. Microbiol. 2020; 86:e00442-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Citorik R.J., Mimee M., Lu T.K.. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014; 32:1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yosef I., Manor M., Kiro R., Qimron U.. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA. 2015; 112:7267–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kilcher S., Loessner M.J.. Engineering bacteriophages as versatile biologics. Trends Microbiol. 2019; 27:355–367. [DOI] [PubMed] [Google Scholar]
- 19. Lu T.K., Collins J.J.. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl Acad. Sci. USA. 2009; 106:4629–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cui J., Schlub T.E., Holmes E.C.. An allometric relationship between the genome length and virion volume of viruses. J. Virol. 2014; 88:6403–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hua J., Huet A., Lopez C.A., Toropova K., Pope W.H., Duda R.L., Hendrix R.W., Conway J.F.. Capsids and genomes of jumbo-sized bacteriophages reveal the evolutionary reach of the HK97 fold. Mbio. 2017; 8:e01579-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edwards K.F., Steward G.F., Schvarcz C.R.. Making sense of virus size and the tradeoffs shaping viral fitness. Ecol. Lett. 2021; 24:363–373. [DOI] [PubMed] [Google Scholar]
- 23. Forster A.C., Church G.M.. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006; 2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang L.Y., Chang S.H., Wang J.. How to make a minimal genome for synthetic minimal cell. Protein Cell. 2010; 1:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Al-Shayeb B., Sachdeva R., Chen L.-X., Ward F., Munk P., Devoto A., Castelle C.J., Olm M.R., Bouma-Gregson K., Amano Y.et al.. Clades of huge phages from across Earth's ecosystems. Nature. 2020; 578:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hutchison C.A. 3rd, Chuang R.Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L.et al.. Design and synthesis of a minimal bacterial genome. Science. 2016; 351:aad6253. [DOI] [PubMed] [Google Scholar]
- 27. Studier F.W. Genetic analysis of non-essential bacteriophage T7 genes. J. Mol. Biol. 1973; 79:227–236. [DOI] [PubMed] [Google Scholar]
- 28. Calendar R.L. The Bacteriophages. 2006; 2nd edn.NY: Oxford University Press. [Google Scholar]
- 29. Moffatt B.A., Studier F.W.. Entry of bacteriophage T7 DNA into the cell and escape from host restriction. J. Bacteriol. 1988; 170:2095–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cerritelli M.E., Cheng N., Rosenberg A.H., McPherson C.E., Booy F.P., Steven A.C.. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997; 91:271–280. [DOI] [PubMed] [Google Scholar]
- 31. Yu B.J., Sung B.H., Koob M.D., Lee C.H., Lee J.H., Lee W.S., Kim M.S., Kim S.C.. Minimization of the Escherichia coli genome using a Tn5-targeted cre/loxP excision system. Nat. Biotechnol. 2002; 20:1018–1023. [DOI] [PubMed] [Google Scholar]
- 32. Luo Z., Yu K., Xie S., Monti M., Schindler D., Fang Y., Zhao S., Liang Z., Jiang S., Luan M.et al.. Compacting a synthetic yeast chromosome arm. Genome Biol. 2021; 22:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reuss D.R., Altenbuchner J., Mader U., Rath H., Ischebeck T., Sappa P.K., Thurmer A., Guerin C., Nicolas P., Steil L.et al.. Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 2017; 27:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mushegian A.R., Koonin E.V.. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA. 1996; 93:10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martel B., Moineau S.. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014; 42:9504–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tao P., Wu X., Tang W.C., Zhu J., Rao V.. Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synth. Biol. 2017; 6:1952–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lemay M.L., Tremblay D.M., Moineau S.. Genome engineering of virulent lactococcal phages using CRISPR-Cas9. ACS Synth. Biol. 2017; 6:1351–1358. [DOI] [PubMed] [Google Scholar]
- 38. Hatoum-Aslan A. Phage genetic engineering using CRISPR-Cas systems. Viruses. 2018; 10:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahler M., Costa A.R., van Beljouw S.P.B., Fineran P.C., Brouns S.J.J.. Approaches for bacteriophage genome engineering. Trends Biotechnol. 2022; 10.1016/j.tibtech.2022.08.008. [DOI] [PubMed] [Google Scholar]
- 40. Springman R., Molineux I.J., Duong C., Bull R.J., Bull J.J.. Evolutionary stability of a refactored phage genome. ACS Synth. Biol. 2012; 1:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bao Z., HamediRad M.. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision. Nat. Biotechnol. 2018; 36:505–508. [DOI] [PubMed] [Google Scholar]
- 42. Chen L., Guan G., Liu Q., Yuan S., Yan T., Tian L., Zhou Y., Zhao Y., Ma Y., Wei T.et al.. Characterization and complete genomic analysis of two Salmonella phages, SenALZ1 and SenASZ3, new members of the genus Cba120virus. Arch. Virol. 2019; 164:1475–1478. [DOI] [PubMed] [Google Scholar]
- 43. Rao V.B., Feiss M.. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol. 2015; 2:351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S.. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015; 81:2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie S., Shen B., Zhang C., Huang X., Zhang Y.. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014; 9:e100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hyman P., Abedon S.T.. Practical methods for determining phage growth parameters. Methods Mol. Biol. 2009; 501:175–202. [DOI] [PubMed] [Google Scholar]
- 48. Kropinski A.M. Practical advice on the one-step growth curve. Methods Mol. Biol. 2018; 1681:41–47. [DOI] [PubMed] [Google Scholar]
- 49. Rousset F., Cui L.. Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet. 2018; 14:e1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Al-Anany A.M., Fatima R., Hynes A.P.. Temperate phage–antibiotic synergy eradicates bacteria through depletion of lysogens. Cell Rep. 2021; 35:109172. [DOI] [PubMed] [Google Scholar]
- 51. Lo H.R., Chao Y.C.. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 2004; 20:354–360. [DOI] [PubMed] [Google Scholar]
- 52. Peng X., Nguyen A., Ghosh D.. Quantification of M13 and T7 bacteriophages by TaqMan and SYBR green qPCR. J. Virol. Methods. 2018; 252:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bull J.J., Molineux I.J.. Predicting evolution from genomics: experimental evolution of bacteriophage T7. Heredity (Edinb.). 2008; 100:453–463. [DOI] [PubMed] [Google Scholar]
- 54. Bull J.J., Heineman R.H., Wilke C.O.. The phenotype-fitness map in experimental evolution of phages. PLoS One. 2011; 6:e27796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Endy D., You L., Yin J., Molineux I.J.. Computation, prediction, and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc. Natl Acad. Sci. USA. 2000; 97:5375–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deatherage D.E., Barrick J.E.. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014; 1151:165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barrick J.E., Colburn G., Deatherage D.E., Traverse C.C., Strand M.D., Borges J.J., Knoester D.B., Reba A., Meyer A.G.. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics. 2014; 15:1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tong J., Liu C., Summanen P., Xu H., Finegold S.M.. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe. 2011; 17:64–68. [DOI] [PubMed] [Google Scholar]
- 59. Wang I.N. Lysis timing and bacteriophage fitness. Genetics. 2006; 172:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lindberg H.M., McKean K.A., Wang I.N.. Phage fitness may help predict phage therapy efficacy. Bacteriophage. 2014; 4:e964081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Springman R., Badgett M.R., Molineux I.J., Bull J.J.. Gene order constrains adaptation in bacteriophage T7. Virology. 2005; 341:141–152. [DOI] [PubMed] [Google Scholar]
- 62. Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y.et al.. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012; 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R.. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020; 13:1194–1202. [DOI] [PubMed] [Google Scholar]
- 64. Tao P., Wu X.. Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci. Adv. 2018; 4:eaar4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hampton H.G., Watson B.N.J., Fineran P.C.. The arms race between bacteria and their phage foes. Nature. 2020; 577:327–336. [DOI] [PubMed] [Google Scholar]
- 66. Miller E.S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Ruger W.. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003; 67:86–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carlson K., Overvatn A.. Bacteriophage T4 endonucleases II and IV, oppositely affected by dCMP hydroxymethylase activity, have different roles in the degradation and in the RNA polymerase-dependent replication of T4 cytosine-containing DNA. Genetics. 1986; 114:669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bryson A.L., Hwang Y., Sherrill-Mix S., Wu G.D., Lewis J.D., Black L., Clark T.A., Bushman F.D.. Covalent modification of bacteriophage T4 DNA inhibits CRISPR-Cas9. Mbio. 2015; 6:e00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chan L.Y., Kosuri S., Endy D.. Refactoring bacteriophage T7. Mol. Syst. Biol. 2005; 1:2005.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shao Y., Wang I.N.. Effect of late promoter activity on bacteriophage lambda fitness. Genetics. 2009; 181:1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reuß D.R., Altenbuchner J., Mäder U., Rath H., Ischebeck T., Sappa P.K., Thürmer A., Guérin C., Nicolas P., Steil L.et al.. Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 2017; 27:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shen J., Zhou J., Chen G.Q., Xiu Z.L.. Efficient genome engineering of a virulent Klebsiella bacteriophage using CRISPR-Cas9. J. Virol. 2018; 92:e00534-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Studier F.W. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J. Mol. Biol. 1975; 94:283–295. [DOI] [PubMed] [Google Scholar]
- 74. Tenaillon O., Barrick J.E., Ribeck N., Deatherage D.E., Blanchard J.L., Dasgupta A., Wu G.C., Wielgoss S., Cruveiller S., Médigue C.et al.. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016; 536:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequences of the phage and the raw sequencing data used in this study have been deposited in the GenBank database with the accession numbers shown in Supplementary Table S10. All data, plasmids and strains used for this study are available from the authors upon request.