Abstract
Pif1 family 5′ → 3′ DNA helicases are important for replication fork progression and genome stability. The budding yeast Saccharomyces cerevisiae encodes two Pif1 family helicases, Rrm3 and Pif1, both of which are multi-functional. Here we describe novel functions for Rrm3 in promoting mutation avoidance during DNA replication. We show that loss of RRM3 results in elevated spontaneous mutations made by DNA polymerases Pols ϵ and δ, which are subject to DNA mismatch repair. The absence of RRM3 also causes higher mutagenesis by the fourth B-family DNA polymerase Pol ζ. By genome-wide analysis, we show that the mutational consequences due to loss of RRM3 vary depending on the genomic locus. Rrm3 promotes the accuracy of DNA replication by Pols ϵ and δ across the genome, and it is particularly important for preventing Pol ζ-dependent mutagenesis at tRNA genes. In addition, mutation avoidance by Rrm3 depends on its helicase activity, and Pif1 serves as a backup for Rrm3 in suppressing mutagenesis. We present evidence that the sole human Pif1 family helicase in human cells likely also promotes replication fidelity, suggesting that a role for Pif1 family helicases in mutation avoidance may be evolutionarily conserved, a possible underlying mechanism for its potential tumor-suppressor function.
INTRODUCTION
Duplication of eukaryotic genomes by DNA replication is a challenging undertaking. A variety of mechanisms ensure that DNA replication is faithful. For instance, multiple DNA repair pathways, such as nucleotide excision repair, base excision repair, and DNA break repair contribute to maintaining the integrity of DNA. Three major processes are responsible for mutation avoidance during and immediately after replication of the undamaged nuclear DNA genome (1,2). First, the replicative DNA polymerases (Pols α, ϵ and δ), also known as replicases, strongly select correct rather than incorrect nucleotides for incorporation (1,2). Second, Pols ϵ and δ possess 3′ → 5′ exonuclease activity that conducts mismatch correction in a process called proofreading (3). Proofreading occurs during ongoing replication, when occasional misinserted bases at the 3′ DNA terminus are excised before DNA synthesis resumes. Pol α, along with primase, synthesizes RNA–DNA primers that initiate both leading strand synthesis and Okazaki fragment synthesis, but it does not contain proofreading activity (4). Because mismatches pose kinetic barriers to polymerase activity (5–7), Pol α likely dissociates from the 3′ terminus when it is unable to extend a mismatch, allowing Pol δ to excise the mismatch in a process we call extrinsic proofreading (8). We recently demonstrated that Pol δ can also extrinsically proofread errors by Pol ϵ and by itself (9), thereby contributing to the high fidelity of DNA replication. Finally, mismatches that are erroneously extended can be recognized and repaired by DNA mismatch repair (MMR) (3,10).
During DNA synthesis in vitro, yeast and human Pols α, ϵ and δ were found to have error rates between 10−3 and 10−5 (11–17). These values are on average ∼100 times higher than our in vivo estimates using proofreading and MMR-defective yeast strains (∼2 × 10−6 for both Pols ϵ and δ) (9). The discrepancy between in vitro and in vivo values suggests that there are additional factors that operate in vivo to promote polymerase fidelity, and/or that there are unknown repair processes that correct errors that escape proofreading and MMR.
Pif1 family DNA helicases are conserved from yeast to humans (18,19). The budding yeast Saccharomyces cerevisiae genome encodes two closely related Pif1 family members, Rrm3 and Pif1. In contrast, most eukaryotes have only one. Rrm3 promotes replication through over 1000 protein associated sites, including telomeric DNA, RNA polymerase III-transcribed genes such as tRNA genes, and centromeres (20). At tRNA genes, Rrm3 also suppresses R-loop mediated DNA damage (20–23). The related Pif1 helicase shares these functions by acting as a backup helicase for Rrm3 during replication at tRNA genes and centromeres (21,22,24). In addition, Pif1 negatively regulates telomere length by inhibiting telomerase activity, promotes replication through G-quadruplex DNA structures, has a key role in replication termination (25) and is required for maintenance of mitochondrial DNA (18,23,26).
The budding yeast Pif1 has 5′ to 3′ DNA helicase activity. This activity can unwind both DNA duplexes and G-quadruplexes and can displace RNA from RNA–DNA hybrids (27–30). Pif1 also displaces proteins from DNA (29). The S. pombe Pif1 family helicase, called Pfh1, has activities similar to those of Pif1(31). Like Pif1 and Pfh1, Rrm3 is a 5′ to 3′ helicase, but detailed biochemical studies of Rrm3 are lacking, due to difficulties in purifying active protein. However, Rrm3 is predicted to have similar activities as Pif1, based both on its in vivo functions and its binding to G-quadruplexes and R-loops (20,21,32).
Rrm3 associates with DNA polymerase ϵ and travels with the replication fork (33), suggesting an unidentified, genome-wide function for Rrm3 throughout DNA replication. Mutations in human PIF1 (hPIF1) are found in some high-risk breast cancer families, suggesting that it may have a tumor suppressor function (34). Here we reveal a novel function of yeast Rrm3 and human PIF1 in promoting mutation avoidance during DNA replication.
MATERIALS AND METHODS
Yeast strains
All S. cerevisiae strains were derived from Δ|(−2)|-7B-YUNI300 (MATa CAN1 his7-2 leu2Δ::kanMX ura3Δ trp1-289 ade2-1 lys2ΔGG2899-2900) (35), known as Δ7 background. Yeast strains used for this study is listed in Supplementary Table S2.
Measuring mutation rates of reporter genes
Mutation rates of reporter genes URA3 and CAN1 genes were measured using fluctuation assays. As previously described (36), at least 20 independent colonies were inoculated in 5 ml YPDA yeast media supplemented with 100 μg/ml of adenine. The liquid cultures were rotated at 30°C for about 3 days till saturation. The cells were washed with sterile water and and diluted for plating on non-selective and selective media. For selection of ura3 or can1 mutants, the cells were plated on 5-fluoroorotic acid or canavanine containing media respectively. The mutation rates are calculated from the individual culture's mutation frequency using Drake's formula (37).
Mutation accumulation
Diploid yeast strains were passaged for mutation accumulation as previously described (38). For each passaging each single clone was allowed to grow into a 2 mm colony on YPDA solid media with 100 μg/ml of adenine supplemented, which was estimated to equal 30 cell divisions. For each genotype, multiple isolates were passaged up to 30 times to accumulate sufficient mutations (summarized in Supplementary Table S1).
Genome sequencing and mutation analysis
Yeast DNA was isolated using the Lucigen MasterPure Yeast DNA Purification kit (MPY80200). Sequencing libraries were prepared using the KAPA HyperPrep kit (KK8504) with low amplification cycles (∼6 cycles). The libraries were sequenced using Illumina HiSeq 2500 or 4000 for paired-end 150 bp reads. Sequencing reads were mapped on to the L03 reference genome built for the Δ7 background (39). Mutation calling was performed using the muver pipeline which does so by comparing the endpoint to the timepoint 0 genome from the mutation accumulation experiment (40). Per basepair per generation mutation rates were calculated by dividing number of mutations by number of generations and by number of queryable bases. Mutation counts of each isolate for mutations analysed for Figures 5 and 6 are provided in Supplementary Table S3.
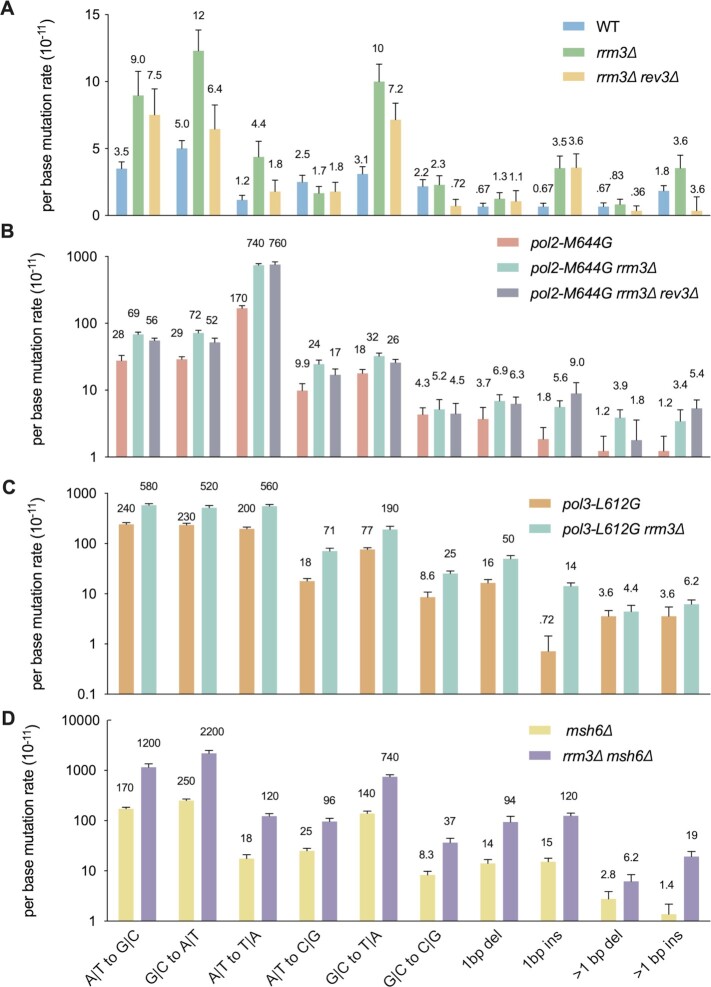
Figure 5.
Mutation spectra of rrm3Δ cells. All mutations were categorized by six substitution types and four insertion/deletion (indel) types. Panels (A) through (D) are aligned vertically by mutation types. The mutation types are indicated at the bottom of the figure. All panels except (A) use a log10 scale. Error bars show standard errors.
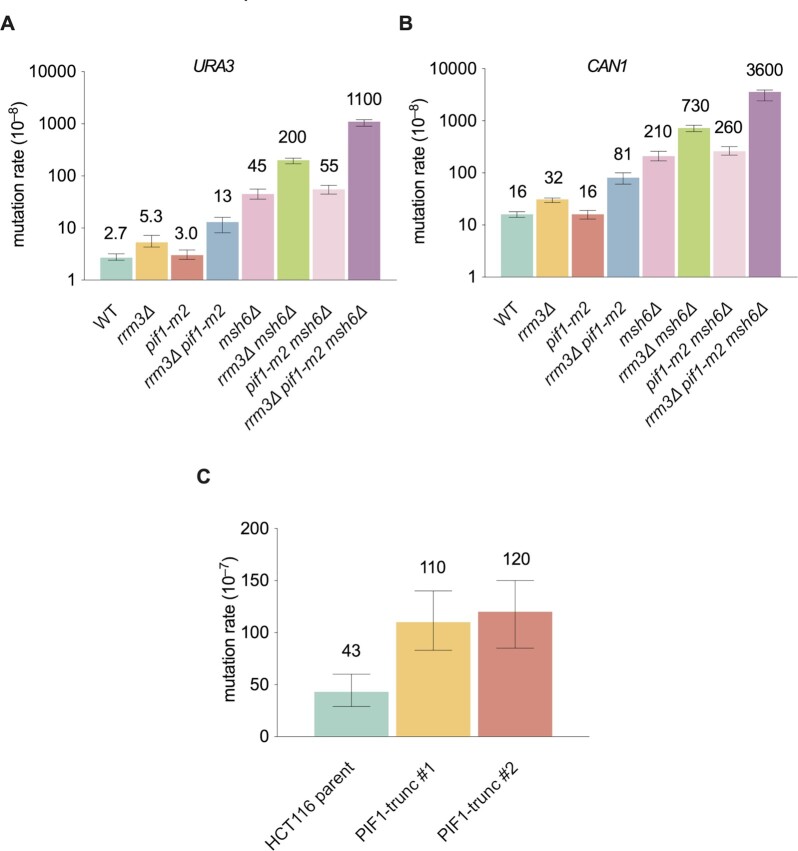
Figure 6.
Evolutionary conservation of Rrm3 function. (A, B) mutation rates of reporter genes (URA3 and CAN1) in pif1-m2 cells and the genetic interaction between rrm3Δ and pif1-m2. Data are presented as median ± 95% confidence interval. (C) Mutation rate of HPRT reporter gene in HCT116 cells with or without PIF1. Data are presented as median ± 95% confidence interval. Twelve independent cultures were used for the fluctuation experiment.
Statistical comparison of mutation rates
In all cases where comparisons are made between two strains (e.g. rates in Figures 4 and 5), P-values were calculated using Welch's t-test (41). The null-hypothesis is that the two data sets have equivalent means. This test assumes normality but allows for data sets with unequal variance. Normality is expected for mutation accumulation in the absence of selection so long as no novel mutator or suppressor phenotypes emerge during the experiment. A null-hypothesis of normality could not be rejected for any strain reported here (overall mutation rates, Shapiro–Wilk test (42) for non-normality P > Šidák corrected threshold (43) of 0.00516 for a family-wise error rate of 0.05), except for the previously published msh6Δ strain. That strain contained one outlier (3.3 standard deviations above the mean). Removal of that outlier drove the Shapiro–Wilk P-value to 0.272. The outlier caused the average msh6Δ error rate to increase by only 6%, insufficient to change any of our conclusions. The outlier was therefore retained in order to remain consistent with previous publications.
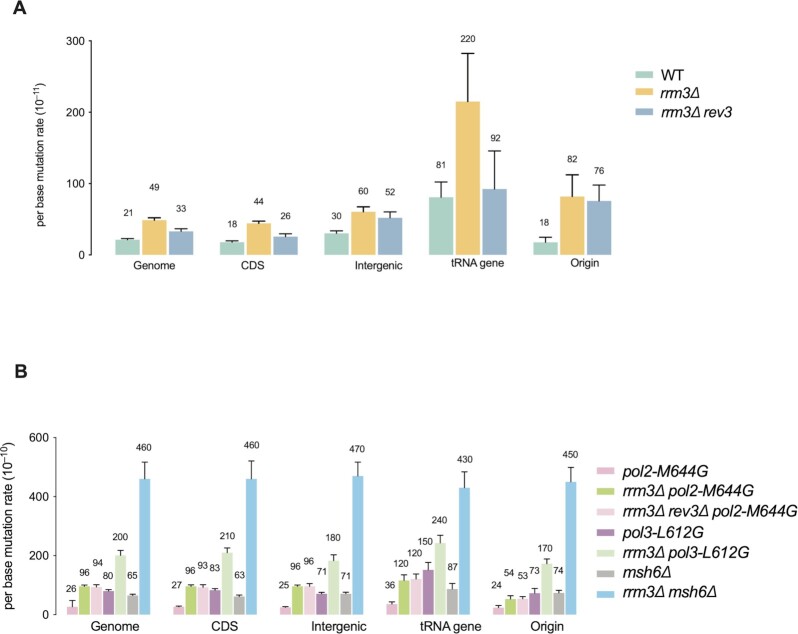
Figure 4.
Mutation rates across the genome and at specific genomic loci. (A, B) Genome average mutation rates. Error bars show standard errors. Also included are average mutation rates within protein-coding genes, intergenic regions, tRNA genes (±300 bp) regions, replication origins (1 kb windows surround the autonomously replicating sequences (ARS)).
Construction of PIF1 truncation mutant HCT116 cell lines
HCT116 cells grown in McCoys 5A media supplemented with 10% FBS with 1% l-glutamine and 1% Pen/Strep were transfected with CRISPR67 gRNA vector and Cas9 vector. Simultaneously, the cells were infected with rAAV-Pif1-CondKO-SEPT-Neo viruses. rAAV viruses were produced using AAV Helper-free system (Stratagene, 240071). On the next day, cells were trypsinized and plated into 96 well plates at a concentration of 2000 cells per well, in medium containing G418 selection medium (0.5 mg/ml). After approximately two weeks, single colonies were expanded to 24-well plates. Once the cells were confluent enough some cells were collected to screen for positive clones by PCR using different pairs of primers (control primers: Fwd-TACCCTCAGGAGCAAGCA, Rev-TCATCCTGGTGGGTGCAGAG; insertion primers (SEPT-seq): Fwd-CCAAATTTTAAGGTACCACTGTGCA, Rev-TCATCCTGGTGGGTGCAGAG). Sequencing showed that the CRISPR67 target site contains insertion of a neomycin expressing cassettes (Supplementary Figure S3a, b). RT-PCR shows that the insertion interrupted hPIF1 transcription (Supplementary Figure S3c). Positive clones were also confirmed by Southern blot that both alleles were modified (Supplementary Figure S3d). Modification at the site of the CRISPR67 gRNA target was verified by PCR-sequencing using the following primers: 5′-CTTCTATCCACTTGCCTCCTAC-3′ and 5′-CGGTTCTGCTTCCAGGTATTA-3′.
Plasmids used for CRISPR mutagenesis
pAAV-Pif1-CondKO-SEPT-Neo was built using the vector pAAV-MCS from AAV Helper-free system (Stratagene, 240071). The PIF1-CondKO-SEPT cassette containing the conditional cassette with 3 LoxP sites and the Neomycin resistant marker was introduced between the two Not1 sites. CRISPR67 gRNA vector was built using the gRNA vector backbone vector (Addgene plasmid #41824) (44). The gRNA sequence GGAAAGGACGAAACACCGACCTTCATCTCTCTACTGCGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCTTTCTTGGCTTTATATATCTTGT was introduced into the vector using Gibson Assembly kit (NEB E2611S). Cas9 vector was also from Addgene (#41815) (44).
Southern blot
Genomic DNA was extracted using the QIAgene Dneasy Blood and Tissue kit and digested by BstEII and EcoRI. DNA was run on a 0.8% agarose gel, gently washed in HCl 0.25 N for 20 min, rinsed with diH2O, washed with NaOH 0.4 M and finally transferred overnight in NaOH 0.4M to Zeta-Probe GT Genomic Tested Blotting Membranes. The blot was then probed using either a genomic probe (CATTGCACATGGGCCAAGAA) or a neomycin probe (GGTGGAAAATGGCCGCTTTT).
Measuring HPRT mutation rates
To prepare for fluctuation assays, HPRT mutants were purged by culturing the HCT116 cell lines in 1x HAT (sodium hypoxanthine, aminopterin and thymidine, Thermo Fisher 21060017) containing DMEM with high glucose media for five passages. As previously described, for fluctuation analysis of each cell line, 500 cells were seeded in each well of two six-well plates for a total of 12 cultures. After cells were grown into confluence, cells harvested from each well were counted. Five hundred cells were plated in each of a triplicate 10 cm dishes. After about 7–10 days, colonies were stained with crystal violet and counted for estimation of viable cells plated. On the other hand, 1 million cells were seeded in each of a triplicate 10 cm dishes containing 5 μg/ml 6-thioguanine (Sigma Aldrich, A4882). After about 14 days, colonies were stained with crystal violet and counted for estimation of viable cells plated. The mutation rates were estimated by Ma-Sandri-Sarkar Maximum method using the FALCOR web tool (45). FALCOR is currently maintained by the Ping Liang lab at https://lianglab.brocku.ca/FALCOR/.
DATA AVAILABILITY
All whole genome sequencing data is available through Sequence Read Archive accession number PRJNA689775.
Code availability
Muver suite is available via GitHub (40) or upon request.
RESULTS
Elevated spontaneous mutations in rrm3Δ cells
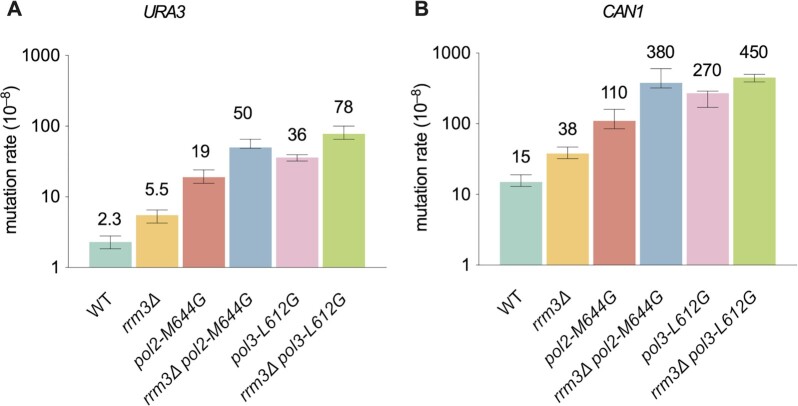
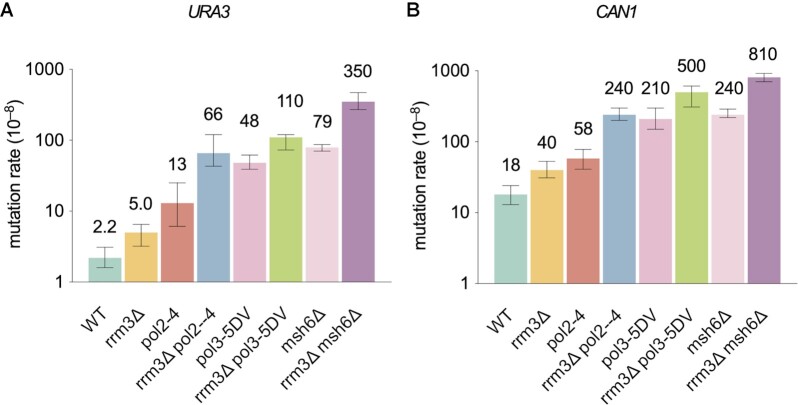
Because Rrm3 moves with the nuclear DNA replication fork and interacts with DNA polymerase ϵ in vivo (33), we considered that it might affect replication fidelity. To test this idea, we determined the effects of its deletion on spontaneous mutation rates using the URA3 and CAN1 reporter genes for forward mutation assays. The results showed 2–3-fold increases in mutation rates in the absence of RRM3 (P < 0.01 for both URA3 and CAN1, Figure 1). A previous study found that spontaneous mutations at the CAN1 locus were increased in rrm3Δ cells (46). To determine if these mutations result from DNA synthesis by the replicative DNA polymerases, we tested the genetic interaction of RRM3 with variants of Pols ϵ and δ that themselves have reduced fidelity. We used polymerase alleles that contain active site mutations (pol2-M644G and pol3-L612G for Pols ϵ and δ, respectively) that show reduced fidelity but otherwise support relatively normal cell growth (47,48). Both rrm3Δ pol2-M644G and rrm3Δ pol3-L612G double mutants had synergistic increases in mutation rates over the corresponding single mutants (P < 0.01 for both reporters, Figure 1). These synergistic increases are higher than expected from the additive effects of the corresponding two single mutants. These increases suggest that the mutations generated in the absence of Rrm3 are likely introduced during DNA replication by Pols ϵ and δ.
Figure 1.
Mutational effect of rrm3Δ and genetic interaction of replicase mutators. Mutation rates of reporter genes (URA3 and CAN1) measured in forward mutation assays. Data are presented as median ±95% confidence interval. n ≥ 20 independent cultures were used for fluctuation analysis for each genotype.
Mutations in rrm3Δ cells are subject to proofreading and MMR
We reasoned that if the mutations in rrm3Δ cells are indeed replication errors, they should be subject to correction by proofreading by Pols ϵ and δ and should also be substrates for MMR. Therefore, we tested the genetic interactions between RRM3 and the exonuclease activities of Pols ϵ or δ, and between RRM3 and MMR. We found that mutations in rrm3Δ cells were synergistically higher in the absence of exonuclease activity of Pol ϵ or Pol δ (pol2-exo− or pol3-exo−) (P < 0.01 for both reporters, Figure 2). MSH6 encodes a subunit of the MutSα heterodimer, deletion of which abolishes the repair of base-base mismatches and reduces the repair of small misalignments (49). Similarly, rrm3Δ was also synergistic with msh6Δ (P < 0.01 for both reporters, Figure 2). These data suggest that at least some of the mutations made in rrm3Δ strains are due to replication errors made by Pols ϵ and Pol δ.
Figure 2.
Genetic interaction between rrm3Δ and exonuclease-defective replicases and MMR mutant. Mutation rates of reporter genes (URA3 and CAN1) measured in forward mutation assays. Data are presented as median ± 95% confidence interval. n ≥ 20 independent cultures were used for fluctuation analysis for each genotype.
Elevated mutations in rrm3Δ cells are due partly to Pol ζ
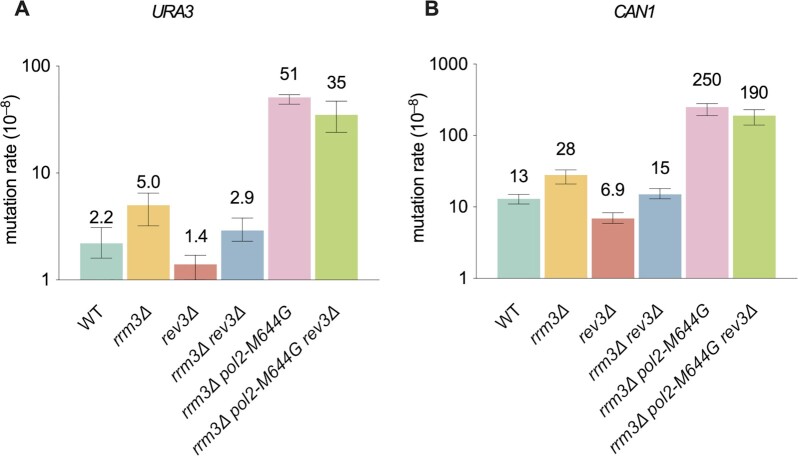
One well-characterized function of Rrm3 is to promote replication progression through hard-to-replicate sites (20,21,33). For example, actively transcribed genes are often obstacles for the replisome (50). The fourth B-family DNA polymerase, Pol ζ, is a translesion DNA polymerase that contributes to 40–85% of spontaneous mutations in normal cycling cells (35,51–54). It affects mutagenesis induced by replication stress and DNA damaging agents, and during DNA repair (55–58). Therefore, we asked whether Pol ζ contributes to mutagenesis in rrm3Δ cells. By deleting REV3, which encodes the catalytic subunit of Pol ζ, we found that Polζ is responsible for 40–50% of the mutations in both WT and rrm3Δ cells (P < 0.05 for all pairwise comparisons, Figure 3). Since rrm3Δ cells have higher spontaneous mutation rates than WT, this result suggests that Pol ζ causes more mutations in rrm3Δ cells. The fact that rrm3Δ rev3Δ cells had >2-fold higher mutation rate than rev3Δ (P < 0.01 for both URA3 and CAN1, Figure 3) suggests that not all of the mutations are due to Pol ζ. This interpretation is consistent with the idea that some of the mutations in rrm3Δ cells are dependent on the replicases Pols ϵ and δ. Pol ζ is not a significant source of mutations in rrm3Δ pol2-M644G cells (P > 0.05, Figure 3).
Figure 3.
Pol ζ contributes to mutagenesis in rrm3Δ cells. Mutation rates of reporter genes (URA3 and CAN1) measured in forward mutation assays. Data are presented as median ± 95% confidence interval. n ≥ 20 independent cultures were used for fluctuation analysis for each genotype.
Genome-wide mutation rates in rrm3Δ mutant cells
Replication of most of the genome does not appear to be Rrm3-sensitive (20,50). Rather it promotes replication at specific sites such as tRNA genes. Particularly, Rrm3 is not required for normal replication progression through RNA Pol II-transcribed protein-coding genes. Yet, Rrm3 is required for mutation avoidance at reporter genes URA3 and CAN1. To better understand mutagenic events in rrm3Δ cells across the genome, we performed genome mutation accumulation in rrm3Δ cells. Briefly, diploid yeast cells were passaged a number of times on solid media to allow mutation accumulation (38). The accumulated mutations were identified by comparing the genomes sequenced before and after the passagings. As shown in Figure 4A, deletion of RRM3 resulted in a 2.4-fold increase (P < 0.001) in the genome-wide average mutation rate, similar to the increase in mutations at the two reporter genes in this same genetic background (Figure 1). Also similar to the reporter genes, mutation rates in rrm3Δ cells were synergistic with polymerase mutants and with msh6Δ (P < 0.001, Figure 4A), while deletion of REV3 resulted in reduced mutations in rrm3Δ cells. Mutation rates are elevated across different substitution types and insertion/deletion (indel) mutations in rrm3Δ cells (Figure 5A). Thus, the presence of Rrm3 suppresses mutagenesis genome-wide.
Different sources of mutagenesis in rrm3Δ mutant cells
Next, we analyzed the mutational effect of rrm3Δ in different regions of the genome. We found that the mutational rate within coding regions in rrm3Δ cells was similar to the genome-wide average (Figure 4A). These genic regions account for ∼70% of the S. cerevisiae genome. Moreover, rrm3Δ cells had a similar increase in mutation rate in intergenic regions, and this increase was synergistic with mutants in DNA polymerases or MMR (P < 0.001, Figure 4B). However, deletion of REV3 resulted in no significant reduction in mutation rate in intergenic regions in rrm3Δ cells (P = 0.22, Figure 4A). This result suggests that mutations in inter-genic regions may be due mostly to replication by the replicases, not Pol ζ.
In WT cells, replication pauses at tRNA genes, especially in genes where replication and transcription moves in the opposite directions through the gene. This pausing depends on the pre-initiation transcription complex and was increased dramatically in rrm3Δ cells (22,59). Likewise, in WT cells, the spontaneous mutation rate at tRNA genes was significantly higher than the genome average (P < 0.01), and this rate was even higher (∼3-fold) in rrm3Δ cells (P < 0.05, Figure 4B). In contrast to other sites, mutations at tRNA genes were reduced by ∼3-fold in rev3Δ rrm3Δ cells (P = 0.08). These results suggest a particularly important role for RRM3 in preventing Pol ζ-related mutagenesis at tRNA genes. Notably, although not statistically significant due to low mutation counts (Supplementary Table S3), in rrm3Δ rev3Δ cells, the mutation rate at tRNA genes (92 × 10−11) is higher than that of the genome average (33 × 10−11, P = 0.14) or protein-coding genes (26 × 10−11, P = 0.12) (Figure 4A). In contrast, in rrm3Δ pol2-M644G or rrm3Δ pol3-L612G, the mutation rate at tRNA genes is similar to the genome average (Figure 4B). These findings suggest that, in addition to the replicases and Pol ζ, there may be additional, as yet unidentified, sources of mutagenesis at tRNA genes.
In addition, rrm3Δ cells had a 5-fold increase in mutation rate in genomic regions surrounding replication origins (1 kb windows), with mutation rates even higher than genome average (82 × 10−11 versus 49 × 10−11 for the genome average, P < 0.0001, Figure 4A). In contrast to tRNA genes, rev3Δ caused no significant decrease in mutation rate at these regions in rrm3Δ cells (P > 0.05, Figure 4A). However, the polymerase mutators do not contribute to more mutagenesis in these regions compared to genome average (Figure 4B, see Supplementary Table S3 for all pairwise comparisons). Consistently, in rrm3Δ pol2-M644G, the mutation rate in these regions is also lower compared to the genome-wide average (P < 0.01, Figure 4B). The rrm3Δ msh6Δ double mutant behaves similarly in these regions compared to the genome-wide average (Figure 4A, B). Therefore, mutagenic proceses that are independent of the replicases and Pol ζ may also contribute to mutations at replication origins in rrm3Δ cells.
Mutation spectra in rrm3Δ mutant cells
We also examined the mutational effect of rrm3Δ across different substitution types and indels. We found that most substitutions except for A•T to C•G and G•C to C•G transversions were elevated in rrm3Δ cells compared to WT (Figure 5A), as were indels, particularly so for single base pair insertions. Deletion of REV3 reduced the mutations rate of different substitution types to varying degrees. Particularly significant were A•T to T•A and G•C to C•G transversions (P < 0.01, Figure 5A) which are preferred Pol ζ-dependent mutations observed in WT cells, although the G•C to C•G mutation rate is not elevated in rrm3Δ cells (53). G•C to A•T is reduced to a similar extent with rev3Δ (P < 0.01, Figure 5A), which was not a preferred mutation type by Pol ζ in WT cells (53). Furthermore, the dramatic increase in single base insertions does not depend on Pol ζ (Figure 5A). These findings suggests that Pol ζ-dependent mutagenesis is somehow different in rrm3Δ than in WT cells.
Pol ϵ carrying the pol2-M644G variant mostly makes A•T to T•A transversions via T•T mispair (11,47). We observed a strong synergy between pol2-M644G and rrm3Δ for A•T to T•A mutations (p < 0.01, Figure 5B). Similarly, we observed synergy between pol3-L612G and rrm3Δ across most mutation types (P < 0.01, except for >1 bp indels, Figure 5C), with different specificity compared to pol2-M644G, suggesting that the two replicases respond differently to the loss of RRM3. Moreover, we observed strong synergy between rrm3Δ and msh6Δ (P < 0.01, except for >1 bp deletion, Figure 5D), further highlighting the interpretation that mutations made in rrm3Δ cells are replication errors that are subject to MMR.
Rrm3 promotes replication fidelity via its helicase activity and independently of replication checkpoint
Like all Pif1 family helicases, Rrm3 is an ATP-dependent 5′ to 3′ helicase (60). To determine if its helicase activity is required for its role in suppressing mutagenesis, we introduced a plasmid borne copy of either wild type RRM3 or rrm3-K260A into rrm3Δ cells. The K260A mutation is a Walker A box mutation that eliminates the ATPase activity of Rrm3 (61). While WT RRM3 fully suppressed the increased mutation rate in rrm3Δ cells, rrm3-K260A behaved similarly to rrm3Δ (Supplementary Figure S1). Thus, the helicase activity of Rrm3 is essential for its role in mutation avoidance during replication. Suppression of spontaneous mutations in the CAN1 gene also requires the helicase activity of Rrm3 (46).
The DNA damage checkpoint is moderately activated in rrm3Δ cells (20). Checkpoint activation causes changes in cell cycle and nucleotide pool (62). To test whether the mutagenic consequences of RRM3 loss is due to checkpoint activation, we measure mutation rates in cells lacking Rad53 checkpoint kinase with or without Rrm3. Shown in Supplementary Figure S2, we found that the elevated mutagenesis in rrm3Δ cells is not dependent on checkpoint activation.
Pif1 may serve a backup role in mutation avoidance for Rrm3
In addition to Rrm3, S. cerevisiae encodes a second Pif1 family helicase called Pif1. Although Pif1 has several distinct functions, in some cases, it also acts as a backup for Rrm3 during DNA replication, for example, at tRNA genes and centromeres (21,22,24). Because PIF1 is required for maintenance of mitochondrial function (18,23,26), we used the pif1-m2 allele which is defective only for the nuclear functions of Pif1, even though it is not a null in some in vivo assays (63). Although pif1-m2 cells had a normal mutation rate, pif1-m2 exacerbated the mutational effects of rrm3Δ at both CAN1 and URA3 (Figure 6A, B). We observed similar effects of pif1-m2 in a MMR-defective background (Figure 6A, B). These data are consistent with a backup role for Pif1 in mutation avoidance.
Human PIF1 (hPIF1) suppresses mutations
In humans, hPIF1 is the sole Pif1 family helicase. To test whether the function we observed for Rrm3 and Pif1 is conserved in mammalian cells, we introduced mutations in both copies of the hPIF1 gene using CRISPR-Cas9 in human HCT116 cells. We obtained two independent clones carrying a neomycin expressing casette insertion in the hPIF1 gene induced by CRISPR-Cas9 (Supplementary Figure S3A–C). RT-PCR confirmed that the insertion interrupted hPIF1 transcription in both strains (Supplementary Figure S3D). We then carried out fluctuation assays to measure the mutation rate of the HPRT gene in both WT and the two mutant hPIF cells lines. We found a 2.6–3-fold increase in the HPRT mutation rate in the PIF1 mutant cell lines (Figure 6C, P < 0.001). The fact that HCT116 is MMR-defective, likely due to lack of the MLH1 gene (64), further suggests that these mutations are at least in part replication errors that are substrates for MMR.
DISCUSSION
We show that RRM3 promotes the fidelity of the two major DNA replicases, Pols ϵ and δ. In addition, RRM3 has locus specific functions in preventing Pol ζ-dependent mutagenesis (Figures 3 and 4), particularly at tRNA genes, but also, to a lesser extent, at replication origins. The evidence also suggests that RRM3 prevents other unknown mutagenic processes, at least at tRNA genes and at regions surrounding origins, which could involve DNA break repair, replication fork – initiated recombination or synthesis by other mutagenic polymerases such as Pol η.
Rrm3 and the S. pombe Pfh1 are both required for efficient replication through hard to replicate sites, such as stable protein complexes and R-loops, as deterimined by both 2D gels and DNA polymerase occupancy assays (20,21,33). However, replication fork progression through most of the genome is not affected by reduced Rrm3 or Pfh1, perhaps because Rrm3 acts only rarely at other sites. Unlike S. cerevisiae Pif1, Rrm3 and S. pome Pfh1 are stably associated with replication forks (33,65) (Pif1 is recruited to its sites of action (66)). These results suggest a genome-wide role for Rrm3 and Pfh1 in DNA replication, in addition to their more specific functions at hard-to-replicate loci.
Here, we demonstrate that Rrm3 has an unanticipated function in the fidelity of DNA replication. That is, the mutation rates throughout the genome, at both Rrm3-sensitive and insensitive sites, were elevated in the absence of Rrm3. Our data do not allow us to determine the mechanism by which Rrm3 increases replication fidelity, which will require future experiments. However, we speculate that Rrm3 suppression of mutations is linked to its role in fork progression. It is possible that replication forks are constantly dealing with minor impediments that may require frequent engagement of Rrm3 helicase function during replication. Without Rrm3, the replication template or the fork environment could become less than ideal for accurate polymerase action. For instance, Rrm3 could facilitate movement of replicases by displacing DNA-binding proteins or RNA–DNA hybrids, or unwinding secondary DNA structures. These actions may affect the fidelity of the replicases. Another possibility is that Rrm3 facilitates error removal. Our data clearly shows that Rrm3 does not participate in the base-base mismatch removel by the MutSα pathway. It is also possible that Rrm3 may play a role in an uncharacterized repair pathway that corrects mismatchs and/or polymerase slippage loops.
In addition to influencing the fidelity of Pols ϵ and δ, Rrm3 also suppresses Pol ζ-dependent mutagenesis. We propose that increased replication fork stalling in rrm3 cells increases the chance that Pol ζ engages in DNA synthesis, thereby increasing mutagenesis owing to its fidelity being lower than that of the replicative polymerases. This model is supported by our finding that tRNA genes, which are among the strongest replication pause sites, are also mutation hotspots, even in WT cells. Fork slowing/stalling is exacerbated in rrm3Δ cells, which could facilitate Pol ζ engagement and result in even higher mutagenesis. Pol ζ engagement at these sites could help extend leading strands or fill in gaps on the lagging strand that arise from incomplete Okazaki fragments. In addition, Pol ζ also contributes to a significant portion of mutagenesis in the protein-coding genes in contrast to intergenic regions, consistent with the idea that RNA Pol II-transcribed gene are also replication impediments (50,67). Moreover, our data suggest additional mutagenic processes in rrm3Δ cells at certain genomic loci. For instance, the mutation rate in regions surrounding replication origins increased by ∼5-fold in rrm3Δ compared to WT cells, which were not due to Pol ζ (Figure 4). However, in rrm3Δ and polymerase mutators or msh6Δ double mutants, the mutations in these regions were not higher than elsewhere, suggesting that many of the additional mutations in rrm3Δ are not due to replicase errors. These mutations could come from other error-prone polymerases and/or DNA repair processes.
Unlike most other eukaryotes, S. cerevisiae encodes two Pif1 family helicases, making the functional similarity and divergence of Rrm3 and Pif1 interesting. Our data suggest that Pif1 acts as a backup for Rrm3 in mutation avoidance as it does for several other Rrm3 functions, such as replication of tRNA genes and centromeres (21,22,24). Likewise, Rrm3 is a backup for Pif1 for unwinding G-quadruplexes but only in Pif1-deficient cells (21). The S. pombe Pfh1 combines many of the functions of Pif1 and Rrm3 (18). We note that the pif1-m2 allele used in our analyses is a nuclear-null in some functional assays (30,68). However, there are currently no PIF1 alleles that are a null for all of Pif1’s nuclear activities that do not also eliminate its mitochondrial functions. Mitochondrial deficient cells are hard to work with because of their slow growth., and nuclear defects seen in pif1Δ cells could be a secondary consequence of loss of mitochondrial function.
Pfh1, the sole Pif1 family helicase in fission yeast has most of the functions of Pif1 and Rrm3 (ref a review). Thus, we anticipate that the human PIF1 may also be a functional homolog for both Rrm3 and Pif1 in suppressing spontaneous mutations. To test this idea, we monitored the mutation rates for HPRT in HCT116 human cell lines. Indeed, the HPRT mutation rates in HCT116 cells with disrupted hPIF1 were ∼3-fold higher than in cells expressing WT hPIF1. These data suggest that at least some functions of Pif1 family helicases may be widely conserved. Interestingly, a mutation disrupting PIF1 function was found in certain breast cancer families, suggesting hPIF1 might be a tumor suppressor gene (34). We propose that human PIF1’s role in mutation avoidance provides a possible and novel explanation for its potential tumor suppressor function.
We began this endeavor to examine whether certain non-essential replication fork-associated factors promote replication fidelity. This issue stems partly from the observation that the apparent fidelities of the replicases are 10–100 times higher in vivo than in vitro (9). The data on Rrm3 support this hypothesis. The question now is how many additional, as yet unknown, factors are involved in replication fidelity. As genes involved in mutation avoidance are likely also tumor suppressors, future endeavors in identifying additional such factors may discover novel tumor suppressor genes or provide mechanistic insight into the functions of known tumor suppressor genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thanks J.S. Williams for sharing reagents and helpful discussion. We thank all Kunkel group members for their thoughtful comments on the manuscript. We thank P. Mieczkowski and others from the High Throughput Sequencing Facility of UNC Chapel Hill for performing Illumina sequencing.
Author contributions: Z.-X.Z. and T.A.K. conceived the project. Z.-X.Z. performed most experiments and data analysis. C.F. constructed the PIF1 mutant HCT116 cell lines. S.A.L. and A.B.B. performed mutation calling using the muver pipeline and assisted with the data analysis. Z.-X.Z., V.A.Z and T.A.K. wrote the manuscript.
Contributor Information
Zhi-Xiong Zhou, Genome Integrity & Structural Biology Laboratory, NIH/NIEHS, DHHS, Research Triangle Park, NC 27709, USA.
Cindy Follonier, Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA.
Scott A Lujan, Genome Integrity & Structural Biology Laboratory, NIH/NIEHS, DHHS, Research Triangle Park, NC 27709, USA.
Adam B Burkholder, Integrative Bioinformatics Support Group, NIH/NIEHS, DHHS, Research Triangle Park, NC 27709, USA.
Virginia A Zakian, Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA.
Thomas A Kunkel, Genome Integrity & Structural Biology Laboratory, NIH/NIEHS, DHHS, Research Triangle Park, NC 27709, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Division of Intramural Research of the NIH, NIEHS [Z01 ES065070 to T.A.K.]; work in the Zakian lab was supported by grant 1R35GM118279 to V.A.Z from the National Institutes of Health. Funding for open access charge: Division of Intramural Research of the NIH, NIEHS [Z01 ES065070 to T.A.K.]; work in the Zakian lab was supported by grant 1R35GM118279 to V.A.Z from the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kunkel T.A., Bebenek K.. DNA replication fidelity. Annu. Rev. Biochem. 2000; 69:497–529. [DOI] [PubMed] [Google Scholar]
- 2. Ganai RaisA., Johansson E.. DNA replication—a matter of fidelity. Mol. Cell. 2016; 62:745–755. [DOI] [PubMed] [Google Scholar]
- 3. Kunkel T.A. Exonucleolytic proofreading. Cell. 1988; 53:837–840. [DOI] [PubMed] [Google Scholar]
- 4. Abbotts J., Loeb L.A.. DNA polymerase alpha and models for proofreading. Nucleic Acids Res. 1985; 13:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wingert B.M., Parrott E.E., Nelson S.W.. Fidelity, mismatch extension, and proofreading activity of the plasmodium falciparum apicoplast DNA polymerase. Biochemistry. 2013; 52:7723–7730. [DOI] [PubMed] [Google Scholar]
- 6. Picher A.J., Garcia-Diaz M., Bebenek K., Pedersen L.C., Kunkel T.A., Blanco L.. Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res. 2006; 34:3259–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen F., Zhao Y., Fan C., Zhao Y.. Mismatch extension of DNA polymerases and high-accuracy single nucleotide polymorphism diagnostics by gold nanoparticle-improved isothermal amplification. Anal. Chem. 2015; 87:8718–8723. [DOI] [PubMed] [Google Scholar]
- 8. Pavlov Y.I., Frahm C., McElhinny S.A.N., Niimi A., Suzuki M., Kunkel T.A.. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006; 16:202–207. [DOI] [PubMed] [Google Scholar]
- 9. Zhou Z.X., Lujan S.A., Burkholder A.B., St Charles J., Dahl J., Farrell C.E., Williams J.S., Kunkel T.A.. How asymmetric DNA replication achieves symmetrical fidelity. Nat. Struct. Mol. Biol. 2021; 28:1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iyer R.R., Pluciennik A., Burdett V., Modrich P.L.. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006; 106:302–323. [DOI] [PubMed] [Google Scholar]
- 11. Shcherbakova P.V., Pavlov Y.I., Chilkova O., Rogozin I.B., Johansson E., Kunkel T.A.. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003; 278:43770–43780. [DOI] [PubMed] [Google Scholar]
- 12. Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M., Kunkel T.A.. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005; 280:29980–29987. [DOI] [PubMed] [Google Scholar]
- 13. Thomas D.C., Roberts J.D., Sabatino R.D., Myers T.W., Tan C.K., Downey K.M., So A.G., Bambara R.A., Kunkel T.A.. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991; 30:11751–11759. [DOI] [PubMed] [Google Scholar]
- 14. Schmitt M.W., Matsumoto Y., Loeb L.A.. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009; 91:1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunkel T.A., Hamatake R.K., Motto-Fox J., Fitzgerald M.P., Sugino A.. Fidelity of DNA polymerase i and the DNA polymerase I-DNA primase complex from saccharomyces cerevisiae. Mol Cell Biol. 1989; 9:4447–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korona D.A., Lecompte K.G., Pursell Z.F.. The high fidelity and unique error signature of human DNA polymerase epsilon. Nucleic Acids Res. 2011; 39:1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arana M.E., Seki M., Wood R.D., Rogozin I.B., Kunkel T.A.. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008; 36:3847–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sabouri N. The functions of the multi-tasking pfh1(pif1) helicase. Curr. Genet. 2017; 63:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bochman M.L., Sabouri N., Zakian V.A.. Unwinding the functions of the pif1 family helicases. DNA Repair (Amst.). 2010; 9:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A.. The saccharomyces cerevisiae helicase rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003; 12:1525–1536. [DOI] [PubMed] [Google Scholar]
- 21. Tran P.L.T., Pohl T.J., Chen C.F., Chan A., Pott S., Zakian V.A.. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat Commun. 2017; 8:15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osmundson J.S., Kumar J., Yeung R., Smith D.J.. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat. Struct. Mol. Biol. 2017; 24:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geronimo C.L., Zakian V.A.. Getting it done at the ends: pif1 family DNA helicases and telomeres. DNA Repair (Amst.). 2016; 44:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen C.F., Pohl T.J., Pott S., Zakian V.A.. Two pif1 family DNA helicases cooperate in centromere replication and segregation in saccharomyces cerevisiae. Genetics. 2019; 211:105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deegan T.D., Baxter J., Ortiz Bazan M.A., Yeeles J.T.P., Labib K.P.M.. Pif1-Family helicases support fork convergence during DNA replication termination in eukaryotes. Mol. Cell. 2019; 74:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muellner J., Schmidt K.H.. Yeast genome maintenance by the multifunctional PIF1 DNA helicase family. Genes (Basel). 2020; 11:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boule J.B., Zakian V.A.. The yeast pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007; 35:5809–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T.. Periodic DNA patrolling underlies diverse functions of pif1 on R-loops and G-rich DNA. Elife. 2014; 3:e02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koc K.N., Singh S.P., Stodola J.L., Burgers P.M., Galletto R.. Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase delta. Nucleic Acids Res. 2016; 44:3811–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ribeyre C., Lopes J., Boule J.B., Piazza A., Guedin A., Zakian V.A., Mergny J.L., Nicolas A.. The yeast pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009; 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ryu G.H., Tanaka H., Kim D.H., Kim J.H., Bae S.H., Kwon Y.N., Rhee J.S., MacNeill S.A., Seo Y.S.. Genetic and biochemical analyses of pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 2004; 32:4205–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A.. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azvolinsky A., Dunaway S., Torres J.Z., Bessler J.B., Zakian V.A.. The s. cerevisiae rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006; 20:3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chisholm K.M., Aubert S.D., Freese K.P., Zakian V.A., King M.C., Welcsh P.L.. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS One. 2012; 7:e30748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pavlov Y.I., Shcherbakova P.V., Kunkel T.A.. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001; 159:47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Z.X., Williams J.S., Kunkel T.A.. Studying ribonucleotide incorporation: strand-specific detection of ribonucleotides in the yeast genome and measuring Ribonucleotide-induced mutagenesis. J. Vis. Exp. 2018; 58020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drake J.W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lujan S.A., Clausen A.R., Clark A.B., MacAlpine H.K., MacAlpine D.M., Malc E.P., Mieczkowski P.A., Burkholder A.B., Fargo D.C., Gordenin D.A.et al.. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014; 24:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larrea A.A., Lujan S.A., Nick McElhinny S.A., Mieczkowski P.A., Resnick M.A., Gordenin D.A., Kunkel T.A.. Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17674–17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burkholder A.B., Lujan S.A., Lavender C.A., Grimm S.A., Kunkel T.A., Fargo D.C.. Muver, a computational framework for accurately calling accumulated mutations. BMC Genomics. 2018; 19:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Welch B.L. The generalisation of student's problems when several different population variances are involved. Biometrika. 1947; 34:28–35. [DOI] [PubMed] [Google Scholar]
- 42. Shapiro S.S., Wilk M.B.. An analysis of variance test for normality (Complete samples). Biometrika. 1965; 52:591–661. [Google Scholar]
- 43. Sidak Z. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 1967; 62:626–633. [Google Scholar]
- 44. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hall B.M., Ma C.X., Liang P., Singh K.K.. Fluctuation analysis calculator: a web tool for the determination of mutation rate using luria-delbruck fluctuation analysis. Bioinformatics. 2009; 25:1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Syed S., Desler C., Rasmussen L.J., Schmidt K.H.. A novel rrm3 function in restricting DNA replication via an orc5-binding domain is genetically separable from rrm3 function as an ATPase/Helicase in facilitating fork progression. PLoS Genet. 2016; 12:e1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pursell Z.F., Isoz I., Lundstrom E.B., Johansson E., Kunkel T.A.. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007; 317:127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Clausen A.R., Lujan S.A., Burkholder A.B., Orebaugh C.D., Williams J.S., Clausen M.F., Malc E.P., Mieczkowski P.A., Fargo D.C., Smith D.J.et al.. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 2015; 22:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008; 18:85–98. [DOI] [PubMed] [Google Scholar]
- 50. Azvolinsky A., Giresi P.G., Lieb J.D., Zakian V.A.. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in saccharomyces cerevisiae. Mol. Cell. 2009; 34:722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roche H., Gietz R.D., Kunz B.A.. Specificity of the yeast rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics. 1994; 137:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kraszewska J., Garbacz M., Jonczyk P., Fijalkowska I.J., Jaszczur M.. Defect of dpb2p, a noncatalytic subunit of DNA polymerase varepsilon, promotes error prone replication of undamaged chromosomal DNA in saccharomyces cerevisiae. Mutat. Res. 2012; 737:34–42. [DOI] [PubMed] [Google Scholar]
- 53. Garbacz M.A., Cox P.B., Sharma S., Lujan S.A., Chabes A., Kunkel T.A.. The absence of the catalytic domains of saccharomyces cerevisiae DNA polymerase strongly reduces DNA replication fidelity. Nucleic Acids Res. 2019; 47:3986–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garbacz M., Araki H., Flis K., Bebenek A., Zawada A.E., Jonczyk P., Makiela-Dzbenska K., Fijalkowska I.J.. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair (Amst). 2015; 29:23–35. [DOI] [PubMed] [Google Scholar]
- 55. Morrison A., Christensen R.B., Alley J., Beck A.K., Bernstine E.G., Lemontt J.F., Lawrence C.W.. REV3, a saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 1989; 171:5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holbeck S.L., Strathern J.N.. A role for REV3 in mutagenesis during double-strand break repair in saccharomyces cerevisiae. Genetics. 1997; 147:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., Mathur R., Chabes A., Malkova A.. Break-induced replication is highly inaccurate. PLoS Biol. 2011; 9:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siebler H.M., Lada A.G., Baranovskiy A.G., Tahirov T.H., Pavlov Y.I.. A novel variant of DNA polymerase zeta, Rev3DeltaC, highlights differential regulation of pol32 as a subunit of polymerase delta versus zeta in saccharomyces cerevisiae. DNA Repair (Amst). 2014; 24:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yeung R., Smith D.J.. Determinants of replication-fork pausing at tRNA genes in <em>Saccharomyces cerevisiae</em>. Genetics. 2020; 214:825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A.. Saccharomyces rrm3p, a 5' to 3' DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002; 16:1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ivessa A.S., Zhou J.Q., Zakian V.A.. The saccharomyces pif1p DNA helicase and the highly related rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000; 100:479–489. [DOI] [PubMed] [Google Scholar]
- 62. Zhao X., Rothstein R.. The dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor sml1. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schulz V.P., Zakian V.A.. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994; 76:145–155. [DOI] [PubMed] [Google Scholar]
- 64. Papadopoulos N., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D.et al.. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994; 263:1625–1629. [DOI] [PubMed] [Google Scholar]
- 65. McDonald K.R., Sabouri N., Webb C.J., Zakian V.A.. The pif1 family helicase pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening. DNA Repair (Amst). 2014; 24:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Paeschke K., Capra J.A., Zakian V.A.. DNA replication through G-quadruplex motifs is promoted by the saccharomyces cerevisiae pif1 DNA helicase. Cell. 2011; 145:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aria V., Yeeles J.T.P.. Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. Mol. Cell. 2018; 73:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Myung K., Chen C., Kolodner R.D.. Multiple pathways cooperate in the suppression of genome instability in saccharomyces cerevisiae. Nature. 2001; 411:1073–1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All whole genome sequencing data is available through Sequence Read Archive accession number PRJNA689775.
Code availability
Muver suite is available via GitHub (40) or upon request.