Abstract
STUDY QUESTION
Does an estradiol-based combined oral contraceptive (COC) have a milder effect on the serum proteome than an ethinylestradiol (EE)-based COC or dienogest (DNG) only?
SUMMARY ANSWER
The changes in serum proteome were multifold after the use of a synthetic EE-based COC compared to natural estrogen COC or progestin-only preparation.
WHAT IS KNOWN ALREADY
EE-based COCs widely affect metabolism, inflammation, hepatic protein synthesis and blood coagulation. Studies comparing serum proteomes after the use of COCs containing EE and natural estrogens are lacking.
STUDY DESIGN, SIZE, DURATION
This was a spin-off from a randomized, controlled, two-center clinical trial. Women (n = 59) were randomized to use either EE + DNG, estradiol valerate (EV) + DNG or DNG only continuously for 9 weeks.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Participants were healthy, young, white volunteer women. Serum samples were collected before and after 9 weeks of hormonal exposure. Samples from 44 women were available for analysis (EE + DNG n = 14, EV + DNG n = 16 and DNG only n = 14). Serum proteins were analyzed by quantitative, discovery-type label-free proteomics.
MAIN RESULTS AND THE ROLE OF CHANCE
Altogether, 446 proteins/protein families with two or more unique peptides were detected and quantified. The number of proteins/families that altered over the 9-week period within the study groups was 121 for EE + DNG and 5 for EV + DNG, while no changes were detected for DNG only. When alterations were compared between the groups, significant differences were detected for 63 proteins/protein families, of which 58 were between the EE + DNG and EV + DNG groups. The most affected functions during the use of EE + DNG were the complement system, acute phase response signaling, metabolism and the coagulation system. The results were validated by fetuin-B and cortisol-binding globulin ELISA and sex hormone-binding globulin immunoassay.
LARGE SCALE DATA
Data are available via ProteomeXchange with identifiers PXD033617 (low abundance fraction) and PXD033618 (high abundance fraction).
LIMITATIONS, REASONS FOR CAUTION
The power analysis of the trial was not based on the proteomic analysis of this spin-off study. In the future, targeted proteomic analysis with samples from another trial should be carried out in order to confirm the results.
WIDER IMPLICATIONS OF THE FINDINGS
The EE-based COC exerted a broader effect on the serum proteome than the EV-based COC or the DNG-only preparation. These results demonstrate that the effects of EE in COCs go far beyond the established endpoint markers of estrogen action, while the EV combination is closer to the progestin-only preparation. The study indicates that EV could provide a preferable option to EE in COCs in the future and signals a need for further studies comparing the clinical health outcomes of COCs containing EE and natural estrogens.
STUDY FUNDING/COMPETING INTEREST(S)
Funding for this researcher-initiated study was obtained from the Helsinki University Hospital research funds, the Hospital District of Helsinki and Uusimaa, the Sigrid Juselius Foundation, the Academy of Finland, the Finnish Medical Association, the University of Oulu Graduate School, the Emil Aaltonen Foundation, the Swedish Cultural Foundation in Finland, the Novo Nordisk Foundation, Orion Research Foundation and the Northern Ostrobothnia Regional Fund. The funders had no role in study design, data collection and analysis, publishing decisions or manuscript preparation. T.P. has received honoraria for lectures, consultations and research grants from Exeltis, Gedeon Richter, MSD, Merck, Pfizer, Roche, Stragen and Mithra Pharmaceuticals. O.H. occasionally serves on advisory boards for Bayer AG and Gedeon Richter and has designed and lectured at educational events for these companies. The other authors have nothing to disclose. O.H. occasionally serves on advisory boards for Bayer AG and Gedeon Richter and has designed and lectured at educational events for these companies. The other authors have nothing to disclose.
TRIAL REGISTRATION NUMBER
ClinicalTrials.gov NCT02352090
TRIAL REGISTRATION DATE
27 January 2015
DATE OF FIRST PATIENT’S ENROLMENT
1 April 2015
Keywords: estradiol valerate, ethinylestradiol, combined contraceptive, proteome, randomized controlled trial, complement, acute phase signaling, metabolism, coagulation
Introduction
In addition to the contraceptive effect, combined oral contraceptives (COCs) have vast effects on female physiology. In the liver, COCs stimulate the synthesis of steroid-binding globulins, such as sex hormone-binding globulin (SHBG), thereby affecting circulating, free steroid levels (Mashchak et al., 1982; Wiegratz et al., 2003; Sitruk-Ware et al., 2007a,b; Piltonen et al., 2012). COCs also increase low-grade inflammation, alter lipid metabolism and affect the coagulation system, resulting in an increased risk for thromboembolic events (van Rooijen et al., 2006; Morin-Papunen et al., 2008; Haarala et al., 2009; Lidegaard et al., 2009, 2012; Piltonen et al., 2012; Wang et al., 2016; Kangasniemi et al., 2020). Even though millions of women worldwide use COCs, some of the effects and health outcomes, as well as their mode of action, remain poorly understood.
The net effect of COCs results from both estrogen and progestin action and their interplay (Sitruk-Ware and Nath, 2013). In COCs, ethinylestradiol (EE) is the most commonly used estrogen. It is a potent estrogen having up to 600 times greater effect on hepatic protein synthesis than natural estradiol (E2) (Mashchak et al., 1982). EE provides efficient bleeding control in combination with different progestins; however, its oral dose is directly proportional to the risk of venous thrombosis (Sitruk-Ware and Nath, 2013; de Bastos et al., 2014; Sitruk-Ware, 2016; Abou-Ismail et al., 2020). Over recent decades, the dose of EE in COCs has been reduced to lower this risk.
Recently, natural estrogens have been included in COCs to reduce EE-related adverse events. New combinations containing E2, its ester estradiol valerate (EV), and estetrol (E4) have proven effective in combined contraceptives. They induce good cycle control and provoke less marked changes in metabolic and endocrine parameters compared with EE (Junge et al., 2011; Sitruk-Ware and Nath, 2013; Stanczyk et al., 2013; Mawet et al., 2015; Grandi et al., 2016; Kluft et al., 2017; Haverinen et al., 2021; Klipping et al., 2021). However, most trials comparing COCs with different estrogens have included preparations that also differ in the progestin component; hence, knowledge of the specific estrogen effects in COCs is lacking. As the progestin component of COCs modulates the estrogen effects, the net effect of a COC depends on both the type of estrogen and the properties of the progestin (Sitruk-Ware and Nath, 2013; Abou-Ismail et al., 2020).
Serum proteomic analysis provides an option to investigate the broad pharmacodynamic effects during medication use. Instead of specific, planned targets, this shotgun method analyzes all proteins and enables a broad overview and novel findings. To our knowledge, this is the first study to utilize such discovery-type proteomic analysis on hormonal contraceptives.
This study compared changes induced in the serum proteome during the use of COCs containing different estrogen components. A progestin-only preparation (containing the same progestin) was included as an active control. This study is a spin-off of a larger randomized controlled trial comparing the metabolic effects of EE + dienogest (DNG), EV + DNG and DNG only. We hypothesized that the EV + DNG preparation would have a milder impact on the serum proteome than that of EE + DNG and that the effect of DNG only would be neutral.
Materials and methods
This randomized, controlled, investigator-initiated clinical trial was conducted at Helsinki and Oulu University Hospitals, Finland, between April 2015 and January 2018. The study protocol has been described previously (Haverinen et al., 2021), and it was approved by the independent Ethics Committee of Helsinki University Central Hospital and the Finnish Medicines Agency. The regional ethics committee of the Northern Ostrobothnia Hospital District was informed of the approval. The study was registered with ClinicalTrials.gov (NCT02352090; https://clinicaltrials.gov/) and the EU Clinical Trials Register (EudraCT Number 2014-001243-20; https://www.clinicaltrialsregister.eu). All participants signed an informed consent form. The sample size calculation of the study was based on glucose metabolism, which was the trial’s primary outcome (Haverinen et al., 2021).
Subjects
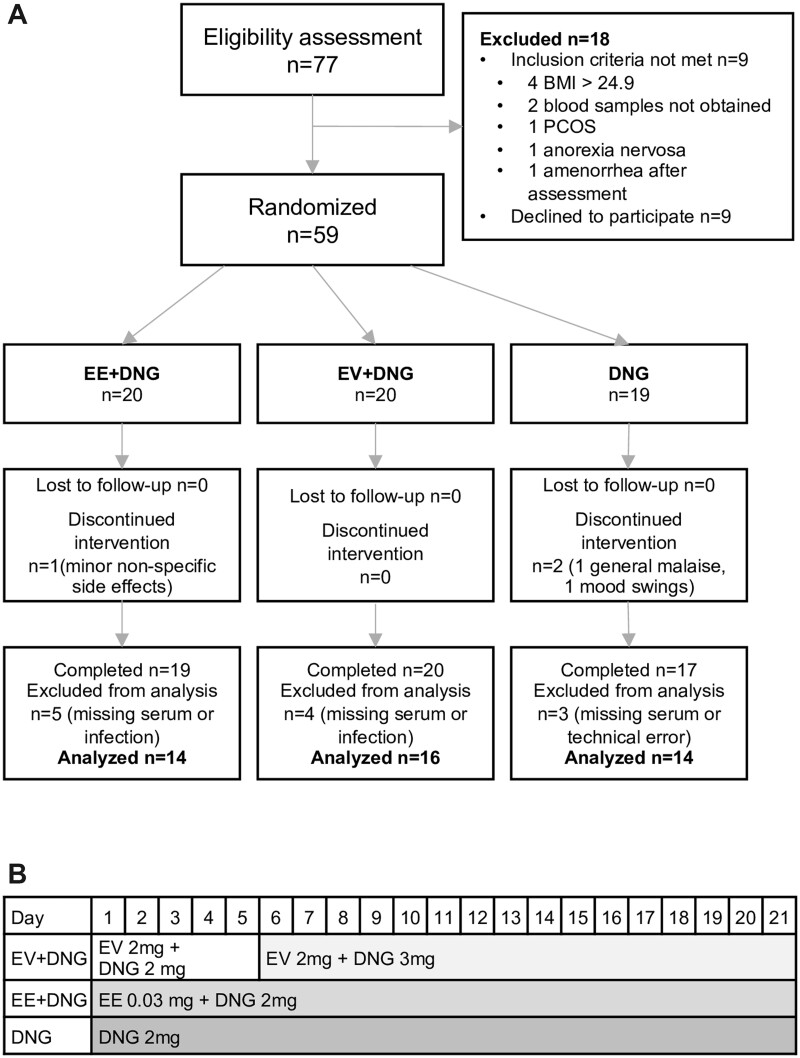
Seventy-seven healthy women volunteered for the study (Fig. 1). After the eligibility assessment, 59 women were randomized. The participants had regular menstrual cycles and a minimum wash-out period of 2 months from hormonal medications and 3 months from breastfeeding. Exclusion criteria were age over 35 years, BMI ≥25 kg/m2, blood pressure ≥140/90 mmHg, smoking, alcohol or drug abuse, or any contraindication to COC use. Also, abnormalities in the 2-h oral glucose tolerance test or gynecological ultrasound examination led to exclusion from the study.
Figure 1.
Flow chart and study preparations. Flow chart of the study (A) and hormone contents of the study preparations (B). The regimens were used continuously for 9 weeks. DNG, dienogest; EE, ethinylestradiol; EV, estradiol valerate.
Intervention
Participants were randomized to use either EE + DNG (Valette®, Bayer AG, Germany), EV + DNG (Qlaira®, Bayer AG, Germany) or DNG only (Visanne®, Jenapharm, Bayer AG, Germany) for 9 weeks continuously. Original packages were modified by removing placebo pills and pills containing only estrogen to minimize the differences in hormonal contents. The study flow chart and the exact hormonal contents of the study preparations are shown in Fig. 1. Serum samples were collected after a 12-h fast before the intervention on menstrual cycle days 1–5 and during the 9th week of treatment. After analysis for the primary and secondary outcomes of the original trial, serum samples for this spin-off study were available for 44 patients (EE + DNG n = 14, EV + DNG n = 16 and DNG n = 14).
Serum proteomic analysis
Serum samples were stored at −70°C until analysis. High-abundance proteins and their protein complexes were affinity-purified from the samples, and the samples were divided into two pools: high- and low-abundance proteins. Human serum albumin was further removed from the high-abundance samples. Proteins were digested with trypsin and applied via a reverse-phase nano-liquid chromatograph column to a high-resolution mass spectrometer. Data were collected in a data-independent manner, and peptides were identified and quantified.
Affinity chromatography
An Agilent Technologies (Santa Clara, CA, USA) Multiple Affinity Removal Column Human 14 (Cat No 5188-6557) was used in the affinity chromatography, according to the manufacturer’s instructions. Samples were diluted four-fold with buffer A. Fourteen proteins and their complexes (i.e. ‘high-abundance proteins’) were retained in the column: albumin, IgG, IgA, transferrin, haptoglobin, antitrypsin, fibrinogen, alpha2-macroglobulin, alpha1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3 and transthyretin. Low-abundance proteins were collected as a flow-through fraction. High-abundance proteins were eluted with buffer B and collected.
Desalting and albumin removal from high-abundance samples
All samples were desalted using Thermo Fisher Scientific (Waltham, MA, USA) Zeba Spin desalting 96-well plates (Cat No 89808) to change the buffer to 25 mM Trizma BASE + 75 mM NaCl pH 7–8. The high-abundance samples were albumin-depleted with a Thermo Fisher Scientific Pierce Albumin Depletion Kit (Cat No 85160). Protein concentrations were measured in all samples using the Bradford method; 120 μg of proteins were aliquoted, dried, and stored at −70°C until digested. A SpeedVac vacuum concentrator (Model DNA120; Savant Systems LLC, MA, USA) was used for drying the samples.
Trypsin digestion
Proteins in the samples were trypsin-digested with the aid of Waters Corporation (Milford, MA, USA) 0.2% surfactant RapiGest™ SF. Samples were boiled for 10 min, and proteins were reduced by adding 0.5 M 1,4-dithiothreitol to a final concentration of 5 mM, vortexed and incubated for 30 min in 60°C water bath. In the next step, the proteins were alkylated with a 15 mM final concentration of iodoacetamide and incubated for 30 min in the dark at room temperature. Sequence grade trypsin Promega Biotech AB (Madison, WI, USA) Trypsin Gold (Cat No V5280) was added to the samples in a 1:100 enzyme-to-protein ratio. Samples were incubated at 37°C overnight. Samples were acidified to a final concentration of 0.5% trifluoroacetic acid and incubated for 45 min at 37°C, after which cleaved RapiGest™ SF was removed by a 10-min centrifugation at 16 000 g at 8°C. The supernatant was transferred to a new vial, and half of it was purified with Thermo Fisher Scientific Pierce C18 Spin Columns (Cat No 89870), according to the manufacturer’s instructions. The samples were dried and resuspended in 50 µl of 0.1% formic acid (FA) + 2% acetonitrile (ACN); 10 µl were used for peptide quantitation. Peptide concentrations were measured with a Thermo Fisher Scientific Pierce™ Quantitative Fluorometric Peptide Assay (Cat No 23290). The mass spectrometry samples were spiked in with a Waters Corporation Hi3 E. Coli Standard (Cat No 186006012), using 50 fmol/500 ng injected peptide mixture.
Mass spectrometry
Five hundred nanograms of the Hi3 spiked peptide mixture was injected to the Waters Corporation Synapt G2-Si HDMS and nanoACQUITY system. The system was equipped with Trap Column Symmetry C18, nanoACQUITY 10K 2G V/M Trap Column, 100 Å, 5 µm, 180 µm × 20 mm (Cat No 186006527) and analytical ultra-performance liquid chromatography (UPLC) column BEH C18 nanoACQUITY 10 K psi, 300 Å, 1.7 µm, 75 µm × 250 mm (Cat No 1886003815).
Samples were loaded to trapping column 8 µl/min for 2 minutes with 2% B buffer. The analytical gradient used was as follows: 0–1 min 2% B buffer, 1–65 min 55% B buffer, 65–80 min 80% B buffer, 80–87 min 80% B buffer, 87–90 min 2% B buffer and 90–100 min 2% B buffer. The gradient curve between the time points was linear. Analytical gradient flow rate was 200 nl/min. Buffer A was 0.1% FA in water, and buffer B was 0.1% FA in ACN. The gradient curve between the time points was linear. Buffer A was 0.1% FA in water, and buffer B was 0.1% FA in ACN. All solvents were mass spectrometry–grade from Merck Life Science (Darmstadt, Germany).
Data acquisition was performed with UPLC–ultra definition mass spectrometry (UPLC-UDMSE). Data were collected in the range of 100–2000 m/z, scan time 1 s, and ion mobility mass spectrometry wave velocity 650 m/s. Collision energy was ramped to fragment the peptides in the high-energy mode from 20 to 60 V. Calibration was performed with sodium iodide clusters over a mass range of 50–2500 m/z by infusing 2 µg/µl sodium iodide solution in 50/50 2-propanol/water into the mass spectrometer. At the same time, a leucine–enkephalin function for post-acquisition mass correction was collected (M+H+ 556.2771 m/z). In all, 10% of the samples were acquired as triplicates: the percentage coefficient of variation for the high-abundance proteins was 1.82, and for the low-abundance proteins 8.11.
Peptide identification and quantitation
The raw data were imported to Progenesis QI for proteomics (Nonlinear Dynamics, La Jolla, CA, USA), and simultaneously mass corrected with the leucine–enkephalin function. Proteinlynx Global Server (Waters Corporation) was used to identify peptides from the UDMSE-type data (Silva et al., 2006a). In Progenesis, we used default parameters for peak picking and alignment, and the peptides were identified against Uniprot human FASTA sequences release 2019_06. A ClpB protein sequence (CLPB_ECOLI (P63285)) was inserted for label-free quantification to match the spike-in Hi3 standard. We used fixed modification at cysteine (carbamidomethyl) and variable at methionine (oxidation), and trypsin with two miss cleavages as digestion agents. The maximal false discovery rate was set to 1%, and fragment and peptide errors were set as automatic. The default parameters for ion matching were used: one or more ion fragments per peptide, three or more fragments per protein, and one or more peptides per protein. The proteins were assembled from the peptides by using the parsimony principle, and at least two unique peptides per protein were required. Progenesis used the method of Silva (Silva et al., 2006b) for relative quantitation.
Hierarchical clustering and pathway analysis
Hierarchical clustering analysis was performed with MetaboAnalyst (https://www.metaboanalyst.ca/). QIAGEN Ingenuity Pathway Analysis (IPA) was used for pathway enrichment and network analysis. The desired protein accessions and fold changes were uploaded to the IPA, and canonical pathway enrichment was carried out with IPA Core Analysis. Default parameters were used for the analysis. The most significant pathway proteins were transferred to the network, and the proteins were connected where data were available in the Ingenuity Knowledge Base.
Validation
For proteomic analysis validation, fetuin-B measurements were performed using ELISA. Serum samples were diluted 1:10 000 with DPBS (Corning #20-031-CVR, Mediatech, Inc., Manassas, VA, USA) and kit-specific dilution buffer, and analyzed with a FETUB ELISA Kit (Cat No EKX-LDKNLJ-96; Nordic BioSite AB, Täby, Sweden), according to the manufacturer’s instructions. Graphpad Prism 9 (GraphPad Software, San Diego, CA, USA) was used for interpolation of the data. Corticosteroid-binding globulin (CBG) was measured as part of the other sub-study (Kangasniemi et al., 2022) using ELISA (Cat. No. RD192234200R, BioVendor, Brno, Czech Republic) and SHBG in another sub-study of the same trial (Haverinen et al., 2022a) using a chemiluminescent microparticle immunoassay (Abbott Cat No 8K26, RRID: AB_2895255, Abbot Architect i2000SR analyzer, Abbot Diagnostics, IL, USA).
Statistics
The distributions of protein changes were examined with the Shapiro–Wilk test. Most changes were skewed; thus, non-parametric tests were used for the analyses. The changes within the study groups were analyzed with the Wilcoxon test using the Benjamini–Hochberg procedure to control false discovery rates.
To investigate the differences between the study groups, we calculated the difference between the baseline and 9 weeks for all proteins. These differential variables were then analyzed with the Kruskal–Wallis test and the Benjamini–Hochberg procedure. Statistically significant results were further compared with the Mann–Whitney post hoc test, in which a value of P < 0.05 was considered significant. Wilcoxon test was performed using Python script from SciPY (Virtanen et al., 2020) and other statistics using R package stats (R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria).
Results
Study subjects
The baseline characteristics of the participants are shown in Table I. The randomization resulted in balanced study groups regarding age, BMI, blood pressure, glucose and lipid measurements.
Table I.
Baseline characteristics of the study groups taking a 9-week course of combined oral contraceptive.
| EE + DNG |
EV + DNG |
DNG |
Anova P-value* | |
|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | ||
| Number of patients | 14 | 16 | 14 | |
| Age (years) | 26 ± 4 | 24 ± 3 | 24 ± 3 | 0.17 |
| BMI (kg/m2) | 22.74 ± 1.78 | 22.53 ± 1.55 | 21.88 ± 2.07 | 0.42 |
| WHR | 0.79 ± 0.05 | 0.77 ± 0.04 | 0.78 ± 0.04 | 0.44 |
| Systolic BP (mmHg) | 118.93 ± 9.77 | 118.31 ± 7.59 | 111.57 ± 10.70 | 0.08 |
| Diastolic BP (mmHg) | 74.50 ± 8.26 | 74.06 ± 7.45 | 72.86 ± 7.85 | 0.85 |
| Fasting glucose (mmol/l) | 5.09 ± 0.35 | 5.21 ± 0.47 | 4.91 ± 0.38 | 0.14 |
| HbA1c (mmol/mol) | 32.79 ± 2.52 | 33.13 ± 2.66 | 32.14 ± 2.54 | 0.77 |
| Fasting total cholesterol (mmol/l) | 4.08 ± 0.60 | 3.93 ± 0.71 | 4.04 ± 0.47 | 0.19 |
| HDL (mmol/l) | 1.80 ± 0.41 | 1.59 ± 0.33 | 1.59 ± 0.26 | 0.58 |
| LDL (mmol/l) | 2.14 ± 0.51 | 2.17 ± 0.67 | 2.35 ± 0.49 | 0.95 |
| Triglycerides (mmol/l) | 0.67 ± 0.16 | 0.66 ± 0.25 | 0.64 ± 0.18 | 0.58 |
| hs-CRP (mg/l) | 0.98 ± 0.92 | 0.65 ± 0.54 | 0.56 ± 0.39 | 0.21 |
EE, ethinylestradiol; DNG, dienogest; EV, estradiol valerate; WHR, waist–hip ratio; BP, blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein; hs-CRP, high sensitivity C-reactive protein.
Comparison between groups by one-way ANOVA.
Proteomic analysis
Protein identification
A total of 446 proteins or protein families with two or more unique peptides were quantified. Some of the protein sequences are shared by many proteins (protein families), and some are unique in the human proteome. Depending on which peptides are identified, one can detect either a distinct protein or a protein family. Certain proteins were therefore identified and analyzed together as a family. Some proteins were detected both in high- and low-abundance analyses. The complete list of detected proteins is shown in Supplementary Table SI.
Hierarchical clustering
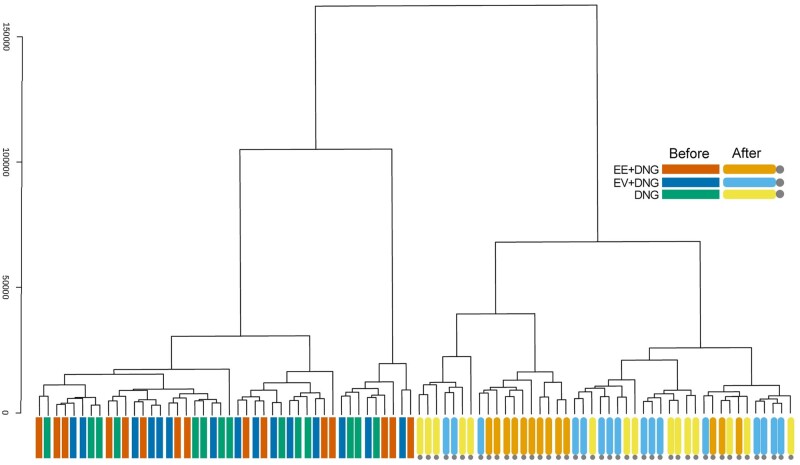
Successful randomization was further confirmed by hierarchical clustering based on protein compositions (Fig. 2). The 9-week intervention resulted in apparent clustering of the participants, especially the women using the EE + DNG preparation.
Figure 2.
Hierarchical clustering analysis based on the protein compositions of serum samples during a 9-week use of combined oral contraceptive. Hierarchical clustering dendrogram of samples using the Euclidean distance measure (Y-axis) and the Ward clustering algorithm. The analysis shows that samples from the three study groups were mixed at baseline but after 9 weeks of use of the combined oral contraceptive preparations, the serum proteome profile clustered the participants according to preparation. The EE + DNG group in particular showed distinct clustering. The cluster was produced using MetaboAnalyst. DNG, dienogest; EE, ethinylestradiol; EV, estradiol valerate.
Proteome alterations within groups
A total of 122 proteins/protein families changed significantly during the 9-week trial within at least one group: 121 in the EE + DNG and 5 in the EV + DNG group. No changes were observed in the DNG-only group. Some proteins were found in both low- and high-abundance fractions, indicating that proteins appeared both alone and in complexes with some high-abundance proteins, such as albumin. After consideration of these duplicate findings, the numbers of changing, unique proteins/families are shown in Fig. 3. All the proteins with relevant statistical data are compiled in Supplementary Table SI. The changes during EE + DNG use are further described in the ‘Pathway analysis’ section. Most of the changing proteins were related to inflammation, metabolism and coagulation. The five proteins that showed significant change during EV + DNG use were ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ICOS ligand & Ig-like domain-containing protein, fetuin-B, angiotensinogen and interleukin-1 receptor accessory protein (IL1RAP).
Figure 3.
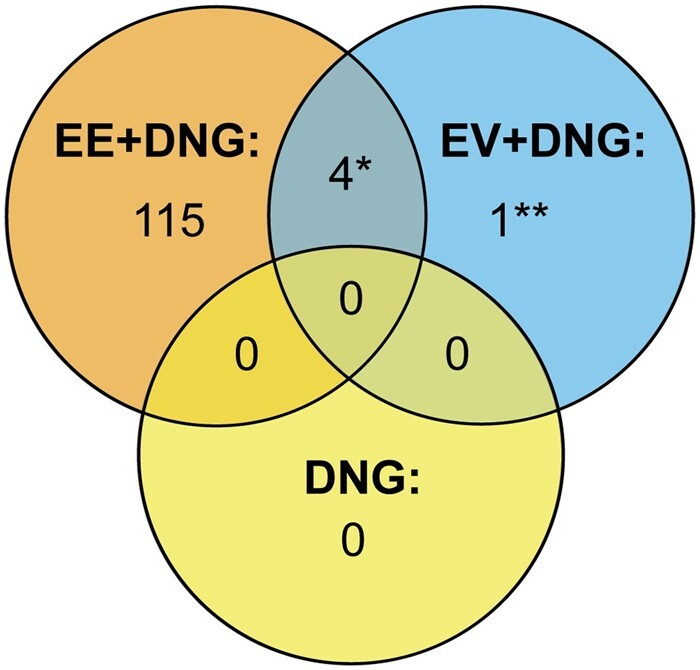
Amounts of changing and unique proteins/protein families in serum from women taking a 9-week course of combined oral contraceptive. Venn diagram showing the numbers of changed proteins after within-group analysis. DNG, dienogest; EE, ethinylestradiol; EV, estradiol valerate. *Interleukin1 receptor accessory protein, fetuin-B, angiotensinogen and ICOS ligand and Ig-like domain-containing protein. **Ectonucleotide pyrophosphatase/phosphodiesterase family member 2.
Change in protein abundances between study groups
The change in protein abundance from baseline to 9 weeks was significantly different between the study groups for 63 proteins/protein families. In all, 58 proteins/protein families altered differently between the EE + DNG and EV + DNG groups, 61 between EE + DNG and DNG only and 17 between EV + DNG and DNG only. All significantly altered proteins between the groups are listed in Table II. Among the 10 most differently altered proteins, the highest fold changes were detected in CBG, SHBG, IL1RAP, angiotensinogen and fetuin-B.
Table II.
Proteins with significantly differing alterations between study groups during the 9-week trial.
| Fold changes |
Group comparison |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fraction | Accession | Gene | Protein name | EE + DNG | EV + DNG | DNG | A | B | C |
| LOW | Q9NPH3 | IL1RAP | Interleukin-1 receptor accessory protein | 3.0* | 1.7* | 1.2 | S | S | S |
| LOW | Q9UGM5 | FETUB | Fetuin-B | 2.0* | 1.4* | 1.2 | S | S | S |
| LOW | P01019 | AGT | Angiotensinogen | 2.6* | 1.7* | 1.2 | S | S | S |
| LOW | P04278 | SHBG | Sex hormone-binding globulin | 3.2* | 1.4 | 1.2 | S | S | NS |
| LOW | O75144 | ICOSLG | ICOS ligand | 2.2* | 1.5* | 1.1 | S | S | S |
| A0A087X1L8 | LOC102723996 | Ig-like domain-containing protein | |||||||
| LOW | P08185 | SERPINA6 | Corticosteroid-binding globulin | 3.2* | 1.2 | 1.3 | S | S | NS |
| LOW | Q9HD23 | MRS2 | Magnesium transporter MRS2 homolog, mitochondrial | 2.1* | 1.2 | 1.1 | S | S | NS |
| LOW | P08294 | SOD3 | Extracellular superoxide dismutase [Cu-Zn] | 0.5* | 0.8 | 1.0 | S | S | NS |
| LOW | Q96PD5 | PGLYRP2 | N-acetylmuramoyl-L-alanine amidase | 0.5* | 0.9 | 1.1 | S | S | S |
| LOW | Q562R1 | ACTBL2 | Beta-actin-like protein 2 | 1.7* | 1.2 | 1.1 | S | S | NS |
| LOW | Q5TCS8 | AK9 | Adenylate kinase 9 | 0.5* | 0.9 | 1.1 | S | S | S |
| LOW | P04196 | HRG | Histidine-rich glycoprotein | 0.6* | 0.9 | 1.1 | S | S | NS |
| LOW | Q8N6C8 | LILRA3 | Leukocyte immunoglobulin-like receptor subfamily A member 3 | 0.6* | 0.9 | 1.2 | S | S | S |
| LOW | Q04756 | HGFAC | Hepatocyte growth factor activator | 0.5* | 0.8 | 1.0 | S | S | S |
| LOW | P01008 | SERPINC1 | Antithrombin-III | 0.6* | 0.9 | 1.1 | S | S | S |
| LOW | P07225 | PROS1 | Vitamin K-dependent protein S | 0.7* | 1.0 | 1.1 | S | S | NS |
| HIGH | P04083 | ANXA1 | Annexin A1 | 1.5* | 1.1 | 1.0 | S | S | NS |
| LOW | P05155 | SERPING1 | Plasma protease C1 inhibitor | 1.1* | 1.5 | 2.0 | S | S | NS |
| HIGH | Q9NRF9 | POLE3 | DNA polymerase epsilon subunit 3 | 1.3* | 1.0 | 1.0 | S | S | NS |
| HIGH | Q9Y4X3 | CCL27 | C-C motif chemokine 27 | 1.3* | 1.0 | 1.0 | S | S | NS |
| LOW | P05452 | CLEC3B | Tetranectin | 0.6* | 0.8 | 1.0 | S | S | S |
| LOW | P07357 | C8A | Complement component C8 alpha chain | 0.8* | 0.9 | 1.1 | S | S | NS |
| LOW | P20851 | C4BPB | C4b-binding protein beta chain | 5.6* | 6.1 | 7.0 | S | S | NS |
| HIGH | A4FU69 | EFCAB5 | EF-hand calcium-binding domain-containing protein 5 | 0.9* | 1.0 | 1.1 | S | S | NS |
| HIGH | P20742 | PZP | Pregnancy zone protein | 1.5* | 1.0 | 1.0 | S | S | NS |
| LOW | P06396 | GSN | Gelsolin | 0.7* | 0.9 | 1.1 | S | S | NS |
| P22460 | KCNA5 | Voltage-gated potassium channel HK2 | |||||||
| Q8N1V2 | CFAP52 | Cilia- and flagella-associated protein 52 | |||||||
| Q99607 | ELF4 | ETS-related transcription factor Elf-4 | |||||||
| Q9UH62 | ARMCX3 | Armadillo repeat-containing X-linked protein 3 | |||||||
| Q9Y2M5 | KLHL20 | Kelch-like protein 20 | |||||||
| HIGH | P01009 | SERPINA1 | Alpha-1-antitrypsin | 1.4* | 1.0 | 1.0 | S | S | NS |
| LOW | A5A3E0 | POTEF | POTE ankyrin domain family member F | 0.6* | 0.8 | 1.0 | S | S | S |
| LOW | P02747 | C1QC | Complement C1q subcomponent subunit C | 0.8* | 0.9 | 1.1 | NS | S | S |
| LOW | Q92835 | INPP5D | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | 0.8* | 0.9 | 1.1 | S | S | NS |
| HIGH | P01880 | IGHD | Immunoglobulin heavy constant delta | 0.8* | 1.0 | 1.0 | S | S | NS |
| LOW | P04003 | C4BPA | C4b-binding protein alpha chain | 1.9* | 2.1 | 2.8 | S | S | NS |
| LOW | P51884 | LUM | Lumican | 0.6* | 0.8 | 1.0 | S | S | NS |
| LOW | P02749 | APOH | Beta-2-glycoprotein 1 | 331.5* | 443.3 | 589.0 | S | S | NS |
| LOW | P07737 | PFN1 | Profilin-1 | 0.7* | 0.9 | 1.1 | S | S | NS |
| LOW | P09871 | C1S | Complement C1s subcomponent | 0.8* | 0.9 | 1.1 | S | S | NS |
| LOW | P13671 | C6 | Complement component C6 | 0.8* | 0.9 | 1.0 | S | S | NS |
| LOW | P02748 | C9 | Complement component C9 | 0.7* | 0.9 | 1.0 | S | S | NS |
| LOW | P08697 | SERPINF2 | Alpha-2-antiplasmin | 0.8* | 1.0 | 1.1 | S | S | NS |
| LOW | P01011 | SERPINA3 | Alpha-1-antichymotrypsin | 0.7* | 0.9 | 1.1 | NS | S | NS |
| Q9HCY8 | S100A14 | Protein S100-A14 | |||||||
| LOW | P06727 | APOA4 | Apolipoprotein A-IV | 10.7* | 13.0 | 17.2 | NS | S | S |
| LOW | Q16473 | TNXA | Putative tenascin-XA | 0.6* | 0.8 | 1.0 | S | S | NS |
| LOW | Q13822 | ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | 1.5 | 0.8* | 1.0 | S | NS | S |
| LOW | P02745 | C1QA | Complement C1q subcomponent subunit A | 0.7* | 0.9 | 1.1 | S | S | NS |
| LOW | P00450 | CP | Ceruloplasmin | 1.5* | 1.1 | 1.2 | S | S | NS |
| Q96L14 | CEP170P1 | Cep170-like protein | |||||||
| LOW | Q9BWP8 | COLEC11 | Collectin-11 | 0.7* | 1.0 | 1.1 | S | S | NS |
| LOW | P35442 | THBS2 | Thrombospondin-2 | 0.7* | 0.9 | 1.0 | S | S | NS |
| LOW | Q9NQ79 | CRTAC1 | Cartilage acidic protein 1 | 0.7* | 0.9 | 1.0 | S | S | NS |
| LOW | P00736 | C1R | Complement C1r subcomponent | 0.8* | 0.9 | 1.1 | S | S | NS |
| LOW | P02746 | C1QB | Complement C1q subcomponent subunit B | 0.7* | 0.9 | 1.1 | S | S | NS |
| LOW | P07360 | C8G | Complement component C8 gamma chain | 0.8* | 1.0 | 1.1 | S | S | NS |
| LOW | P02776 | PF4 | Platelet factor 4 | 1.1 | 0.9 | 1.1 | S | NS | S |
| HIGH | P08185 | SERPINA6 | Corticosteroid-binding globulin | 1.4* | 1.0 | 1.0 | S | S | NS |
| LOW | P09172 | DBH | Dopamine beta-hydroxylase | 0.7* | 0.9 | 1.1 | S | S | NS |
| LOW | P10643 | C7 | Complement component C7 | 0.8* | 0.9 | 1.1 | S | S | NS |
| LOW | Q8WXW3 | PIBF1 | Progesterone-induced-blocking factor 1 | 0.7* | 1.0 | 1.0 | S | S | NS |
| LOW | Q8N531 | FBXL6 | F-box/LRR-repeat protein 6 | 0.8* | 1.0 | 1.0 | S | S | NS |
| LOW | P05019 | IGF1 | Insulin-like growth factor I | 0.8* | 0.9 | 1.1 | S | S | NS |
| LOW | P05543 | SERPINA7 | Thyroxine-binding globulin | 1.4* | 1.1 | 1.1 | S | S | NS |
| LOW | Q6EMK4 | VASN | Vasorin | 0.7* | 0.9 | 1.1 | S | S | NS |
| LOW | Q9UBP9 | GULP1 | PTB domain-containing engulfment adapter protein 1 | 0.8 | 0.9 | 1.1 | NS | S | S |
| LOW | Q96QU8 | XPO6 | Exportin-6 | 0.7* | 0.9 | 1.1 | S | S | NS |
| HIGH | Q9H078 | CLPB | Caseinolytic peptidase B protein homolog | 0.6* | 0.9 | 1.0 | NS | S | NS |
Proteins with a statistically different change between the groups after the Kruskal–Wallis test and Benjamini–Hochberg method (in order of significance). The Mann–Whitney U test was used for pairwise analysis. Proteins shown in italics belong to the family of proteins with similar peptides with shared statistical analysis. The high-abundance fraction indicates that the protein co-eluted with high-abundance proteins, i.e. it had probably formed a complex with highly concentrated proteins in the serum. A fold change >1 indicates an increase in protein amount, whereas a fold change between 0 and 1 reflects reduced quantity.
Statistically significant change within the group.
A: ethinylestradiol (EE) + dienogest (DNG) versus estradiol valerate (EV) + DNG. B: EE + DNG versus DNG. C: EV + DNG versus DNG. S: Mann–Whitney U test P < 0.05; NS, non-significant.
Pathway analysis
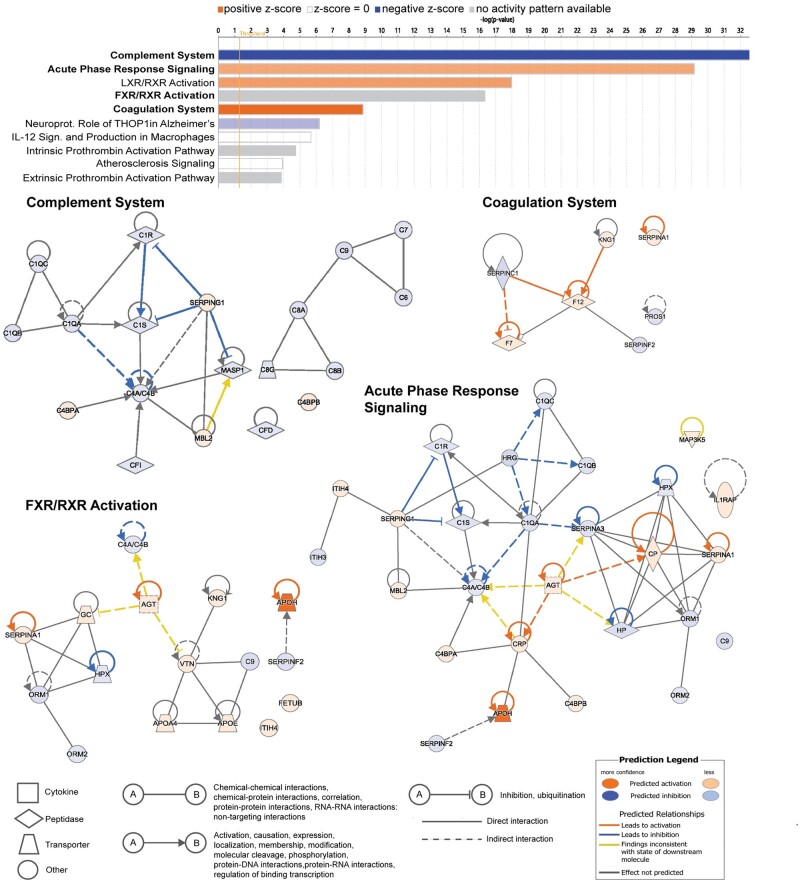
Most changes in the serum proteins were detected in the EE + DNG group; therefore, the functionality analysis was based on the proteins with a significant change during this treatment. The 10 most significantly affected pathways are listed in Fig. 4. The use of EE + DNG led to predicted inhibition of the complement system and activation of acute phase response signaling, liver X receptor/retinoid X receptor (LXR/RXR) and the coagulation system. The detected proteins of the farnesoid X receptor (FXR)/RXR activation pathway were 88% the same as in the LXR/RXR activation pathway. The complement system was the most-affected pathway, with 19 significantly changed proteins. The proteins belonging to the five pathways most changed during EE + DNG treatment are shown in Supplementary Table SII.
Figure 4.
Pathway and network analyses of changing proteins during EE + DNG use. Pathway analysis of the 10 pathways most affected during 9-week use of ethinylestradiol + dienogest (EE + DNG). Graph shows category scores; ‘threshold’ indicates the minimum significance level (scored as −log(P-value) from Fisher’s exact test, set here to 1.25). A positive z-score (orange bar) predicts activation of the pathway, whereas a negative (blue bar) predicts inhibition. The analysis was performed with QIAGEN Ingenuity Pathway Analysis (IPA) software. The four pathways in bold text are depicted below as networks. FXR, farnesoid X receptor; LXR, liver X receptor; RXR, retinoid X receptor.
Full lists of normalized abundance data are included in Supplementary Tables SIII (high-abundance protein fraction) and SIV (low-abundance protein fraction).
Fetuin-B, CBG and SHBG
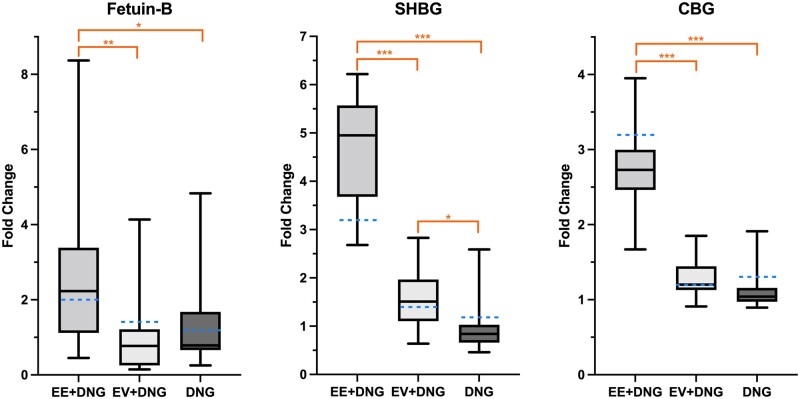
After proteomic analysis, fetuin-B (a novel finding with a significant difference between the study groups) was chosen for additional validation. We previously reported the changes in CBG (Kangasniemi et al., 2022) and SHBG (Haverinen et al., 2022a) and those analyses were also used for validation of proteomic analysis (Fig. 5). For fetuin-B, the median change (95% CI, P-value for change within the group) during the trial was the following: EE + DNG 206.04 ng/ml ((92.86–410.6) P = 0.02), EV + DNG −52.20 ng/ml ((−77.92–22.83) P = 0.112) and DNG −12.62 ng/ml ((−81.84–185.44) P = 0.73). In the proteomics analysis, EE + DNG induced a higher increase in serum CBG and SHBG compared with the other study preparations. These findings are in line with our previous ELISA and immunoassay results (Kangasniemi et al., 2022; Haverinen et al., 2022a); the fold changes derived from these data are shown in Fig. 5.
Figure 5.
Validation of proteomic analysis of serum from women taking a 9-week course of combined oral contraceptive. The fold changes in serum Fetuin-B, CBG and SHBG during the trial, as measured by ELISA and immunoassay. Box blots present the fold changes gained from the validation analyses, whereas the blue dotted lines show the fold changes from the proteomic analysis. The fetuin-B concentration increased especially during use of the combined oral contraceptive containing ethinylestradiol + dienogest (EE + DNG). One major fetuin-B outlier was removed from the EE + DNG group, which did not affect the statistical analysis. CBG and SHBG concentrations have been reported previously (Kangasniemi et al., 2022; Haverinen et al., 2022a). DNG, dienogest; EE, ethinylestradiol; EV, estradiol valerate; CBG, cortisol-binding globulin; SHBG, sex hormone-binding globulin. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
This study showed that 9 weeks of EE-based COC use had a multifold effect on the serum proteome compared with EV-based COC or DNG-only preparation; the number of affected proteins after exposure to EE + DNG was 24-fold the number for EV + DNG and 120-fold that for DNG only. Few proteins changed during the use of EV + DNG and no changes were detected during the use of DNG only. The most-affected pathways were the complement system, acute phase response signaling, metabolism-related LXR&FXR/RXR activation and the coagulation system. The findings of the proteomic analysis were validated with fetuin-B ELISA and our previously reported CBG ELISA and SHBG immunoassay, showing that treatment with EE + DNG increased serum levels of fetuin-B, CBG and SHBG more than EV + DNG or DNG only.
To our knowledge, this is the first study to explore the effects of hormonal contraceptives on serum proteins with an untargeted, discovery-type proteomic analysis. Previously, only one study has utilized a proteomic analysis method during contraceptive use (Josse et al., 2012). However, that cross-sectional study had selected targets for analysis and compared hormonal contraceptive use to non-use but did not specify the treatments. Similar to our data, differences were observed in proteins related to inflammation, innate immunity, coagulation, and blood pressure regulation (Josse et al., 2012). Quantitation in the discovery-type analysis is not as accurate as in the targeted method, but it offers a wider, indicative insight into the matter in question. The next validating step for the discovery-type analysis would be targeted analysis based on the results of this study with a new sample set from other subjects. However, many of the effects seen in this study have been previously observed with other targeted methods, which supports the reliability of the results.
EE-containing COCs have been shown to increase low-grade inflammation, reflected by increased levels of C-reactive protein (CRP) (van Rooijen et al., 2006; Morin-Papunen et al., 2008; Haarala et al., 2009; Piltonen et al., 2012; Wang et al., 2016), which is a well-known marker of the acute phase. Activation of the acute phase response signaling pathway by EE + DNG was also detected in the present study (Fig. 4), exemplified by the increase in IL1RAP and angiotensinogen, a precursor of the vasoconstrictor angiotensin. EV + DNG also increased these proteins, but the increase was significantly milder compared with EE + DNG. Similarly, in our earlier study, EE + DNG had a greater effect on the inflammatory markers CRP and pentraxin 3 compared with EV + DNG (Kangasniemi et al., 2020). These inflammatory changes are relevant as regards the cardiovascular morbidity related to COC use compared to progestin-only preparations, as shown in a recent review (Okoth et al., 2020). To support this, moderate effects of natural estrogens were also shown in a study comparing EE + drospirenone (DRSP) and E4 + DRSP, which found a greater increase in angiotensinogen during the use of EE + DRSP (Klipping et al., 2021). In the same study, EE + DRSP also increased CRP, although the difference compared to E4 + DRSP did not reach statistical significance (Klipping et al., 2021).
EE is a highly potent estrogen known to have a major effect on hepatic protein synthesis: EE increases SHBG, CBG and angiotensinogen multifold compared with estradiol (Mashchak et al., 1982). Previous studies have demonstrated increases in angiotensinogen and binding globulins during the use of EE-based combined contraceptives, regardless of the administration route (Sitruk-Ware et al., 2007b; Piltonen et al., 2012; Westhoff et al., 2013; Klipping et al., 2021). However, the impact of COCs containing natural estrogens on hepatic protein synthesis appears to be lower than that of EE-based COCs. This was shown in a comparative study in which E4 + DRSP promoted SHBG and CBG synthesis less than EE + DRSP (Klipping et al., 2021). The lower impact of natural estrogens on hormone-binding globulins has also been observed comparing EV + DNG with EE + levonorgestrel (LNG) (Junge et al., 2011), and E2 + nomegestrol acetate with EE + LNG (Ågren et al., 2011). These studies indicate a lesser hepatic impact of natural estrogens, supporting the results of the present study. It is also well known that COCs increase the risk of thromboembolic events (Sitruk-Ware et al., 2007a; Lidegaard et al., 2009, 2012; de Bastos et al., 2014). Interestingly, new data suggest that the effects on coagulation and the risk for thromboembolism might be lower with natural estrogen combinations (Sitruk-Ware, 2016; Kluft et al., 2017; Heikinheimo et al., 2022; Haverinen et al., 2022b), which may be related to the lesser impact on the liver.
In the pathway analysis, as many as 19 proteins related to the complement system were altered during EE + DNG treatment, which predicts inhibition of this pathway. The complement system is a complex immunological system involving ∼30 interacting proteins that compose a cascade of activation steps leading to activation of inflammation, opsonization of pathogens and cells for destruction, and direct killing of target cells or microbes by lysis (Oksjoki et al., 2007). The complement system has a central role in the immune system by discriminating healthy tissues from apoptotic cells and microbes, but its imbalance and uncontrolled activation is associated with various diseases, such as glomerulonephritis, sepsis and cardiovascular diseases (Oksjoki et al., 2007; Ricklin et al., 2010). Complement components C3 and C4 in serum increase during pregnancy (He et al., 2020), which is a high estrogen state. During pregnancy, complement dysregulation may lead to miscarriage, pre-eclampsia or pre-term birth (Girardi et al., 2020). As shown above, COCs are known to alter the inflammatory milieu, but data are limited on their effects on the complement system. A study from 1975 showed increases in C3 and decreases in CH50, C6, and C7 during estrogen–progestogen contraceptive use compared with women not using hormonal contraceptives (Tedder et al., 1975). Estrogen-regulated complement action has recently been demonstrated by showing that estrogen promotes an adaptation response in Candida albicans: through inhibition of complement-mediated opsonophagocytosis, C. albicans can evade the innate immune system (Kumwenda et al., 2022). In rheumatoid arthritis, lower circulating levels of properdin, one complement component, were measured in women who had ever used COCs compared with never-users (Bemis et al., 2019). Inflammatory activation of EE is unknown, but the effects on the complement cascade probably contribute to this process and should be further studied.
Untargeted proteomic analysis also enables the identification of interesting novel targets for future research. Fetuin-B is a relatively understudied hepatokine associated with insulin resistance, dyslipidemia, and non-alcoholic fatty liver disease (Meex and Watt, 2017; Mokou et al., 2020; Olkowicz et al., 2021). Serum levels of fetuin-B increase during pregnancy but normalize after delivery (Šimják et al., 2018), possibly related to hormonal changes of pregnancy. One study investigated fetuin-B levels in four women taking COCs containing EE in combination with LNG, DRSP, or DNG, all of which increased fetuin-B levels (Floehr et al., 2016). In a cross-over model, one woman used DNG only first and continued with EE + DNG: fetuin-B levels remained unchanged during DNG only but increased after initiation of EE + DNG treatment (Floehr et al., 2016). These data are in line with our results. Fetuin-B is part of the FXR/RXR activation pathway (Fig. 4), which is closely related to the LXR/RXR activation pathway (Redinger, 2003). Both of these pathways, also affected by EE + DNG in the present study, play critical roles in regulating lipid and glucose metabolism (Edwards et al., 2002; Redinger, 2003; Wang et al., 2008). EE-containing contraceptives alter lipid metabolism, increasing especially high-density lipoprotein and triglycerides, but also affect glucose metabolism (Morin-Papunen et al., 2008; Piltonen et al., 2012; Wang et al., 2016). Whether the activations of the FXR/RXR and LXR/RXR pathways are mediators of these changes remains to be studied. If this is confirmed, natural estrogens might also be less detrimental in this aspect (Junge et al., 2011; Mawet et al., 2015; Kangasniemi et al., 2020; Klipping et al., 2021).
The main strength of the present study is that this randomized trial is the first comparison of EE- and EV-containing contraceptives combined with the same progestin. Randomization was successful regarding the clinical characteristics but also at the proteome level, as reflected in the mixed clustering of baseline samples. Therefore, the changes observed during the trial are likely related to the intervention and not to chance. However, we acknowledge that the 9-week study period is relatively short for concluding long-term effects. Therefore, these findings should be further examined in longer trials with specifically targeted measurements. Proteomic analysis was not considered in the original sample size calculation, given that the analysis was not the primary outcome. On the other hand, as no previous data exist, estimating power for a discovery-type setup is extremely challenging. However, in the field of discovery-type proteomics, the groups of 14 are quite extensive, especially considering the trial with repeated measurements. Despite these limitations, our results give valuable insight into the extent of the differences between the estrogen types in COCs.
To conclude, EE + DNG had a greater impact on the serum proteome than that of EV + DNG or the DNG-only preparation. The differences between EE and EV go far beyond the well-known, traditional estrogen actions. Therefore, EV could provide a preferable option to EE in COCs in the future. Moreover, although our findings need to be validated in further studies utilizing targeted methods, this study provides a valuable basis for future research and guides the development of hormonal contraceptives.
Supplementary Material
Acknowledgements
We thank M.Sc. Elisa Korhonen for statistical assistance and Mrs Maarit Haarala, Ms Elina Huikari and Mrs Pirjo Ikonen for expert technical assistance.
Authors’ roles
The study was designed by J.S.T. and O.H. in collaboration with T.T.P. and A.H. M.H.K., A.H., K.L., O.H., J.S.T. and T.T.P. contributed to the data collection. S.J., T.T., R.R., R.K.A. and M.H.K. performed the laboratory analyses and data interpretation. M.H.K. wrote the first draft of the manuscript; all authors contributed to revision and approved the final version of the manuscript. T.T.P., J.S.T. and O.H. supervised the project.
Funding
Funding was obtained from Helsinki University Hospital research funds (J.S.T. and A.H.), the Hospital District of Helsinki and Uusimaa (K.L., T.T. and O.H.), the Sigrid Juselius Foundation (J.S.T. and T.T.P.), the Academy of Finland (J.S.T. and T.T.P. 315921 and 321763), the Finnish Medical Association (M.H.K., A.H. and T.T.P.), the University of Oulu Graduate School (M.H.K.), the Emil Aaltonen Foundation (M.H.K.), the Swedish Cultural Foundation in Finland (A.H.), the Novo Nordisk Foundation (R.K.A.), Orion Research Foundation (A.H.) and the Northern Ostrobothnia Regional Fund (R.K.A.). The funders had no role in study design, data collection and analysis, publishing decisions or manuscript preparation.
Conflict of interest
T.P. has received honoraria for lectures, consultations and research grants from Exeltis, Gedeon Richter, MSD, Merck, Pfizer, Roche, Stragen and Mithra Pharmaceuticals. O.H. occasionally serves on advisory boards for Bayer AG and Gedeon Richter and has designed and lectured at educational events for these companies. The other authors have nothing to disclose. O.H. occasionally serves on advisory boards for Bayer AG and Gedeon Richter and has designed and lectured at educational events for these companies. The other authors have nothing to disclose.
Contributor Information
M H Kangasniemi, Department of Obstetrics and Gynecology, Clinical Research Unit, Medical Research Center, Oulu University Hospital, University of Oulu, Oulu, Finland.
R K Arffman, Department of Obstetrics and Gynecology, Clinical Research Unit, Medical Research Center, Oulu University Hospital, University of Oulu, Oulu, Finland.
S Joenväärä, Transplantation Laboratory, Haartman Institute, University of Helsinki, Helsinki, Finland; HUS Diagnostic Center, Helsinki University Hospital, Helsinki, Finland.
A Haverinen, Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
K Luiro, Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
T Tohmola, Transplantation Laboratory, Haartman Institute, University of Helsinki, Helsinki, Finland; HUS Diagnostic Center, Helsinki University Hospital, Helsinki, Finland.
R Renkonen, Transplantation Laboratory, Haartman Institute, University of Helsinki, Helsinki, Finland; HUS Diagnostic Center, Helsinki University Hospital, Helsinki, Finland.
O Heikinheimo, Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
J S Tapanainen, Department of Obstetrics and Gynecology, Clinical Research Unit, Medical Research Center, Oulu University Hospital, University of Oulu, Oulu, Finland; Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
T T Piltonen, Department of Obstetrics and Gynecology, Clinical Research Unit, Medical Research Center, Oulu University Hospital, University of Oulu, Oulu, Finland.
Data Availability
Normalized abundance data of high- and low-abundance fractions are included in Supplementary Tables SIII and SIV. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifiers PXD033617 and 10.6019/PXD033617 for the low-abundance proteins and PXD033618 and 10.6019/PXD033618 for the high fraction.
References
- Abou-Ismail MY, Citla Sridhar D, Nayak L.. Estrogen and thrombosis: a bench to bedside review. Thromb Res 2020;192:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ågren UM, Anttila M, Mäenpää-Liukko K, Rantala ML, Rautiainen H, Sommer WF, Mommers E.. Effects of a monophasic combined oral contraceptive containing nomegestrol acetate and 17β-oestradiol in comparison to one containing levonorgestrel and ethinylestradiol on markers of endocrine function. Eur J Contracept Reprod Health Care 2011;16:458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis EA, Norris JM, Seifert J, Frazer-Abel A, Okamoto Y, Feser ML, Demoruelle MK, Deane KD, Banda NK, Holers VM.. Complement and its environmental determinants in the progression of human rheumatoid arthritis. Mol Immunol 2019;112:256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bastos M, Stegeman BH, Rosendaal FR, van HVA, Helmerhorst FM, Stijnen T, Dekkers OM.. Combined oral contraceptives: venous thrombosis. Cochrane Database Syst Rev 2014;2014:CD010813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA, Kennedy MA, Mak PA.. LXRs; Oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vascul Pharmacol 2002;38:249–256. [DOI] [PubMed] [Google Scholar]
- Floehr J, Dietzel E, Neulen J, Rösing B, Weissenborn U, Jahnen-Dechent W.. Association of high fetuin-B concentrations in serum with fertilization rate in IVF: a cross-sectional pilot study. Hum Reprod 2016;31:630–637. [DOI] [PubMed] [Google Scholar]
- Girardi G, Lingo JJ, Fleming SD, Regal JF.. Essential role of complement in pregnancy: from implantation to parturition and beyond. Front Immunol 2020;11:1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi G, Napolitano A, Cagnacci A.. Metabolic impact of combined hormonal contraceptives containing estradiol. Expert Opin Drug Metab Toxicol 2016;12:779–787. [DOI] [PubMed] [Google Scholar]
- Haarala A, Eklund C, Pessi T, Lehtimäki T, Huupponen R, Jula A, Viikari J, Raitakari O, Hurme M.. Use of combined oral contraceptives alters metabolic determinants and genetic regulation of C‐reactive protein. The Cardiovascular Risk in Young Finns Study. Scand J Clin Lab Invest 2009;69:168–174. [DOI] [PubMed] [Google Scholar]
- Haverinen A, Kangasniemi MH, Luiro K, Piltonen T, Heikinheimo O, Tapanainen JS.. Ethinyl estradiol vs estradiol valerate in combined oral contraceptives—effect on glucose tolerance: a randomized, controlled clinical trial. Contraception 2021;103:53–59. [DOI] [PubMed] [Google Scholar]
- Haverinen A, Luiro K, Kangasniemi MH, Piltonen TT, Hustad S, Heikinheimo O, Tapanainen JS.. Estradiol valerate vs ethinylestradiol in combined oral contraceptives: effects on the pituitary-ovarian axis. J Clin Endocrinol Metab 2022a;107:e3008–e3017. [DOI] [PubMed] [Google Scholar]
- Haverinen A, Luiro KM, Szanto T, Kangasniemi MH, Hiltunen L, Sainio S, Piltonen TT, Lassila R, Tapanainen JS, Heikinheimo O.. Combined oral contraceptives containing estradiol valerate vs ethinylestradiol on coagulation: a randomized clinical trial. Acta Obstet Gynecol Scand 2022b;101:1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YD, Xu BN, Song D, Wang YQ, Yu F, Chen Q, Zhao MH.. Normal range of complement components during pregnancy: a prospective study. Am J Reprod Immunol 2020;83:e13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikinheimo O, Toffol E, Partonen T, But A, Latvala A, Haukka J.. Systemic hormonal contraception and risk of venous thromboembolism. Acta Obstet Gynecol Scand 2022;101:846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse AR, Garcia-Bailo B, Fischer K, El-Sohemy A.. Novel effects of hormonal contraceptive use on the plasma proteome. PLoS One 2012;7:e45162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge W, Mellinger U, Parke S, Serrani M.. Metabolic and haemostatic effects of estradiol valerate/dienogest, a novel oral contraceptive: a randomized, open-label, single-centre study. Clin Drug Investig 2011;31:573–584. [DOI] [PubMed] [Google Scholar]
- Kangasniemi MH, Arffman RK, Haverinen A, Luiro K, Hustad S, Heikinheimo O, Tapanainen JS, Piltonen TT.. Effects of estradiol- and ethinylestradiol-based contraceptives on adrenal steroids: a randomized trial. Contraception 2022;https://doi.org/10.1016/j.contraception.2022.08.009. [DOI] [PubMed] [Google Scholar]
- Kangasniemi MH, Haverinen A, Luiro K, Hiltunen JK, Komsi EK, Arffman RK, Heikinheimo O, Tapanainen JS, Piltonen TT.. Estradiol valerate in COC has more favorable inflammatory profile than synthetic ethinyl estradiol: a randomized trial. J Clin Endocrinol Metab 2020;105:e2483–e2490. [DOI] [PubMed] [Google Scholar]
- Klipping C, Duijkers I, Mawet M, Maillard C, Bastidas A, Jost M, Foidart JM.. Endocrine and metabolic effects of an oral contraceptive containing estetrol and drospirenone. Contraception 2021;103:213–221. [DOI] [PubMed] [Google Scholar]
- Kluft C, Zimmerman Y, Mawet M, Klipping C, Duijkers IJM, Neuteboom J, Foidart JM, Bennink HC.. Reduced hemostatic effects with drospirenone-based oral contraceptives containing estetrol vs. ethinyl estradiol. Contraception 2017;95:140–147. [DOI] [PubMed] [Google Scholar]
- Kumwenda P, Cottier F, Hendry AC, Kneafsey D, Keevan B, Gallagher H, Tsai HJ, Hall RA.. Estrogen promotes innate immune evasion of Candida albicans through inactivation of the alternative complement system. Cell Rep 2022;38:110183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidegaard Ø, Lokkegaard E, Jensen A, Skovlund CW, Keiding N.. Thrombotic stroke and myocardial infarction with hormonal contraception. N Engl J Med 2012;366:2257–2266. [DOI] [PubMed] [Google Scholar]
- Lidegaard Ø, Løkkegaard E, Svendsen AL, Agger C.. Hormonal contraception and risk of venous thromboembolism: national follow-up study. BMJ 2009;339:b2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashchak CA, Lobo RA, Dozono-Takano R, Eggena P, Nakamura RM, Brenner PF, Mishell DR.. Comparison of pharmacodynamic properties of various estrogen formulations. Am J Obstet Gynecol 1982;144:511–518. [DOI] [PubMed] [Google Scholar]
- Mawet M, Maillard C, Klipping C, Zimmerman Y, Foidart JM, Bennink HJTC.. Unique effects on hepatic function, lipid metabolism, bone and growth endocrine parameters of estetrol in combined oral contraceptives. Eur J Contracept Reprod Health Care 2015;20:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meex RCR, Watt MJ.. Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat Rev Endocrinol 2017;13:509–520. [DOI] [PubMed] [Google Scholar]
- Mokou M, Yang S, Zhan B, Geng S, Li K, Yang M, Yang G, Deng W, Liu H, Liu D et al Elevated circulating fetuin-B levels are associated with insulin resistance and reduced by GLP-1RA in newly diagnosed PCOS women. Mediators Inflamm 2020;2020:2483435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Papunen L, Martikainen H, McCarthy MI, Franks S, Sovio U, Hartikainen A-L, Ruokonen A, Leinonen M, Laitinen J, Järvelin M-R et al Comparison of metabolic and inflammatory outcomes in women who used oral contraceptives and the levonorgestrel-releasing intrauterine device in a general population. Am J Obstet Gynecol 2008;199:529.e1–529.e10. [DOI] [PubMed] [Google Scholar]
- Okoth K, Chandan JS, Marshall T, Thangaratinam S, Thomas GN, Nirantharakumar K, Adderley NJ.. Association between the reproductive health of young women and cardiovascular disease in later life: umbrella review. BMJ 2020;371:m3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksjoki R, Kovanen PT, Meri S, Pentikainen MO.. Function and regulation of the complement system in cardiovascular diseases. Front Biosci 2007;12:4696–4708. [DOI] [PubMed] [Google Scholar]
- Olkowicz M, Czyzynska-Cichon I, Szupryczynska N, Kostogrys RB, Kochan Z, Debski J, Dadlez M, Chlopicki S, Smolenski RT.. Multi-omic signatures of atherogenic dyslipidaemia: pre-clinical target identification and validation in humans. J Transl Med 2021;19:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M et al The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 2022;50:D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piltonen T, Puurunen J, Hedberg P, Ruokonen A, Mutt SJ, Herzig KH, Nissinen A, Morin-Papunen L, Tapanainen JS.. Oral, transdermal and vaginal combined contraceptives induce an increase in markers of chronic inflammation and impair insulin sensitivity in young healthy normal-weight women: a randomized study. Hum Reprod 2012;27:3046–3056. [DOI] [PubMed] [Google Scholar]
- Redinger RN. The coming of age of our understanding of the enterohepatic circulation of bile salts. Am J Surg 2003;185:168–172. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD.. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 2010;11:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel C, Gorenstein M V, Li G-Z, Richardson K, Wall D, Geromanos SJ.. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome. Mol Cell Proteomics 2006a;5:589–607. [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ.. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 2006b;5:144–156. [DOI] [PubMed] [Google Scholar]
- Šimják P, Cinkajzlová A, Anderlová K, Kloučková J, Kratochvílová H, Lacinová Z, Kaválková P, Krejčí H, Mráz M, Pařízek A et al Changes in plasma concentrations and mRNA expression of hepatokines fetuin A, fetuin B and FGF21 in physiological pregnancy and gestational diabetes mellitus. Physiol Res 2018;67:S531–S542. [DOI] [PubMed] [Google Scholar]
- Sitruk-Ware R, Nath A.. Characteristics and metabolic effects of estrogen and progestins contained in oral contraceptive pills. Best Pract Res Clin Endocrinol Metab 2013;27:13–24. [DOI] [PubMed] [Google Scholar]
- Sitruk-Ware R, Plu-Bureau G, Menard J, Conard J, Kumar S, Thalabard JC, Tokay B, Bouchard P.. Effects of oral and transvaginal ethinyl estradiol on hemostatic factors and hepatic proteins in a randomized, crossover study. J Clin Endocrinol Metab 2007a;92:2074–2079. [DOI] [PubMed] [Google Scholar]
- Sitruk-Ware R. Hormonal contraception and thrombosis. Fertil Steril 2016;106:1289–1294. [DOI] [PubMed] [Google Scholar]
- Sitruk-Ware RL, Menard J, Rad M, Burggraaf J, de Kam M, Tokay BA, Sivin I, Kluft C.. Comparison of the impact of vaginal and oral administration of combined hormonal contraceptives on hepatic proteins sensitive to estrogen. Contraception 2007b;75:430–437. [DOI] [PubMed] [Google Scholar]
- Stanczyk FZ, Archer DF, Bhavnani BR.. Ethinyl estradiol and 17β-estradiol in combined oral contraceptives: pharmacokinetics, pharmacodynamics and risk assessment. Contraception 2013;87:706–727. [DOI] [PubMed] [Google Scholar]
- Tedder RS, Nelson M, Eisen V.. Effects on serum complement of normal and pre-eclamptic pregnancy and of oral contraceptives. Br J Exp Pathol 1975;56:389–395. [PMC free article] [PubMed] [Google Scholar]
- van Rooijen M, Hansson LO, Frostegård J, Silveira A, Hamsten A, Bremme K.. Treatment with combined oral contraceptives induces a rise in serum C-reactive protein in the absence of a general inflammatory response. J Thromb Haemost 2006;4:77–82. [DOI] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J et al; SciPy 1.0 Contributors. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 2020;17:261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wurtz P, Auro K, Morin-Papunen L, Kangas AJ, Soininen P, Tiainen M, Tynkkynen T, Joensuu A, Havulinna AS et al Effects of hormonal contraception on systemic metabolism: cross-sectional and longitudinal evidence. Int J Epidemiol 2016;45:1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-D, Chen W-D, Moore DD, Huang W.. FXR: a metabolic regulator and cell protector. Cell Res 2008;18:1087–1095. [DOI] [PubMed] [Google Scholar]
- Westhoff CL, Petrie KA, Cremers S.. Using changes in binding globulins to assess oral contraceptive compliance. Contraception 2013;87:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegratz I, Kutschera E, Lee JH, Moore C, Mellinger U, Winkler UH, Kuhl H.. Effect of four different oral contraceptives on various sex hormones and serum-binding globulins. Contraception 2003;67:25–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Normalized abundance data of high- and low-abundance fractions are included in Supplementary Tables SIII and SIV. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifiers PXD033617 and 10.6019/PXD033617 for the low-abundance proteins and PXD033618 and 10.6019/PXD033618 for the high fraction.