Abstract
Background
Diffuse midline gliomas (DMG) are highly invasive brain tumors with rare survival beyond two years past diagnosis and limited understanding of the mechanism behind tumor invasion. Previous reports demonstrate upregulation of the protein ID1 with H3K27M and ACVR1 mutations in DMG, but this has not been confirmed in human tumors or therapeutically targeted.
Methods
Whole exome, RNA, and ChIP-sequencing was performed on the ID1 locus in DMG tissue. Scratch-assay migration and transwell invasion assays of cultured cells were performed following shRNA-mediated ID1-knockdown. In vitro and in vivo genetic and pharmacologic [cannabidiol (CBD)] inhibition of ID1 on DMG tumor growth was assessed. Patient-reported CBD dosing information was collected.
Results
Increased ID1 expression in human DMG and in utero electroporation (IUE) murine tumors is associated with H3K27M mutation and brainstem location. ChIP-sequencing indicates ID1 regulatory regions are epigenetically active in human H3K27M-DMG tumors and prenatal pontine cells. Higher ID1-expressing astrocyte-like DMG cells share a transcriptional program with oligo/astrocyte-precursor cells (OAPCs) from the developing human brain and demonstrate upregulation of the migration regulatory protein SPARCL1. Genetic and pharmacologic (CBD) suppression of ID1 decreases tumor cell invasion/migration and tumor growth in H3.3/H3.1K27M PPK-IUE and human DIPGXIIIP* in vivo models of pHGG. The effect of CBD on cell proliferation appears to be non-ID1 mediated. Finally, we collected patient-reported CBD treatment data, finding that a clinical trial to standardize dosing may be beneficial.
Conclusions
H3K27M-mediated re-activation of ID1 in DMG results in a SPARCL1+ migratory transcriptional program that is therapeutically targetable with CBD.
Keywords: CBD, DMG, ID1, invasion/migration, ROS
Key Points.
ID1 is found to be a novel therapeutic target in DMG.
ID1 is found to be epigenetically upregulated in H3K27M-DMG.
Preliminary data suggest that therapeutic targeting of ID1 decreases tumor cell invasion/migration and tumor growth in human and murine pHGG models.
Importance of the Study.
Diffuse midline gliomas (DMGs) are incurable pediatric brain tumors, and there is a need to identify novel genetic targets that can be exploited therapeutically. We show that ID1 drives DMG cell invasion and migration. ID1-knockdown (genetic and pharmacologic) in human DMG cells reduces tumor growth and improves survival of mice. Taken together, our data demonstrate that CBD, which inhibits ID1, affects multiple malignant phenotypes of DMG, including proliferation, invasion, and migration. The effects of CBD on DMG are mediated by both ID1-dependent and ID1-independent mechanisms. These results lay the groundwork for additional investigations targeting ID1 and hold potentially major implications for future clinical trials.
Diffuse midline gliomas (DMG) are lethal pediatric brain tumors that originate in the midline brain structures, and have a median survival of 10–11 months.1 Standard of care consists of palliative radiation, and experimental chemotherapies have yet to demonstrate additional benefits. Even with the advent of precision-based medicine, clinical trials targeting specific molecular targets are lacking, highlighting the need to identify novel therapeutic targets.
As many as 80% of DMGs harbor a lysine-to-methionine substitution (H3K27M) in Histone H3.3 (H3.3A) or Histone H3.1 (H3C2).1,2 H3K27M now defines a distinct subgroup in DMG and is associated with worse prognoses.3 The H3K27M mutation represses the polycomb repressive complex 2 (PRC2), resulting in global reduction of H3K27me3 (with focal gains),4 and global increases in acetylation of H3K27 (H3K27ac) with focal losses, associated with upregulation of tumor-driving genes.5
Constitutive expression of Inhibitor of DNA binding (ID) proteins has been shown to inhibit the differentiation of multiple tissues.6 ID proteins dimerize with basic helix-loop-helix (bHLH) transcription factors, preventing DNA binding.7 bHLH transcription factors are key regulators of tissue and lineage-specific gene expression.8 Overexpression of the Inhibitor of DNA binding 1 (ID1) gene has been tied to the pathogenesis and invasiveness of multiple human cancers.8,9 ID1 is associated with downstream activin A receptor type 1 (ACVR1) signaling, which itself is mutated/activated in 25% of DMGs,10–12 indicating a potential role for ID1. Prior studies have shown H3K27M and ACVR-mutation upregulate ID1 in cultured human astrocytes and murine models of DMG.10,11 Invasion into normal brain tissue is a pathognomonic feature of DMG, but its regulation remains poorly understood. Further, analysis of ID1 in human DMG preclinical models, and its regulation and targetability, have not been previously investigated.
In the present study, we show that DMGs demonstrate epigenetic activation and increased expression of ID1, influenced most strongly by H3K27M mutational status and brain location. ID1-high DMG tumor cells most closely resemble developing human and murine prenatal pontine astrocyte precursors with an activated “migratory” transcriptional program. Genetic and pharmacologic (CBD) suppression of ID1 decreases invasion, and to a greater extent, migration, and improves survival in multiple preclinical DMG models. These findings represent an exciting new direction for understanding the regulation and targetability of invasion in DMG, with broad implications for therapeutic targeting of solid tumors with ID1 upregulation.
Materials and Methods
PPK-IUE Model of pHGG (Pediatric High-grade Gliomas)
All animal studies were conducted according to guidelines approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan. In-utero electroporation (IUE) was performed using sterile technique on isoflurane/oxygen anesthetized pregnant CD1 females at embryonic stage E13.5, using established methodology.13,14 In this study, we injected the following four plasmids together: [1] PBase, [2] PB-CAG-DNp53-Ires-Luciferase (dominant negative TP53 or TP53 hereafter), [3] PB-CAG-PdgfraD842V (PDGFRA D842V), and [4] PB-CAG-H3.3 K27M- (H3K27M), referred to as the “PPK”-IUE pHGG model15 (Supplementary Material).
Whole Exome and Transcriptome Sequencing (Sick Kids, Toronto)
Use of patient tissues was approved by the Hospital for Sick Children (Toronto) Research Ethics Board. WES/WGS (accession EGAS00001000575) from 34 DMG samples plus 18 matched normal brains were used. The total RNA Library Prep with Ribo-Zero Gold Kit (Illumina, CA, USA) was used and paired end sequencing generated with Illumina HiSeq 2500 machines (accession EGAD00001006450)16 (Supplementary Material).
MiNT-ChIP-sequencing of Tumor Tissue
Analyses for the two classical histone modifications H3K27ac and H3K27me3, representing accessible and repressed chromatin states, respectively, were performed as part of a MiNT-ChIP analysis for 9 tumor samples of DMG patients in comparison to a control tissue sample of healthy pons according to the protocol published by Buenstro et al. (Supplementary Material).
Single-cell RNA Sequencing (scRNA-seq) Analysis from Developing Brain and H3K27M-mutant DMGs
Single-cell gene expression data and their clusters in the developing brain were obtained from GSE133531 (mouse pons), GSE120046 (human pons, gestational week 8–28), and GSE144462 (human cortex, gestational week 21–26) (Supplementary Material).
Native ChIP-qPCR
Native ChIP-qPCR was performed on post-mortem tissue using antibodies against H3K27ac (2 μl, Cat#07360, Millipore Sigma), H3K27me3 (1 μg, Cat#07449, Millipore Sigma), and control IgG (2 μg Cat#12370, Millipore Sigma) (Supplementary Material).
Invasion Assay
Invasion assays were performed using growth factor-reduced Matrigel invasion chambers with 8 μm pores (Cat#354483, Corning) as described in previously published work.17
Migration (Scratch) Assay
Migration assays were performed following a previously published protocol with slight modifications.18 Cells were seeded in 6-well plates and grown to approximately 80% confluence. Scratches were made using a 200 μl pipette tip and migration was then monitored using the IncuCyte® live-cell analysis system.
CBD Treatment Studies In In Vivo PPK-IUE and DIPGXIIIP* Mouse Model
Mice harboring IUE-generated PPK HGG tumors were treated with cannabidiol (CBD) when tumors reached logarithmic growth phase (minimum 2 × 106 photons/s via bioluminescent imaging). Mice litters from each experimental group were randomized to treatment with: (A) 15 mg/kg CBD (10% CBD suspended in Ethanol, 80% DPBS, 10% Tween-80) and (B) control treatment (10% Ethanol, 80% DPBS, 10% Tween-80). Mice were treated via intraperitoneal injection 5 days/week until morbidity all control mice (Supplementary Material).
Results
ID1 Elevated in DMG and PPK-IUE pHGG with H3K27M Mutation
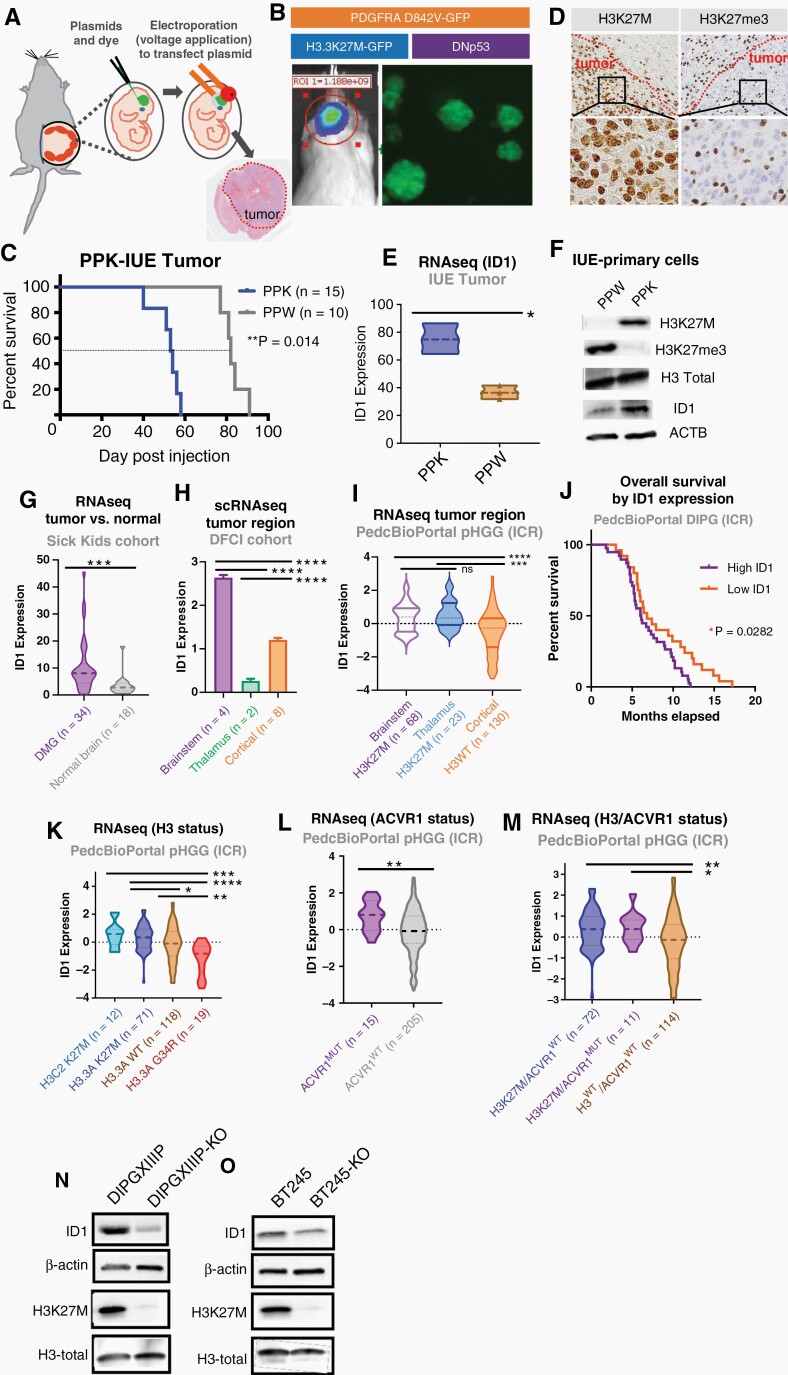
We first sought to confirm whether ID1 expression is affected by H3K27M. We adopted a PPK-IUE model of pHGG, as previously described.15 Mice developed tumors [mutant TP53, mutant PDGFRA (D842V) with H3.3A K27M mutation (“PPK”) or H3.3A wildtype (“PPW”)] via plasmid injection into the lateral ventricles of E13.5 embryonic CD1 mice (Figure 1A–B). Primary neurosphere cell cultures were generated by tumor dissociation (Figure 1B). Survival analysis revealed that H3.3A K27M-mutant PPK tumor bearing mice (n = 15) had significantly reduced survival compared to PPW (n = 10) (Figure 1C). Additionally, immunohistochemistry (IHC) analyses of murine tumors showed tumor-specific expression of H3K27M and global loss of H3K27me3 expression, a salient feature of H3K27M-DMG (Figure 1D).19 RNA-sequencing of PPK and PPW showed elevated ID1 expression in PPK tumors (Figure 1E). Further, western blot analysis confirmed increased ID1 expression in a PPK tumor-derived cell line (Figure 1F).
Fig. 1.
Elevated expression of ID1 in DIPG population. (A) IUE-mediated H3K27M-tumor model. (B) PDGFRAMUT-p53MUT-H3WT (“PPW”) and PDGFRAMUT-p53MUT-H3K27M (“PPK”) tumors generated by IUE. Tumor growth monitored by in vivo bioluminescence imaging and primary neurosphere cell cultures generated by dissociation of tumor tissue. (C) Survival curve for PPW and PPK mice; **P = .014, log-rank test. (D) IHC-stained images of PPK tumor for H3K27M and H3K27me3 expression (representative of n = 3 tumors). Magnification = 10× (top row); 40× (bottom row). (E) ID1 expression by RNA-sequencing from PPK and PPW from IUE mouse tumors, *P < .01, Welch’s t-test. (F) Western blot (WB) of PPW and PPK primary neurospheres for assessment of H3K27M, H3K27ac, and ID1 expression by H3-K27M mutational status. (G) ID1 expression of DIPG tissue (n = 34) compared to matched normal brain tissue (n = 18) from SickKids cohort; ***P < .001, unpaired parametric t-test. (H) ID1 expression by scRNA-seq from DFCI cohort, including brainstem, thalamus, and cortex; ****P< .0001, one-way ANOVA t-test. (I) ID1 expression of DMG tissue compared to hemispheric pHGG tissue from ICR cohort; ****P < .0001, unpaired t-test. (J) Kaplan–Meier survival curve of DIPG patients (n = 66) grouped by high and low ID1 expression. *P = .0282, Mantel–Cox test. (K) ID1 expression across H3.1 (H3C2) K27M (n = 12), H3.3 (H3.3A) K27M (n = 71), H3WT (n = 118), and H3.3 (H3.3A) G34R/V (n = 19) DIPG tumors from ICR cohort, presented in Mackay et al.; *P < .05, **P < .01, ***P < .001, ****P < .0001, one-way ANOVA t-test. (L) ID1 expression of pHGG tissue by ACVR1 mutational status (n = 15 ACVR1MUT; n = 205 ACVR1WT). Data from ICR cohort; **P < .01, unpaired parametric t-test. (M) ID1 expression of pHGG tumors with H3K27M only (n = 72), H3K27M, and ACVR1 mutations (n = 11) and neither mutation (H3WT/ACVR1 WT; n = 114) from ICR cohort; *P < .05, **P < .01, one-way ANOVA t-test. (N–O) WB showing removal of H3K27M in two human DIPG cell lines (DIPGXIIIP* and BT-245) decrease ID1 expression.
ID1 Expression is Influenced by H3 Mutational Status and Associated with Lower Overall Survival in DMG
In whole transcriptome sequencing from 34 DMG and 18 normal brain samples (cortex), DMG tissue showed significantly higher ID1 expression (Figure 1G). ScRNA-seq data from H3K27M-mutant DMGs [Dana-Farber Cancer Institute (DFCI) cohort20] confirmed that malignant cells display significantly higher ID1 expression compared to nonmalignant cells within these tumors (Fig. S1A-B). ScRNA-seq data from H3K27M-DMG patients (n = 4) revealed higher ID1 expression in brainstem H3K27M-DMG cells compared to thalamic and cortical tumor cells (Figure 1H), although the number of thalamic tumors (n = 2) is very low. Further analysis of bulk RNA-seq [ICR cohort (Institute for Cancer Research), n = 22121], in which both brainstem and thalamic DMG tumors showed significantly higher ID1 expression than cortical pHGGs (Figure 1I). Further, H3K27M-DMG patients with higher ID1 expression (ICR cohort) have lower OS22 (Figure 1J).
Analysis of bulk tumor RNA-seq (ICR cohort) revealed that ID1 expression is significantly increased in DMGs harboring H3K27M (H3.3A or H3C2) compared to H3.3WT and H3.3G34R/V (Figure 1K).21ACVR1-mutant tumors have significantly higher ID1 expression compared to WT tumors (Figure 1L), but the addition of mutated ACVR1 to H3 doesn’t significantly increase ID1 expression (Figure 1M). Further, western blot analysis of two isogenic cell lines DIPGXIIIP K27M-KO vs DIPGXIIIP and BT245 K27M-KO vs K27M shows that ID1 expression decreases when H3K27M is knocked out (Figure 1N–O).
Epigenetic State of ID1 Loci in DMG Tumor Cells by H3 Mutational Status
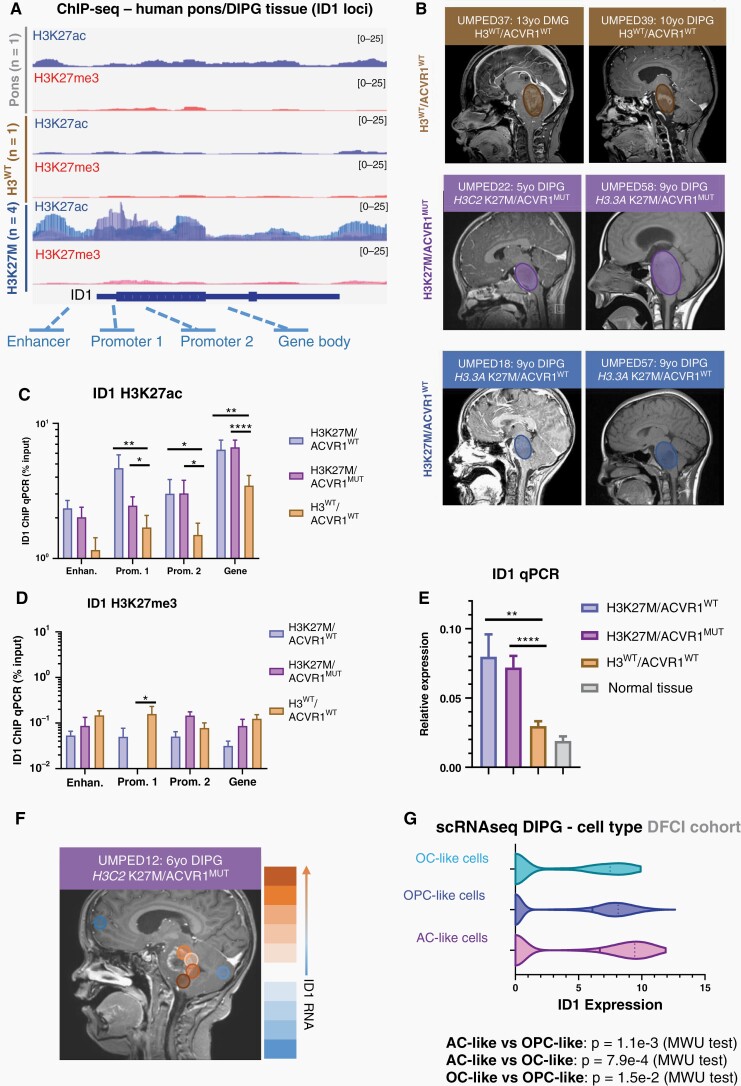
As DMGs are driven by epigenetic dysregulation, we sought to assess H3K27ac and H3K27me3 marks at regulatory regions of the ID1 gene in human DMG tumors. ChIP-sequencing on normal pediatric pontine tissue (n = 1), H3WT DMG (n = 1), and H3K27M DMG (n = 4) tumor samples revealed a marked increase in H3K27ac deposition at ID1 gene body elements in H3K27M DMG tissue (Figure 2A). Subsequent ChIP-qPCR analysis confirmed significantly elevated H3K27ac at predicted promotor and gene body regions of the ID1 locus compared to H3WT/ACVR1WT DMG samples (Figure 2B–C) (Supplementary Table 1). Changes in H3K27me3 were less notable (Figure 2D). Further, qPCR of tumor tissue demonstrated ID1 expression to be highest in H3K27M tumors (n = 4 sites) regardless of ACVR1 status (Figure 2E and Fig. S2A-C). We noted differences in ID1 expression (qPCR) between multi-focal autopsy samples, leading us to analyze multi-focal (n = 6) bulk RNA-sequencing of a single H3K27M/ACVR1-mutant DMG patient (UMPED12), which revealed varying levels of ID1 expression across different regions of the tumor (Figure 2F). Assessment of ID1 expression across all malignant cell types in scRNA-seq performed on four DMG patients showed that ID1 is most highly expressed in DMG cells with an astrocytic differentiation program [“AC-like cells” 20], followed by oligodendrocyte precursor cell-like (“OPC-like”) cells (Figure 2G and Fig. S3A). We observed higher levels of ID1 expression in cycling compared to noncycling malignant cells (Fig. S3B).
Fig. 2.
ID1 is epigenetically active in H3K27M-DIPG. (A) ChIP-sequencing of H3K27ac and H3K27me3 deposition at ID1 gene locus in H3K27M human DIPG tumor tissue (n = 4), H3WT DIPG tumor tissue (n = 1), and normal human pontine tissue (n = 1). (B) Multifocal DIPG tumor samples were obtained at autopsy from n = 2 patients with H3K27M mutation and wildtype ACVR1 (ACVR1WT), n = 2 patients with H3K27M mutation and ACVR1 mutation (ACVR1MUT), and n = 3 patients with wildtype H3 and ACVR1. Circles over MRI images represent approximate region of tumor. (C–D) ChIP-qPCR quantification of deposited (C) H3K27ac, and (D) H3K27me3 marks at gene body elements identified in part A for ID1. Data represent samples from patients in (B), mean ± SEM; *P < .05, **P < .01, ****P < .0001, one-way ANOVA t-test. (E) ID1 expression (qPCR) for multifocal samples collected from patients in (B) showed highest ID1 expression in H3K27M tumors. Data represent mean ± SEM; **P < .01, ****P < .0001, one-way ANOVA t-test. (F) MRI of H3K27M/ACVR1MUT DIPG patient with circles representing regions where samples were obtained at autopsy. Color scale on right displays relative level of ID1 expression by qPCR (orange = higher ID1 expression; blue = lower ID1 expression). (G) ScRNA-seq data (DFCI, n = 4 DIPGs) of malignant DIPG cells plotted to show ID1 expression across varying subtypes of cells [oligodendrocyte-like (OC-like); OPC-like; AC-like]. AC-like vs OPC-like: p = 1.1e-3; AC-like vs OC-like: p = 7.9e-4; OC-like vs OPC-like: p = 1.5e-2, Mann–Whitney U (MWU) test.
ID1+ Malignant DMG Cells Share Transcriptional Program with OAPC-like Cells in Developing Human Brain
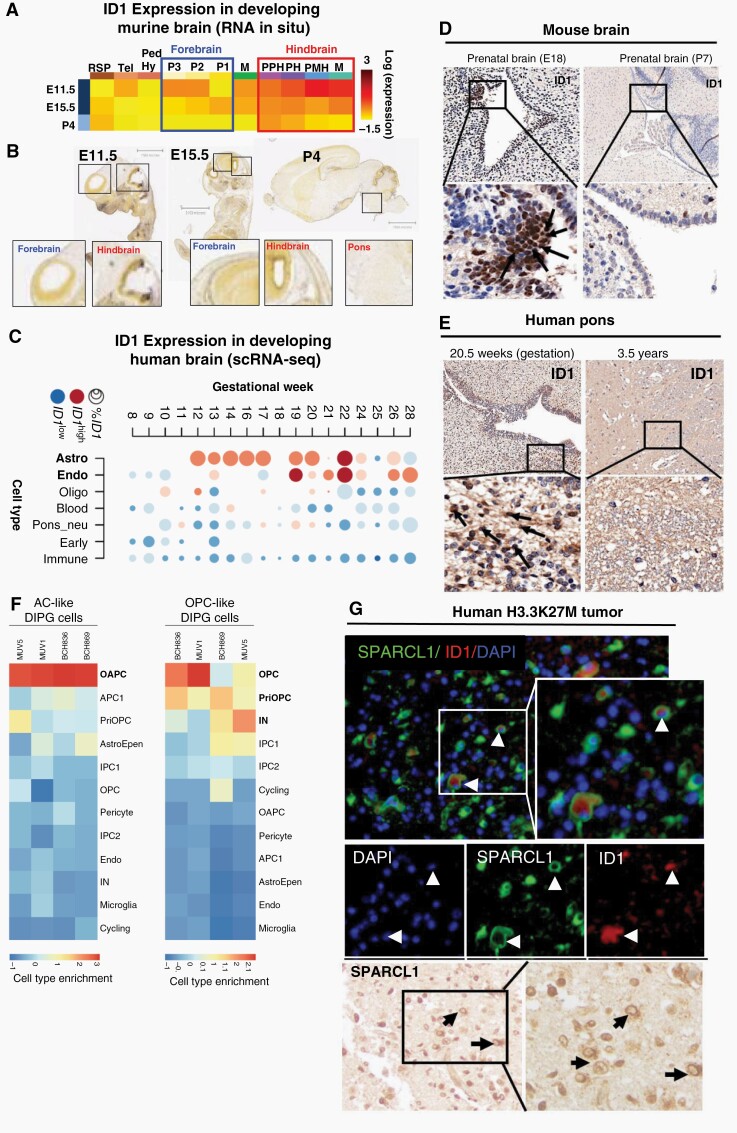
We next assessed ID1 expression and deposition of H3K27ac at the ID1 gene locus across pre and postnatal mouse brain developmental stages. RNA in-situ hybridization data (Allen Brain Atlas) demonstrated that ID1 RNA was higher in the developing prenatal mouse hindbrain (including developing pons) compared to forebrain or midbrain, and postnatal mouse brain (Figure 3A–B and Fig. S4). ChIP-sequencing in E15.5 mouse brains revealed elevated H3K27ac in the hindbrain compared to midbrain and forebrain,23 including at ID1 enhancer sites (Fig. S5A-B).
Fig. 3.
ID1 expression is elevated in developing astrocyte cells in prenatal human and murine hindbrain. (A) Heat map showing relative ID1 expression by in situ hybridization (ISH) in murine brain across development. Available from: http://developingmouse.brain-map.org/. (B) ISH of sagittal developing murine brain sections showing higher ID1 RNA in embryonic hindbrain than forebrain, and minimal ID1 RNA in all postnatal brain [Allen Developing Mouse Brain Atlas. Available from: http://developingmouse.brain-map.org/]. (C) Heatmap of ID1 expression across varying cell types during normal human pontine development [data from Fan et al.24]. Circle size indicates percentage of cells that express ID1 and color indicates expression level in ID1+ cells (red = high expression; blue = low expression). (D) ID1 IHC staining of normal human pontine tissue across development. (E) ID1 IHC of normal murine pontine tissue across development. Magnification = 10× (top row); 40× (bottom row). (F) Overlap of genes expressed by cell types in the developing human pons [Fu et al.] in DIPG tumor cell subsets. (Red = cell type marker genes enriched in DIPG cells; blue = cell type marker genes not enriched in DIPG cells). (G) Immunostaining of SPARCL1 (green) and ID1 (red) in human DIPG tissue showing colocalization of ID1 and SPARCL1 in a subset of cells (white arrow). Scale bar, 20 μm. Tumor nuclei stained with DAPI (blue). IHC staining of SPARCL1in human DIPG tissue (black arrow) (representative of n = 3 human DIPG tumors).
Analysis of developing human24 and mouse25 brain scRNA-seq data showed that ID1 expression peaks at gestational week (GW) 12–22 in the human pons (Figure 3C) and early postnatal mouse pons (P0; Fig. S6A-D), and is most highly expressed in astrocytes. IHC analyses of pre and postnatal brains confirmed elevated ID1 in the murine embryonic brain (E18; Figure 3D) and human GW20.5 brain (Figure 3E) in subventricular regions lining the 4th ventricle.
We next sought to assess whether ID1+ sub-populations of malignant DMG cells share a transcriptional program with ID1+ developing brain cells. Interestingly, AC-like cells from all four DMG tumors show the strongest overlap with the transcriptional program of the recently defined OAPC cell population in the developing human brain (Figure 3F). OAPCs express both astrocyte (GFAP) and oligodendrocyte (OLIG1, OLIG2) marker genes as well as SPARCL1, which is involved in regulation of cell adhesion/migration. We found ID1 to be a marker gene for both AC-like DMG cells and OAPCs. Immunofluorescence and IHC of human H3K27M-DMG samples revealed colocalization of ID1 and SPARCL1 expression in cell sub-populations (Figure 3G).
In vitro ID1-knockdown Decreases Invasion and Migration of DMG Cells
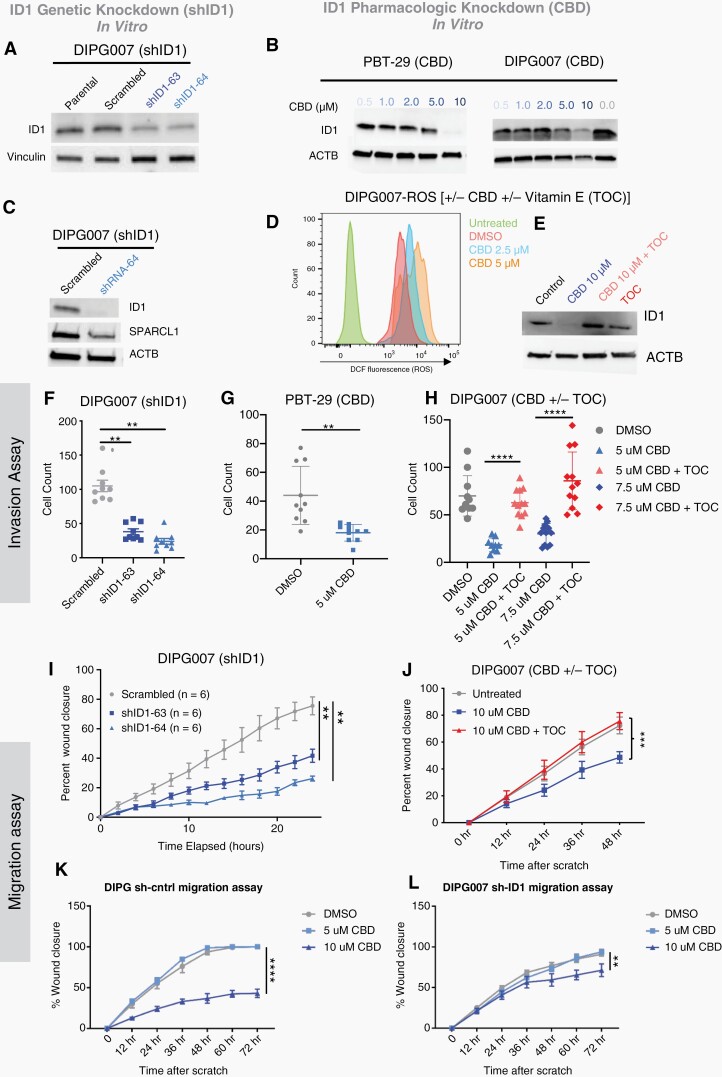
Single cell RNA-seq analysis of ID1-expressing AC-like DMG cells from human tumors and analysis of bulk RNA-seq from mouse IUE-PPK tumors revealed elevated SPARCL1 expression (Fig. S7A-B), and enrichment of gene sets involved in regulation of cell adhesion and migration (Fig. S7C). To explore ID1’s role in DMG cell migration and invasion, we pursued genetic (shRNA) and pharmacologic knockdown of ID1 in patient-derived H3K27M-mutant DMG cells. CBD (the nonpsychoactive compound found in Cannabis sativa26–28) has been shown to reduce ID1 expression in multiple preclinical solid tumor models,9,29 potentially through increased intracellular levels of reactive oxygen species (ROS).30 Genetic knockdown (shID1) in DIPG007 resulted in significant ID1 reduction (Figure 4A). CBD reduced ID1 protein levels in two K27M-DMG cell cultures (Figure 4B) and murine H3.3-PPK cell culture (Fig. S8A), but not in two control [NHA (normal human astrocytes) and KNS42 (hemispheric HGG)] cell lines without K27M mutation (Fig. S8B). ID1 knockdown resulted in SPARCL1 reduction in DIPG007 cells (Figure 4C), further indicating that SPARCL1 functions downstream of ID1. CBD treatment increased ROS levels in DIPG007 cells treated with 2.5 or 5.0 μM of CBD (Figure 4D). Additional treatment with Vitamin E [α-tocopherol (TOC)], an established ROS scavenger,29,30 consistently rescued the downregulation of ID1 expression seen with CBD treatment (Figure 4E).
Fig. 4.
Genetic and pharmacological inhibition of ID1 decreases DIPG cell invasion and migration. (A) Western blot (WB) confirming ID1-knockdown with ID1 specific shRNA in DIPG007 cells. (B) WB shows reduced ID1 expression in PBT-29 and DIPG007 cells treated with increasing concentrations of CBD. (C) WB depicting reduction in SPARCL1 expression along with decreased ID1 expression in ID1-knockdown DIPG007 cells. (D) Histogram showing increase in reactive oxygen species (ROS) with increasing CBD doses (0–5 μM) over two days in DIPG007 cells measured by 2′,7′-Dichlorodihydrofluorescein (DCF) using flow cytometry. (E) WB of DIPG007 cells treated with vehicle (control) or 10 μM of CBD for 14 h in the absence or presence of 50 μM TOC. (F–G) Effect of ID1 knockdown on invasion as measured by Matrigel-coated Boyden chamber assay. shRNA ID1-knockdown in DIPG007 or CBD-mediated ID1 knockdown in PBT-29 showed reduced invasion, **P < .01, unpaired parametric t-test. (H) Effect of CBD and α-tocopherol (TOC) (50 μM) on DIPG007 invasion. DIPG007 cells were treated for 2 days with DMSO (control) and 5 μM or 7.5 μM CBD ± TOC (50 μM). Invasion was measured by Matrigel-coated Boyden chamber. **P < .01, unpaired parametric t-test. (I) Effect of ID1-knockdown on DIPG007 migration and (J) Effect of CBD and TOC (50 μM) on DIPG007 migration. (K–L) Effect of CBD on migration of DIPG007-sh-control and DIPG007-sh-ID1, as measured by scratch assay and quantified as percent wound closure. Experiments were conducted in triplicate; data points represent mean ±- SEM, **P < .01; images taken with Incucyte; area measured by ImageJ.
Genetic- (Sh-ID1) and CBD-mediated ID1-inhibition significantly reduced invasion of multiple DMG cell lines including DIPG007 (Figure 4F and Fig. S9A-B), PBT-29 (Figure 4G), and JUMP4 (Fig. S11A-B) but CBD did not reduce invasion of control cell line (NHA and KNS42) to the same extent (Fig. S10A-B). ID1 knockdown decreased the efficacy of CBD-mediated invasion reduction (Fig. S9C-E). Proliferation rates were unchanged by shID1, but CBD did impact cell proliferation of multiple human DMG (DIPG007, PBT-29, DIPGXIIIP*, and JUMP4) and mouse PPK cells (Fig. S8C-E and S11A,C,E). Rescue of ID1 with the addition of TOC blunted the CBD-induced invasion reduction (Figure 4H). ID1-knockdown significantly reduced DIPG007 migration using a “scratch assay,” (Figure 4I) and CBD treatment significantly reduced migration in three DMG cell lines (Figure 4J, Fig. S11D,F,G) when compared to control cells (NHA and KNS42) (Fig. S10C). Similar to CBD-induced invasion reduction, rescue of ID1 with TOC also blunted the CBD-induced migration reduction (Figure 4J). Genetic knockdown of ID1 (shID1) in DIPG007 cells decreased sensitivity to CBD treatment in a scratch assay (Figure 4K–L), indicating that CBD, in part, mediates cell migration through ID1.
In vivo Genetic Knockdown of ID1 Increase Survival of Mice with pHGG in IUE Model
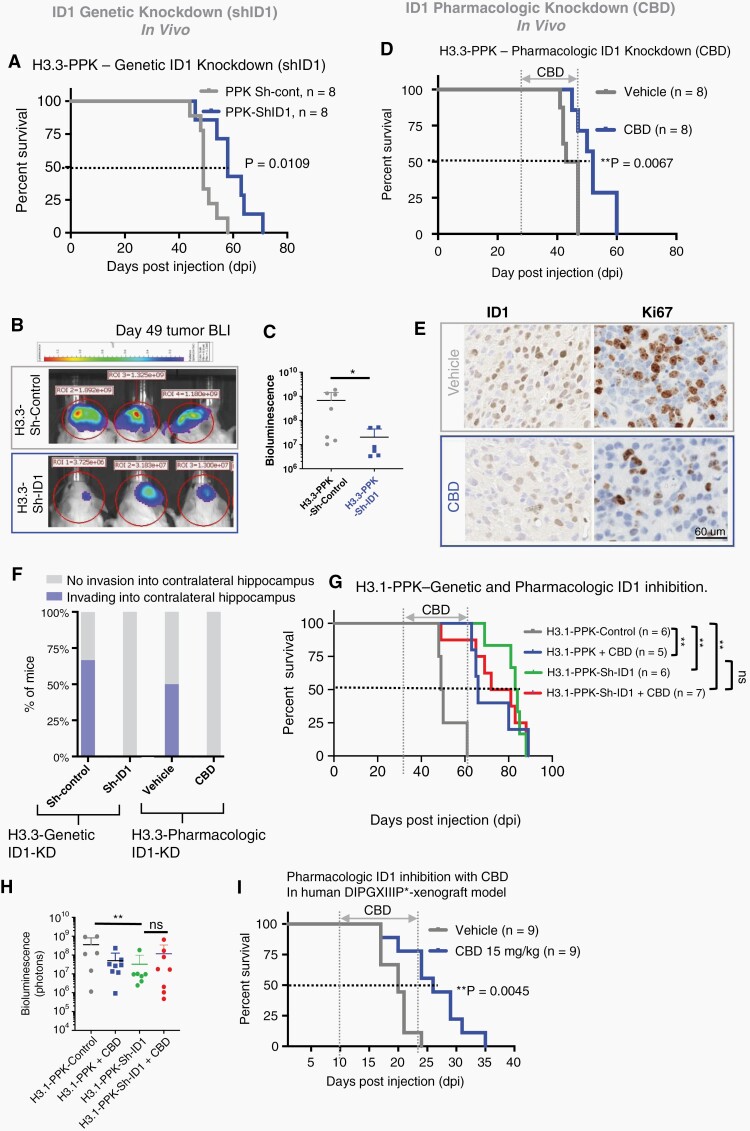
To assess whether ID1 suppression would impede in vivo tumor growth in PPK tumor mice, we knocked down ID1 expression in our both H3.3-PPK and H3.1-PPK-IUE tumor model using PiggyBac transposon ID1-shRNA plasmids and scrambled short hairpins (“Sh-control”). Both H3.3-PPK-ShID1 and H3.1-PPK mice exhibited significantly prolonged survival and reduced luminescent tumor signals when compared to PPK-control (Figure 5A–C and 5G-H and Fig. S13C). H3.3-PPK-Sh-ID1 tumors demonstrated reductions in ID1 and Ki67 expression by IHC (Fig. S12A-B). In a human patient-derived intracranial xenograft in vivo mouse model, orthotopic implantation of DIPG007 cells with ID1-knockdown into the brainstem of NSG mice also demonstrated reduced tumor growth compared to DIPG007-Sh-control cells, as shown by bioluminescent imaging (Fig. S12C-D) and demonstrated reductions in ID1 and Ki67 expression by IHC (Fig. S12E-F). Orthotopic implantation of human DIPGXIIIP* cells showed increased survival in ID1 KD tumor bearing mice when compared to controls (Fig. S12G).
Fig. 5.
Genetic and pharmacologic (CBD) targeting of ID1 decreases migration and slows in vivo tumor growth in pHGG with H3K27M mutation. (A) Standard Kaplan–Meier survival plot for H3.3-PPK-Sh-ID1 (median survival = 58 days) and H3.3-PPK-Sh-control mice (median survival = 49 days); P = .01, Log-rank test. (B) Representative bioluminescence images of H3.3-PPK-Sh-control and H3.3-PPK-Sh-ID1 tumors (representative from n = 8) 49 days post-IUE injection. (C) H3.3-PPK bioluminescence tumor growth for H3.3-PPK-Sh-control and H3.3-PPK-Sh-ID1 mice 49 days post-IUE injection. *P < .05, one-way ANOVA t-test. (D) Survival analysis of mice bearing H3.3-PPK tumor treated with CBD. Median survival for control condition was 45 days post-IUE injection and 55 days for CBD condition (15 mg/kg). **P < .005, Log-rank test. (E) IHC analysis of tumor images for ID1 and Ki67 expression (representative of n = 3 tumors); N = 3 animals/treatment group and 4 images/animal. Data represent mean ± SEM. Magnification = 10×. (F) Analysis of tumor invasion in tumor-bearing mice (n = 3 mice/group) with genetic (H3.3-PPK-Sh-ID1) or pharmacologic (CBD) ID1 knockdown. Invasion was defined as tumor infiltration into the contralateral hippocampus. (G) Survival analysis of mice bearing H3.1-PPK tumor with ID1 genetic KD (H3.1-PPK-ShID) or treated with CBD. Median survival for control = 45 days, CBD = 66 days, for Sh-ID183 days, and for Sh-ID1 + CBD = 76.5 days, respectively **P < .005, Log-rank test. (H) H3.1-PPK tumor bearing mice bioluminescence tumor growth for control, CBD, Sh-ID1, and for Sh-ID1 + CBD. *P < .05, one-way ANOVA t-test. (I) Survival analysis of NSG mice bearing human DIPGXIIIP* xenograft tumor treated with CBD. Median survival for control condition was 22 and 26 days for CBD condition (15 mg/kg). **P < 0.005, Log-rank test.
We then assessed the impact of CBD treatment on in vivo tumor growth using our H3.3- and H3.1-PPK-IUE model. Both PPK-IUE mice received daily treatment with CBD (15 mg/kg) or vehicle control. CBD treatment significantly improved median survival compared to vehicle control (Figure 5D and G–H). Further Moribund PPK tumors treated with CBD showed reductions in ID1 and Ki67 expression by IHC (Figure 5E) and more distinct tumor borders, indicating reduced invasion into normal brain (Figure 5F and Fig. S13A-B). ID1-KD, H3.1-PPK tumor mice (H3.1-PPK-shID1) showed no further improvement in survival when treated with CBD (Figure 5G–H and Fig. S13C), consistent with the majority of CBD impact on survival being ID1-mediated in this model. Further, CBD treatment showed significantly improved survival of NSG mice with human patient-derived DIPGXIIIP* cells orthotopically implanted into the brainstem (Figure 5I).
CBD Dose Reporting in DMG Patients
CBD is a supplemental nonprescribed therapy gaining use among patients with pHGG.31 We gathered patient-reported CBD alongside tetrahydrocannabinol (THC) dosing from families of DMG patients at two institutions (n = 14 total, n = 11, DMG with K27M, n = 3, H3WT), including a medical marijuana therapy registry study (NCT03052738) at Children’s Hospital of Colorado and retrospective interviews with families of patients who underwent research autopsy at the University of Michigan. CBD was given orally in all but one case (suppository) one to three times per day with a wide range of dosing (0.07 mg/kg to 25 mg/kg/day) (Fig. S14). This CBD dosing data represents feasibility of CBD treatment in DMG, with the clear need for further investigation and prospective therapeutic clinical trial.
Discussion
Overexpression of the Inhibitor of DNA binding 1 (ID1) gene has been tied to the pathogenesis of multiple human cancers.32–34 A role for ID1 in DMG has been proposed, based on its association with activating mutations in ACVR1, which increase ID1 expression through the activation of the BMP signaling pathway.10,12,13 Our data show that in DMG tissue, ID1 expression is increased with H3K27M mutation with minimal additional impact from concurrent ACVR1 mutation. This raises the possibility that the epigenetic activation of the ID1 locus may be the primary driver of ID1 signaling in human DMG tumors. Additionally, our data support that ID1+/SPARCL1+ expressing AC-like DMG cells share a migratory expression transcriptional profile with a subset of developing brain cells, the recently identified OAPCs. This may indicate new and exciting developmental origins for this subset of DMG cells. Further, our data indicate that ID1 promotes migration and invasion in DMG cells, which are disease-defining features of this infiltrative tumor.
We propose a model by which ID1 is upregulated through multiple mechanisms to “re-activate” prenatal brain developmental signaling (Figure 6, upper panel). Further, our data suggest that CBD inhibits DMG cell proliferation, invasion, and migration, through both ID1-dependent and ID1-independent mechanisms, with inhibition of migration being the most ID1-dependent and inhibition of proliferation being the most ID1-independent (Figure 6, lower panel). Our studies implicate an active epigenetic state at the ID1 locus shared between H3K27M tumor cells and prenatal precursor brain cells. This is consistent with prior studies focused on H3K27M mutations that have associated changes in H3K27ac/H3K27me3 with differential regulation of key DMG-associated genes.35,36 Additionally, we provide evidence that postnatal activation of ID1 in tumor cells replicates a prenatal “migratory” transcriptional state seen in a recently discovered subset of developing OAPC brain cells. These OAPCs (Olig2+SPARCL1+ glial progenitor cells) were identified as astrocyte-like at the molecular and transcriptional levels. In line with this, we found AC-like DMG cells to transcriptionally mimic the program of OAPCs, with the OAPC-marker SPARCL1 and ID1 colocalizing in a subset of H3K27M tumor cells. Interestingly, previous work has suggested a role for SPARCL1 in promoting DMG cell invasion into the subventricular zone (SVZ).37 Our data demonstrate that ID1 is most highly expressed by cycling AC-like cells in DMG tumors and SPARCL1 is one of the strongest expression markers of these cells.
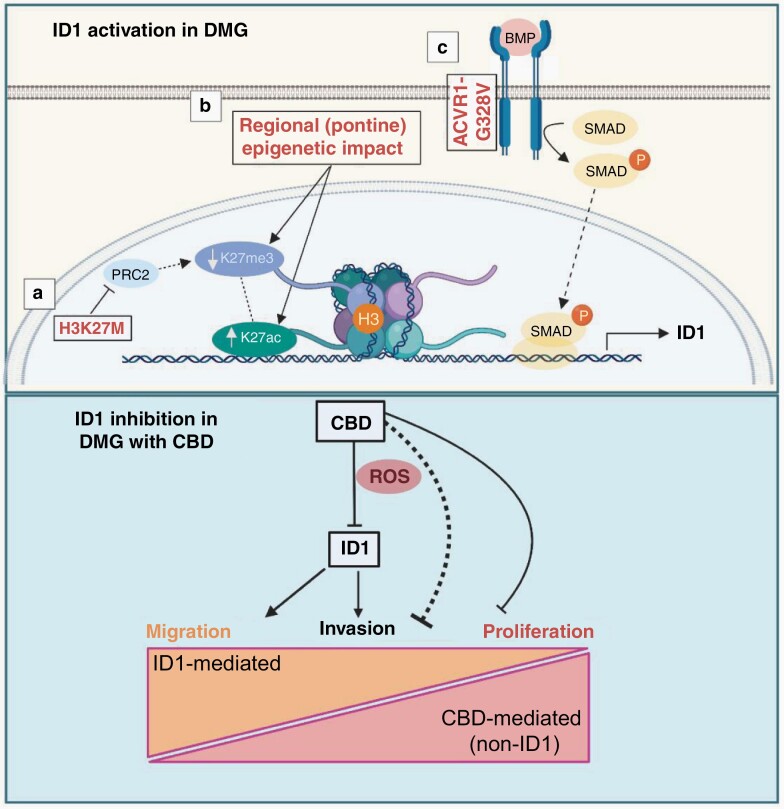
Fig. 6.
Proposed model of ID1 activation in DMG (upper panel) and its inhibition with CBD and downstream phenotypic impact (lower panel). The proposed activation model is made up of the following sub-sections: (a) H3K27M inhibits PRC2, leading to global decreases in H3K27me3 and subsequently allowing for increased H3K27ac. (b) Regional or tissue-specific factors and/or (c) constitutively activating ACVR1 mutations increase ID1 expression via SMAD protein signaling. ID1 inhibition by CBD treatment, which partially acts through increasing intracellular levels of reactive oxygen species (ROS), regulating migration and invasion in an ID1-dependent manner, while inhibiting proliferation in an ID1-independent manner. Image created with BioRender.
Our data show that ID1-knockdown has the potential to severely impede DMG tumor cell migration and invasion in preclinical models. These phenotypes are consistent with the inherent invasion into normal brainstem tissue that is observed histologically in DMGs, and with the role of ID1 in other cancers.8,34 In our experiments involving both genetically-engineered and intracranial implantation models, H3K27M-mutant tumor cells with ID1 reduction show decreased tumor growth and invasion. Our data raise important insights into the mechanisms underlying one of the most critical and problematic features of DMG tumors.
In the present study, CBD reduced ID1 expression in DMG cells at concentrations that are likely clinically achievable in the human brain. CBD is an often-unregulated supplemental therapy already being used for palliative treatment in pediatric oncology. It is commonly taken alongside THC, though the effects of coadministration are unknown in DMG patients. Our data show that CBD treatment reduces ID1 expression and DMG cell invasion and migration through increasing intracellular levels of ROS. Additionally, CBD increases expression of ATF3 (data not shown), which could be another possible mechanism by which CBD inhibits ID1 expression in DMG cells.38
The present study makes significant strides in establishing the mechanism of this controversial and popular off-trial supplemental compound in high-risk brain tumor patients. Given that patients currently take a variety of doses, it may be important to standardize both dosage and treatment length, as well as looking into potential toxicities in a future clinical trial. A recent CBD formulation (Epidiolex) is FDA-approved for epilepsy treatment,39 opening the door to a future clinical trial in DMG (and other ID1-driven tumors).
Our data support a model in which multifactorial genetic and epigenetic processes promote ID1-driven prenatal development transcriptional programs, which also promote the invasive features of DMG. These results improve our understanding of the pathogenesis of DMG tumors and provide a strong argument for the inclusion of ID1-targeting therapies into future treatments.
Supplementary Material
Acknowledgments
The authors thank the patients and their families for participation in this study.
Contributor Information
Dana Messinger, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Micah K Harris, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Jessica R Cummings, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Chase Thomas, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Tao Yang, Department of Neurology, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Stefan R Sweha, Department of Pathology, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Rinette Woo, Cancer Research, California Pacific Medical Center Research Institute; San Francisco, California, USA.
Robert Siddaway, Arthur and Sonia Labatt Brain Tumour Research Centre and Division of Pathology, Hospital for Sick Children, Toronto, Canada.
Martin Burkert, Centre for Molecular Medicine Norway (NCMM), Nordic EMBL Partnership, University of Oslo and Oslo University Hospital, Oslo, Norway.
Stefanie Stallard, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Tingting Qin, Department of Computational Medicine and Bioinformatics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA; Rogel Cancer Center, University of Michigan, Ann Arbor, USA.
Brendan Mullan, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Ruby Siada, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Ramya Ravindran, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Michael Niculcea, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Abigail R Dowling, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Joshua Bradin, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Kevin F Ginn, Department of Pediatrics, Children’s Mercy Kansas City, Kansas City, Missouri, USA.
Melissa A H Gener, Department of Pathology and Laboratory Medicine, Children’s Mercy Kansas City, Kansas City, Missouri, USA.
Kathleen Dorris, Department of Pediatrics, University of Colorado School of Medicine, Aurora, Colorado, USA.
Nicholas A Vitanza, Department of Pediatrics, Seattle Children’s, Seattle, Washington, USA.
Susanne V Schmidt, Institute of Innate Immunity, AG Immunogenomics, University Bonn, Bonn, Germany.
Jasper Spitzer, Institute of Innate Immunity, AG Immunogenomics, University Bonn, Bonn, Germany.
Jiang Li, Dana-Farber Boston Children’s Cancer and Blood Disorders Center, Department of Pediatric Oncology, Boston, Massachusetts, USA.
Mariella G Filbin, Dana-Farber Boston Children’s Cancer and Blood Disorders Center, Department of Pediatric Oncology, Boston, Massachusetts, USA.
Xuhong Cao, Department of Pathology, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Maria G Castro, Department of Neurosurgery, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Pedro R Lowenstein, Department of Neurosurgery, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA; Department of Cell and Developmental Biology, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Rajen Mody, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Arul Chinnaiyan, Department of Pathology, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Pierre-Yves Desprez, Cancer Research, California Pacific Medical Center Research Institute; San Francisco, California, USA.
Sean McAllister, Cancer Research, California Pacific Medical Center Research Institute; San Francisco, California, USA.
Matthew D Dun, Cancer Signalling Research Group, School of Biomedical Sciences and Pharmacy, College of Health, Medicine and Wellbeing, University of Newcastle, Callaghan NSW, Australia.
Cynthia Hawkins, Arthur and Sonia Labatt Brain Tumour Research Centre and Division of Pathology, Hospital for Sick Children, Toronto, Canada.
Sebastian M Waszak, Centre for Molecular Medicine Norway (NCMM), Nordic EMBL Partnership, University of Oslo and Oslo University Hospital, Oslo, Norway; Division of Pediatric and Adolescent Medicine, Department of Pediatric Research, Rikshospitalet, Oslo University Hospital, Oslo, Norway.
Sriram Venneti, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Carl Koschmann, Division of Pediatric Hematology-Oncology, Department of Pediatrics, University of Michigan Medical School (UMMS), Ann Arbor, Michigan, USA.
Viveka Nand Yadav, Department of Pediatrics at Children’s Mercy Research Institute, Kansas City, Missouri, USA.
Funding
This work was supported by ChadTough Defeat DIPG Foundation, Joan and David Evans Prosper Road Foundation (V.N.Y.); NIH/NINDS Grant [K08-NS099427, R01-NS119231] and Department of Defense Grant [CA201129P1], University of Michigan Chad Carr Pediatric Brain Tumor Center, ChadTough Defeat DIPG Foundation, DIPG Collaborative, Catching Up With Jack, The Pediatric Brain Tumor Foundation, The Morgan Behen Golf Classic, and Michael Miller Memorial Foundation (C.K.); National Institutes of Health [T32CA009676 to D.M.]; National Institutes of Health Clinical Sequencing Exploratory Research Award [1UM1HG006508 to A.C.], National Institutes of Health [R44CA206723 to S.M., P.Y.D.], The Research Council of Norway [187615 to S.M.W.], The South-Eastern Norway Regional Health Authority [S.M.W.], The University of Oslo [S.M.W.]. The Cure Starts Now Foundation, The Julian Boivin Courage for Cures Foundation, ChadTough Foundation, The Cure Starts Now Australia, Love Chloe Foundation, Aidan’s Avengers, Austin Strong, Brooke Healey Foundation, Cure Brain Cancer, Jeffrey Thomas Hayden Foundation, Musella Foundation, Pray Hope Believe, Reflections Of Grace, Storm the Heavens Fund, Ryan’s Hope, Benny’s World, The Isabella and Marcus Foundation, Lauren’s Fight for Cure, Robert Connor Dawes Foundation, Whitley’s Wishes, Aubreigh’s Army, Grant’s Ginormous Gift, Wayland Villars Foundation, Danni Kemp Cancer Support Fund, The Gold Hope Project, Smile for Brooklyn, Julia Barbara Foundation, Lily Larue Foundation, American Childhood Cancer Organization, RUN DIPG, Gabriella’s Smile Foundation, The DIPG Collaborative, and Snapgrant.com and and The Yuvaan Tiwari Foundation (C.K.).
Conflict of interest statement. The authors declare no potential conflicts of interest.
Authorship statement. Conception and design of the study: D.M, M.K.H., C.K., and V.N.Y. Acquisition, analysis, and/or interpretation of data: D.M., M.K.H., J.R.C., C.T., T.Y., R.W., Ro.S., M.B., S.S., T.Q., B.M., Ru.S., R.R., M.N., K.F.G., M.A.H.G., K.D., N.A.V., J.L., M.G.F., X.C., M.G.C., P.R.L., R.M., A.C., P.Y.D., S.M., C.H., S.M.W., S.V., C.K., and V.N.Y. Drafting and revision of the written manuscript: D.M., M.K.H., J.R.C., Ru.S., C.K., and V.N.Y. All authors discussed and reviewed the manuscript and approved the manuscript for publication.
References
- 1. Schroeder KM, Hoeman CM, Becher OJ. Children are not just little adults: recent advances in understanding of diffuse intrinsic pontine glioma biology. Pediatr Res. 2014; 75(1-2):205–209. [DOI] [PubMed] [Google Scholar]
- 2. Castel D, Philippe C, Calmon R, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015; 130(6):815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012; 124(3):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan KM, Fang D, Gan H, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013; 27(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mohammad F, Weissmann S, Leblanc B, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017; 23(4):483–492. [DOI] [PubMed] [Google Scholar]
- 6. Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998; 8(2):58–65. [PubMed] [Google Scholar]
- 7. Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990; 61(1):49–59. [DOI] [PubMed] [Google Scholar]
- 8. Soroceanu L, Murase R, Limbad C, et al. Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2013; 73(5):1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McAllister SD, Christian RT, Horowitz MP, Garcia A, Desprez PY. Cannabidiol as a novel inhibitor of Id-1 gene expression in aggressive breast cancer cells. Mol Cancer Ther. 2007; 6(11):2921–2927. [DOI] [PubMed] [Google Scholar]
- 10. Buczkowicz P, Hoeman C, Rakopoulos P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014; 46(5):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoeman CM, Cordero FJ, Hu G, et al. ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat Commun. 2019; 10(1):1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fortin J, Tian R, Zarrabi I, et al. Mutant ACVR1 arrests glial cell differentiation to drive tumorigenesis in pediatric gliomas. Cancer Cell. 2020; 37(3):308–323 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patel SK, Hartley RM, Wei X, et al. Generation of diffuse intrinsic pontine glioma mouse models by brainstem-targeted in utero electroporation. Neuro-Oncology. 2020; 22(3):381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pathania M, De Jay N, Maestro N, et al. H3.3(K27M) Cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell. 2017; 32(5):684–700 e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miklja Z, Yadav VN, Cartaxo RT, et al. Everolimus improves the efficacy of dasatinib in PDGFRα-driven glioma. J Clin Invest. 2020; 130(10):5313–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pajovic S, Siddaway R, Bridge T, et al. Epigenetic activation of a RAS/MYC axis in H3.3K27M-driven cancer. Nat Commun. 2020; 11(1):6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assays. J Vis Exp. 2014; 88:e51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007; 2(2):329–333. [DOI] [PubMed] [Google Scholar]
- 19. Lewis PW, Muller MM, Koletsky MS, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013; 340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filbin MG, Tirosh I, Hovestadt V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 2018; 360(6386):331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 2017; 32(4):520–537 e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schoppmann SF, Schindl M, Bayer G, et al. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int J Cancer. 2003; 104(6):677–682. [DOI] [PubMed] [Google Scholar]
- 23. Davis CA, Hitz BC, Sloan CA, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46(D1):D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan X, Fu Y, Zhou X, et al. Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci Adv. 2020; 6(34):eaaz2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jessa S, Blanchet-Cohen A, Krug B, et al. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet. 2019; 51(12):1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sachdeva R, Wu M, Smiljanic S, et al. ID1 is critical for tumorigenesis and regulates chemoresistance in glioblastoma. Cancer Res. 2019; 79(16):4057–4071. [DOI] [PubMed] [Google Scholar]
- 27. Tada K, Kawahara K, Matsushita S, et al. MK615, a Prunus mume Steb. Et Zucc (“Ume”) extract, attenuates the growth of A375 melanoma cells by inhibiting the ERK1/2-Id-1 pathway. Phytother Res. 2012; 26(6):833–838. [DOI] [PubMed] [Google Scholar]
- 28. Wilkie G, Sakr B, Rizack T. Medical Marijuana use in oncology: a review. JAMA Oncol. 2016; 2(5):670–675. [DOI] [PubMed] [Google Scholar]
- 29. McAllister SD, Murase R, Christian RT, et al. Pathways mediating the effects of cannabidiol on the reduction of breast cancer cell proliferation, invasion, and metastasis. Breast Cancer Res Treat. 2011; 129(1):37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singer E, Judkins J, Salomonis N, et al. Reactive oxygen species-mediated therapeutic response and resistance in glioblastoma. Cell Death Dis. 2015; 6:e1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilkie G, Sakr B, Rizack T. Medical marijuana use in oncology: a review. JAMA Oncol. 2016; 2(5):670–675. [DOI] [PubMed] [Google Scholar]
- 32. Tzeng SF, de Vellis J. Id1, Id2, and Id3 gene expression in neural cells during development. Glia. 1998; 24(4):372–381. [DOI] [PubMed] [Google Scholar]
- 33. Ling MT, Wang X, Zhang X, Wong YC. The multiple roles of Id-1 in cancer progression. Differentiation. 2006; 74(9-10):481–487. [DOI] [PubMed] [Google Scholar]
- 34. Sachdeva R, Wu M, Smiljanic S, et al. ID1 is critical for tumorigenesis and regulates chemoresistance in glioblastoma. Cancer Res. 2019; 79(16):4057–4071. [DOI] [PubMed] [Google Scholar]
- 35. Piunti A, Hashizume R, Morgan MA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017; 23(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larson JD, Kasper LH, Paugh BS, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019; 35(1):140–155 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin EY, Cooper DD, Abbott KL, et al. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell. 2017; 170(5):845–859.e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003; 11(4):915–926. [DOI] [PubMed] [Google Scholar]
- 39. Devinsky O, Verducci C, Thiele EA, et al. Open-label use of highly purified CBD (Epidiolex(R)) in patients with CDKL5 deficiency disorder and Aicardi, Dup15q, and Doose syndromes. Epilepsy Behav. 2018; 86:131–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.