Abstract
Background
The role of intestinal microbiota in inflammatory bowel diseases is intensively researched. Pediatric studies on the relation between microbiota and treatment response are sparse. We aimed to determine whether absolute abundances of gut microbes characterize the response to infliximab induction in pediatric inflammatory bowel disease.
Methods
We recruited pediatric patients with inflammatory bowel disease introduced to infliximab at Children’s Hospital, University of Helsinki. Stool samples were collected at 0, 2, and 6 weeks for microbiota and calprotectin analyses. We defined treatment response as fecal calprotectin value <100 µg/g at week 6. Intestinal microbiota were analyzed by 16S ribosomal RNA gene amplicon sequencing using the Illumina MiSeq platform. We analyzed total bacterial counts using quantitative polymerase chain reaction and transformed the relative abundances into absolute abundances based on the total counts.
Results
At baseline, the intestinal microbiota in the treatment responsive group (n = 10) showed a higher absolute abundance of Bifidobacteriales and a lower absolute abundance of Actinomycetales than nonresponders (n = 19). The level of inflammation according to fecal calprotectin showed no statistically significant association with the absolute abundances of fecal microbiota. The results on relative abundances differed from the absolute abundances. At the genus level, the responders had an increased relative abundance of Anaerosporobacter but a reduced relative abundance of Parasutterella at baseline.
Conclusions
High absolute abundance of Bifidobacteriales in the gut microbiota of pediatric patients reflects anti-inflammatory characteristics associated with rapid response to therapy. This warrants further studies on whether modification of pretreatment microbiota might improve the outcomes.
Keywords: children, Crohn’s disease, ulcerative colitis
Key Messages.
What is already known?
- Therapeutic response to a tumor necrosis α antagonist in pediatric inflammatory bowel disease associates with the pretreatment gut microbiota composition.
What is new here?
- We conducted the first study to examine the quantitative—instead of relative—fecal microbiota composition in patients with pediatric inflammatory bowel disease introduced to tumor necrosis α antagonist infliximab.
How can this study help patient care?
- Increased understanding of the association of gut microbiota composition and therapeutic response may lead to improved care in the future.
Introduction
The incidence of inflammatory bowel diseases (IBDs), comprising ulcerative colitis (UC), Crohn’s disease (CD), and IBD–unclassified (IBD-U), is rising steadily across the globe.1 A systematic review including 139 studies in 32 countries found globally a more consistent increase in the incidence of pediatric CD than that of UC.2 Thus, pediatric IBD (PIBD) is an increasing health care burden.
PIBD presents with similar symptoms at diagnosis as adult IBD, which may include diarrhea, abdominal pain, bloody stools, and fever. The disease, however, is more extensive and its course more severe in PIBD, which is reflected on the higher need of surgeries compared with the adult patients.3,4 Additionally, PIBD (especially CD) may cause malnourishment, leading to growth impairment, delayed puberty, and even psychosocial problems.3,5,6 Therefore, to avoid long-term sequelae, effective treatments are especially important among pediatric patients.
The etiology and the precise pathogenesis behind PIBD remain unclear. Up to 200 genes potentially associated with IBD have been indicated in large, genome-wide studies.7 Several environmental risk factors for IBD have been identified, including a family history of IBD and urban living.8 Protective factors include breastfeeding, large family size, early-life exposure to pets, and farm animals.9 Many of these may influence the development of the immune system directly or via changes in the host–microbe interactions. The current hypothesis for the pathogenesis of IBD is that the combination of genetic, environmental, and microbial factors leads to altered permeability of the gut, the alteration of intestinal microbiota composition with predominance of pathobionts that contribute to inflammation and prolonged immune responses.5
Anti-tumor necrosis factor α (anti-TNF) treatment has proven effective in PIBD.5 Infliximab and adalimumab are the anti-TNF antibodies approved for pediatric patients by the Food and Drug Administration and the European Medicines Agency.5 In a guideline by European Crohn’s and Colitis Organization and European Society of Paediatric Gastroenterology, Hepatology and Nutrition for pediatric CD, anti-TNF therapy was the first-line treatment for children with high risk of poor outcome, children with fistulizing perianal disease, if severe growth delay is present or if therapy with corticosteroids and dietary therapy with exclusive enteral nutrition do not induce remission.10 The European Crohn’s and Colitis Organization/European Society of Paediatric Gastroenterology, Hepatology and Nutrition guideline for pediatric UC recommended anti-TNF treatment for steroid-dependent or continuously active disease, if 5-aminosalicylic acid and thiopurines have failed.11 Although most patients benefit first from anti-TNF therapy, secondary loss of response happens often; in a recent study on PIBD patients treated with anti-TNF medication, up to 83% of patients needed therapy enhancement.12
The gut microbiota in IBD are characterized by aberrant composition and decreased bacterial diversity.13 Additionally, there are reports on specific microbial signatures associating with IBD, including a decreased abundance of anti-inflammatory bacteria, including taxa belonging to Roseburia, Bifidobacterium, and Faecalibacterium.14,15 We previously examined the gut microbiota in relation to the induction of infliximab in a few patients with PIBD, discovering several bacteria associating with the treatment response. Of note, a high abundance of bacteria related to Eubacterium rectale, a known butyrate-producer, and Bifidobacterium spp. known to have anti-inflammatory properties, at introduction of the therapy predicted attenuation of inflammation.16 Another previous study linked sustaining response to anti-TNF in PIBD with an increase in Blautia and Lachnospira spp. as well as several known butyrate producers, such as Faecalibacterium and Roseburia in postinfliximab samples.17 Additionally, a PIBD study reported that response to infliximab associated with the evolution of microbiota closer to healthy control subjects.18 A recent review regarding gut microbiota profiles in PIBD, encompassing 41 studies, reported a gain in Enterococcus and significant decrease in Anaerostipes, Blautia, Coprococcus, Faecalibacterium, Roseburia, Ruminococcus, and Lachnospira in PIBD compared with healthy control subjects, and a decrease in microbial diversity within majority of the included articles.19
Previous reports on microbiota and therapeutic response are based on assessment of relative abundances of the most prevalent bacteria. The relative abundance of a particular microbe is influenced by abundances of other microbe(s), as it is evident that when the relative abundance of one microbe increases, the relative abundance of another one decreases. This bias could be tackled by analyzing the absolute abundances rather than relative ones. To our knowledge, there are no quantitative data on microbiota related to response to a given therapy in PIBD. The aim of this study was to determine whether the absolute profiles of intestinal microbiota characterize the response to infliximab in PIBD. We hypothesized that there would be substantial differences in bacterial profiles between treatment response groups at baseline.
Methods
Study design
PIBD patients introduced to infliximab at Children’s Hospital, University of Helsinki were recruited to the study between March 21, 2014, and March 2, 2016. The participants were requested to take a stool sample for the study purpose at 0, 2, and 6 weeks at the same time when the routine sample for fecal calprotectin measurement was obtained. Samples were immediately frozen at home in -20°C and transported to the laboratory for storage in -80°C until processing.
Clinical disease activity was routinely assessed with a validated tool including a symptom score and a visual analog scale.20 The symptom score contains 5 questions (scale 0-15 points, with a higher score being worse) regarding general well-being, abdominal pain, nocturnal and daily bowel movements, and the presence of blood in the stools.20 Treatment outcome was assessed in chart review. All decisions regarding management were made at the physicians’ discretion.
We evaluated disease activity using fecal calprotectin as a marker of inflammation. A cutoff of <100 µg/g defines a low level of inflammation.21 The cutoff value of <100 µg/g has a specificity of 0.92 and negative prediction value of 95% to predict deep healing.22 The fecal calprotectin level was measured in a routine clinical laboratory using a quantitative enzyme immunoassay (PhiCal Test; Calpro AS, Oslo, Norway). This level of calprotectin can be considered deep biochemical remission. The patients who reached final calprotectin <100 µg/g were considered the group with remission, and patients with final calprotectin ≥100 µg/g were considered the group with no remission.
DNA extraction and preparation sequencing libraries
Fecal samples were analyzed for microbiota composition by 16S ribosomal RNA (rRNA) amplicon sequencing. DNA was extracted from Circa 250 mg of fecal samples using a repeated bead-beating method with the following modification for automated DNA purification: 250 μL of the lysates was transferred to a 96-well deep plate and stored on ice. The cell pellet was subjected for second round of beat beating after addition of 340 μL of fresh lysis buffer. The pooled supernatants were then heated at +95°C for 15 minutes with shaking at 400 rpm, centrifuged at room temperature for 5 minutes at 13 000 rpm, and 200 μL of the clarified supernatant was used for DNA extraction with KingFisher Flex automated purification system (Thermo Fisher Scientific, Waltham, MA, USA) and Ambion Magma Total Nucleic Acid Isolation Kit (Life Technologies, Carlsbad, CA, USA) using the MagMAX Pathogen High Vol Duo program (Thermo Fisher Scientific). DNA was quantified using Quanti-iT Pico Green dsDNA Assay (Invitrogen, San Diego, CA, USA).
The samples were subjected to 16S rRNA gene V3-V4 amplicon sequencing (primers 341F/785R) with Illumina MiSeq (BaseClear, Leiden, the Netherlands). A few samples (a total of 11 out of 80) failed, and those samples were reanalyzed with the same primers at the Biomedicum Functional Genomics Unit, University of Helsinki, Finland. After reanalyzing the failed samples, all samples met the threshold of 10 000 reads (median 28 106 [range, 14 194-73 132]). The reanalyzed samples were evenly distributed within the treatment response groups (Table 1).
Table 1.
Characteristics of the study population of patients with pediatric IBD introduced to infliximab
| Baseline Characteristic | Remission (n = 10) | No Remission (n = 19) | Total (n = 29) | P Value |
|---|---|---|---|---|
| Male | 7 (70) | 9 (47) | 16 (55) | .43 |
| Age, y | 14 (9.4-18) | 14 (6.3-20) | 14 (6.3-20) | .82 |
| Disease duration, y | 1.5 (0.09-3.8) | 1.7(0-8.4) | 1.7 (0-8.4) | .96 |
| <1 y | 2 (20) | 8 (42) | 10 (34) | .41 |
| Crohn’s disease | 7 (70) | 10 (53) | 17 (59) | .44 |
| Ileal (L1) | 1 (10) | 0 (0) | 1 (3) | .34 |
| Colonic(L2) | 2 (20) | 2 (11) | 4 (14) | .59 |
| Ileocolonic (L3) | 4 (40) | 7 (37) | 11 (38) | 1 |
| Other (L4BP) | 0 | 1 (5) | 1 (3) | .59 |
| UC/IBD-U | 3 (30) | 9 (47) | 12 (41) | .45 |
| UC | 1 (10) | 5 (26) | 6 (21) | .63 |
| Extensive colitis (E3) | 0 (0) | 2 (11) | 2 (7) | .53 |
| Pancolitis (E4) | 1 (10) | 3 (16) | 4 (14) | 1 |
| IBD-U | 2 (20) | 4 (21) | 6 (21) | 1 |
| Fecal calprotectin, µg/g | 326 (38-1317) | 1038 (186-6293) | 915 (38–6293) | .002a |
| Elevated C-reactive protein (≥4 mg/L) | 2 (20) | 10 (53) | 12(41) | .13 |
| Hemoglobin, g/L | 134 (98-145) | 118 (94-139) | 119 (94–145) | .27 |
| Erythrocyte sedimentation rate, mm/h | 8.5 (2-33) | 18 (5-44) | 15 (2–44) | .052 |
| Serum albumin, g/L | 37 (33-43) | 35 (30-37) | 35 (30–43) | .005a |
| Serum albumin below age-adjusted reference value | 2 (20) | 13 (68) | 15 (52) | .021a |
| Previously treated with IFX | 2 (20) | 3 (16) | 5 (17) | 1 |
| Days since last IFX treatment | 788 (777-799) | 1008 (497–1335) | 799 (497–1335) | 1 |
| Previous IBD surgery | 1 (10) | 3 (16) | 4 (14) | 1 |
| Sample analyzed in BaseClear | 45 (87) | 24 (86) | 69 (86) | 1 |
| Concomitant medication | ||||
| Azathioprine | 6 (60) | 6 (32) | 12 (41) | .23 |
| Glucocorticoid | 6 (60) | 10 (53) | 15 (51) | 1 |
| Antibioticsb | 4 (40) | 8 (42) | 12 (41) | 1 |
| 5-aminosalicylic acid | 4 (40) | 6 (32) | 10 (35) | .69 |
| Methotrexate | 1 (10) | 0 (0) | 1 (3) | .34 |
| Cyclosporine | 1 (10) | 0 (0) | 1 (3) | .34 |
| Lactic acid bacteria supplement (on a weekly basis) | 2 (20) | 2 (11) | 4 (14) | .59 |
Values are n (%) or mean (range).
Abbreviations: IBD, inflammatory bowel disease; IBD-U, inflammatory bowel disease–unclassified; IFX, infliximab; UC, ulcerative colitis.
statistically significant at p < .05.
3 metronidazole, 2 amoxicillin + doxycycline + metronidazole, 1 cefuroxime + metronidazole, 1 ciprofloxacin, 1 metronidazole, 1 amoxicillin + metronidazole, 1 amoxicillin, 1 cefuroxime + metronidazole, and 1 unknown.
Statistical analysis
Analysis of the 16S rRNA gene amplicon sequence data was conducted in R software (version 1.3.1093) (R Foundation for Statistical Computing, Vienna, Austria) using the package mare.23 Preprocessing, quality filtering, and taxonomic annotation of the reads was performed using USEARCH24 by mapping the reads to the SILVA 16S rRNA reference database version 115,25 restricted to gut-associated taxa (available through the R package mare).23 Only high-quality forward reads trimmed to 150 nt were used for analysis. Comparative analyses for the differences in the relative and relative absolute abundances of common bacteria genera, families, order, class, and phyla were performed with mare functions “GroupTest,” “CovariateTest,” and “ChangeTest” that implement generalized linear models using negative binomial distribution from MASS,26 with tools from packages vegan,27 MASS,26 and nlme.28
Quantification of total bacteria was performed with quantitative polymerase chain reaction using the universal primers and assay conditions described previously.29 This method reflects the total bacterial abundances as reliably as taxon-specific quantitative polymerase chain reaction.29 The relative bacterial abundances were translated into relative absolute abundances by multiplying with the total bacterial count in the sample and correcting for differences in 16S rRNA gene copy numbers.29 This “relative absolute abundance” is shortened into “absolute abundance” in the text for the sake of clarity.
All categorical variables between the group with remission and the group with no remission were tested for statistical significance using Fisher’s exact test and numerical variables using the Mann-Whitney U test (2-tailed) (Table 1). We also tested for the association between intestinal inflammation (as defined with fecal calprotectin) and microbiota composition. All analyses were adjusted for the sequencing place (BaseClear or FuGu), type of IBD (CD, UC/IBD-U), sex, ongoing corticosteroid treatment, and antibiotics within 1 month prior to sample. In all analyses, 0.1 was used as minimum prevalence and 0.01 as minimum relative abundance, analyzing only taxa that exceeded these cutoffs. P values <.05 were considered significant. The diversity was calculated as the inverse Simpson diversity index and richness as the number of operational taxonomic units.
Results
Patient characteristics and response to infliximab
A total of 30 patients with PIBD introduced to infliximab were sampled for fecal calprotectin and gut microbiota at 0, 2, and 6 weeks. One patient was excluded due to missing calprotectin values. One patient did not return the 2-week sample, and 6 patients did not provide the 6-week samples (Supplementary Figure 1). Out of the remaining 29 patients, 10 achieved remission and 19 did not achieve remission. A total of 17 were diagnosed with CD, 6 were diagnosed with UC, and 6 were diagnosed with IBD-U. Infliximab induction therapy was started to 14 patients due to extensive disease (CD = 11, IBD-U = 3) and in 15 patients because oral medication had failed (CD = 6, UC = 6, IBD-U = 3). The given dose of infliximab ranged from 4.5 to 5.5 mg/kg, and 1 patient received 7.6 mg/kg. All background characteristics according to treatment response are listed in Table 1.
Fecal microbiota composition
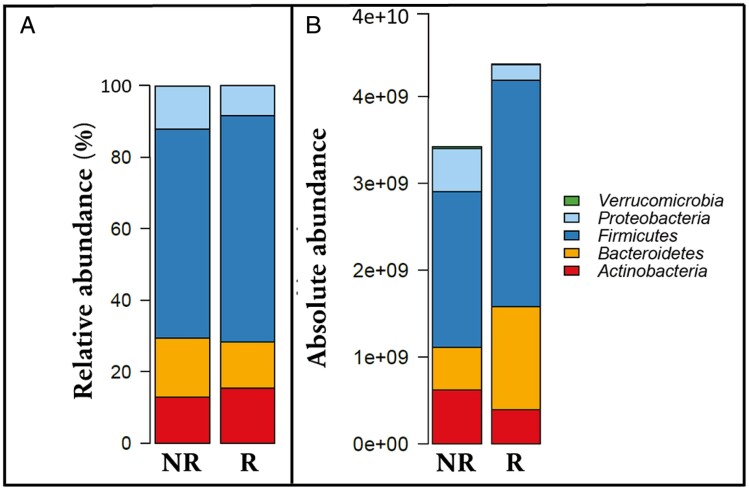
The most abundant phyla across all samples were Firmicutes (68%), Bacteroidetes (8.4%), Actinobacteria (7.0%), and Proteobacteria (1.3%). Taxa in the phyla Fusobacteria and Verrucomicrobia were less abundant and almost absent from the samples except 2 (relative abundances 8.0% and 13%). In all other samples, the 4 major phyla—Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria—accounted for over 95% of all reads (Figure 1). Similar percentages were calculated from the total copy number–corrected abundances: Firmicutes (median 77%), Bacteroidetes (median 11%), Actinobacteria (median 10%), and Proteobacteria (median 2%).
Figure 1.
A, The relative abundance of the most abundant phyla in all samples according to treatment response. B, The absolute abundance of the most abundant phyla in all samples according to treatment response (logarithmic scale), as the number of cells. NR, no remission; R, remission.
The microbial diversity varied from 1.1 to 22 (median 6.6) and richness varied from 30 to 126 (median 72).
Response to infliximab
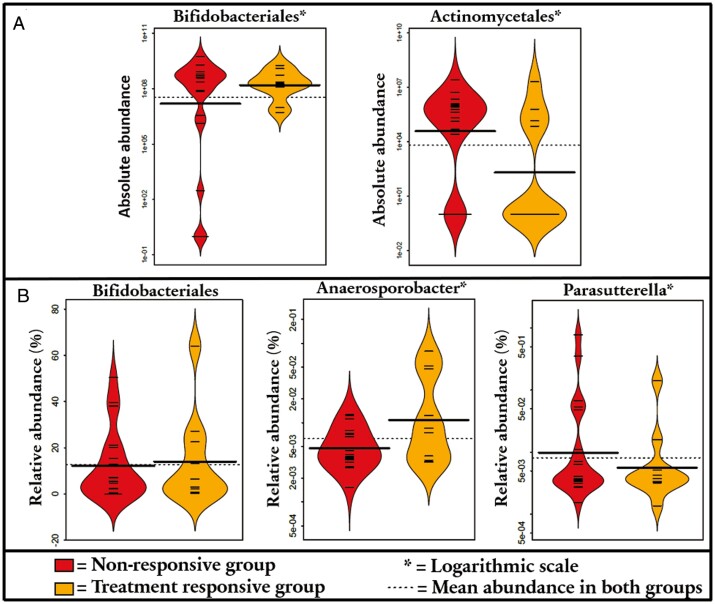
Regarding absolute data and infliximab response, we discovered that the remission group had an increased abundance of order Bifidobacteriales (1.7-fold; P = .039) and a reduced abundance of order Actinomycetales (3.4-fold; P = .018) at baseline (Figure 2A). Additionally, 1 species of genus Actinomyces was increased the no remission group at baseline. All differences between groups from baseline to 2- and 6-week samples are listed in Supplementary Table 1.
Figure 2.
A, Order-level differences between treatment response groups: absolute abundance. B, Order- and genus-level differences between treatment response groups: relative abundance.
In the relative abundance analysis, the remission group had an increased abundance of order Bifidobacteriales (by 2.2-fold; P = .026) at baseline, but there was no significant finding related to Actinomycetales, unlike with absolute abundances (see previous). On the other hand, on the genus level, the remission group had an increased relative abundance of Anaerosporobacter (15-fold; P = .0013) at baseline, whereas the no remission group had a significant increase in the relative abundance of Parasutterella (144-fold; P < .001) at baseline (Figure 2B). All differences between groups on other taxonomic levels and changes in the microbiota on 2- and 6-week samples are listed in Supplementary Table 2.
Regarding the differences in the changes of microbiota during treatment, we discovered that at 2 weeks the absolute abundances of the genera Phascolarctobacterium (difference between remission group and no remission group: 4 700 000 genomes; P = .048) and Morganella (difference between remission group and no remission group: 1 100 000 genomes; P = .024) increased in the remission group, whereas they decreased in the no remission group. At 6 weeks, Anaerosporobacter (difference between remission group and no remission group: 100 000 reads; P = .0062) increased more in the remission group. Changes of other taxonomic levels samples are listed in Supplementary Table 3.
Regarding changes of relative abundances during treatment, the findings differed from the absolute counts. At 2 weeks, the class Negativicutes increased more in the no remission group than in the remission group (fold change = 0.053; P = .032), as did order Selomonadales (fold change = 0.053; P = .032), family Veillonellaceae (fold change = 0.058; P = .012), and genus Anaerostipes (fold change = 0.006; P = .0049). At 6 weeks, the genus Streptococcus was increased in the remission group, whereas it was decreased in the no remission group (fold change = -0.32; P = .005). Changes of other taxonomic levels are listed in Supplementary Table 4.
The bacterial richness or diversity did not differ statistically significantly at baseline, 2 weeks, or 6 weeks between the treatment response groups.
Disease subtypes
Next, we examined whether disease subtype or the use of antibiotics or steroids had an impact on the microbiota at baseline. When examining absolute bacterial abundances at baseline, we found only 1 minor species belonging to genus Lachnospira to be increased in CD when compared with UC and IBD-U (Supplementary Table 5). Regarding the relative abundances (Supplementary Table 6), patients with UC had an increase in genera Barnesiella (14-fold; P = .042) and Anaerotruncus (14-fold; P = .030) and a decrease in Catenibacterium (7.0-fold; P = .032) at baseline. None of these bacteria associated with therapeutic response to infliximab.
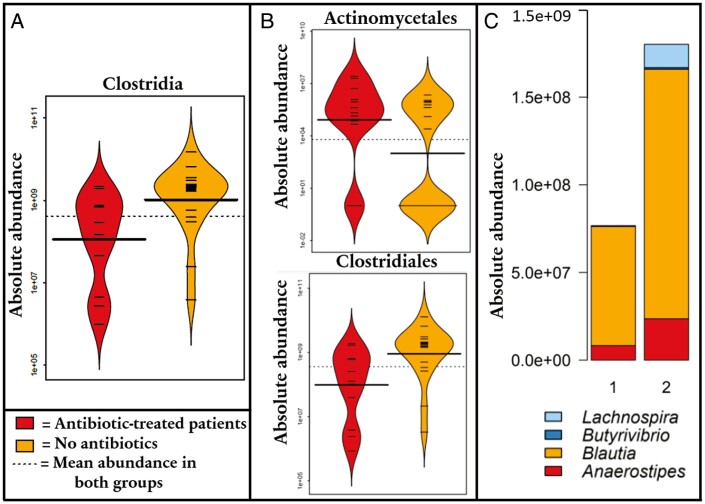
Antibiotics
We observed that the patients who had received antibiotics during the month prior to introducing infliximab had a decreased absolute abundance of class Clostridia (8.3-fold; P = .022) and order Clostridiales (8.4-fold; P = .022) but significantly increased abundances of orders Actinomycetales (120-fold; P = .042) and Bacillales (130-fold; P = .049) at baseline (Figure 3A, 3B). At the genus level, patients who received antibiotics had decreased abundances of Anaerostipes (33-fold; P = .0055), Blautia (20-fold; P = .043), Butyrivibrio (120-fold; P = .048), and Lachnospira (620-fold; P = .015) (Figure 3C). All differences of other taxonomic levels are displayed in Supplementary Table 7. However, these findings showed no significant association with the response to infliximab.
Figure 3.
A, Difference in absolute abundance of class Clostridia in antibiotic-treated patients and patients not receiving antibiotics. B, Order-level differences of absolute abundances between antibiotic-treated patients and patients not receiving antibiotics (the Bacillales order was too sparse to be made into a graph). C, Boxplot depicting the genus-level differences of absolute abundances between antibiotic-treated patients and patients not receiving antibiotics.
Regarding relative data, there were several significant findings that were not found in absolute data. The antibiotic-treated patients had a reduced relative abundance of classes Clostridia (2.1-fold; P = .0088), Verrucomicrobiae (34-fold; P = .027), and Negativicutes (1.6-fold; P = .015) and orders Clostridiales (2.1-fold; P = .0088), Selomonadales (1.6-fold; P = .015), and Verrucomicrobiales (34-fold; P = .027) but an increased relative abundance of Actinomycetales (4.5-fold; P = .0080) at baseline. At the genus level, antibiotics-treated patients had a decreased relative abundance of Akkermansia (34-fold; P = .027), Barnesiella (80-fold; P = .0006), Parabacteroides (13-fold; P = .022), uncultured Christensenellaceae (9.0-fold; P = .026), Anaerosporobacter (6.0-fold; P = .042), Coprococcus (6.5-fold; P = .034), and Catenibacterium (7.5-fold; P = .021) but an increased relative abundance of Actinomyces (4.5-fold; P = .0080). Taxa of other taxonomic levels are listed in Supplementary Table 8. However, none of these findings associated with analyses on absolute abundances and response to infliximab.
Systemic steroid
We discovered that the patients who were on steroids had an increased absolute abundance of order Pasteurellales (30-fold; P = .0021) at baseline. Taxa of other taxonomic levels are listed in Supplementary Table 9. When observing relative abundances, these patients had an increase in class Negativicutes (4.2-fold; P = .0014), orders Selenomonadales (4.2-fold; P = .0014) and Pasteurellales (6.9-fold; P = .014) and genera Anaerosporobacter (8.5-fold; P = .021) and Haemophilus (6.9-fold; P = .014) at baseline when compared with patients not receiving oral steroid treatment. Results of other taxonomic levels are listed in Supplementary Table 8. However, none of these findings associated with analyses on absolute abundances and response to infliximab.
The effect of inflammation
When analyzing absolute abundances, we gained no statistically significant results associating with the level of fecal calprotectin (analyzed as a continuous variable) in any taxonomic level. In the relative abundance analysis, class Erysipelotrichia decreased as the level of inflammation progressed (slope = -0.00087; P = .013). At the family level, Erysipelotrichiae (slope = -0.00087; P = .013) and Acidaminococcaceae (slope = -0.0051; P = .024) decreased as inflammation progressed, whereas Enterobacteriaceae increased along with inflammation (slope = 0.0013; P = .033). At the genus level, the relative abundance of Faecalibacterium (slope = = -0.0017; P = .008) and Blautia (slope = -0.00097; P = .0004) decreased as inflammation progressed. All data are presented in Supplementary Table 11. None of these bacteria associated with therapeutic response to infliximab.
Discussion
We examined the microbiota of patients with PIBD introduced to induction therapy with infliximab assessing the relative and relative absolute data of the gut microbiota. To our knowledge, this is the first study regarding absolute abundances of fecal microbiota in relation to response to infliximab induction on a pediatric cohort. Treatment response was defined by low level of fecal calprotectin (<100 µg/g) mirroring deep healing.22 In absolute analyses, we report bacteria associating with response to infliximab. Importantly, the results in absolute and relative microbiota analyses differed to some extent. This difference is explained by the influence of microbial density, eg, total bacterial loads, and its considerable variability between individuals.30
The phyla Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria composed over 95% of the microbiota in 98% of the samples. This is in line with previous studies on adults, which have reported these 4 phyla to account for 95% to 99% of all reads.31,32
Total bacterial counts in the remission group indicated an increased absolute abundance of order Bifidobacteriales and a reduced absolute abundance of order Actinomycetales when compared with the no remission group at baseline. In relative abundance analyses, order Bifidobacteriales was also more prominent in the remission group. These findings are in line with our previous study showing an increased relative abundance of Bifidobacterium in the remission group to infliximab.16 When compared with healthy control subjects, Wang et al17 reported a reduced relative abundance of family Bifidobacteriaceae and an increased relative abundance of the family Actinomycetaceae in pediatric patients with CD. It has been speculated that the main metabolic pathways of Bifidobacterium spp. are involved in decreasing inflammation by contributing to intestinal cell renewal, participating in building an epithelial barrier against pathological organisms by adhering to the mucosa, and producing short-chain fatty acids, which are important for enterocyte homeostasis.14,33Actinomyces spp. are facultatively pathogenic commensals that in some cases have been implied in intestinal diseases, while they are known to produce antimicrobial compounds that may affect the gut microbiota and its activity.34 Its direct effects on the gut immune system are not well characterized. Taken together, our previous and current findings show that the response to infliximab is associated with an intestinal microbiota closer to that in healthy control subjects. This emphasizes the role of intestinal microbiota in reaching remission.
The remission group had a lower level of fecal calprotectin and a higher level of serum albumin at the start of treatment, indicating toward milder disease. Therefore, it is rational that the remission group had more beneficial microbiota at the start of treatment. However, most patients within the remission group had an elevated level of fecal calprotectin at the baseline (70%), showing active inflammation. Intriguingly, the remission group also had slightly higher concentration of baseline serum albumin. This could imply that the no remission group had increased loss of albumin and other proteins, such as infliximab the intestine. Previously, low blood albumin concentrations correlated with low anti-TNF levels in blood during induction therapy in CD.5 The higher abundance of Bifidobacteriales could indeed play a role in the integrity of the epithelial barrier that would result in less loss of albumin and infliximab.14
The absolute abundance of Phascolarctobacterium and Morganella increased in the remission group but decreased in the no remission group 2 weeks after the first dose of infliximab. At week 6, the absolute abundance of Anaerosporobacter and the relative abundances of several bacteria varied between the study groups, but unexpectedly the differences were so small that the impact to the outcome of the treatment is unlikely. However, the absolute increase in Phascolarctobacterium in the remission group was toward the microbiota profile in healthy peers.35 This gut commensal is known to produce propionate from succinate.36
We discovered that the patients treated with antibiotics prior to infliximab induction had a lower absolute and relative abundance of Clostridia and Clostridiales compared with non–antibiotic-treated patients, but these findings did not associate with the response to infliximab. The absolute and relative abundance of order Actinomycetales and the relative abundance of Actinomyces was increased in antibiotic-treated patients. Previously, the relative abundances of Clostridiales and Clostridiaceae have been reported to be decreased and Actinomycetaceae and Actinomyces increased in pediatric CD when compared with healthy control subjects.17,37 These results are expected, as the antibiotics used largely target Gram-positive anaerobic bacteria, of which Clostridia represent the dominant group.
Furthermore, the antibiotic-treated patients had a reduced absolute abundance of the dominant and anti-inflammatory Clostridial taxa Anaerostipes, Blautia, Butyrivibrio, and Lachnospira paralleling published data on their decreased relative abundances (except Lachnospira) in pediatric CD when compared with healthy control subjects.16,17,37 Regarding relative abundances, the genera Akkermansia, Parabacteroides, Coprococcus, and Catenibacterium were decreased in antibiotic-treated patients, while all of these have been reported reduced in PIBD.18,37–39 Our results provide further evidence of the importance of antibiotics when assessing dysbiosis in IBD. However, we did not find any of these genera to predict therapeutic response to infliximab.
Regarding concomitant medication, we assessed the association of corticosteroid use to the microbiota profiles but found no predictive markers for response to infliximab. Patients on corticosteroids at baseline had an increased absolute abundance of Pasteurellales when compared with patients who were off steroids, but this was not detected when studying relative abundances. On the other hand, in relative abundances but not in absolute abundances, the steroid-treated patients had an increase in Negativicutes and Haemophilus. Negativicutes has been reported increased in adult IBD, and Haemophilus in pediatric CD when compared with healthy control subjects.37,40 Based on these observations, the steroid-treated patients seem to have more aberrant microbiota, further away from healthy peers mirroring the overall severity of their disease.
When examining the effect of inflammation on microbiota (ie, association with levels of fecal calprotectin), we gained no statistically significant results in absolute abundances. This was unexpected, as the level of inflammation is considered to impact the microbiota based on reports on relative abundances16–18,37 Accordingly, the relative abundance of Erysipelotrichaceae, Blautia, and Faecalibacterium decreased but the relative abundance of Enterobacteriaceae increased as inflammation progressed.16–18,34
The discovery that the relative and absolute bacterial analyses gave different results is an important finding. The relative abundance of a particular microbe is influenced by abundances of other microbe(s), as it is evident that when the relative abundance of one microbe increases another one decreases respectively. Therefore, analyses made on absolute bacterial abundances should be used in parallel in future studies to distinguish true biological signals from noise created by compositional data. Importantly, more published data on absolute abundances in PIBD are warranted.
As a limitation, we had to combine various IBD subtypes into one group for most analyses for gaining improved statistical power, even though it is well established that these disease conditions differ from each other. Analyses comparing patients with CD and UC could not be made due to the limited number of patients. However, we adjusted for the disease subtype in the analyses of microbiota changes during infliximab induction therapy and the treatment response. Notably, we focused on response to induction therapy. As 16S RNA sequencing does not detect fungi or viruses and depicts metabolic functions, future work with metagenomics could alleviate the functional differences in the microbiota between responders and nonresponders. As a strength, the fecal samples for microbiota analyses were immediately frozen, and the samples were transported frozen from home to our laboratory. Also, the use of antibiotics and corticosteroids was controlled. In addition, the definition of therapeutic response was defined with a strict criterion of fecal calprotectin value <100 µg/g mirroring transmural healing of the gut.22
Conclusions
We discovered that treatment response to infliximab was associated with higher absolute and relative abundance of short-chain fatty acid–producing Bifidobacteriales at baseline. Also, a decreased absolute abundance of Actinomycetales was associated with response to infliximab. During induction, the development of microbiota was rather similar in both treatment response groups, indicating that the microbiota changes in a similar pattern during infliximab induction therapy regardless of the microbiota composition prior to treatment. Therefore, our results highlight the importance of beneficial microbiota composition prior to treatment in reaching remission. In the future, intervention studies are warranted to research whether it is possible to improve therapeutic outcomes by modifying microbiota prior to introduction of infliximab.
Supplementary Material
Contributor Information
Miikka Höyhtyä, Departmentof Pediatrics, Tampere University Hospital, Faculty of Medicine and Health Technology, University of Tampere, Tampere, Finland.
Katri Korpela, Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Schahzad Saqib, Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Sofia Junkkari, Departmentof Pediatrics, Tampere University Hospital, Faculty of Medicine and Health Technology, University of Tampere, Tampere, Finland.
Eija Nissilä, Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Anne Nikkonen, Children’s Hospital, Department of Pediatric Gastroenteroloy, Helsinki University, Helsinki, Finland.
Evgenia Dikareva, Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Anne Salonen, Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Willem M de Vos, Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland; Laboratory of Microbiology, Wageningen University, Wageningen, the Netherlands.
Kaija-Leena Kolho, Departmentof Pediatrics, Tampere University Hospital, Faculty of Medicine and Health Technology, University of Tampere, Tampere, Finland; Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland; Children’s Hospital, Department of Pediatric Gastroenteroloy, Helsinki University, Helsinki, Finland.
Acknowledgments
The study was approved by the ethical committee of the Hospital District of Helsinki and Uusimaa (extension approved in 2014 to study, approved with a diary number 183/13/03/03/2011).
Funding
This work was supported by the Pediatric Research Foundation (Finland), a Helsinki University Hospital Grant and Helsinki University (to K.-L.K.), SIAM Gravitation Grant 024.002.002, and the Spinoza 2008 Award of the Netherlands Organization for Scientific Research (to W.M.d.V.).
Conflicts of Interest
The authors declare no conflicts.
Data Availability
All participants (or their guardians) signed an informed consent. All data available on request.
References
- 1. Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 2. Benchimol EI, Fortinsky KJ, Gozdyra P, et al. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17:423-439. [DOI] [PubMed] [Google Scholar]
- 3. Kelsen J, Baldassano RN.. Inflammatory bowel disease: the difference between children and adults. Inflamm Bowel Dis. 2008;14(Suppl 2):S9-S11. [DOI] [PubMed] [Google Scholar]
- 4. Ruemmele F, Veres G, Kolho K, et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J Crohns Colitis. 2014;8:1179-1207. [DOI] [PubMed] [Google Scholar]
- 5. Aardoom MA, Veereman G, de Ridder L.. A review on the use of anti-TNF in children and adolescents with inflammatory bowel disease. Int J Mol Sci 2019;20:2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diefenbach KA, Breuer CK.. Pediatric inflammatory bowel disease. World J Gastroenterol. 2006;12:3204-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gearry RB, Richardson AK, Frampton CM, et al. Population-based cases control study of inflammatory bowel disease risk factors. J Gastroenterol Hepatol. 2010;25:325-333. [DOI] [PubMed] [Google Scholar]
- 9. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12(4):205-217. doi: 10.1038/nrgastro.2015.34 [DOI] [PubMed] [Google Scholar]
- 10. van Rheenen PF, Aloi M, Assa A, et al. The medical management of paediatric Crohn’s disease: an ECCO-ESPGHAN guideline update. J Crohns Colitis. 2021;15:171-194. [DOI] [PubMed] [Google Scholar]
- 11. Turner D, Ruemmele F M, Orlanski-Meyer E, et al. Management of paediatric ulcerative colitis, part 1: ambulatory care-an evidence-based guideline from European Crohn’s and Colitis Organization and European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2018;67:257-291. [DOI] [PubMed] [Google Scholar]
- 12. Nikkonen A, Kolho KL.. Infliximab and its biosimilar produced similar first-year therapy outcomes in patients with inflammatory bowel disease. Acta Paediatr. 2020;109:836-841. [DOI] [PubMed] [Google Scholar]
- 13. Manichanh C, Borruel N, Casellas F, et al. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599-608. [DOI] [PubMed] [Google Scholar]
- 14. Knox NC, Forbes JD, Peterson C, et al. The Gut microbiome in inflammatory bowel disease: lessons learned from other immune-mediated inflammatory diseases. Am J Gastroenterol. 2019;114:1051-1070. [DOI] [PubMed] [Google Scholar]
- 15. Nishida A, Inoue R, Inatomi O, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11(1):1-10. doi: 10.1007/s12328-017-0813-5 [DOI] [PubMed] [Google Scholar]
- 16. Kolho KL, Korpela K, Jaakkola T, et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol. 2015;110:921-930. [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Gao X, Ghozlane A, et al. Characteristics of faecal microbiota in paediatric Crohn’s disease and their dynamic changes during infliximab therapy. J Crohns Colitis. 2018;12:337-346. [DOI] [PubMed] [Google Scholar]
- 18. Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2017;22:247. [DOI] [PubMed] [Google Scholar]
- 19. Zhuang X, Liu C, Zhan S, et al. Gut microbiota profile in pediatric patients with inflammatory bowel disease: a systematic review. Front Pediatr. 2021;9:626232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puolanne AM, Kolho KL, Alfthan H, et al. Rapid fecal calprotectin test and symptom index in monitoring the disease activity in colonic inflammatory bowel disease. Dig Dis Sci. 2017;62:3123-3130. [DOI] [PubMed] [Google Scholar]
- 21. Kolho KL, Raivio T, Lindahl H, et al. Fecal calprotectin remains high during glucocorticoid therapy in children with inflammatory bowel disease. Scand J Gastroenterol. 2006;41:720-725. [DOI] [PubMed] [Google Scholar]
- 22. Weinstein-Nakar I, Focht G, Church P, et al. Associations among mucosal and transmural healing and fecal level of calprotectin in children with Crohn’s disease. Clin Gastroenterol Hepatol. 2018;16:1089-1097.e4. [DOI] [PubMed] [Google Scholar]
- 23. Korpela K. mare: Microbiota analysis in R Easily. R package version 1.0. 2016. Accessed April 10, 2022. https://github.com/katrikorpela/mare
- 24. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460-2461. [DOI] [PubMed] [Google Scholar]
- 25. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Venables WN, Ripley BD. Modern Applied Statistics with S, Fourth edition. Springer; 2022. ISBN 0-387-95457-0. https://www.stats.ox.ac.uk/pub/MASS4/. [Google Scholar]
- 27. Oksanen J, Blanchet FG, Kindt R, et al. vegan: Community Ecology Package. R package version 2.0-2. 2012. Accessed April 10, 2022.
- 28. Pinheiro J, Bates D, DebRoy S, et al. nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-145. 2020. Accessed April 10, 2022. https://svn.r-project.org/R-packages/trunk/nlme/
- 29. Jian C, Luukkonen P, Yki-Järvinen H, et al. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS One. 2020;15:e0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jian C, Salonen A, Korpela K.. Commentary: How to count our microbes? The effect of different quantitative microbiome profiling approaches. Front Cell Infect Microbiol. 2021;11:627910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Y, Xu ZZ, He Y, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 2018;3:e00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frank DN, Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Connell Motherway M, Houston A, O’Callaghan G, et al. A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol Microbiol. 2019;111:287-301. [DOI] [PubMed] [Google Scholar]
- 34. Li J, Li Y, Zhou Y, et al. Actinomyces and alimentary tract diseases: a review of its biological functions and pathology. Biomed Res Int. 2018;2018:3820215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dot T, Osawa R, Stackebrandt E.. Phascolarctobacterium faecium gen. nov, spec. nov., a novel taxon of the Sporomusa group of bacteria. Syst Appl Microbiol. 1993;16:380-384. doi: 10.1016/S0723-2020(11)80269-9 [DOI] [Google Scholar]
- 37. Ren B, Schwager E, Knights D, et al. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2015;15:382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang M, Molin G, Ahrné S, et al. High proportions of proinflammatory bacteria on the colonic mucosa in a young patient with ulcerative colitis as revealed by cloning and sequencing of 16S rRNA genes. Dig Dis Sci. 2007;52:620-627. [DOI] [PubMed] [Google Scholar]
- 39. Olbjørn C, Småstuen MC, Thiis-Evensen E, et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease – associations with disease phenotype, treatment, and outcome. Clin Exp Gastroenterol. 2019;12:37-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alam MT, Amos GCA, Murphy ARJ, et al. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathogens. 2020;12:1-8. doi: 10.1186/s13099-019-0341-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All participants (or their guardians) signed an informed consent. All data available on request.