Abstract
BACKGROUND
Modern reproductive behavior in most developed countries is characterized by delayed parenthood. Older gametes are generally less fertile, accumulating and compounding the effects of varied environmental exposures that are modified by lifestyle factors. Clinicians are primarily concerned with advanced maternal age, while the influence of paternal age on fertility, early development and offspring health remains underappreciated. There is a growing trend to use assisted reproductive technologies for couples of advanced reproductive age. Thus, the number of children born from older gametes is increasing.
OBJECTIVE AND RATIONALE
We review studies reporting age-associated epigenetic changes in mammals and humans in sperm, including DNA methylation, histone modifications and non-coding RNAs. The interplay between environment, fertility, ART and age-related epigenetic signatures is explored. We focus on the association of sperm epigenetics on epigenetic and phenotype events in embryos and offspring.
SEARCH METHODS
Peer-reviewed original and review articles over the last two decades were selected using PubMed and the Web of Science for this narrative review. Searches were performed by adopting the two groups of main terms. The first group included ‘advanced paternal age’, ‘paternal age’, ‘postponed fatherhood’, ‘late fatherhood’, ‘old fatherhood’ and the second group included ‘sperm epigenetics’, ‘sperm’, ‘semen’, ’epigenetic’, ‘inheritance’, ‘DNA methylation’, ‘chromatin’, ‘non-coding RNA’, ‘assisted reproduction’, ‘epigenetic clock’.
OUTCOMES
Age is a powerful factor in humans and rodent models associated with increased de novo mutations and a modified sperm epigenome. Age affects all known epigenetic mechanisms, including DNA methylation, histone modifications and profiles of small non-coding (snc)RNA. While DNA methylation is the most investigated, there is a controversy about the direction of age-dependent changes in differentially hypo- or hypermethylated regions with advanced age. Successful development of the human sperm epigenetic clock based on cross-sectional data and four different methods for DNA methylation analysis indicates that at least some CpG exhibit a linear relationship between methylation levels and age. Rodent studies show a significant overlap between genes regulated through age-dependent differentially methylated regions and genes targeted by age-dependent sncRNA. Both age-dependent epigenetic mechanisms target gene networks enriched for embryo developmental, neurodevelopmental, growth and metabolic pathways. Thus, age-dependent changes in the sperm epigenome cannot be described as a stochastic accumulation of random epimutations and may be linked with autism spectrum disorders. Chemical and lifestyle exposures and ART techniques may affect the epigenetic aging of sperm. Although most epigenetic modifications are erased in the early mammalian embryo, there is growing evidence that an altered offspring epigenome and phenotype is linked with advanced paternal age due to the father’s sperm accumulating epigenetic changes with time. It has been hypothesized that age-induced changes in the sperm epigenome are profound, physiological and dynamic over years, yet stable over days and months, and likely irreversible.
WIDER IMPLICATIONS
This review raises a concern about delayed fatherhood and age-associated changes in the sperm epigenome that may compromise reproductive health of fathers and transfer altered epigenetic information to subsequent generations. Prospective studies using healthy males that consider confounders are recommended. We suggest a broader discussion focused on regulation of the father’s age in natural and ART conceptions is needed. The professional community should be informed and should raise awareness in the population and when counseling older men.
Keywords: sperm, epigenetics, advanced paternal age, age, DNA methylation, non-coding RNA, epigenetic clock, assisted reproductive technologies, delayed parenthood, pregnancy success
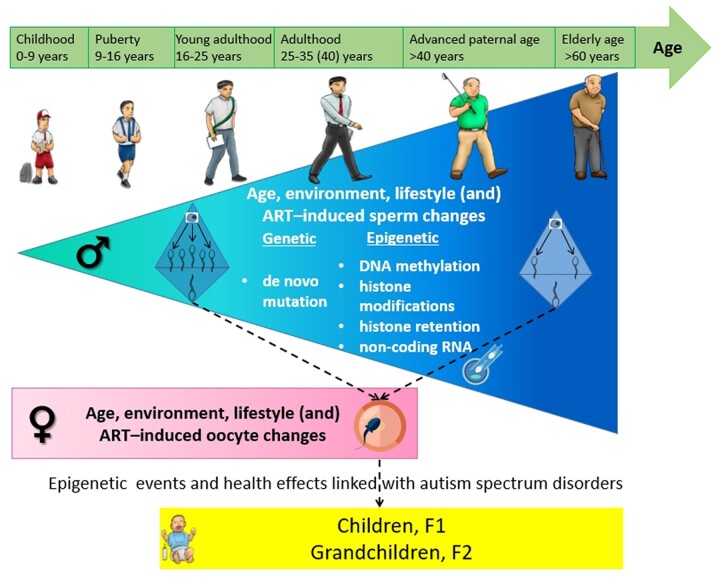
Graphical Abstract
Graphical Abstract.
Age is a driver of sperm epigenetic changes, including DNA methylation, histone modifications and small non-coding RNA profiles that may be linked with neurodevelopmental disorders in offspring.
Introduction
Delayed parenthood and advanced paternal age
Modern reproductive behavior in most developed countries is characterized by delayed parenthood due to increased life expectancy, contraception, socioeconomic status and pressures and overall changes in society, rates of divorce and remarriage and increased access to ART (Schmidt et al., 2012; Cedars, 2015; Goisis et al., 2020). Until recently, clinicians and researchers were mainly concerned about the advanced age of prospective mothers which known to reduce the ovarian reserve (Richardson et al., 2014) and increase the risks of miscarriages and aneuploid offspring presenting, for example Down syndrome (Nugent and Balen, 2001). The influence of paternal age on reproductive outcomes has largely been ignored. However, the age of fathers at the time of conception has increased worldwide, primarily in high-income countries (Martin et al., 2018; Brandt et al., 2019). For example, 15% of newborns were fathered by men older than 40 in England and Wales in 2016 (Office for National Statistics, 2019; Morris et al., 2021), and in the USA, newborns fathered by men over 40 doubled from 4.1% in 1972–1975 to 8.9% in 2011–2015 (Khandwala et al., 2017).
In the past decade, data accumulating from epidemiological studies showed that the paternal age at conception is associated with an increased risk of de novo mutations (Smits et al., 2022, reviewed in Goldmann et al., 2019), which have significant effects on pregnancy outcomes and offspring health (Belloc et al., 2014; Sharma et al., 2015; Khandwala et al., 2018). As noted in recent reviews (Brandt et al., 2019; Couture et al., 2021), there is no consensus for the definition of advanced paternal age (APA), although some professional societies define APA as greater than 40 years of age.
Age and natural fertility
Decreased female fertility with age is associated with decreased ovarian reserve, oocyte function and oocyte ploidy, followed by menopause. In contrast, men maintain lifelong reproductive function although it also declines over time (Ford et al., 2000; Hassan and Killick, 2003).
Age-associated physiological changes in the male reproductive system affect the testis, the seminal vesicles, the prostate and the epididymis (Sharma et al., 2015; Gunes et al., 2016). Some studies have shown that APA is associated with alterations in reproductive hormone levels, declining testicular and sexual function and sperm production (Eskenazi et al., 2003; Belloc et al., 2014; Avellino et al., 2017; Paoli et al., 2019). Semen quality declines with advancing age and is associated with increased sperm DNA damage (Evenson et al., 2020), altered sperm protamination and seminal plasma miRNA profiles (Paoli et al., 2019), increased oxidative stress (Aitken, 2018), decreased spermatogenic efficiency and increased proliferation of spermatogonia (Pohl et al., 2019).
Clinicians underestimate male age as a factor in assessing the reproductive potency of couples. This is exasperated given that men are less likely to seek evaluation of their reproductive health and fertility, and the assessment is rather rudimentary compared to that for women. For example, female age is a key factor in when treating infertility (Petok, 2015; Turner et al., 2020), while male age is typically not considered (Jungwirth et al., 2019). Although APA has been associated with an increased risk of pregnancy loss, birth defects and offspring diseases (Belloc et al., 2014; du Fossé et al., 2020), in clinical guidelines of professional andrology societies, there is a gap in the understanding of the importance of APA and its potentially harmful influence on early development and short- and long-term offspring health. As an example, recent European Association of Urology Guidelines for sexual and reproductive health focuses on an association of APA with infertility only (Salonia et al., 2020).
Environment and lifestyle-induced changes in gametes
Increasing evidence indicates that the interaction between aging and environmental impacts contributes to the epigenetic programming of gametes (Jenkins et al., 2018; Suvorov et al., 2020; Oluwayiose et al., 2021; Pilsner et al., 2021). As one ages, it is not surprising that the potential adverse effects of environmental conditions on gametes are compounded. Men may be more sensitive to these accumulating effects due to the fundamental differences in gametogenesis among men and women. In particular, the mitotic divisions in spermatogonia, the undifferentiated male germ cell, occur throughout the life span of men (Rosenwaks and Wassarman, 2014). Spermatogonia are characterized by their inherent self-renewal, with around 23 divisions every year (Goriely, 2016), that results in sperm production throughout man’s life span. While appearing to be evolutionary beneficial for men, the self-renewal properties of male germ cells allow for the accumulation of genetic and epigenetic errors throughout the life course.
Sperm are essential vehicles with specific molecular and epigenetic markers connecting paternal experiences with the phenotypes of their offspring (Chen et al., 2016). It has been shown that the preconception period is a sensitive developmental window in which a variety of environmental conditions such as toxicants, nutrition, drugs, stress and exercise affect sperm epigenetics (reviewed by Marcho et al., 2020).
Assisted reproductive technologies prolong fertile age
Some older couples use natural conception. Up to 1–3% of women and 7% of men who conceive naturally are older than 40 years in the USA (Wesselink et al., 2017; Bertoncelli Tanaka et al., 2019). However, there is a growing trend toward the use of ART for older men and women who have may be subfertile or fertile. Among couples seeking ART in the USA, approximately 20% are older than 40 years (American Society for Reproductive Medicine, Society for Assisted Reproductive Technology, 2017; Bertoncelli Tanaka et al., 2019; Centers for Disease Control and Prevention, 2019, 2021). Although ART increases the chances for older men to become fathers, fertilization, pregnancy and live birth rates decrease with increasing paternal age when using in vitro fertilization or intracytoplasmic sperm injection (Cheung et al., 2019; Marsidi et al., 2021; McCarter et al., 2021; Morris et al., 2021; Oluwayiose et al., 2021). This can be confounded by the reproductive biological age of the female (Ntostis et al., 2021).
The number of children born using gametes from older men by ART is increasing (Goisis et al., 2020). While ART is considered generally safe, it is associated with an increased risk of adverse perinatal outcomes, including low birthweight, preterm birth and birth defects, compared to spontaneous gestations (reviewed in Berntsen et al., 2019). Thus, the negative effects of delayed fatherhood raise concerns for the health and development of the next generation (Bertoncelli Tanaka et al., 2019). Despite this, the characterization of the molecular effects of APA, such as epigenetics, is not considered standard care in ART clinics. Another point for consideration is that ART allows and promotes the use of sperm for research. For example, most human sperm epigenetics findings come from ART clinics. However, the investigated subjects are men from couples seeking fertility treatment or donors of sperm, who do not represent the general population, and thus, the research findings may not be generalizable to non-ART populations.
In this review, we summarize the current knowledge of age-associated sperm epigenetic changes in mammals and humans, including DNA methylation and epigenetic clocks, histone modifications and sncRNA profiles. The interplay between environment, lifestyle and fertility and age-related epigenetic markers is explored.
Germ cells’ fate and epigenetic mechanisms of the sperm
The germline transmits genetic and epigenetic information to the subsequent generation (Hackett et al., 2012; Seisenberger et al., 2012; Kagiwada et al., 2013; Xavier et al., 2019). The mammalian germline generates critical epigenetic information for embryo development through gene imprinting (Kelsey and Feil, 2013; Li, 2013; Tucci et al., 2019) as well as through some RNAs (Sendler et al., 2013; Zhang et al., 2019; Cecere, 2021; Santiago et al., 2021).
Although each parental genome contribute equivalent genetic information to the zygote, their epigenetic contributions are different and depend on the parental origin. For example, paternally imprinted genes only express the allele inherited from the mother, while maternally imprinted genes only express the allele inherited from the father. Erasure and reestablishment of imprints occur in each germline cell (GC) generation (Hayashi and Surani, 2009).
Male GCs undergo two waves of nearly complete erasure of methylated DNA. In the zygote, shortly after fertilization, the first wave of erasure is targeted to the paternal genome (Oswald et al., 2000; Smith et al., 2012). It affects the majority of the genome while sparing paternal imprints (Hajkova, 2011; Smith et al., 2012; Hackett and Surani, 2013). Thus, most DNA methylation gained by GCs during the father’s lifetime is removed before the embryo starts development (Hajkova et al., 2002). At the point at which primordial germ cells (PGCs) are specified from post-implantation epiblast cells, they are epigenetically indistinguishable from their neighbors (Hackett et al., 2012). Therefore, nascent PGCs inherit stable epigenetic states, including DNA methylation and X-inactivation, constituting an epigenetic barrier against the eventual acquisition of totipotency (Surani et al., 2007; Tang et al., 2016). An essential early step in PGC development is reprogramming that erases these stable epigenetic blocks. The second and dynamic wave of genome-wide erasure of DNA methylation and extensive chromatin remodeling occurs after entry into the genital ridges at approximately E10.5 in mice. By E13.5, this almost yields complete erasure of DNA methylation with complete stripping of parental imprints and promoter CpG methylation of germline-specific genes. Male GCs retain about 7% of DNA methylation in mice when they reach the gonads (Seisenberger et al., 2012).
Spermatogenesis begins at puberty, and during mitosis and meiosis, sperm DNA is packaged in nucleosomes, comprised of histones susceptible to covalent modifications that influence gene expression. Monomethylation, dimethylation and trimethylation of H3K4, H3K9 and H3K27 display tightly controlled temporal expression and ensure proper progression through spermatogenesis (Khalil et al., 2004; Godmann et al., 2007). The strict timing of establishing and removing histone methylation marks is critical to spermatogenesis, as demonstrated in numerous transgenic animal models. Dynamic regulation of histone lysine acetylation is essential for spermatogenesis (Lahn et al., 2002; Fenic et al., 2004, 2008; Kurtz et al., 2007). On the one hand, histone acetylation relaxes chromatin and promotes RNA polymerase II gene transcription, on the other hand, deacetylation causes gene silencing.
A hallmark of spermiogenesis is the widespread change in chromatin structure, including the exchange of most canonical histones for protamines (Hao et al., 2019). Protamines are small basic proteins that bind DNA to form toroids, tightly packed structures that compact the genome to approximately 1/13 the size occupied by a human oocyte (Martins and Krawetz, 2007), which is well beyond that attainable with nucleosomes. The high level of compaction is an essential attribute for genome transport in the mature sperm head. The histone to the protamine exchange process is incomplete, with a small percentage (10% and 1% of histones in humans and mice, respectively) of the genome remaining bound to nucleosomes (Wykes and Krawetz, 2003; Hammoud et al., 2009; Brykczynska et al., 2010). The replacement of somatic histones by protamines is important for nuclear chromatin compaction, sperm maturation and fertility (Kramer et al., 1997; Hammoud et al., 2009). In humans, the relative proportion of protamine-1 (P1) to protamine-2 (P2) is strictly regulated at an approximately 1:1 ratio (Carrell and Hammoud, 2010). Changes in the P1/P2 ratio are common in infertile men and are associated with altered sperm quality, decreased embryo quality and reduced IVF outcomes (Aoki et al., 2006). It was shown that nucleosomes retained in sperm are not randomly distributed remnants of inefficient protamine replacement but are significantly enriched at many loci important for embryo development, including gene regulatory sequences (Arpanahi et al., 2009) and genes of key embryonic transcription factors and signaling pathway proteins (Hammoud et al., 2009). Histones are also significantly enriched at the promoters of miRNAs and imprinted genes. However, some studies contradict these findings showing that nucleosomes are not enriched in promoter regions including those of developmental genes but are rather in gene-poor regions (Carone et al., 2014; Samans et al., 2014).
In mice and humans, most protein-coding genes and long non-coding RNAs (lncRNAs) are transcribed in spermatogenic cells with strict temporal regulation (Chen et al., 2018; Hermann et al., 2018; Sohni et al., 2019). Multiple stages of spermatogenesis are defined by specific gene expression profiles (Estill et al., 2019). Mature sperm cells are transcriptionally inactive (Grunewald et al., 2005; Goodrich et al., 2013). The vast majority of RNA molecules in sperm are fragments of longer transcripts (Johnson et al., 2011; Sendler et al., 2013; Soumillon et al., 2013). Cessation of transcription and fragmentation of existing sperm mRNAs may be one of the several safety mechanisms to ensure that, upon fertilization, the highly differentiated sperm gives rise to the totipotent zygote. However, other transcripts remain intact and may have a post-fertilization function, affecting the embryo and offspring phenotype (Jodar et al., 2013; Sendler et al., 2013; Santiago et al., 2021). Mature sperm express a large number of sncRNAs, including microRNAs (miRNAs) (Krawetz et al., 2011; Hammoud et al., 2014), transfer RNA-derived (tsRNA) (Sharma et al., 2016) and PIWI-interacting RNAs (piRNAs) (Fu and Wang, 2014). The composition of different sncRNAs changes dynamically throughout the spermatogenesis cycle, which indicates specific roles for specific subtypes at specific timepoints (Sharma et al., 2018). miRNAs and tsRNA in sperm are involved in intergenerational epigenetic inheritance (Rassoulzadegan et al., 2006; Chen et al., 2016; Wang et al., 2021). Interestingly, this intergenerational transmission of epigenetic information via miRNAs depends on the activity of the DNA methyltransferase 2 (Dnmt2) gene (Kiani et al., 2013). This feature is likely reflective of miRNA stabilization by the known cytosine RNA methyltransferase activity of Dnmt2 (Goll et al., 2006; Ashapkin et al., 2016).
Age and genetic events in gametes
A meta-analysis has shown a direct correlation between age and nuclear DNA damage (Soares et al., 2014). An increase in DNA fragmentation with age has been confirmed in sperm (Wyrobek et al., 2006). Genetic and epigenetic changes in somatic cells related to aging have been extensively studied, including their connection with age-related diseases (reviewed in Ashapkin et al., 2017, 2019; Booth and Brunet, 2016; Jones et al., 2015; Kane and Sinclair, 2019; Sen et al., 2016; Xiao et al., 2019). In comparison, age-related genetic and epigenetic changes in gametes are understudied.
Goldmann et al. reviewed recent findings based on whole-genome sequencing of parent-offspring trios to characterize de novo mutations of the human germline associated with age (Goldmann et al., 2019). They concluded that both the father's age and the mother's age are positively correlated with the number of de novo mutations in offspring, with a larger effect size for APA (Goldmann et al., 2019). The paternal age effect could reflect that, unlike oogenesis, spermatogenesis is a life-long process. The number of spermatogonial cell divisions preceding spermiogenesis increases each year starting at puberty (given that a spermatogonial cell divides once every 16 days, it is estimated that the spermatogonia of a 75-year-old man have 1500 total mitotic cell divisions (Goriely, 2016; Laurentino et al., 2020)) with a concomitant accumulation of mutations (Goldmann et al., 2019).
Recently, Paoli et al. (2019) showed increased sperm DNA damage and altered sperm protamination, as assessed by PRM1 and PRM2 expression, in sperm during aging. However, in contrast with somatic tissues, telomere length in human sperm increases with age (Aston et al., 2012; Eisenberg et al., 2019; Laurentino et al., 2020). Even though telomeres are longer in the offspring of older fathers, paternal age effects are still detectable in grandchildren (Eisenberg et al., 2019). Yatsenko and Turek reviewed the effects of APA on sperm genetic changes. They found that APA is associated with accumulated damage to sperm DNA and mitotic and meiotic quality control mechanisms (mismatch repair) during spermatogenesis. In turn, this causes well-delineated abnormalities in sperm chromosomes, both numerical and structural, with increased sperm DNA fragmentation (3%/year of age) and single-gene mutations (relative risk, RR 10.0) (Yatsenko and Turek, 2018).
Accumulation of de novo mutations explains only a small part of the increased disease risk in children of older fathers. Since the DNA methylation maintenance at replication is much more error-prone than DNA replication (Wigler et al., 1981; Stein et al., 1982), aging somatic cells likely accumulate many more changes in DNA methylation than genetic mutations. It is likely true for gametes as well.
Age-associated epigenetic markers in sperm and fertility
From a clinician’s perspective, paternal age is a factor primarily associated with male infertility. Research has mainly focused on searching for fertility or infertility biomarkers and biomarkers of the efficacy of ART procedures. Epigenetic markers are among the promising biomarkers of fertility or infertility, particularly for idiopathic (“unexplained”) infertility (Jenkins and Turek, 2020).
Selected imprinted genes were investigated in some of the earliest sperm epigenetics and semen parameter studies among normozoospermic and oligozoospermic men. Abnormal methylation patterns at both paternal (hypomethylated) and maternal (hypermethylated) imprinted sites have been found in men with low sperm counts (Kobayashi et al., 2007; Marques et al., 2008; Boissonnas et al., 2010). High-throughput technologies for detecting DNA methylation across the genome allow the investigation of hundreds of DNA methylation targets. Houshdaran et al. (2007) were the first to find significant correlations between methylation levels in dozens of genes and sperm concentration, motility or morphology. Aston et al. (2015) compared genome-wide methylation patterns in the sperm of 54 men with normal semen quality and proven fertility, 55 infertile men whose partners produced high-quality embryos at IVF along with many confirmed pregnancies and 72 infertile men whose partners generally produced poor-quality embryos with far fewer pregnancie. Patterns of sperm methylation highly predictive of male fertility status (fertile or infertile) with 82% sensitivity and 99% positive predictive value were identified. Other types of epigenetic change recently investigated as a biomarker of fertility and better ART outcomes are sncRNA and other RNA profiles (Ostermeier et al., 2005; Jodar et al., 2015; Hua et al., 2019).
To our knowledge, no associations of age, fertility status and epigenetic marker data are available in sperm. At the moment, epigenetic changes associated with altered semen quality and/or infertility seem to be pathological. Causal mechanisms linking the sperm epigenome with sperm quality or fertility remain unknown. For example, it is unclear whether abnormal DNA methylation among imprinted and other genes arises from de novo methylation or failure to erase pre-existing methylation marks (Houshdaran et al., 2007). A large-scale well-designed longitudinal study among healthy men could shed light on the interplay of age-associated epigenetic changes and changes in fertility. One example of a longitudinal retrospective study aiming to construct sperm epigenetic clocks used 17 donors with biobank sperm samples 9–19 years apart with a mean age of 37.7 and 50.3 years (Jenkins et al., 2014; Jenkins and Turek, 2020). However, the relatively small sample size of that study limited the evaluation of the effects of age on fertility over time.
Age-dependent changes in the sperm epigenome: animal data
DNA methylation
Animal studies of age-dependent changes in the sperm epigenome have recently started to emerge, focusing mostly on DNA methylation and small RNA. Unfortunately, these studies have employed different strategies, including different animal models, different timepoints and different bioinformatic pipelines. Thus, it is not surprising that a consensus has not been reached. We do not yet have a good understanding of age-dependent dynamics in sperm epigenome.
The earliest study failed to identify differences in global methylation levels in sperm and livers of 6- and 21–24-month-old Brown Norway rats using thin-layer chromatography (Oakes et al., 2003). Using a different approach (restriction landmark genomic scanning), the authors identified age-dependent hypermethylation of repeated ribosomal DNA (rDNA) loci. Later studies used more sensitive methods of DNA methylation analysis.
In our recent study, we also used a rat model (Wistar rats) to examine the effect of aging on DNA methylation in spermatozoa (Pilsner et al., 2021). Profiles of DNA methylation were compared in 65- and 120-day-old animals, corresponding approximately to young (20–25 years old) and mature men (40–45 years old), respectively (Robb et al., 1978; Zanato et al., 1994). DNA methylation was assessed by reduced representation bisulfite sequencing (RRBS). In control animals, 5319 age-dependent differentially methylated regions (aDMRs) were identified, of which 99.3% of the aDMRs were hypermethylated in mature animals compared to young pubertal rats. These aDMRs were mostly associated with regions with low methylation levels and were enriched for functional categories essential for embryonic development, such as pattern specification, forebrain and sensory organ development and Hippo and Wnt pathways.
The earliest mouse study compared sperm DNA methylation in 3- and 12–24-month-old 129SvEv/Tac mice, corresponding to approximately 25 and 40–60-year-old men, using the methyl-MAPS approach (Milekic et al., 2015). This study did not find global changes in methylation and hydroxymethylation of sperm DNA. However, it reported an age-dependent loss of sperm DNA methylation in regions associated with transcriptional regulation.
Another RRBS study revealed a 3% higher level of sperm DNA methylation in introns, intergenic regions and retrotransposons of 17-month-old and 18-week-old C57BL/6N mice as compared with 8-week-old animals. This corresponds to approximately 60-, 30- and 20-year-old men, respectively (Kobayashi et al., 2016). Specifically, methylation levels of long interspersed nuclear elements (LINEs) increased with age from 80.0% at 8 weeks, 82.0% at 18 weeks, to 83.1% at 17 months of age. It is not clear whether a 3% change in LINE is biologically meaningful. Given the critical role of retrotransposons in zygotic genome activation (Fu et al., 2019), even small change in their methylation in germ cells may hypothetically shift the balance of retrotransposon activity, where excessive activity can cause insertional mutagenesis or chromosomal abnormality, while decreased activity can cause developmental arrest at the stage of two cell embryo (Huang et al., 2017). Additionally, Kobayashi et al. (2016) identified 126 promoters that showed statistically significant absolute methylation changes ≥5% between different age groups. Promoters undergoing age-dependent hypomethylation were significantly enriched for gene ontology (GO) categories related to spermatogenesis. Promoters undergoing age-dependent hypermethylation were enriched for cell projection-related genes. Interestingly, most of the significant changes were observed between the youngest and two older groups, suggesting that sperm DNA methylation changes may occur at variable rates at different age periods.
Another study using RRBS compared DNA methylation in sperm of 4- and >21-month-old C57BL/6J mice, corresponding to approximately 25- and >60-year-old men (Xie et al., 2018). This study identified 484 aDMRs in promoters, with 62% hypomethylation and 38% hypermethylation in older mice. Enrichment analysis of genes associated with aDMRs identified several aging-related pathways, including mTOR, PTEN, IGF1 and p53 signaling and immune-regulatory pathways. Contrary to the previous study, Xie et al. report overall loss of methylation across Alu and LINE-1 repetitive elements in aged relative to young sperm.
In a mouse model of autism spectrum disorder (ASD), the APA effect was manifested by vocal communication deficits in the offspring at infancy (Yoshizaki et al., 2021). Targeted sperm methylome analysis identified 16 hypermethylated and 96 hypomethylated DMRs in aged (12 months old) compared to young (3 months old) mice. Binding motifs for the neuron-restrictive silencer factor (NRSF) were detected in 19 out of 96 hypo-DMRs, while none were found in hyper-DMRs. These changes in sperm DNA methylation appeared to lead to the upregulation of NRSF target genes in the brain of the offspring embryos at E14.5, the peak stage of neuronal production.
In a study of DNA methylation patterns in spermatozoa from bulls at 10 months old (early puberty), 12 months old (late puberty) and 16 months old (adult), no statistically significant differences were detected between 16- and 12-month animals (Lambert et al., 2018). However, about 2600 aDMRs were found when the 16- and 10-month bulls were compared. Furthermore, in blastocysts obtained from these bulls’ spermatozoa by IVF of same-age cows, more than 250 genes were differentially expressed, and nearly 7000 aDMRs were found (Wu et al., 2020a).
In a study of age-dependent DNA methylation in the Japanese Black bull, semen samples were collected from animals of five different ages (between 14 and 162 months) and analyzed using human Infinium EPIC BeadChip array (Takeda et al., 2019). This analysis identified 77 CpGs undergoing an age-dependent change in methylation: 56 with higher methylation levels and 21 with lower methylation levels in older animals. Targeted analysis of 13 age-dependent CpG sites using combined bisulfite restriction analysis (COBRA) demonstrated rapid changes in methylation levels in animals younger than 4 years of age and less pronounced changes in older animals (Takeda et al., 2019).
Small non-coding RNAs
Several animal studies have addressed age-dependent changes in small RNA (sRNA) of spermatozoa. For example, when RNA-seq data were compared for 4- and >21-month-old C57BL/6J mice, 428 differentially expressed sRNAs were identified (Xie et al., 2018). A total of 43% were miRNAs and 53% were piRNAs. The majority of age-dependent miRNAs (98%) were down-regulated in the sperm of old males. Targets of age-dependent miRNAs were enriched for signaling pathways, including mTOR, insulin and growth factor signaling (Xie et al., 2018). Age-dependent differentially methylated promoters enriched for similar sets of molecular pathways as those discussed above were identified.
In a recent study, our group showed that the distribution of small RNA fractions is different in 65- and 120-day-old Wistar rats, corresponding approximately to young (20–25 years old) and mature men (40–45 years old), with fractions of reads mapping to rRNA and lncRNA decreasing and fractions of reads mapping to tsRNA and miRNA increasing with age (Suvorov et al., 2020). Furthermore, we identified 1384 sncRNA, including 249 miRNA, 908 piRNA and 227 tsRNA-derived RNAs significantly differentially expressed between two age groups of rats. Differentially expressed miRNA and piRNA were enriched for protein-coding targets involved in development and metabolism. Additionally, piRNA was enriched for long terminal repeat targets. Interestingly, our results on age-dependent changes in miRNA and piRNA were concordant with changes in DNA methylation in the same rat model (Pilsner et al., 2021). Genes associated with aDMRs were significantly enriched with gene targets of age-dependent miRNA and piRNA. In addition, genes (n = 1052) overlapping as miRNA targets and as aDMR-associated genes were highly enriched for categories related to embryonic development.
Another recent study also reports age-dependent changes in the distribution of fractions of sncRNA (Ma et al., 2020). The piRNA fraction was significantly lower and the miRNA fraction was higher in 12-month-old C57BL/6 mice than in 6–8-week-old animals. There were 162 miRNAs differentially expressed between age groups with 160 miRNA up-regulated in the sperm of older male mice. Gene targets of 33 age-dependent and spermatozoa-specific miRNA (not expressed in the oocyte but expressed in embryo) were enriched for fatty acid metabolism, protein processing, mTOR signaling, Hippo signaling and steroid biosynthesis (Ma et al., 2020).
Notably, age-associated changes in sncRNA profiles of epididymal sperm observed in rats (Suvorov et al., 2020) and in mice (Ma et al., 2020) were similar to maturation-associated changes of sncRNA patterns in sperm collected along the bovine male reproductive tract from testis through the caput, corpus, cauda epididymis to ejaculated sperm (Sellem et al., 2021). miRNA and tsRNA were enriched, while piRNA were expressed at lower levels in ejaculates and cauda compared to testis parenchyma and caput epididymis. Similar small RNA dynamics in sperm maturation were observed in mice (Sharma et al., 2016, 2018). Thus, for sncRNA sperm profiles, it is reasonable to hypothesize similar mechanisms for the maturation-associated changes in the cauda epididymis over the spermatogenesis cycle and age-associated changes over the life span.
In a complex recent study of the role of aging-related changes of sperm tsRNA in the intergenerational transmission of behavioral traits in mice, Guo et al. (2021) found 1202 up-regulated tsRNA and 408 down-regulated tsRNA in the older sperm (14–18 months old) compared to young sperm (3–4 months old). Enrichment analysis showed that up-regulated tsRNA target genes were involved in nervous system-related signaling pathways, including neurotrophin signaling, cholinergic synapses and axon guidance and in the metabolic mTOR signaling pathway.
In a recent study of age-dependent changes in sncRNA profiles in bull spermatozoa, semen was collected and analyzed from the same four animals at the ages of 10, 12 and 16 months, representing the pre-, peri- and post-pubertal stages, respectively (Wu et al., 2020b). Six miRNA were differentially expressed at 16 months compared to 12 months of age, and eight miRNA and one tsRNA were differentially expressed at 16 months compared to 10 months of age. Gene targets of 10 age-dependent and spermatozoa-specific miRNA (not expressed in the oocyte, but expressed in the embryo) were enriched for metabolic and developmental pathways, including TGF-β, Rho family GTPase, PI3K/AKT and IGF-1 signaling.
Histone modifications
Only two studies are available where histone modifications in spermatozoa are analyzed in relation to age. Xie et al. (2018) used the ChIP-seq approach with antibodies for H3K4me3 and H3K27me3 to identify retention of major activating and suppressive histone marks, respectively, in 4- and >21-month-old C57BL/6J mice. They identified a small region on chromosome 5 enriched with spermatogenesis-related genes (Speer), which contained 90% of the differential histone modifications (Xie et al., 2018). Tatehana et al. (2020) used a histochemical approach to analyze the patterns of histone modifications throughout spermatogenesis in young (3 months) and aged (12 months) mice. Among the transcription-repressive marks, H3K9me3 decreased, while H3K27me2/3 increased with age. The activating mark H3K4me2 drastically decreased in the GCs of the aged males.
Technical challenges in the assessment of nucleosome locations and their histone modifications exist due to the structural constraints of highly compact genome of mature sperm. Additional methodological advances are warranted to resolve the findings before a defining role of sperm nucleosomes, as well as their histone modifications, on embryo development or determining how age impacts nucleosome localization and histone modifications.
Interaction between age and environmental factors
There is a significant knowledge gap in understanding how environmental and lifestyle factors interact with the epigenetic aging of sperm. Two examples of such research were published recently by our group, which documented changes in the age-dependent profile of DNA methylation (Pilsner et al., 2021) and small RNA (Suvorov et al., 2020) in spermatozoa of rats exposed perinatally to environmental brominated flame retardant 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47). Surprisingly, perinatal exposure to BDE-47 had very similar effects as age-dependent changes in DNA methylation and sncRNA expression. At first glance, exposure eliminated most of the aging effects on both epigenetic markers. Further analysis revealed convergence between the epigenomes of the younger and older animal. This convergence consisted of opposite effects of exposure on age-dependent changes in profiles of sncRNA and DNA methylation in young and old animals. In young animals, exposure accelerated age-dependent changes while in older animals, exposure resulted in a slowing down of age-dependent changes. As a result, the epigenomes of exposed younger and older animals were much more similar than those of control animals.
Lessons from animal studies
A review of animal data shows that we do not clearly understand the age-dependent dynamics of different epigenetic mechanisms in spermatozoa. Comparison of published studies indicate that these changes may be species and even strain specific or may have more complex dynamics than just linear increases or decreases in methylation. If the later statement is true, then selection of different timepoints for comparison may result in identification of opposite time-trends in age-dependent epigenetic changes. Finally, most methods for both DNA methylation and sncRNA analysis today are prone to selection bias of specific genomic regions (DNA methylation) and specific transcripts (sncRNA). Therefore, results of different studies may be not directly comparable as they focus on somewhat different populations of nucleic acids. Some common findings that can be derived from animal studies include the following (Table I): (i) age is a powerful factor that affects the sperm epigenome; (ii) it is likely that pubertal life is associated with more significant changes in the sperm epigenome than are later stages; (iii) age-dependent changes of DNA methylation and sncRNA expression occur synchronously or are linked causatively and affect the same biological functions; (iv) age-dependent epigenetic changes include changes in fractions of sncRNA subtypes; and (v) genes associated with aDMRs or genes targets of age-dependent sncRNA are enriched with metabolic pathways and major developmental pathways, including nervous system-related signaling, Wnt, Hippo, mTOR and Igf1.
Table I.
Sperm epigenome and age: findings from animal studies.
| Findings | Studies | Species | Ages compared (PND) | N per age group | Methods |
|---|---|---|---|---|---|
| Age significantly affects sperm epigenome | Pilsner et al. (2021) | Rat | 65/120 | 6 | RRBS* |
| Kobayashi et al. (2016) | Mouse | 56/126/510 | 3–7 | RRBS | |
| Xie et al. (2018) | Mouse | 120/630 | Pool of 5 | RRBS/RNA-seq/ChIP-seq# | |
| Yoshizaki et al. (2021) | Mouse | 90/360 | 4 or 9 | SureSelect** | |
| Ma et al. (2020) | Mouse | 50/360 | ? | RNA-seq | |
| Guo et al. (2021) | Mouse | 90–120/420–540 | 3–4 | RNA-seq | |
| Lambert et al. (2018) | Bull | 300/360/480 | 4 | EDMA*** | |
| Takeda et al. (2019) | Bull | 420/570/840/1620/4860 | ∼7 | EPIC##, COBRA$ | |
| Tatehana et al. (2020) | Mouse | 90/360 | ? | Histone immunostaining | |
| More changes in sperm epigenome occur during pubertal life than at older age | Lambert et al. (2018) | Bull | 300/360/480 | 4 | EDMA |
| Takeda et al. (2019) | Bull | 420/570/840/1620/4860 | ∼7 | EPIC, COBRA | |
| Age-dependent changes in DNA methylation and sncRNA affect the same biological functions | Pilsner et al. (2021) /Suvorov et al. (2020) | Rat | 65/120 | 6 | RRBS/RNA-seq |
| Xie et al. (2018) | Mouse | 120/630 | Pool of 5 | RRBS/RNA-seq | |
| Fractions composition of sncRNA subtypes change with age in sperm | Suvorov et al. (2020) | Rat | 65/120 | 6 | RNA-seq |
| Ma et al. (2020) | Mouse | 50/360 | ? | RNA-seq | |
| Genes associated with age-dependent DMRs or genes targets of age-dependent sncRNA are enriched with metabolic and major developmental (including neurodevelopmental) pathways | Pilsner et al. (2021) | Rat | 65/120 | 6 | RRBS |
| Suvorov et al. (2020) | Rat | 65/120 | 6 | RNA-seq | |
| Xie et al. (2018) | Mouse | 120/630 | Pool of 5 | RRBS/RNA-seq | |
| Ma et al. (2020) | Mouse | 50/360 | ? | RNA-seq | |
| Wu et al. (2020b) | Bull | 300/360/480 | 4 | RNA-seq | |
| Yoshizaki et al. (2021) | Mouse | 90/360 | 4 or 9 | SureSelect | |
| Guo et al. (2021) | Mouse | 90–120/420–540 | 3–4 | RNA-seq |
sncRNA, small non-coding RNA; DMRs, differentially methylated regions; RNA-seq, RNA sequencing; RRBS, reduced representation bisulfite sequencing.
ChIP-seq, chromatin immunoprecipitation sequencing.
SureSelect, SureSelect Methyl‐Seq technique (Agilent).
EDMA, EmbryoGENE DNA methylation array.
EPIC, Infinium MethylationEPIC BeadChip (Illumina).
COBRA, combined bisulfite restriction analysis.
Age-associated epigenetic changes and epigenetic clocks in human sperm
Research began on age-associated epigenetic changes in human sperm epigenome by examining the differences in DNA methylation of the BRCA1, BRCA2, HD, DM1, PSEN1 and PSEN2 promoter regions. DNA methylation was quite variable between individual spermatozoa in healthy individuals at 22–56 years of age and between individuals (Flanagan et al., 2006). Some of the methylated loci displayed a clear correlation with age.
A longitudinal study of sperm DNA methylation in fertile human donors at timepoints 9–19 years apart showed that 139 aDMRs lose methylation, while only eight aDMRs gain methylation with age (Jenkins et al., 2014). Of the 139 hypomethylated aDMRs, 112 were associated with a gene body or promoter; of the 8 hypermethylated aDMRs, 7 were associated with a gene. In total, 110 genes showed hypomethylation with age. The majority of hypermethylation events, six of eight, occurred at promoter CpG islands (regions with a high frequency of CpG sites), whereas hypomethylation events were preferentially associated with CpG shores (regions immediately flanking CpG islands, up to 2 kb). Interestingly, most (∼88%) hypomethylated aDMRs were found within 1 kb of known nucleosome retention sites in the mature sperm. When validated in an analysis on an independent cohort of young (<25 years) and aged (>45 years) individuals, the age-associated methylation differences in the same regions were higher by about 2-fold, in accordance with the larger age difference (27.2 years on average) compared with that in the longitudinal study (12.6 years on average). This notion suggested a linear relationship between methylation alterations and age. However, these changes were relatively subtle, considering their possible impacts on gene activity. Although these age-associated changes in sperm methylation are highly significant and consistent in their location, more direct evidence of their biological implications is needed. Of the genes associated with aDMRs, the most relevant was DRD4, which is implicated in schizophrenia and bipolar disorder.
Nine genes known to be associated with neuropsychiatric and other disorders were selected among those reported by Jenkins et al. (2014) to test their correlation with age by Atsem et al. Their methylation levels were measured by bisulfite pyrosequencing in 162 IVF/ICSI sperm samples that led to pregnancy (Atsem et al., 2016). Four of these genes, DMPK, FOXK1, KCNA7 and NCOR2, showed a significant negative correlation with donor age. In DMPK, the age-hypomethylated sites were within a gene-body CpG island, in FOXK1, they were within a gene-body CpG shore, in KCNA7, they were within a promoter CpG shore, while in NCOR2, they were within a distal CpG island. Single allele methylation profiles of FOXK1 (5 CpGs) and KCNA7 (12 CpGs) were then studied in 13 sperm samples from young (25–35 years) and 13 sperm samples from older (40–55 years) donors by deep bisulfite sequencing. In sperm samples from younger donors, both genes showed highly variable (0–100%) methylation levels, with most alleles exhibiting mixed methylation profiles (50–90% CpGs methylated). In the sperm samples from older donors, methylation variation was much lower, and methylation profiles showed hypomethylation (0–20% for FOXK1 and 10–40% for KCNA7) relative to young sperm. Average methylation levels showed a significant negative correlation with paternal age (correlation coefficients −0.63 for FOXK1 and −0.77 for KCNA7).
Sperm DNA methylation data obtained on the Illumina Infinium HumanMethylation450 BeadChip microarray platform from three previous studies (Jenkins et al., 2014, 2017; Aston et al., 2015) were used to generate a DNA methylation sperm age prediction model (Jenkins et al., 2018). It should be noted that the model was constructed mainly using samples from men of typical paternity ages (20–45 years). Of several design strategies tested, the model trained on a subset of 51 most robust aDMRs from Jenkins et al. (2014) was chosen. It performed well in the entire data set with an R2 = 0.89 and a mean absolute error (MAE) of 2.04 years. Testing the model on independent sperm samples (n = 10) showed high accuracy of age prediction, with an MAE of 2.37. When the epigenetic sperm ages were compared between individuals who had never smoked, smokers and long-term (>10 years) smokers of similar chronological ages, an increase of epigenetic age was observed of around 1.5% in all smokers and 2.5% in long-term smokers relative to never smokers. The authors suggested that their model is capable not only of correctly predicting age based on sperm DNA methylation but also of predicting the aging-acceleration effects of smoking.
Another recent study used a customized methylC-capture sequencing approach to characterize DNA methylation in spermatozoa from 94 fertile and infertile men, who were categorized as young (48 men between 18 and 38 years) or old (46 men between 46 and 71 years) (Cao et al., 2020). Among the 2.65 million CpGs covered, 21 971 were associated with aging, most of them (>99.5%) in regions not reported previously. Interestingly, hypomethylation was less common (38.3%; 8409 CpGs) than hypermethylation (61.7%; 13 562 CpGs) with age. The hypomethylated CpGs were frequently located near gene regions, whereas the hypermethylated CpGs were mostly in the distal regions. Furthermore, 26% of age-associated CpGs were clustered in DMRs where ≥5 CpGs in a 500-bp window were either hypomethylated (315 hypoDMRs) or hypermethylated (483 hyperDMRs) with age. These aDMRs showed stronger associations with subgenomic regions. Clusters of hyperDMRs were found in chromosomes 4 and 16. The chr4 hyperDMR cluster overlaps the PGC-1α locus, known to be involved in metabolic aging (Halling and Pilegaard, 2020), and the chr16 hyperDMR cluster overlaps the RBFOX1 gene, implicated in neurodevelopmental disease (Hamada et al., 2016). Overall, both hypoDMRs and hyperDMRs were enriched for developmental pathway genes, including a strong association of hyperDMRs with the nervous system. An age predictor was built by linear regression using the top 5000 age-related CpGs (>99% were not reported previously). It showed high accuracy with an average error of 2.68 years. The authors concluded that genome-wide DNA methylation changes in sperm from a wide range of male ages could be used to create more accurate epigenetic clocks.
A recent prospective study was the first to link male age-related declines in ART outcomes with sperm DNA methylation changes among 47 male participants (21–45 years of age) of couples seeking infertility treatment from the Sperm Environmental Epigenetics and Development Study (Oluwayiose et al., 2021). Adjusting for male BMI, infertility status, smoking and female age, male age was significantly associated with diminished odds of fertilization, high embryo quality (at both Days 3 and 5) and live birth. Male age was also associated with sperm methylation at 1698 CpGs and 1146 regions via the 450k array. These were associated with >750 genes enriched in embryonic development, behavior and neurodevelopment. High-dimensional mediation analyses identified four genes (DEFB126, TPI1P3, PLCH2 and DLGAP2) with age-related sperm differential methylation that accounted for 64% of the effect of male age on the lower fertilization rate. Interestingly, DEFB126 is a member of the beta-defensin protein family that has been shown to be important for sperm to penetrate oocytes and linked to lower clinical pregnancy rates (Tollner et al., 2011; Boroujeni et al., 2019). DLGAP2 functions in synapse organization and signaling and has been implicated as a candidate gene for autism and schizophrenia and linked to male infertility (Rasmussen et al., 2017; Sujit et al., 2018).
Since most sperm epigenome studies have used cohorts of men recruited from fertility clinics, changes in sperm DNA methylation caused by aging may be difficult to differentiate from those due to age- and environment-associated comorbidities. A recent whole-genome bisulfite sequencing (WGBS) study used 198 variously aged (18–84 years) healthy men (Laurentino et al., 2020). A total of 236 aDMRs were identified in sperm DNA between young (18–25 years) and old (>65 years) men, about equal numbers of them hyper- or hypomethylated with age. GO analysis of neighboring genes revealed significant enrichment for homeobox, DNA binding, nucleus and transcription categories in the hypomethylated aDMRs. In contrast, no significant enrichments were found for the hypermethylated aDMRs. Furthermore, 10 aDMRs overlapped with those known to escape the genome-wide demethylation after fertilization. The six aDMRs with the lowest P-values selected from 11 aDMRs validated by deep bisulfite sequencing were used to derive an age predictor for sperm. Its validation in an independent set of sperm samples showed a good correlation between calculated and chronological age with an MAE of 9.8 years. The precision of this age predictor is lower than that of the sperm DNA methylation clock described above (Jenkins et al., 2018; Cao et al., 2020). While all clocks could be used in various experimental settings, they should be confirmed by other studies, and a prospective design using men from a healthy population is preferable.
Most recently, Pilsner et al. (2022) developed a novel sperm epigenetic clock utilizing a state-of-the-art ensemble machine learning algorithm, Super Learner (van der Laan et al., 2007), to predict age from the sperm DNA methylation data from 379 semen samples from male partners from male participants of the Longitudinal Investigation of Fertility and the Environment (LIFE) Study, a prospective pregnancy cohort from the general population. Remarkable male age was associated with 85 434 CpGs and 22 397 CpGs after Bonferroni correction and regional analyses identified >12 000 DMRs. Ontology analyses revealed that sperm DMRs were enriched in numerous biological processes including multiple terms related to signaling, morphogenesis, brain development and learning or memory. Next, they developed two independent biological clocks by applying Super Learner to predict chronological age from the previously identified 22 397 Bonferroni age-associated individual CpGs and the 2364 age-associated sperm DMRs and then calculated sperm epigenetic aging (SEA). The CpG-based clock (SEACpG) model had the highest predictive performance of chronological age within sample (r = 0.91, MAE = 1.6) and out of sample (r = 0.81, MAE = 2.2). Furthermore, SEACpG showed a strong performance in an independent IVF cohort (n = 173; r = 0.83) (Pilsner et al., 2022).
Pilsner et al. examined the association of SEACpG with pregnancy outcomes. In adjusted discrete Cox models, SEACpG was negatively associated with time to pregnancy (TTP), with fecundability odds ratios = 0.83, indicating a longer TTP with advanced SEACpG. Furthermore, advanced SEACpG was associated with a shorter gestational age. Finally, current smokers also displayed advanced SEACpG (Pilsner et al., 2022), which corroborates the previous findings from Jenkins et al. (2018).
A highly significant positive correlation was found between sperm rDNA methylation and age using bisulfite pyrosequencing of 53 CpGs of the upstream control element (UCE), core promoter, 18S rDNA and 28S rDNA sequences in variously aged (25–66 years) men (Potabattula et al., 2020). Sperm samples from old donors consistently displayed significantly higher numbers of hypermethylated (potentially silenced) alleles compared with those from young men. An epigenetic clock model was built by ElasticNet regression using methylation values of the 53 CpG sites. A model involving 15 of the most robust CpGs showed a correlation between the predicted epigenetic age and chronological age (r = 0.67, median absolute difference between predicted and chronological age 2.91). In a similar study using 60 couples who underwent IVF, a positive correlation was found between the methylation of 21 CpG sites in sperm rDNA promoter and donor age, but no statistically significant correlation was observed between rDNA methylation and IVF outcomes (successful fertilization, Day 3 good-quality embryos, clinical pregnancies) (Li et al., 2020). It should be noted that the rDNA methylation clock gives a lower correlation compared with the epigenetic clock of Jenkins et al. described above (Jenkins et al., 2018). Possibly, the precision of the rDNA clock could be higher if all ∼1500 CpGs in the rDNA were analyzed.
Only one study using a small sample size (n = 3 for each age group) and semen samples that led to clinical pregnancy during in vitro fertilization procedures has been published with results of an age-related tsRNA profile in human sperm. Guo et al. found 34 up-regulated and 11 down-regulated tsRNA in older sperm (45–50 years old) compared to young sperm (25–27 years old). Analysis of GO terms suggested that up-regulated tsRNA target genes were involved in neurogenesis and nervous system development (Guo et al., 2021). For non-coding RNA in seminal plasma, one study of 40 older men (50–81 years) and 40 younger men (20–40 years) demonstrated lower expression of three selected miRNAs, miR-371, miR-122 and miR-146a, of 756 analyzed miRNA (Paoli et al., 2019) in older men.
Some discrepancies in results discussed in this section may be due to specific technical challenges of human research. Among others, these include differences in the source, methods of purification and fractionation of sperm, the methylation/sncRNA analysis methodology and the type of bioinformatic analysis (Donkin and Barrès, 2018; Jenkins and Turek, 2020). In addition, the epidemiological design of the study may affect results. While a longitudinal design is time intensive and expensive, the cross-sectional design is limited by an inability to assess intra-individual age-related epigenetic changes, possible selection bias and insufficient control of confounders.
Age-induced epigenetic sperm changes and their influence on fertilization, embryo development and offspring health
Recent reviews have shown that APA is associated with increased miscarriage (RR = 2) and fetal loss (RR = 2) as well as rare single-gene disorders (RR = 1.3–12), congenital anomalies (RR = 1.2), autism, schizophrenia and other forms of ‘psychiatric morbidity’ (RR = 1.5–5.7) in human offspring (Sharma et al., 2015; Yatsenko and Turek, 2018). Using 1057 embryos from 87 couples undergoing IVF treatment, a negative association of APA has been reported with the chance of an optimal eight-cell embryo on the third day of development: couples with older men were less likely to produce an embryo of eight blastomeres, compared to those with younger men (Van Opstal et al., 2021).
The effect of APA and related epigenetic sperm changes on offspring health is still underappreciated, despite growing evidence that an association exists. If APA is considered a harmful factor, sperm epigenetic changes can be markers and mechanisms through which adverse effects are transmitted to offspring. Some authors have suggested the role of DNA methylation profiles in inheritance (Jenkins et al., 2018). Others have highlighted the potential role of extracellular vesicles and their small RNA cargo (Chen et al., 2016; Morgan et al., 2019; Guo et al., 2021).
The direct role of age-associated sperm epigenetic changes in offspring outcomes has been studied in better controlled rodent models. A genome-wide methylation study of epididymal sperm DNA from young (3 months old) and old (12–14 months old) mice showed that age-associated DNA methylation alterations that occur in sperm are passed to the offspring (Milekic et al., 2015). The offspring of older fathers had reduced exploratory and startle behaviors and exhibited similar brain DNA methylation abnormalities as those observed in the paternal sperm. Offspring from old fathers also exhibited transcriptional dysregulation of developmental genes implicated in autism and schizophrenia.
In addition, in a recent study of age-related changes in sperm tsRNA, using the injection of sperm tsRNAs from aged and young male mice into zygotes, Guo et al. investigated behavioral traits of F1 progeny and transcriptome of the cerebral cortex and hippocampus of male mice and the embryo at two-cell and blastocyst stage (Guo et al., 2021). They found that injection of sperm tsRNAs from aged male mice into zygotes induced anxiety-like behaviors in F1 males and altered expression of nerve development genes in embryos and nerve tissue in F1 males.
Xie et al. found a significant reduction of the median life spans in the offspring of old fathers (≥21-month age) compared with the offspring of young fathers (4-month age), 825 and 883.5 days, respectively (Xie et al., 2018). These effects were independent of maternal age. Examination of old (19-month age) offspring of old and young fathers showed that many aging dysfunctions were more pronounced in the progeny of old mice, indicating that APA aggravated specific aging-associated tissue changes. Specifically, RRBS analysis of hippocampus tissue in the offspring of old and young fathers at 4-week age revealed 189 promoter regions that were significantly hypomethylated and 33 promoter regions that were significantly hypermethylated in the offspring of old fathers. Among these differentially methylated promoters, 14 were also aDMR in sperm. Pathway analyses showed enrichment for genes encoding components of mTOR and immune-regulatory pathways, the same pathways enriched with aDMRs and age-dependent miRNAs in paternal sperm.
Zhao et al. (2020) characterized changes in glucose and cholesterol metabolism and also ASD-like behaviors in first and second generations from 12- and 18-month-old male mice, respectively (Zhao et al., 2020). WGBS of sperm from APA mice identified aDMRs within the whole genome and aDMRs within promoter regions, suggesting that specific genes and relevant pathways might be associated with autism and aberrant glucose metabolism in the offspring from APA males. Whether this and the above-mentioned mechanisms are involved in the intergenerational and transgenerational inheritance of paternal age-associated DNA methylation abnormalities remains to be explored.
A few human studies in this field have used fathers' sperm and either fetal cord blood (Atsem et al., 2016) or blastocysts (Denomme et al., 2020) to investigate the transmission of sperm epigenetics to the next generation. Atsem et al. have analyzed by bisulfite pyrosequencing the DNA methylation in fetal cord blood samples of >190 children conceived by IVF/ICSI. Of genes known to display a correlation between paternal age and sperm methylation (Jenkins et al., 2014), several (FOXK1 and KCNA7) were associated with neuropsychiatric and other disorders, and methylation levels in their regions have a negative correlation with age. SNPs in the FOXK1 and KCNA7 target regions were used to distinguish paternal and maternal alleles in the offspring's methylation profiles. For FOXK1, a significant correlation was observed between methylation of the paternal allele in fetal cord blood and paternal age, whereas no correlation was found between the maternal allele methylation and maternal age. Thus, there was a strong tendency for hypomethylation to be inherited between fathers and their offspring (Atsem et al., 2016). Collectively, these results suggest that paternal epigenome reprogramming in early embryogenesis is incomplete, and some features of the sperm epigenome can persist in the offspring’s differentiated somatic cells.
Recently, the possibility of aberrant DNA methylation being inherited via aged sperm was approached by analyses of global methylomes in sperm of younger (∼30 years) and older (>50 years) fathers and blastocysts donated from young fertile donor oocyte IVF cycles (Denomme et al., 2020). More than 3000 sperm aDMRs and nearly 4000 blastocyst aDMRs were detected. A statistical comparison of genes associated with aDMRs in the sperm and blastocysts revealed 218 common genes: 167 were hypomethylated and 61 were hypermethylated in samples from older aged individuals. Ten genes contained both hypo- and hypermethylated aDMRs. Statistically significant enrichment for the known nucleosome retention regions was found in the sperm aDMRs. This colocalization with nucleosomes was even more pronounced for aDMRs common between sperm and blastocysts. Several neurological signaling pathway genes were associated with aDMRs, including multiple genes from the opioid pathway. Furthermore, a highly significant enrichment for genes implicated in ASDs, schizophrenia and bipolar disorder was found in both sperm and blastocyst aDMRs. Notably, all three candidate gene lists for these neurological disorders share genes with the opioid signaling pathway (Denomme et al., 2020) that is a neurotransmitter system in the brain involved in mood regulation and genesis of neurodevelopmental pathologies (Pellissier et al., 2018). In the sperm methylome, 19 hypomethylated and 7 hypermethylated aDMRs, while in blastocysts, 22 hypomethylated and 10 hypermethylated aDMRs were associated with imprinted genes, and 6 imprinted genes were shared between sperm and blastocyst aDMRs. The authors concluded that some changes of genome methylation associated with neurodevelopmental pathways could be directionally induced in sperm of aged fathers and partially transferred to the preimplantation embryo.
However, a comparison of sperm DNA methylation patterns between similar-aged grandsons of younger (<25 years of age at the time of conception of the father) versus older (older than 40 years of age at the time of conception of the father) grandfathers did not reveal any significant differences (Jenkins et al., 2019). The authors suggested that aging signals appeared to be erased and reestablished with each generation. Only a weak trend for the subjects of the older grandfather group to display reduced methylation at the 140 aDMRs known to undergo hypomethylation with age was found. These remnants of age-related DNA methylation signals were too small to have any biological consequences.
For humans, studies of the association of APA, epigenetic changes and health risk in offspring remain a challenge. The one crucial challenge involves the use of ART in the study design as it contributes additional confounders to investigated outcomes.
Age, ART procedures and additional epigenetic risks
APA is associated with increased use of ART (Bertoncelli Tanaka et al., 2019; American Society for Reproductive Medicine, Society for Assisted Reproductive Technology, 2017) due to a significant decrease in age-related female fertility for a couple and the slightly decreased male fertility, and because ART is considered generally safe. A father’s age for ART is rarely regulated in guidelines, except in Germany, where the age limit for ART is 50 years (Belaisch-Allart et al., 2016; Couture et al., 2021). ART spans the periconception period from gametogenesis, fertilization and early embryonic development, coinciding with an intense epigenetic reprogramming in germ cells. ART-induced epigenetic changes may provide additional epigenetic changes and risks for the next generation (Vrooman and Bartolomei, 2017; Chen et al., 2020). ART-related epigenetic changes (disruption of DNA methylation in imprinted control regions in mouse models were mainly investigated) occur during the following ART procedures: ovarian stimulation by gonadotropins, intracytoplasmic sperm injection and embryo culture and transfer (de Waal et al., 2015; Vrooman and Bartolomei, 2017). Possible male-associated epigenetic risks linked to ART techniques may result from either use of sperm with incomplete reprogramming or from the ART procedures themselves.
Among the two most common types of ART, conventional IVF and ICSI (69% of ART cycles in the USA utilized ICSI), ICSI and round spermatid injection seem to be more harmful because they may increase the incidence of imprinting disorders and adversely affect embryonic development by using immature spermatozoa that may not have proper imprints or global methylation established (Kishigami et al., 2006; Rajender et al., 2011). One recent study aimed to clarify epigenetic risks attributable to ARTs (Chen et al., 2020) using high-throughput methods to assess profiles of DNA methylation from 137 umbilical cord blood (UCB) and 158 parental peripheral blood samples, profiles of histone modifications (H3K4me3, H3K4me1, H3K27me3 and H3K27ac) from 33 UCB samples and transcriptomes of 32 UCB samples. They found that H3K4me3 was the most profoundly impacted by ICSI and freeze-thawing operation compared with the other three types of histone modifications. In that study, IVF-fresh embryo transfer seemed to introduce fewer disturbances into infant epigenomes than IVF-frozen embryo transfer or ICSI-embryo transfer did. Another study, however, suggested that cryopreservation may temper some of the epigenetic aberrations induced by IVF or ICSI (Estill et al., 2016). ARTs also decreased the similarity of DNA methylome within twin pairs, and researchers suggested that ART processes potentially caused conservative epigenetic changes distributed widely throughout the genome (Chen et al., 2020). Notably, those unique and common alterations induced by different ART procedures were, for example, highly enriched in the processes related to the nervous system, cardiovascular system and glycolipid metabolism. Given the available data already in hand, a meta-analysis may begin to provide a consensus or at least indicate the variables that should be considered. However, this can only be established from a life course study.
Research associating ART with epigenetic changes in the offspring is still limited and accompanied by methodological challenges, including a large number of confounders related to male and female infertility, environment and lifestyle and ethical restrictions in the use human embryos. Our knowledge and understanding of the association of APA, ART procedures and epigenetic changes is lacking as many gaps remain.
Discussion
Age-dependent changes in sperm epigenome: stochastic errors or program targeting offspring fitness
Today, age-dependent changes in the sperm epigenome and changes induced by different environmental or lifestyle stressors are usually explained by the accumulation of random epigenetic errors: epimutations. Indeed both extended periods of time and unfavorable conditions are likely factors that can increase rates of such stochastic errors. However, this hypothesis contradicts the patterns of enriched biological functions usually identified when sperm epigenome changes are analyzed in relation to time or other factors, as discussed above.
Findings from many independent studies suggest that different epigenetic mechanisms undergo non-random and concerted age-dependent changes. For example, in the mouse study by Xie et al. (2018), both age-dependent changes in DNA methylation and sncRNA were associated with major developmental and aging pathways included mTOR, insulin and growth factor signaling. In our experiments with rats, age-dependent changes in sncRNA (Suvorov et al., 2020) and DNA methylation (Pilsner et al., 2021) targeted the same set of developmental pathways, including those identified by Xie et al. Thus, emerging evidence suggests that a purely stochastic error model cannot explain age-dependent changes in animal sperm epigenome. Instead, these changes follow well-defined patterns.
Findings from several epidemiologic studies have shown that aDMRs were enriched for genes involved in embryonic development, behavior and neurodevelopment pathway (Cao et al., 2020; Denomme et al., 2020; Laurentino et al., 2020; Oluwayiose et al., 2021; Pilsner et al., 2022). Human studies are usually limited by the effect of confounders (ART, men from infertility clinics, concomitant influence of environment and lifestyle); however, construction of epigenetic clocks based on sperm DNA methylation with relatively high precision (Jenkins et al., 2018; Cao et al., 2020; Pilsner et al., 2022) demonstrate clear age-related impacts on the sperm epigenome. The novel recent findings that SEA is associated with TTP in the general population underscores the importance of biological aging of male germ cells to pregnancy success and may enhance our understanding of idiopathic infertility (Pilsner et al., 2022).
Today we do not know what mechanisms determine selective age-associated changes in the sperm epigenome. However, one can propose that these patterns may be partly a result of natural selection, which enabled fine-tuning of offspring development based on the actual paternal state and experience. Age-dependent changes in sperm epigenome may be a part of normal developmental program shaped by natural selection to inform better fitness of offspring phenotypes. One recent study demonstrated that paternal dominance status determines the growth rate in offspring in mice, where offspring of dominant males grow faster than the offspring of subordinate fathers even if available food resources are the same (Cauceglia et al., 2020). One interpretation of this intriguing finding is that in nature, dominance usually ensures better access to resources and may increase the fitness of faster-growing offspring. Fine-tuning offspring growth as a function of paternal dominance status could result from natural selection. Similarly, natural selection may shape epigenetic programs transferred to the new generation by fathers of different ages.
The alternative explanation of clear patterns in age-dependent changes of sperm epigenome refers to the chromatin structure in spermatozoa. One can assume that open areas of chromatin accumulate stochastic epigenetic errors faster than densely packed sites. Several animal studies have demonstrated that genes encoding most key embryonic developmental and morphogenesis genes are associated with modified nucleosomes in mature sperm (Arpanahi et al., 2009; Hammoud et al., 2009; Brykczynska et al., 2010). Interestingly, the majority of regions in fertile human males corresponding to regions coincident with sperm RNAs genes were H3K4me3 and H3K27me3 enriched within hypomethylated DNA regions (Sendler et al., 2013). In one study, most hypomethylated aDMRs were found within 1 kb of known nucleosome retention sites in human sperm (Jenkins et al., 2014). In another study, sperm aDMRs were also significantly enriched for the known nucleosome retention regions (Denomme et al., 2020). Hence, a bias of age-dependent epigenetic changes toward primary developmental pathways in sperm may be due to the loose packaging of corresponding genes in spermatozoa.
Finally, both explanations may be valid, and in fact, they do not contradict each other and may act together to fine tune the response. Additional research on sperm chromatin is needed to identify if age-dependent changes are often associated with loose chromatin segments.
Future perspectives characteristics of sperm epigenetic markers: dynamic/stability and reversibility
When epigenetic markers are detected in sperm, we should first know ‘what is stable and what is dynamic, and what is consistent within and between men over time?’ (Ostermeier et al., 2005; Morgan et al., 2020). ‘Over time’ for males means a period that extends from puberty when spermatogenesis occurs till late adulthood (60 years and older). ‘Over time’ for males also could mean either a short period, within one to several spermatogenesis cycles, 3–12 months or a long period, over the life span.
Based on data of epigenetic clocks in human sperm, we suggest that observed epigenetic markers related to aging are naturally dynamic and appear to be irreversible, as data has yet to suggest otherwise. For environment-modulated sperm epigenetic changes, a few recent epidemiological studies using a cross-over cross-back binary study design demonstrated the reversible fashion after exercise training (Ingerslev et al., 2018) and chemical exposure (Estill et al., 2019), although the period for full recovery is likely to be longer than one spermatogenic cycle. In some cases of phthalate exposure, it appears that a point of no return was reached with high doses (Estill et al., 2019). We hypothesized that environmental and lifestyle epigenetic changes in sperm are rather naturally dynamic over days or months and seem to be reversible if exposure occurs for short periods in adults when spermatogenesis is ongoing. Some observed environment-induced epigenetic changes in sperm could be irreversible if the duration of exposure is chronic or coincides with sensitive windows of development.
Critical issues of age-associated sperm epigenetic changes still remain (Table II). In Fig. 1, we summarize our vision of current and future research in sperm epigenetics and related fields. We propose some recommendations for future longitudinal studies in animal models and human epidemiological studies. Experimental and epidemiological studies using a prospective design, healthy males from peripuberty and multi-omics approaches, are required. They should extend across age periods and encompass all types of genomic elements and epigenetic markers. Only then, may they explain the lifetime dynamics of epigenetic changes, the role of lifestyle and environmental factors on SEA, the modulation by infertility and ART techniques and the relationship with embryo and offspring characteristics.
Table II.
Critical issues of age-associated sperm epigenetic changes (AASEC) that still poorly addressed in human and animals.
| Critical issues | Description |
|---|---|
| Genomic elements | Which sperm genomic elements are affected by age? |
| Lifetime dynamics | Are AASEC linear? Or are changes that occur during peripubertal period more significant? What is the direction of DNA methylation change? |
| Species specificity | Are AASEC the same in humans and animals? |
| Cofactors and vulnerable periods | What factors affect AASEC in different sensitive windows? |
| Inheritance | Do AASEC survive epigenome remodeling in early embryo? |
| Interventions | What critical knowledge gaps need to be addressed to develop therapeutic interventions? |
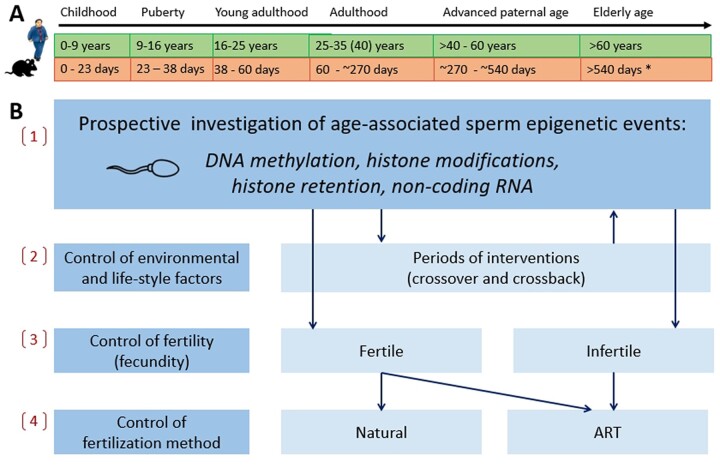
Figure 1.
Proposed future research on age, environment and lifestyle and ART-associated sperm epigenetic changes in humans and mice over their life span. The use of prospective design for human studies and animal models for investigation of age, epigenetic changes and consequent events in offspring among the next generation(s) looks promising and is recommended to be implemented. (A) Although longitudinal human studies over life span stages (childhood, puberty, young adulthood, adulthood, APA, elderly age) are time intensive, expensive and difficult to do, they are more conclusive and, therefore, this needs to be proposed. Animal models have an invaluable advantage of removing the environmental and lifestyle factors from the design and they can be conducted much faster. (B) Involvement of healthy population including twins (human), prospective follow up, controls and use of environmental and lifestyle factors, fertility status and method of fertilization in designs are recommended to distinguish (1) age-associated changes; (2) environment and lifestyle changes; (3) fertility-associated changes; and (4) ART-induced changes of the sperm epigenome. *Equivalency of mouse reproductive ages to these of humans was determined based on previously published data (Flurkey et al., 2007; Brust et al., 2015; Bell, 2018) as well as information from Charles River Laboratories that male mice breeders are usually retired by 9 months of age (270 days) due to the decline of their reproductive efficiency. ART, assisted reproductive technologies.
Conclusions
Throughout a man’s life span, spermatogonia self-renew by undergoing approximately 23 mitotic divisions per year with each cycle yielding millions of spermatozoa. Although male fertility declines over age, older men are fertile or subfertile and a single spermatozoon from the millions of spermatozoa produced, is selected to fertilize an oocyte either from natural sperm competition, or in vitro, with the selection of what appears to be the best sperm. Increasing evidence indicates that delayed fatherhood is associated with the accumulation of changes in male gametes induced by various factors, including aging, environmental exposures, lifestyle factors and in some cases, ART. Age-associated genetic and epigenetic changes affect the developmental trajectories of offspring and may result in adverse health outcomes, including psychiatric and neurodevelopmental disorders. Yet, paternal age is rarely considered an important factor for the fertility of couples’ fertility and the health of offspring.
Although documented in many studies, age-associated changes in the sperm epigenome, including DNA methylation, histone modifications and profiles of sncRNA, are yet poorly understood. For example, some humans and rodent studies suggest that most sperm aDMRs undergo hypermethylation with age, whereas other studies do not. Several animal studies suggest that the rate of DNA methylation changes over the life span is not uniform, and significant changes mainly occur during puberty. However, the successful development of the human sperm epigenetic clock indicates that at least some aDMRs follow linear relations between methylation levels and paternal age.
The few rodent studies in which age-dependent DNA methylation and sncRNA profiles were analyzed show that both age-dependent epigenetic mechanisms target the same genes and biological functions. In addition, most animal and human studies identify similar lists of biological categories enriched by age-dependent epigenetic mechanisms in sperm, including embryonic developmental, neurodevelopmental, growth and metabolic pathways. Thus, age-dependent changes in the sperm epigenome cannot be described by the model of stochastic accumulation of epimutations.
An emerging body of studies indicate that chemical exposures and lifestyle factors may affect the normal process of epigenetic aging of sperm. Future experimental and epidemiological studies using a prospective design, healthy males and multi-omics approaches across all types of genomic elements and epigenetic markers are needed to further understand the mechanisms of age-related genetic and epigenetic changes in sperm, their interplay with environmental and lifestyle factors and their association with epigenetic events after fertilization and the health of the successive generation. We suggest a broader discussion focused on the father’s age in natural and ART conceptions is needed. Professional community should be informed and raise awareness of the general population, particularly older men and couples seeking reproductive care. Future research on SEA may open doors for the development of paternal therapeutic interventions to protect the health of the next generations.
Contributor Information
Vasily Ashapkin, Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia.
Alexander Suvorov, Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia; Department of Environmental Health Sciences, School of Public Health and Health Sciences, University of Massachusetts Amherst, Amherst, MA, USA.
J Richard Pilsner, Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Stephen A Krawetz, Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA; Center for Molecular Medicine and Genetics, Wayne State University School of Medicine, Detroit, MI, USA.
Oleg Sergeyev, Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia.
Data availability
The data that support the findings of our conclusions are available from the corresponding author upon reasonable request.
Authors’ roles
S.A.K., J.R.P., O.S. and A.S. conceptualized study. V.A., O.S. and A.S. set methodology, reviewed included studies, analyzed data and prepared the first draft. V.A., S.A.K., J.R.P., O.S. and A.S. revised the article critically. All authors approved the final version of the article.
Funding
Russian Science Foundation (18-15-00202) (https://rscf.ru/en/project/18-15-00202/).
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Aitken RJ. Not every sperm is sacred; a perspective on male infertility. Mol Hum Reprod 2018;24:287–298. [DOI] [PubMed] [Google Scholar]
- Aoki V, Liu L, Jones K, Hatasaka H, Gibson M, Peterson C, Carrell D.. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil Steril 2006;86:1408–1415. [DOI] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D.. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 2009;19:1338–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI, Vanyushin BF.. Dnmt2 is the most evolutionary conserved and enigmatic cytosine DNA methyltransferase in eukaryotes. Russ J Genet 2016;52:237–248. [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI, Vanyushin BF.. Aging as an epigenetic phenomenon. Curr Genomics 2017;18:385–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI, Vanyushin BF.. Epigenetic clock: just a convenient marker or an active driver of aging? In: Guest PC (ed). Reviews on Biomarker Studies in Aging and Anti-Aging Research, Vol 1178. Cham: Springer International Publishing, 2019, 175–206. [DOI] [PubMed] [Google Scholar]
- Aston KI, Hunt SC, Susser E, Kimura M, Factor-Litvak P, Carrell D, Aviv A.. Divergence of sperm and leukocyte age-dependent telomere dynamics: implications for male-driven evolution of telomere length in humans. Mol Hum Reprod 2012;18:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, Carrell DT.. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 2015;104:1388–1397.e5. [DOI] [PubMed] [Google Scholar]
- Atsem S, Reichenbach J, Potabattula R, Dittrich M, Nava C, Depienne C, Böhm L, Rost S, Hahn T, Schorsch M. et al. Paternal age effects on sperm FOXK1 and KCNA7 methylation and transmission into the next generation. Hum Mol Genet 2016;25:4996–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avellino G, Theva D, Oates RD.. Common urologic diseases in older men and their treatment: how they impact fertility. Fertil Steril 2017;107:305–311. [DOI] [PubMed] [Google Scholar]
- Belaisch-Allart J, Ouazana M, Bailly M, Selva J, Kulski O, Boitrelle F.. [Male age in assisted reproductive technologies: is there a limit?]. Gynecol Obstet Fertil 2016;44:712–715. [DOI] [PubMed] [Google Scholar]
- Bell MR. Comparing postnatal development of gonadal hormones and associated social behaviors in rats, mice, and humans. Endocrinology 2018;159:2596–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloc S, Hazout A, Zini A, Merviel P, Cabry R, Chahine H, Copin H, Benkhalifa M.. How to overcome male infertility after 40: influence of paternal age on fertility. Maturitas 2014;78:22–29. [DOI] [PubMed] [Google Scholar]
- Berntsen S, Söderström-Anttila V, Wennerholm U-B, Laivuori H, Loft A, Oldereid NB, Romundstad LB, Bergh C, Pinborg A.. The health of children conceived by ART: “the chicken or the egg?” Hum Reprod Update 2019;25:137–158. [DOI] [PubMed] [Google Scholar]
- Bertoncelli Tanaka M, Agarwal A, Esteves SC.. Paternal age and assisted reproductive technology: problem solver or trouble maker? Panminerva Med 2019;61:138–151. [DOI] [PubMed] [Google Scholar]
- Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, Vaiman D, Jouannet P, Tost J, Jammes H.. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet 2010;18:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth LN, Brunet A.. The aging epigenome. Mol Cell 2016;62:728–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroujeni PB, Ebrahimian S, Abedini M, Chayjan MR, Hassani M, Gourabi H, Sadighi-Gilani MA, Sabbaghian M, Meybodi AM.. The role of DEFB126 variation in male infertility and medically assisted reproduction technique outcome. Reprod Biomed Online 2019;39:649–657. [DOI] [PubMed] [Google Scholar]
- Brandt JS, Cruz Ithier MA, Rosen T, Ashkinadze E.. Advanced paternal age, infertility, and reproductive risks: a review of the literature. Prenat Diagn 2019;39:81–87. [DOI] [PubMed] [Google Scholar]
- Brust V, Schindler PM, Lewejohann L.. Lifetime development of behavioural phenotype in the house mouse (Mus musculus). Front Zool 2015;12 Suppl 1:S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AHFM.. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 2010;17:679–687. [DOI] [PubMed] [Google Scholar]
- Cao M, Shao X, Chan P, Cheung W, Kwan T, Pastinen T, Robaire B.. High-resolution analyses of human sperm dynamic methylome reveal thousands of novel age-related epigenetic alterations. Clin Epigenetics 2020;12:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Hung J-H, Hainer SJ, Chou M-T, Carone DM, Weng Z, Fazzio TG, Rando OJ.. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 2014;30:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell DT, Hammoud SS.. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod 2010;16:37–47. [DOI] [PubMed] [Google Scholar]
- Cauceglia JW, Nelson AC, Rubinstein ND, Kukreja S, Sasso LN, Beaufort JA, Rando OJ, Potts WK.. Transitions in paternal social status predict patterns of offspring growth and metabolic transcription. Mol Ecol 2020;29:624–638. [DOI] [PubMed] [Google Scholar]
- Cecere G. Small RNAs in epigenetic inheritance: from mechanisms to trait transmission. FEBS Lett 2021;595:2953–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedars MI. Introduction: childhood implications of parental aging. Fertil Steril 2015;103:1379–1380. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. 2017 Assisted Reproductive Technology Fertility Clinic Success Rates Report. Atlanta, GA: U.S. Department of Health and Human Services, 2019. https://ftp.cdc.gov/pub/Publications/art/ART-2017-Clinic-Report-Full.pdf (22 August 2022, date last accessed). [Google Scholar]
- Centers for Disease Control and Prevention. 2019 Assisted Reproductive Technology Fertility Clinic and National Summary Report. Atlanta, GA: U.S. Department of Health and Human Services, 2021. https://www.cdc.gov/art/reports/2019/pdf/2019-Report-ART-Fertility-Clinic-National-Summary-h.pdf (22 August 2022, date last accessed). [Google Scholar]
- Centers for Disease Control and Prevention, American Society for Reproductive Medicine, Society for Assisted Reproductive Technology. 2015 Assisted Reproductive Technology Fertility Clinic Success Rates Report. Atlanta, GA: U.S. Department of Health and Human Services, 2017. https://www.cdc.gov/art/pdf/2015-report/ART-2015-National-Summary-Report.pdf (22 August 2022, date last accessed). [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng G-h, Peng H, Zhang X, Zhang Y. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397–400. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan W, Duan E.. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 2016;17:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Peng Y, Ma X, Kong S, Tan S, Wei Y, Zhao Y, Zhang W, Wang Y, Yan L. et al. Integrated multi-omics reveal epigenomic disturbance of assisted reproductive technologies in human offspring. EBioMedicine 2020;61:103076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Gao Y, Lin Z, Yang S, Wang T, Wang Q, Xie N, Hua R, Liu M. et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res 2018;28:879–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung S, Parrella A, Rosenwaks Z, Palermo GD.. Genetic and epigenetic profiling of the infertile male. PLoS One 2019;14:e0214275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture V, Delisle S, Mercier A, Pennings G.. The other face of advanced paternal age: a scoping review of its terminological, social, public health, psychological, ethical and regulatory aspects. Hum Reprod Update 2021;27:305–323. [DOI] [PubMed] [Google Scholar]
- de Waal E, Vrooman LA, Fischer E, Ord T, Mainigi MA, Coutifaris C, Schultz RM, Bartolomei MS.. The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum Mol Genet 2015;24:6975–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denomme MM, Haywood ME, Parks JC, Schoolcraft WB, Katz-Jaffe MG.. The inherited methylome landscape is directly altered with paternal aging and associated with offspring neurodevelopmental disorders. Aging Cell 2020;19:e13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Fossé NA, van der Hoorn M-LP, van Lith JMM, le Cessie S, Lashley EELO.. Advanced paternal age is associated with an increased risk of spontaneous miscarriage: a systematic review and meta-analysis. Hum Reprod Update 2020;26:650–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin I, Barrès R.. Sperm epigenetics and influence of environmental factors. Mol Metab 2018;14:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DTA, Lee NR, Rej PH, Hayes MG, Kuzawa CW.. Older paternal ages and grandpaternal ages at conception predict longer telomeres in human descendants. Proc Biol Sci 2019;286:20190800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Wyrobek AJ, Sloter E, Kidd SA, Moore L, Young S, Moore D.. The association of age and semen quality in healthy men. Hum Reprod 2003;18:447–454. [DOI] [PubMed] [Google Scholar]
- Estill M, Hauser R, Nassan FL, Moss A, Krawetz SA.. The effects of di-butyl phthalate exposure from medications on human sperm RNA among men. Sci Rep 2019;9:12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estill MS, Bolnick JM, Waterland RA, Bolnick AD, Diamond MP, Krawetz SA.. Assisted reproductive technology alters deoxyribonucleic acid methylation profiles in bloodspots of newborn infants. Fertil Steril 2016;106:629–639.e10. [DOI] [PubMed] [Google Scholar]
- Estill MS, Hauser R, Krawetz SA.. RNA element discovery from germ cell to blastocyst. Nucleic Acids Res 2019;47:2263–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson DP, Djira G, Kasperson K, Christianson J.. Relationships between the age of 25,445 men attending infertility clinics and sperm chromatin structure assay (SCSA®) defined sperm DNA and chromatin integrity. Fertil Steril 2020;114:311–320. [DOI] [PubMed] [Google Scholar]
- Fenic I, Hossain HM, Sonnack V, Tchatalbachev S, Thierer F, Trapp J, Failing K, Edler KS, Bergmann M, Jung M. et al. In vivo application of histone deacetylase inhibitor trichostatin-A impairs murine male meiosis. J Androl 2008;29:172–185. [DOI] [PubMed] [Google Scholar]
- Fenic I, Sonnack V, Failing K, Bergmann M, Steger K.. In vivo effects of histone-deacetylase inhibitor trichostatin-A on murine spermatogenesis. J Androl 2004;25:811–818. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang S-C. et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet 2006;79:67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, M. Currer J, Harrison DE.. Chapter 20 - Mouse models in aging research. In: Fox JG, Davisson MT, Quimby FW, Barthold SW, Newcomer CE, Smith AL (eds). The Mouse in Biomedical Research, 2nd edn.Burlington: Academic Press, 2007, 637–672. [Google Scholar]
- Ford WC, North K, Taylor H, Farrow A, Hull MG, Golding J.. Increasing paternal age is associated with delayed conception in a large population of fertile couples: evidence for declining fecundity in older men. The ALSPAC Study Team (Avon Longitudinal Study of Pregnancy and Childhood). Hum Reprod 2000;15:1703–1708. [DOI] [PubMed] [Google Scholar]
- Fu B, Ma H, Liu D.. Endogenous retroviruses function as gene expression regulatory elements during mammalian pre-implantation embryo development. Int J Mol Sci 2019;20:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Wang PJ.. Mammalian piRNAs: biogenesis, function, and mysteries. Spermatogenesis 2014;4:e27889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godmann M, Auger V, Ferraroni-Aguiar V, Sauro AD, Sette C, Behr R, Kimmins S.. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol Reprod 2007;77:754–764. [DOI] [PubMed] [Google Scholar]
- Goisis A, Håberg SE, Hanevik HI, Magnus MC, Kravdal Ø.. The demographics of assisted reproductive technology births in a Nordic country. Hum Reprod 2020;35:1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann JM, Veltman JA, Gilissen C.. De novo mutations reflect development and aging of the human germline. Trends Genet 2019;35:828–839. [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, Golic KG, Jacobsen SE, Bestor TH.. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006;311:395–398. [DOI] [PubMed] [Google Scholar]
- Goodrich RJ, Anton E, Krawetz SA.. Isolating mRNA and small noncoding RNAs from human sperm. In: Carrell DT, Aston KI (eds). Spermatogenesis, Vol. 927. Totowa, NJ: Humana Press, 2013, 385–396. [DOI] [PubMed] [Google Scholar]
- Goriely A. Decoding germline de novo point mutations. Nat Genet 2016;48:823–824. [DOI] [PubMed] [Google Scholar]
- Grunewald S, Paasch U, Glander H-J, Anderegg U.. Mature human spermatozoa do not transcribe novel RNA. Andrologia 2005;37:69–71. [DOI] [PubMed] [Google Scholar]
- Gunes S, Hekim GNT, Arslan MA, Asci R.. Effects of aging on the male reproductive system. J Assist Reprod Genet 2016;33:441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Bai D, Liu W, Liu Y, Zhang Y, Kou X, Chen J, Wang H, Teng X, Zuo J. et al. Altered sperm tsRNAs in aged male contribute to anxiety-like behavior in offspring. Aging Cell 2021;20:e13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Surani MA.. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci 2013;368:20110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA.. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet 2012;28:164–174. [DOI] [PubMed] [Google Scholar]
- Hajkova P. Epigenetic reprogramming in the germline: towards the ground state of the epigenome. Philos Trans R Soc Lond B Biol Sci 2011;366:2266–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA.. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 2002;117:15–23. [DOI] [PubMed] [Google Scholar]
- Halling JF, Pilegaard H.. PGC-1α-mediated regulation of mitochondrial function and physiological implications. Appl Physiol Nutr Metab 2020;45:927–936. [DOI] [PubMed] [Google Scholar]
- Hamada N, Ito H, Nishijo T, Iwamoto I, Morishita R, Tabata H, Momiyama T, Nagata K-I.. Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep 2016;6:30805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Low DHP, Yi C, Carrell DT, Guccione E, Cairns BR.. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 2014;15:239–253. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR.. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009;460:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S-L, Ni F-D, Yang W-X.. The dynamics and regulation of chromatin remodeling during spermiogenesis. Gene 2019;706:201–210. [DOI] [PubMed] [Google Scholar]
- Hassan MAM, Killick SR.. Effect of male age on fertility: evidence for the decline in male fertility with increasing age. Fertil Steril 2003;79:1520–1527. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Surani MA.. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell 2009;4:493–498. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen I-C, Gildersleeve H, Lehle JD, Mayo M, Westernströer B et al The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep 2018;25:1650–1667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ.. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One 2007;2:e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua M, Liu W, Chen Y, Zhang F, Xu B, Liu S, Chen G, Shi H, Wu L.. Identification of small non-coding RNAs as sperm quality biomarkers for in vitro fertilization. Cell Discov 2019;5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kim JK, Do DV, Lee C, Penfold CA, Zylicz JJ, Marioni JC, Hackett JA, Surani MA.. Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. eLife 2017;6:e22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerslev LR, Donkin I, Fabre O, Versteyhe S, Mechta M, Pattamaprapanont P, Mortensen B, Krarup NT, Barrès R.. Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clin Epigenet 2018;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Cairns B, Smith A, Carrell DT.. Paternal germ line aging: DNA methylation age prediction from human sperm. BMC Genomics 2018;19:763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT.. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet 2014;10:e1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, James ER, Alonso DF, Hoidal JR, Murphy PJ, Hotaling JM, Cairns BR, Carrell DT, Aston KI.. Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 2017;5:1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, James ER, Aston KI, Salas-Huetos A, Pastuszak AW, Smith KR, Hanson HA, Hotaling JM, Carrell DT.. Age-associated sperm DNA methylation patterns do not directly persist trans-generationally. Epigenetics Chromatin 2019;12:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Turek PJ.. Epigenetics and male infertility. In: Parekattil SJ, Esteves SC, Agarwal A (eds). Male Infertility: Contemporary Clinical Approaches, Andrology, ART and Antioxidants. Cham: Springer International Publishing, 2020, 139–146. [Google Scholar]
- Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA, Reproductive Medicine Network. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update 2013;19:604–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M, Sendler E, Moskovtsev SI, Librach CL, Goodrich R, Swanson S, Hauser R, Diamond MP, Krawetz SA.. Absence of sperm RNA elements correlates with idiopathic male infertility. Sci Transl Med 2015;7:295re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Sendler E, Lalancette C, Hauser R, Diamond MP, Krawetz SA.. Cleavage of rRNA ensures translational cessation in sperm at fertilization. Mol Hum Reprod 2011;17:721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MJ, Goodman SJ, Kobor MS.. DNA methylation and healthy human aging. Aging Cell 2015;14:924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth A, Diemer T, Kopa Z, Krausz C, Minhas S, Tournaye H.. EAU guidelines on male infertility 2020. In: European Association of Urology Guidelines, 2019 edn., presented at the EAU Annual Congress Barcelona 2019. Arnhem, The Netherlands: European Association of Urology Guidelines Office, 2019. https://uroweb.org/guideline/male-infertility/#8 (25 February 2021, date last accessed). [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M.. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 2013;32:340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane AE, Sinclair DA.. Epigenetic changes during aging and their reprogramming potential. Crit Rev Biochem Mol Biol 2019;54:61–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G, Feil R.. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 2013;368:20110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Boyar FZ, Driscoll DJ.. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc Natl Acad Sci USA 2004;101:16583–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandwala YS, Baker VL, Shaw GM, Stevenson DK, Lu Y, Eisenberg ML.. Association of paternal age with perinatal outcomes between 2007 and 2016 in the United States: population based cohort study. BMJ 2018;363:k4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandwala YS, Zhang CA, Lu Y, Eisenberg ML.. The age of fathers in the USA is rising: an analysis of 168 867 480 births from 1972 to 2015. Hum Reprod 2017;32:2110–2116. [DOI] [PubMed] [Google Scholar]
- Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M.. RNA–mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet 2013;9:e1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Van Thuan N, Hikichi T, Ohta H, Wakayama S, Mizutani E, Wakayama T.. Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Dev Biol 2006;289:195–205. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T.. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet 2007;16:2542–2551. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Okae H, Hiura H, Chiba H, Shirakata Y, Hara K, Tanemura K, Arima T.. Genome-scale assessment of age-related DNA methylation changes in mouse spermatozoa. PLoS One 2016;11:e0167127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JA, Zhang S, Yaron Y, Zhao Y, Krawetz SA.. Genetic testing for male infertility: a postulated role for mutations in sperm nuclear matrix attachment regions. Genet Test 1997;1:125–129. [DOI] [PubMed] [Google Scholar]
- Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP.. A survey of small RNAs in human sperm. Hum Reprod 2011;26:3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz K, Martínez-Soler F, Ausió J, Chiva M.. Acetylation of histone H4 in complex structural transitions of spermiogenic chromatin. J Cell Biochem 2007;102:1432–1441. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, Page DC.. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci USA 2002;99:8707–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Blondin P, Vigneault C, Labrecque R, Dufort I, Sirard M-A.. Spermatozoa DNA methylation patterns differ due to peripubertal age in bulls. Theriogenology 2018;106:21–29. [DOI] [PubMed] [Google Scholar]
- Laurentino S, Cremers J, Horsthemke B, Tüttelmann F, Czeloth K, Zitzmann M, Pohl E, Rahmann S, Schröder C, Berres S. et al. A germ cell‐specific ageing pattern in otherwise healthy men. Aging Cell 2020;19:e13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li H, Tian Y, Hu M, Le F, Wang L, Liu X, Jin F.. Sperm ribosomal DNA promoter methylation levels are correlated with paternal aging and may relate with in vitro fertilization outcomes. Front Genet 2020;11:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Genomic imprinting is a parental effect established in mammalian germ cells. Curr Top Dev Biol 2013;102:35–59. [DOI] [PubMed] [Google Scholar]
- Ma J, Chen Q, Wang S, Ma R, Jing J, Yang Y, Feng Y, Zou Z, Zhang Y, Ge X. et al. Mitochondria-related miR-574 reduces sperm ATP by targeting ND5 in aging males. Aging 2020;12:8321–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcho C, Oluwayiose OA, Pilsner JR.. The preconception environment and sperm epigenetics. Andrology 2020;8:924–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M.. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod 2008;14:67–74. [DOI] [PubMed] [Google Scholar]
- Marsidi AM, Kipling LM, Kawwass JF, Mehta A.. Influence of paternal age on assisted reproductive technology cycles and perinatal outcomes. Fertil Steril 2021;116:380–387. [DOI] [PubMed] [Google Scholar]
- Martin JA, Hamilton BE, Osterman MJK, Driscoll AK, Drake P.. Births: final Data for 2017. Natl Vital Stat Syst 2018;67:1–50. [PubMed] [Google Scholar]
- Martins RP, Krawetz SA.. Nuclear organization of the protamine locus. Soc Reprod Fertil Suppl 2007;64:1–12. [DOI] [PubMed] [Google Scholar]
- McCarter K, Setton R, Chung A, An A, Rosenwaks Z, Spandorfer S.. Is increasing paternal age negatively associated with donor oocyte recipient success? A paired analysis using sibling oocytes. Fertil Steril 2021;116:373–379. [DOI] [PubMed] [Google Scholar]
- Milekic MH, Xin Y, O'Donnell A, Kumar KK, Bradley-Moore M, Malaspina D, Moore H, Brunner D, Ge Y, Edwards J. et al. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry 2015;20:995–1001. [DOI] [PubMed] [Google Scholar]
- Morgan CP, Chan JC, Bale TL.. Driving the next generation: paternal lifetime experiences transmitted via extracellular vesicles and their small RNA Cargo. Biol Psychiatry 2019;85:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CP, Shetty AC, Chan JC, Berger DS, Ament SA, Epperson CN, Bale TL.. Repeated sampling facilitates within- and between-subject modeling of the human sperm transcriptome to identify dynamic and stress-responsive sncRNAs. Sci Rep 2020;10:17498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G, Mavrelos D, Odia R, Viñals Gonzalez X, Cawood S, Yasmin E, Saab W, Serhal P, Seshadri S.. Paternal age over 50 years decreases assisted reproductive technology (ART) success: a single UK center retrospective analysis. Acta Obstet Gynecol Scand 2021;100:1858–1867. [DOI] [PubMed] [Google Scholar]
- Ntostis P, Swanson G, Kokkali G, Iles D, Huntriss J, Pantou A, Tzetis M, Pantos K, Picton HM, Krawetz SA. et al. The effects of aging on molecular modulators of human embryo implantation. iScience 2021;24:102751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent D, Balen AH.. The effects of female age on fecundity and pregnancy outcome. Hum Fertil 2001;4:43–48. [DOI] [PubMed] [Google Scholar]
- Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B.. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci USA 2003;100:1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office for National Statistics. Conceptions in England and Wales: 2017. Great Britain: ONS, 2019. https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/conceptionandfertilityrates/bulletins/conceptionstatistics/2017 (22 August 2022, date last accessed). [Google Scholar]
- Oluwayiose OA, Wu H, Saddiki H, Whitcomb BW, Balzer LB, Brandon N, Suvorov A, Tayyab R, Sites CK, Hill L. et al. Sperm DNA methylation mediates the association of male age on reproductive outcomes among couples undergoing infertility treatment. Sci Rep 2021;11:3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Diamond MP, Dix DJ, Krawetz SA.. Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil Steril 2005;83:1687–1694. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J.. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000;10:475–478. [DOI] [PubMed] [Google Scholar]
- Paoli D, Pecora G, Pallotti F, Faja F, Pelloni M, Lenzi A, Lombardo F.. Cytological and molecular aspects of the ageing sperm. Hum Reprod 2019;34:218–227. [DOI] [PubMed] [Google Scholar]
- Pellissier LP, Gandía J, Laboute T, Becker JAJ, Le Merrer J.. μ opioid receptor, social behaviour and autism spectrum disorder: reward matters. Br J Pharmacol 2018;175:2750–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petok WD. Infertility counseling (or the lack thereof) of the forgotten male partner. Fertil Steril 2015;104:260–266. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Saddiki H, Whitcomb BW, Suvorov A, Buck Louis GM, Mumford SL, Schisterman EF, Oluwayiose OA, Balzer LB.. Sperm epigenetic clock associates with pregnancy outcomes in the general population. Hum Reprod Oxf Engl 2022;37:1581–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Shershebnev A, Wu H, Marcho C, Dribnokhodova O, Shtratnikova V, Sergeyev O, Suvorov A.. Aging-induced changes in sperm DNA methylation are modified by low dose of perinatal flame retardants. Epigenomics 2021;13:285–297. [DOI] [PubMed] [Google Scholar]
- Pohl E, Höffken V, Schlatt S, Kliesch S, Gromoll J, Wistuba J.. Ageing in men with normal spermatogenesis alters spermatogonial dynamics and nuclear morphology in Sertoli cells. Andrology 2019;7:827–839. [DOI] [PubMed] [Google Scholar]
- Potabattula R, Zacchini F, Ptak GE, Dittrich M, Müller T, Hajj NE, Hahn T, Drummer C, Behr R, Lucas Hahn A. et al. Increasing methylation of sperm rDNA and other repetitive elements in the aging male mammalian germline. Aging Cell 2020;19:e13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajender S, Avery K, Agarwal A.. Epigenetics, spermatogenesis and male infertility. Mutat Res 2011;727:62–71. [DOI] [PubMed] [Google Scholar]
- Rasmussen AH, Rasmussen HB, Silahtaroglu A.. The DLGAP family: neuronal expression, function and role in brain disorders. Mol Brain 2017;10:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F.. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006;441:469–474. [DOI] [PubMed] [Google Scholar]
- Richardson MC, Guo M, Fauser BCJM, Macklon NS.. Environmental and developmental origins of ovarian reserve. Hum Reprod Update 2014;20:353–369. [DOI] [PubMed] [Google Scholar]
- Robb GW, Amann RP, Killian GJ.. Daily sperm production and epididymal sperm reserves of pubertal and adult rats. J Reprod Fertil 1978;54:103–107. [DOI] [PubMed] [Google Scholar]
- Rosenwaks Z, Wassarman PM.. Human Fertility: Methods and Protocols. New York: Springer, 2014. https://books.google.ru/books?id=OGXonQEACAAJ (22 August 2022, date last accessed). [Google Scholar]
- Salonia A, Bettochi C, Carvalho J, Corona G, Jones TH, Kadioğlu A, Martinez-Salamanca JI, Minhas S, Serefoğlu EC, Verze P.. EAU guidelines on sexual and reproductive health 2020. In: European Association of Urology Guidelines, 2020 edn. presented at the EAU Annual Congress Amsterdam 2020 Arnhem, The Netherlands: European Association of Urology Guidelines Office, 2020.. https://uroweb.org/guideline/sexual-and-reproductive-health/ (22 August 2022, date last accessed). [Google Scholar]
- Samans B, Yang Y, Krebs S, Sarode GV, Blum H, Reichenbach M, Wolf E, Steger K, Dansranjavin T, Schagdarsurengin U.. Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev Cell 2014;30:23–35. [DOI] [PubMed] [Google Scholar]
- Santiago J, Silva JV, Howl J, Santos MAS, Fardilha M.. All you need to know about sperm RNAs. Hum Reprod Update 2021;28:67–91. [DOI] [PubMed] [Google Scholar]
- Schmidt L, Sobotka T, Bentzen JG, Nyboe Andersen A, ESHRE Reproduction and Society Task Force. Demographic and medical consequences of the postponement of parenthood. Hum Reprod Update 2012;18:29–43. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W.. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 2012;48:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellem E, Marthey S, Rau A, Jouneau L, Bonnet A, Le Danvic C, Guyonnet B, Kiefer H, Jammes H, Schibler L.. Dynamics of cattle sperm sncRNAs during maturation, from testis to ejaculated sperm. Epigenetics Chromatin 2021;14:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Shah PP, Nativio R, Berger SL.. Epigenetic mechanisms of longevity and aging. Cell 2016;166:822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendler E, Johnson GD, Mao S, Goodrich RJ, Diamond MP, Hauser R, Krawetz SA.. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res 2013;41:4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Agarwal A, Rohra VK, Assidi M, Abu-Elmagd M, Turki RF.. Effects of increased paternal age on sperm quality, reproductive outcome and associated epigenetic risks to offspring. Reprod Biol Endocrinol 2015;13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ.. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell 2018;46:481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A.. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012;484:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits RM, Xavier MJ, Oud MS, Astuti GDN, Meijerink AM, de Vries PF, Holt GS, Alobaidi BKS, Batty LE, Khazeeva G. et al. De novo mutations in children born after medical assisted reproduction. Hum Reprod 2022;37:1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares JP, Cortinhas A, Bento T, Leitão JC, Collins AR, Gaivão I, Mota MP.. Aging and DNA damage in humans: a meta‐analysis study. Aging 2014;6:432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohni A, Tan K, Song H-W, Burow D, de Rooij DG, Laurent L, Hsieh T-C, Rabah R, Hammoud SS, Vicini E. et al. The neonatal and adult human testis defined at the single-cell level. Cell Rep 2019;26:1501–1517.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A. et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 2013;3:2179–2190. [DOI] [PubMed] [Google Scholar]
- Stein R, , GruenbaumY, , PollackY, , RazinA, , Cedar H.. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci U S A 1982;79:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujit KM, Sarkar S, Singh V, Pandey R, Agrawal NK, Trivedi S, Singh K, Gupta G, Rajender S.. Genome-wide differential methylation analyses identifies methylation signatures of male infertility. Hum Reprod 2018;33:2256–2267. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P.. Genetic and epigenetic regulators of pluripotency. Cell 2007;128:747–762. [DOI] [PubMed] [Google Scholar]
- Suvorov A, Pilsner JR, Naumov V, Shtratnikova V, Zheludkevich A, Gerasimov E, Logacheva M, Sergeyev O.. Aging induces profound changes in sncRNA in rat sperm and these changes are modified by perinatal exposure to environmental flame retardant. Int J Mol Sci 2020;21:8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Kobayashi E, Nishino K, Imai A, Adachi H, Hoshino Y, Iwao K, Akagi S, Kaneda M, Watanabe S.. Age-related changes in DNA methylation levels at CpG sites in bull spermatozoa and in vitro fertilization-derived blastocyst-stage embryos revealed by combined bisulfite restriction analysis. J Reprod Dev 2019;65:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA.. Specification and epigenetic programming of the human germ line. Nat Rev Genet 2016;17:585–600. [DOI] [PubMed] [Google Scholar]
- Tatehana M, Kimura R, Mochizuki K, Inada H, Osumi N.. Comprehensive histochemical profiles of histone modification in male germline cells during meiosis and spermiogenesis: comparison of young and aged testes in mice. PLoS One 2020;15:e0230930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollner TL, Venners SA, Hollox EJ, Yudin AI, Liu X, Tang G, Xing H, Kays RJ, Lau T, Overstreet JW. et al. A common mutation in the defensin DEFB126 causes impaired sperm function and subfertility. Sci Transl Med 2011;3:92ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Tucci V, Bartolomei MS, Benvenisty N, Bourc’his D, Charalambous M, Dulac C, Erice Imprinting Group et al. Genomic imprinting and physiological processes in mammals. Cell 2019;176:952–965. [DOI] [PubMed] [Google Scholar]
- Turner KA, Rambhatla A, Schon S, Agarwal A, Krawetz SA, Dupree JM, Avidor-Reiss T.. Male infertility is a women’s health issue-research and clinical evaluation of male infertility is needed. Cells 2020;9:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan MJ, Polley EC, Hubbard AE.. Super learner. Stat Appl Genet Mol Biol 2007;6:Article 25. [DOI] [PubMed] [Google Scholar]
- Van Opstal J, Fieuws S, Spiessens C, Soubry A.. Male age interferes with embryo growth in IVF treatment. Hum Reprod 2021;36:107–115. [DOI] [PubMed] [Google Scholar]
- Vrooman LA, Bartolomei MS.. Can assisted reproductive technologies cause adult-onset disease? Evidence from human and mouse. Reprod Toxicol 2017;68:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen Z-P, Hu H, Lei J, Zhou Z, Yao B, Chen L, Liang G, Zhan S, Zhu X. et al. Sperm microRNAs confer depression susceptibility to offspring. Sci Adv 2021;7:eabd7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselink AK, Rothman KJ, Hatch EE, Mikkelsen EM, Sørensen HT, Wise LA.. Age and fecundability in a North American preconception cohort study. Am J Obstet Gynecol 2017;217:667.e1-667–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M, , LevyD, , Perucho M.. The somatic replication of DNA methylation. Cell 1981;24:33–40. [DOI] [PubMed] [Google Scholar]
- Wu C, Blondin P, Vigneault C, Labrecque R, Sirard M-A.. The age of the bull influences the transcriptome and epigenome of blastocysts produced by IVF. Theriogenology 2020a;144:122–131. [DOI] [PubMed] [Google Scholar]
- Wu C, Blondin P, Vigneault C, Labrecque R, Sirard M-A.. Sperm miRNAs-potential mediators of bull age and early embryo development. BMC Genomics 2020b;21:798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA.. The structural organization of sperm chromatin. J Biol Chem 2003;278:29471–29477. [DOI] [PubMed] [Google Scholar]
- Wyrobek AJ, Eskenazi B, Young S, Arnheim N, Tiemann-Boege I, Jabs EW, Glaser RL, Pearson FS, Evenson D.. Advancing age has differential effects on DNA damage, chromatin integrity, gene mutations, and aneuploidies in sperm. Proc Natl Acad Sci USA 2006;103:9601–9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier MJ, Roman SD, Aitken RJ, Nixon B.. Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health. Hum Reprod Update 2019;25:519–541. [DOI] [PubMed] [Google Scholar]
- Xiao F-H, Wang H-T, Kong Q-P.. Dynamic DNA methylation during aging: a “prophet” of age-related outcomes. Front Genet 2019;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Ryan DP, Pearson BL, Henzel KS, Neff F, Vidal RO, Hennion M, Lehmann I, Schleif M, Schröder S. et al. Epigenetic alterations in longevity regulators, reduced life span, and exacerbated aging-related pathology in old father offspring mice. Proc Natl Acad Sci USA 2018;115:E2348–E2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko AN, Turek PJ.. Reproductive genetics and the aging male. J Assist Reprod Genet 2018;35:933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki K, Kimura R, Kobayashi H, Oki S, Kikkawa T, Mai L, Koike K, Mochizuki K, Inada H, Matsui Y. et al. Paternal age affects offspring via an epigenetic mechanism involving REST/NRSF. EMBO Rep 2021;22:e51524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanato VF, Martins MP, Anselmo-Franci JA, Petenusci SO, Lamano-Carvalho TL.. Sexual development of male Wistar rats. Braz J Med Biol Res 1994;27:1273–1280. [PubMed] [Google Scholar]
- Zhang Y, Shi J, Rassoulzadegan M, Tuorto F, Chen Q.. Sperm RNA code programmes the metabolic health of offspring. Nat Rev Endocrinol 2019;15:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W-L, Gu N-H, Li Z-Z, Wang G-S, Cheng CY, Sun F.. Autism-like behaviors and abnormality of glucose metabolism in offspring derived from aging males with epigenetically modified sperm. Aging 2020;12:19766–19784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of our conclusions are available from the corresponding author upon reasonable request.