Abstract
Pasteurella multocida is the causative agent of infectious diseases of economic importance such as fowl cholera, bovine hemorrhagic septicemia, and porcine atrophic rhinitis. However, knowledge of the molecular mechanisms and determinants that P. multocida requires for virulence and pathogenicity is still limited. To address this issue, we developed a genetic expression system, based on the in vivo expression technology approach first described by Mahan et al. (Science 259:686–688, 1993), to identify in vivo-expressed genes of P. multocida. Numerous genes, such as those encoding outer membrane lipoproteins, metabolic and biosynthetic enzymes, and a number of hypothetical proteins, were identified. These may prove to be useful targets for attenuating mutation and/or warrant further investigation for their roles in immunity and/or pathogenesis.
Pasteurella multocida is an opportunistic veterinary and human pathogen with worldwide distribution. Certain serotypes are the etiologic agents of severe types of pasteurellosis, such as fowl cholera in avian species, hemorrhagic septicemia in cattle and buffalo, and atrophic rhinitis in swine. Despite considerable research into the mechanisms of immunity, virulence, and pathogenesis, safe and effective vaccines against pasteurellosis are still lacking and little is known of the molecular mechanisms of pathogenesis.
Mahan et al. (35) first described a system to identify in vivo-expressed genes and termed this “in vivo expression technology” (IVET). Various IVET systems have since been designed and used in a number of different organisms (reviewed in references 9, 24, and 25). Information gained from these research efforts has identified a number of known virulence factors, metabolic and biosynthetic genes, and, interestingly, many genes with no known function. IVET systems provide an insight into the genes which are required for survival and multiplication in vivo, and the gene products identified may represent new targets for attenuating mutations, antimicrobial agents, or recombinant vaccines. The inactivation of genes identified by IVET systems has, in many cases, resulted in the attenuation of virulence, indicating an important role for these in vivo-expressed genes in pathogenesis (34, 52). In addition, the in vivo promoters themselves could be utilized for heterologous antigen expression in vivo.
Outer membrane protein preparations from in vivo-grown P. multocida cells protect birds from heterologous serotypes, whereas in vitro-grown bacteria provide protection only against the homologous somatic serotype (17, 22, 23). The in vivo-expressed antigens involved in providing heterologous protection have been termed the cross-protective factors. Much interest has been focused on identifying the cross-protective factors of P. multocida fowl cholera strains, yet none has been isolated and characterized to date. The IVET system provides a new approach for identifying such genes and overcomes the limitations of using in vitro media and conditions to mimic the host factors responsible for triggering bacterial gene expression in vivo. The IVET approach is designed to identify simultaneously a number of genes expressed in vivo.
This report describes an IVET system for use in P. multocida, termed PmIVET, to identify genes that are expressed exclusively or preferentially during infection. A plasmid-based promoter-probe system which relies on the expression of an antibiotic resistance marker, kan, was designed and constructed for use in P. multocida. A plasmid carrying a promoterless kan gene, pMKΩ, was constructed, and fragments were cloned upstream of this gene to generate transcriptional fusions which were then introduced into the virulent P. multocida strain X-73. After infection with P. multocida, mice were treated with kanamycin (KAN), and in vivo-expressed genes were identified by analyzing the bacteria that survived in vivo but were KAN sensitive (Kans) in vitro.
MATERIALS AND METHODS
Media.
P. multocida strains were cultured at 37°C on nutrient agar (NA) or in nutrient broth (NB). Escherichia coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar at 37°C. Antibiotics were added when required at the following concentrations: ampicillin, 100 μg ml−1; KAN, 5, 10, 20, 30, 40, or 50 μg ml−1; streptomycin (STR), 25 μg ml−1; and spectinomycin (SPE), 25 μg ml−1.
Individual P. multocida strains for mouse in vivo selection assays were grown overnight in NB, diluted 1:100 in 10 ml of fresh NB, and incubated with shaking at 37°C for 4 to 6 h. The absorbance at 600 nm of the cultures was determined, after which the cultures were diluted in sterile phosphate-buffered saline, pH 7.2, to provide the required number of CFU.
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| P. multocida X-73 | Serogroup A: serotype 1 chicken isolate; reference strain | 23 |
| PmIVET clones | ||
| MHS6 | X-73 harboring three noncontiguous X-73 genomic Sau3AI fragments, 355, 253, and 127 bp, in pMKΩ | This study |
| MHS7 | X-73 harboring three noncontiguous X-73 genomic Sau3AI fragments, 143, 149, and 967 bp, in pMKΩ | This study |
| MHS17 | X-73 harboring two noncontiguous X-73 genomic Sau3AI fragments, 339 and 191 bp, in pMKΩ | This study |
| MHS23 | X-73 harboring a 604-bp X-74 genomic Sau3AI fragment in pMKΩ | This study |
| MHS25 | X-73 harboring four noncontiguous X-73 genomic Sau3AI fragments, 94, 98, 351, and 144 bp, in pMKΩ | This study |
| MHS27 | X-73 harboring a 465-bp X-73 genomic Sau3AI fragment in pMKΩ | This study |
| MHS30 | X-73 harboring three noncontiguous X-73 genomic Sau3AI fragments, 123, 276, and 387 bp, in pMKΩ | This study |
| MHT6 | X-73 harboring a 267-bp X-73 genomic Sau3AI fragment in pMKΩ | This study |
| MHW32 | X-73 harboring three noncontiguous X-73 genomic Sau3AI fragments, 666, 267, and 129 bp, in pMKΩ | This study |
| MHW39 | X-73 harboring two noncontiguous X-73 genomic Sau3AI fragments, 198 and 909 bp, in pMKΩ | This study |
| MHX6 | X-73 harboring two noncontiguous X-73 genomic Sau3AI fragments, 431 and 549 bp, in pMKΩ | This study |
| MHY6 | X-73 harboring four noncontiguous X-73 genomic Sau3AI fragments, 171, 1,111, 158, and 876 bp, in pMKΩ | This study |
| MHY29 | X-73 harboring two noncontiguous X-73 genomic Sau3AI fragments, 119 and 261 bp, in pMKΩ | This study |
| MHY40 | X-73 harboring six noncontiguous X-73 genomic Sau3AI fragments, 444, 295, 465, 092, 021, and 123 bp, in pMKΩ | This study |
| MHZ12 | X-73 harboring a 66-bp X-73 genomic Sau3AI fragment in pMKΩ | This study |
| Plasmids | ||
| pUC4-KIXX | Ampr, Kanr, ColE1 origin | 51 |
| pUCΩ | Ampr, Sper, Strr, ColE1 origin | 46 |
| pPMK1 | 4.9-kb Kanr, P. multocida origin | 6 |
| pPBA844 | 2.0-kb BamHI SPE/STR cassette of pUCΩ cloned into BglII-digested pPMK1, removing kan gene promoter | This study |
| pMKΩ | SPE and STR selection in vitro; 6.5 kb Expand PCR product generated using primers BAP-03 and BAP-04 from pPBA844 template, digested with BamHI and religated on itself; used for selection of in vivo-expressed genes in PmIVET | This study |
DNA manipulations.
P. multocida genomic DNA was prepared using the method of Ausubel et al. (4), and plasmid DNA from E. coli and P. multocida strains was prepared as described by Le Gouill et al. (32). DNA was digested using restriction endonucleases supplied by Roche Molecular Biochemicals (Basel, Switzerland) or New England Biolabs Inc. (Beverly, Mass.) under conditions recommended by the manufacturer.
Library construction.
P. multocida strain X-73 genomic DNA was digested to completion with Sau3AI, and the resulting fragments were purified using a Qiaex II gel extraction kit (Qiagen, Hilden, Germany) before ligation into BamHI-digested, dephosphorylated pMKΩ (Table 1). The ligation mix was used to transform cells of X-73 (49), which were allowed to recover at 37°C for 2 h and then spread onto NA containing 25-μg ml−1 concentrations of both SPE and STR to select transformants containing pMKΩ. Transformants were used in mouse infection experiments to select for recombinant clones expressing KAN resistance (Kanr) in vivo.
Estimation of the in vivo KAN level.
Female BALB/c mice were weighed, and KAN doses of 50, 100, or 200 μg g of body weight−1 were injected intraperitoneally (i.p.). To measure the circulating KAN concentration, 100-μl volumes of heparinized blood obtained from the orbital plexus were added to 5-mm-diameter wells cut in diagnostic sensitivity test agar plates which had been seeded with sufficient E. coli to produce a confluent lawn after subsequent overnight incubation at 37°C. Zones of inhibition of E. coli growth were then measured, and the circulating concentration of KAN in the mouse was determined by comparison with KAN standards. All measurements were performed at least in triplicate.
In vivo library selection.
An X-73 Sau3AI genomic DNA library in the vector pMKΩ was expressed in X-73 cells grown in NB containing 25 μg of SPE and STR ml−1 in vitro. Female 6- to 8-week-old BALB/c outbred mice were injected i.p. with 2.9 × 106 CFU of the library, and the infection was allowed to progress for a minimum of 2 h before an appropriate volume of KAN, calculated by determining the mouse weight, was injected i.p. to give an approximate in vivo level in blood of 100 μg ml−1. Mice were bled from the orbital plexus immediately prior to the injection of KAN and then periodically over 13 h. Blood was plated onto NA containing 25 μg of SPE and STR ml−1 and incubated at 37°C overnight. Bacterial colonies appearing after overnight incubation were patched or replica plated onto NA containing 50 μg of KAN ml−1. Plasmids purified from Kans clones were analyzed further by restriction digestion, and the nucleotide sequence of the cloned inserts was determined to identify putative in vivo-expressed genes.
Nucleotide sequence analysis of the PmIVET clones.
Synthetic oligonucleotides were designed that were homologous to the 5′ and 3′ ends (BAP-1056, 5′-ATCTAGCGAGGGCTTTAC-3′, and BAP-503, 5′-ACCGAATAGCCTCTCCAC-3′, respectively) of the BamHI cloning site of pMKΩ. These were used in either Taq DyeDeoxy terminator cycle kit or Taq Big DyeDeoxy terminator kit (Applied Biosystems Inc., Foster City, Calif.) sequencing reactions, which typically contained 200 to 500 ng of purified template DNA, 6 to 8 μl of Taq DyeDeoxy or Taq Big DyeDeoxy Terminator mix, and 3.2 pmol of the required oligonucleotide in a 15- to 20-μl volume. Cycle sequencing was performed on a Perkin-Elmer GeneAmp PCR system 2400 thermocycler, and the DNA products were purified according to the manufacturer's instructions and analyzed with an Applied Biosystems Inc. automated DNA sequencer, model 373. The Sequencher 3.0 program (Gene Codes Corp., Ann Arbor, Mich.) was used to align and assemble individual sequences. Comparison of sequences with those in GenBank, EMBL, and unfinished genome databases was performed using the BLAST (3) and FASTA (44) programs through the Australian National Genomic Information Service (ANGIS) (http://www.angis.org.au) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Computer analysis of deduced protein sequences was carried out through ANGIS using programs within the Genetics Computer Group sequencing analysis software package (GCG, Inc., Madison, Wis.).
Colony PCR.
Colonies picked from a plate were resuspended in 50 μl of distilled water, boiled for 10 min, then centrifuged at 12,000 × g for 10 min. A 2-μl portion was then used as template DNA in a 20-μl PCR mixture containing 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 0.01% (wt/vol) gelatin, 200 μM (each) dATP, dCTP, dGTP, and dTTP, 1 μM concentrations each of forward and reverse oligonucleotide primers, and 1 μl of Taq DNA polymerase (Roche Molecular Biochemicals). Reactions were performed using a Perkin-Elmer GeneAmp PCR system 2400 thermocycler and 0.6-ml thin-walled tubes. Thermocycling conditions were 94°C for 15 s, 50°C for 30 s, and 72°C for 2 min, using 25 cycles.
RESULTS
Construction of the promoter probe vector pMKΩ for selection of in vivo-expressed genes.
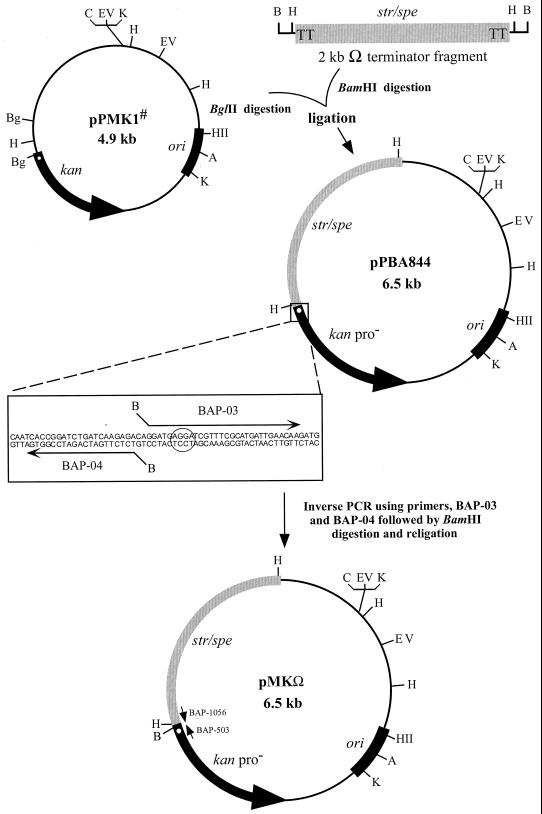
The 4.9-kb plasmid pPMK1 (Table 1; Fig. 1) was constructed by cloning the KAN resistance cartridge from pUC4KIXX (51) into a native cryptic P. multocida plasmid (6). This plasmid was used to generate the promoterless kan gene promoter-probe vector for use in PmIVET. A putative ribosome binding site, AGGA, was present 10 bp upstream of the ATG start codon of the kan gene. The native kan promoter was identified in the region 100 to 160 bp upstream of the start codon. Two BglII sites that could be used to remove the kan gene promoter were identified in pPMK1. The 2-kb BamHI omega (Ω) terminator fragment (46), which carries two transcriptional terminators and translational stop codons in all three frames at either end of the element, in addition to encoding STR and SPE resistance, was cloned into BglII-digested pPMK1 upstream of the promoterless kan gene, resulting in the 6.5-kb plasmid pPBA844 (Fig. 1).
FIG. 1.
Construction of the PmIVET in vivo promoter-probe vector pMKΩ. The P. multocida plasmid origin of replication region (ori) is shown on each plasmid as a black box. The open circle at the 5′ end of the kan gene indicates the ribosome binding site, AGGA. The oligonucleotide primers BAP-03 and BAP-04, used for inverse PCR to introduce a unique BamHI restriction site, are shown as bent arrows above and below the sequence in the expanded boxed section. The ribosome binding site preceding the start of the kan gene is circled. The positions of two sequencing primers, BAP-503 and BAP-1056, are denoted by the small arrows. Restriction site abbreviations: A, AvaI; B, BamHI; Bg, BglII; C, ClaI; EV, EcoRV; H, HindIII; HII, HindII; K, KpnI. #, the approximate size of pPMK1, published as 4.5 kb by Bills et al. (6), has been subsequently reestimated to be 4.9 kb as shown here.
To allow the cloning of P. multocida genomic DNA Sau3AI fragments upstream of the promoterless kan gene in pPBA844, inverse PCR was employed, using outward-facing primers carrying BamHI sites at their 5′ ends, to introduce a unique BamHI cloning site (Fig. 1). The resulting vector, carrying a promoterless kan gene, an Ω terminator fragment, a P. multocida plasmid origin of replication, and a unique BamHI cloning site upstream of the kan gene, was designated pMKΩ (Fig. 1).
P. multocida carrying pMKΩ was unable to grow in NB or on NA plates containing >5 μg of KAN ml−1, whereas P. multocida carrying the vector pPMK1 grew in NB containing up to 400 μg of KAN ml−1.
Survival of P. multocida harboring either pPMK1 or pMKΩ in KAN-treated mice.
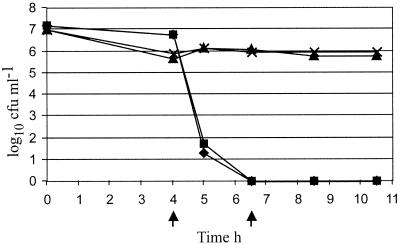
A KAN-treated mouse infection model that selected for bacteria expressing Kanr in vivo but did not permit the survival of detectable levels of Kans bacteria was established. A KAN dose of 100 μg g of body weight−1 was found to remove Kans organisms in vivo while allowing Kanr organisms to multiply and infect mice. Pairs of mice were given either 1 × 107 CFU of X-73(pPMK1) or 2 × 107 CFU of X-73(pMKΩ) and then given a KAN dose (100 μg g of body weight−1) after 4 and 6.5 h (Fig. 2). The effectiveness of even a single KAN dose was demonstrated by the significant reduction in bacterial numbers observed in the mice infected with X-73(pMKΩ), where within 1 h of the first KAN dose, the numbers had decreased from 6 × 106 CFU ml−1 to 20 to 50 CFU ml−1 (Fig. 2). No reduction in numbers was seen for the mice infected with X-73(pPMK1) after the first KAN dose. A second KAN dose was given after 2.5 h, and the circulating bacteria were enumerated after a further 2 and 4 h. No X-73(pMKΩ) bacteria were detected, whereas X-73(pPMK1) continued to be detected in high numbers (Fig. 2).
FIG. 2.
Survival of P. multocida strain X-73(pPMK1) (▴, ×) and X-73(pMKΩ) (▪, ♦) in KAN-treated mice. The arrows indicate the times at which a KAN dose of 100 μg g of body weight−1 was injected. The value given on the y axis at 0 h is the total CFU administered to each mouse to initiate infection. All other values plotted are CFU milliliter of blood−1. Where a y value of 100 is given, no bacteria were recovered.
Generation of an X-73 genomic DNA Sau3AI library in pMKΩ.
The genomic library contained a total of 10,300 transformants, of which approximately 61% (6,300) contained insert DNA with an average size of 0.8 kb. Using the Clarke and Carbon (10) formula and the approximate genome size of 2,350 kb of another A:1 strain (26) (the size of the X-73 genome is not known), this library was estimated to cover 46% of the genome. Replica plating of 500 colonies onto NA plates containing 50 μg of KAN ml−1 demonstrated that the library contained approximately 4.4% in vitro Kanr bacteria prior to in vivo selection. This experiment also confirmed the function of the kan gene in pMKΩ.
Selection of in vivo-active promoters.
The transformants were scraped off plates and pooled into 200 ml of NB containing SPE and STR before incubation for 1.5 h at 37°C. Aliquots of this suspension (representing 3 × 106 CFU) were injected i.p. into four pairs of mice for in vivo selection of clones carrying promoter elements expressed during infection. A fifth pair of mice was injected i.p. with 105 CFU of X-73(pMKΩ) as a control for the in vivo clearance of Kans bacteria. KAN was administered to this control group at 2 h postinfection. The four pairs of mice, used for the in vivo selection of the X-73 library, were treated with KAN at various time points in an attempt to maximize the recovery of in vivo-expressed genes by allowing the infection to progress to different stages before KAN treatment.
Blood was taken from the mice at various times over a 13-h period and bacteria were selected on NA plates containing SPE and STR at 37°C. Of the resulting colonies recovered from the mice, 2,900 were transferred to NA plates containing 50 μg of KAN ml−1 to determine the in vitro KAN resistance phenotype. Of these, 292 (10%) were found to be Kans, indicating that a promoter functioning in vitro was not present upstream of the promoterless kan gene of pMKΩ in these clones.
Plasmid DNA was prepared from the 292 Kans clones, and 159 of these (54.5%) were shown to contain insert DNA in pMKΩ. The nucleotide sequence of insert DNA was obtained using the two oligonucleotide primers, BAP-503 and BAP-1056. Analysis of the individual nucleotide sequences and comparison with sequences in GenBank and other databases identified 76 clones which could potentially contain promoter regions driving the expression of kan in vivo.
Repassaging potential in vivo-expressed clones through KAN-treated mice.
To confirm the specificity of in vivo selection, clones were repassaged individually using pairs of mice. In these experiments each clone (105 to 106 CFU) was injected i.p. into pairs of mice together with X-73(pMKΩ) (105 to 106 CFU) as an internal control for in vivo clearance of Kans organisms. The mice were treated with KAN (100 μg g of body weight−1) at 2 and 4 h postinfection. Bacteria were recovered from blood at 8 h, and Kanr bacteria were plated on NA containing 25 μg of SPE and STR ml−1. Measurement of blood KAN levels using this regimen indicated that concentrations of 80 to 100 μg ml of blood−1 were achieved 15 min after each KAN injection in mice. Random colonies were patched onto NA containing 50 μg of KAN ml−1 to check in vitro sensitivity. Where possible, 12 colonies were screened by PCR from each plate using the primers BAP-503 and BAP-1056 to differentiate between clones containing plasmids with inserts and those containing pMKΩ vector only. Plasmids were selected as potentially carrying an in vivo-expressed promoter only if they were recovered without corecovery of bacteria carrying pMKΩ. Repassaging the clones through mice with the internal pMKΩ control greatly improved the specificity of the PmIVET system.
Of the 76 clones with potential in vivo promoters, only 43 contained unique fragments (Table 2) and one representative of each of these gene fusions was used for repassaging. After repassaging, 17 of the 43 clones were recovered from mice without coisolation of X-73 carrying pMKΩ. These clones were designated as carrying an in vivo-expressed promoter element. As a number of these clones contained noncontiguous Sau3AI fragments, definite assignment of some in vivo-expressed genes is not yet possible. However, from nucleotide sequence analysis of the individual Sau3AI fragments of each clone, the promoter region of the gene most likely to be driving kan expression in vivo was predicted. The analysis of these clones is detailed in Table 2 and in Discussion. Of the remaining 25 repassaged clones, 14 were not recovered and 11 clones were reisolated but with the concurrent isolation of X-73(pMKΩ), indicating that KAN selection in these mice was insufficient to remove Kans bacteria.
TABLE 2.
P. multocida genes identified as in vivo expressed
| Category | Straina | CFU recovered after repassage in vivob | In vitro KAN MIC (μg/ml) | Gene identified as in vivo expressed | % Amino acid identity (similarity)c | Function or role of homolog | Pm70 no.d |
|---|---|---|---|---|---|---|---|
| Lipoproteins | MHS25 (3) | +++ | 30 | hpd | 65 (78) to protein D | Glycerol metabolism, surface-exposed lipoprotein | PM1444 |
| MHS30 (6) | ++ | 10 | pcp | 73 (88) to PCP | Outer membrane-associated lipoprotein | PM0554 | |
| Pyrimidine synthesis and salvage functions | MHS17 (1) | +++ | 20 | dcd | 91 (97) to Dcd | Deoxycytidine deaminase | PM0951 |
| Biosynthetic and metabolic functions | MHS23 (6) | +++ | 10 | dsbD | 43 (58) to DsbD | Thiol-disulfide interchange protein | PM0221 |
| MHT6 | + | 10 | speF | 56 (77) to SpeF | Inducible ornithine decarboxylase, maintenance of cellular polyamines | PM0806 | |
| MHZ12 | + | 20 | ackA-pta | Only putative promoter region of pta present | Fermentation of acetyl-CoA; regulation of virulence gene expression | PM0704 and PM705 | |
| MHW40 | + | 10 | srlD | 81 (87) to SrlD of E. amylovora | Sorbitol-6-phosphate dehydrogenase | PM1968 | |
| MHS7 (2) | ++ | 10 | nrfE | Only putative promoter region present | Formate-dependent nitrite reduction protein | PM0027 | |
| MHX6 | ++ | 10 | yiaK | 71 (83) to YiaK | Putative dehydrogenase | PM1256 | |
| Hypothetical or unknown proteins | MHS6 | + | 40 | ycbK | 64 (82) to YcbK | Unknown | PM0271 |
| MSH27 (10) | + | 10 | ycbL | 90 (100) to YcbL | Unknown | PM0272 | |
| MHW39 | + | 5 | ychN | 44 (76) to YchN of E. coli | Unknown | PM0514 | |
| MHW32 | + | 0 | orfX | No database match | Unknown | ||
| MHY6 | ++ | 10 | No database match | Unknown | |||
| MHY29 | + | 5 | H10894 | Only putative promoter region present | Putative membrane protein | PM1135 | |
| MHY40 | +++ | 20 | yeeX | 80 (89) to YeeX of E. coli | Putative alpha-helix protein | PM0836 |
Strain isolated multiple times from different mice during primary in vivo selection. The numbers in parentheses indicate the number of other identical clones found.
Approximate numbers recovered after repassaging strains in mice, +, 10 to 102 CFU; ++, 102 to 103 CFU; +++, >103 CFU.
All database matches were to proteins of H. influenzae unless otherwise noted.
Gene designation scheme for the P. multocida Pm70 genome sequence (http://www.cbc.umn.edu/ResearchProjects/AGAC/Pm/pmhome.html).
In vitro KAN MIC.
Clones containing in vivo-expressed promoter regions upstream of the pMKΩ kan gene were grown in vitro on NA plates containing 5, 10, 20, 30, or 40 μg of KAN ml−1 to determine if the clones demonstrated any detectable KAN resistance in vitro. Various levels of KAN resistance were demonstrated by the clones, indicating different basal levels of expression from the cloned regions upstream of the promoterless kan gene of pMKΩ (Table 2). In vitro KAN resistance levels appeared to have no relation to bacterial numbers recovered from mice, indicating that survival in vivo was independent of the in vitro KAN resistance.
DISCUSSION
A system for the identification of in vivo-expressed genes of P. multocida was constructed and tested in mice. As this PmIVET system is plasmid based, it is readily transferable to any P. multocida strain provided that the plasmid can be maintained and KAN resistance can be expressed. The method of selection is straightforward, as it does not require complex media or the generation of P. multocida mutants. Importantly, the inherent problems of variability between individual animals were effectively addressed for PmIVET by the use of an internal Kans X-73(pMKΩ) control during repassaging.
Using various IVET systems, researchers have noted significant in vivo selection in animal models, with approximately 86 to 95% of the recovered population expressing the phenotype required for survival in vivo (35, 52, 56). Significant selective pressure for Kanr bacteria appears to have been exerted in vivo when the prepassaged Kanr level in the library, measured at 4.4%, was compared to the 90% Kanr bacteria recovered from mice after the first in vivo selection of the library pool. However, of the 292 Kans clones recovered after the initial in vivo library selection, 45.5% contained pMKΩ without any insert DNA. Variability in the circulating KAN concentrations between mice may play a role in the survival of some Kans bacteria in vivo. Measurement of the blood KAN levels showed that the concentration of KAN reached the required level 15 min after injection. To address the problem of variability, repassaging was performed using duplicate KAN doses 2 h apart to maintain KAN selection for a longer period of time. In addition, mice were injected simultaneously with both the clone to be tested and a similar dose of X-73(pMKΩ). Clones were then selected as potentially carrying an in vivo-expressed gene only if after in vivo KAN selection the clone demonstrated an in vitro Kans phenotype and was recovered without corecovery of bacteria carrying pMKΩ. Multiple isolations of identical clones from mice with different treatment regimens and the occurrence of identical clones isolated from more than one mouse during the first round of in vivo selection also provided confidence in the PmIVET selection procedure. Of the 46 clones that were chosen for repassage through mice, 8 were isolated a number of times. Six of these were recovered after a second round of PmIVET selection. The other two clones were recovered after repassage in mice but in conjunction with X-73(pMKΩ), indicating that selection in these mice was inadequate. Thus, the classification of these clones as potentially expressing Kanr in vivo awaits further testing.
In vivo-expressed genes identified.
Although a number of clones carried noncontiguous genomic Sau3AI fragments and thus we cannot rule out the possibility that some Sau3AI fusions have created fortuitous in vivo-active promoters, we propose that the promoters of the forward-pointing open reading frames (ORFs) in these clones were responsible for the expression of kan. In the following discussion, PM numbers refer to the gene designation scheme for the P. multocida Pm70 genome sequence (http://www.cbc.umn.edu/Research_Projects/AGAC/Pm/pmhome.html).
Lipoproteins.
The identification of potentially in vivo-expressed putative membrane proteins may reflect the change in the bacterial surface properties required during infection. Two clones that contained the forward-pointing ORFs pcp (PM0554) and hpd (PM1444) were identified. The amino acid sequences deduced from these ORFs showed 88 and 78% similarity, respectively, to two membrane lipoproteins of Haemophilus influenzae, PCP and protein D, encoded by the pcp and hpd genes, respectively. Multiple isolations of both clones were obtained (Table 2). In nontypeable H. influenzae, PCP was found to be antigenically conserved and has shown promise for use in recombinant subunit vaccines (12, 18, 19). The immunogenic 42-kDa surface-exposed protein D is widely distributed among H. influenzae strains (1) and has been shown to mediate binding to immunoglobulin D, which may represent a form of immune evasion by the bacteria (47, 48). Protein D is also involved in the metabolism of glycerol, mediating glycerophosphodiester phosphodiesterase activity (40), and has been shown to cause damage to epithelial-cell cilia in a nasopharyngeal tissue culture model (27). Additionally, an H. influenzae hpd mutant was found to be 100 times less virulent than the wild-type strain (28). The identification of an in vivo-expressed P. multocida homologue of protein D raises the possibility that it may play a similar role in immunity and virulence.
Pyrimidine synthesis and salvage.
IVET systems for a number of organisms have identified genes involved in pyrimidine and purine biosynthesis and nucleotide recycling in vivo (24, 25, 36). Mammalian blood contains very low levels of purines and pyrimidines, and thus, blood-borne pathogenic bacteria must synthesize nucleotides de novo. The ORF fused to kan on MHS17 showed sequence similarity to a dCTP deaminase (dcd) of H. influenzae Rd (14). Upstream of P. multocida dcd (PM0951), a gene encoding the enzyme uridine kinase (udk) involved in pyrimidine salvage was identified (Table 2). A similar arrangement occurs in E. coli and H. influenzae Rd (7, 14). The E. coli and Salmonella enterica serovar Typhimurium dcd genes are involved in the formation of dUTP, a precursor for the de novo synthesis of thymidylate (41, 54). Although the udk and dcd genes on MHS17 were separated by only 20 bp, suggesting that they are part of an operon and thus transcribed from a promoter preceding udk, the possibility exists that sequences upstream of and within the udk gene could allow transcription of dcd in vivo.
Biosynthetic and metabolic genes.
Many in vivo-expressed genes identified to date have been involved in biosynthesis, metabolism, or nutrient acquisition. The identification of such genes reveals information about the environmental stimuli within the host during infection that may act as signals for the induction of bacterial genes to complement nutrient limiting conditions and signal the induction of virulence genes required for immediate survival and spread to other anatomical sites of infection (24).
The P. multocida dsbD homologue (PM0221) was fused to kan on clone MHS23. After the first PmIVET in vivo selection, six clones identical to MHS23 were recovered independently (Tables 1 and 2). In E. coli the integral membrane protein, DsbD, is essential for growth above 42°C, with dsbD transcripts still detectable at 50°C (39). This may contribute to the virulence of X-73, given that the normal body temperature of poultry ranges between 39 and 43°C. The dsbD gene in E. coli is transcribed from its own promoter and is involved in disulfide bond formation in periplasmic proteins. As disulfide bonds are often essential for the proper folding, stability, and activity of many extracellular proteins, the expression of such a gene in vivo may have implications in bacterial pathogenesis for the secretion of toxins or other virulence factors. Recently, Fuller et al. (16) found that mutation of dsbB, also involved in disulfide bond formation, caused attenuation of virulence in P. multocida in a septicemic mouse model. Additionally, DsbD is required for c-type cytochrome biogenesis under anaerobic conditions and plays a role in copper (Cu2+) tolerance in bacteria such as E. coli, S. enterica serovar Typhimurium, and P. aeruginosa (11, 21, 42). Other genes involved in Cu2+ homeostasis have been identified, using IVET systems, from S. enterica serovar Typhimurium (24) and Staphylococcus aureus (34).
A P. multocida homolog of the H. influenzae speF gene (14) (PM0806) that encodes an ornithine decarboxylase, was found to potentially drive kan expression in vivo (MHT6) (Table 2). Similarity to E. coli SpeF, a biodegradative enzyme for the production of putrescine that is inducible at low pH in the presence of ornithine, was also found (30). In E. coli and H. influenzae, speF is the first gene of the speF-potE operon which is involved in the maintenance of cellular polyamines required for normal cell growth (50) and transport of putrescine (30). Induction of this operon is believed to neutralize the extracellular medium via the excretion of putrescine (31). Other genes involved in biosynthesis of the polyamines cadaverine, spermidine, and putrescine have been isolated from S. enterica serovar Typhimurium, Streptococcus pneumoniae, and Vibrio cholerae using IVET and signature-tagged mutagenesis systems (24, 38, 45). These genes are also induced at low pH and are believed to play a role in acid tolerance (38, 43). However, Merrell and Camilli (38) did not find a role for SpeF in acid tolerance of V. cholerae. Further work is required to determine what role speF plays in pathogenesis of pasteurellosis and why this gene is up-regulated in vivo.
A single 66-bp Sau3AI fragment was present in MHZ12. Sequence analysis of this short fragment indicated the presence of a truncated ORF with the same orientation as kan, demonstrating similarity to an acetate kinase, AckA, of H. influenzae Rd (14).
Only 5 bp are present downstream of the P. multocida ackA stop codon on this fragment. Analysis of the P. multocida genome sequence identified a homolog of the H. influenzae Rd phosphotransacetylase gene, pta (14), 78 bp downstream of ackA. Pta (PM0705), together with the acetate kinase AckA (PM0704), is involved in the fermentation of acetyl coenzyme A (acetyl-CoA) to generate ATP and acetate and the reverse process utilizing acetate to produce acetyl-CoA. A potential role for acetyl phosphate, the intermediate of the acetyl-CoA⇄acetate pathway, as an effector of gene regulation through interaction with two component response regulators such as the phosphate regulon of E. coli has been proposed (37, 53). Recently, Chiang and Mekalanos (8) identified an attenuated pta mutant of V. cholerae using signature-tagged mutagenesis. The expression of cholera toxin and the toxin coregulated pilus was also affected by the pta mutation, further suggesting a regulatory relationship between the Pta-AckA metabolic pathway and virulence gene expression (8).
A gene (PM1968) with deduced amino acid sequence similarity to SrlD (also named GutD) of Erwinia amylovora and E. coli (2, 7) involved in the conversion of sorbitol-6-phosphate to fructose-6-phosphate was fused to kan on MHW40 (Table 2). The srlD gene is the third and fourth gene of an operon in E. coli and E. amylovora, respectively (2, 55). Transcription from a promoter upstream of the first gene in the operon, srlA, was found to be sensitive to catabolite repression by glucose and also dependent on repressor and activator proteins (2, 15, 33). However, until further genetic experiments are performed, a promoter present immediately upstream of the srlD gene cannot be ruled out.
An ORF (PM1256) with similarity to a hypothetical protein, HI1031, of H. influenzae Rd (14) and the putative dehydrogenase, YiaK, of E. coli (5) appeared to be the ORF driving kan transcription on MHX6 in vivo. In E. coli yiaK is the first of nine genes in an operon involved in carbohydrate utilization (5). A divergently transcribed repressor of the E. coli yiaK-yiaS operon, yiaJ, was found upstream (5). Analysis of the Pm70 genome sequence also indicated the presence of a yiaJ homologue (PM1257) 217 bp upstream of the P. multocida yiaK gene. The up-regulation of genes involved in the use of alternative carbohydrates may indicate different nutritional requirements in vivo.
Two ORFs with sequence similarity to the formate-dependent nitrate-reducing proteins from E. coli and H. influenzae nrfD and nrfE, respectively, were identified in the clone MHS7. Comparison of MHS7 with the Pm70 genome indicated that the start of nrfD was not present on this clone. However, the nrfE start codon was found 71 bp downstream of nrfD on MHS7, preceding the kan gene. Therefore, a promoter in the intergenic region between nrfD and nrfE may be expressed in vivo. Interestingly, nrfEFG gene products have been implicated as part of a heme lyase that is responsible for attaching heme to cytochrome c552 (nrfA) at a motif that has been shown to be the site of nitrite reduction (13). These data are consistent with previous reports that nrfEFG were essential for nitrite reduction (20).
Hypothetical proteins.
A number of PmIVET-isolated clones contained ORFs which were similar to hypothetical proteins and which appeared to be driving kan expression in vivo. Why these gene products are required and what cellular function they perform in vivo is unknown. It is of interest that two clones contained ORFs not present in the P. multocida Pm70 database (W32 and Y6) which hence may represent genes unique to X-73.
The only forward-pointing ORF of clone MHW32 spanned the entire 129-bp Sau3AI fragment and displayed similarity to an internal region of a hypothetical protein, Sll1723, of Synechocystis sp. strain PCC6803 (29). Amino acid sequence similarity to similar regions of a number of bacterial glycosyl transferases was also seen. Comparison to the Pm70 genome did not reveal any sequence similar to that of MHW32. Thus, this ORF may represent a gene unique to X-73 (Table 2).
A forward-pointing 377-bp ORF was found on a Sau3AI fragment in clone MHY6 that displayed no nucleotide or amino acid sequence similarity to any sequences stored in the databases searched. This ORF may represent a gene unique to X-73, as it did not show similarity to the genome of Pm70.
This study represents the first application of the IVET approach to P. multocida for the identification of genes that are preferentially expressed in vivo. Using a mouse model of infection, a number of genes were isolated that represent in vivo-expressed loci from the fowl cholera-causing isolate X-73. These may prove to be useful targets for attenuating mutation and/or warrant further investigation for their products' roles in immunity and pathogenesis. Additionally, the promoter regions of these in vivo-expressed genes will be useful for the delivery of recombinant antigens in vivo by placing a heterologous gene under the control of an “in vivo” promoter for expression in an attenuated P. multocida strain.
ACKNOWLEDGMENTS
Components of this work were supported by grants from the Australian Research Council, the Australian Centre for International Agricultural Research, and the Rural Industries Research and Development Corporation.
We gratefully acknowledge the excellent technical assistance of Ian McPherson and Vicki Vallance.
REFERENCES
- 1.Akkoyunlu M, Ruan M, Forsgren A. Distribution of protein D, an immunoglobulin D-binding protein, in Haemophilus strains. Infect Immun. 1991;59:1231–1238. doi: 10.1128/iai.59.4.1231-1238.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge P, Metzger M, Geider K. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol Gen Genet. 1997;256:611–619. doi: 10.1007/s004380050609. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- 5.Badia J, Ibanez E, Sabate M, Baldoma L, Aguilar J. A rare 920-kilobase chromosomal inversion mediated by IS1 transposition causes constitutive expression of the yiaK-S operon for carbohydrate utilization in Escherichia coli. J Biol Chem. 1998;273:8376–8381. doi: 10.1074/jbc.273.14.8376. [DOI] [PubMed] [Google Scholar]
- 6.Bills M M, Medd J M, Chappel R J, Adler B. Construction of a shuttle vector for use between Pasteurella multocida and Escherichia coli. Plasmid. 1993;30:268–273. doi: 10.1006/plas.1993.1058. [DOI] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Chiang S L, Mekalanos J J. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 9.Chiang S L, Mekalanos J J, Holden D. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- 10.Clarke L, Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 11.Crooke H, Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 12.Deich R A, Anilionis A, Fulginiti J, Metcalf B J, Quataert S, Quinn-Dey T, Zlotnick G W, Green B A. Antigenic conservation of the 15,000-dalton outer membrane lipoprotein PCP of Haemophilus influenzae and biologic activity of anti-PCP antisera. Infect Immun. 1990;58:3388–3393. doi: 10.1128/iai.58.10.3388-3393.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaves D J, Grove J, Staudenmann W, James P, Poole R K, White S A, Griffiths I, Cole J A. Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine-lysine motif in the cytochrome c552 nitrite reductase from Escherichia coli. Mol Microbiol. 1998;28:205–216. doi: 10.1046/j.1365-2958.1998.00792.x. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Feilds C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Furhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Fratamico P M, Buchanan R L, Cooke P H. Virulence of an Escherichia coli 0157:H7 sorbitol-positive mutant. Appl Environ Microbiol. 1993;59:4245–4252. doi: 10.1128/aem.59.12.4245-4252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuller T E, Kennedy M J, Lowery D E. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb Pathog. 2000;29:25–38. doi: 10.1006/mpat.2000.0365. [DOI] [PubMed] [Google Scholar]
- 17.Glisson J R, Cheng I H. In vivo antigen expression by Pasteurella multocida. Avian Dis. 1991;35:392–396. [PubMed] [Google Scholar]
- 18.Green B A, Farley J E, Quinn-Dey T, Deich R A, Zlotnick G W. The e (P4) outer membrane protein of Haemophilus influenzae: biologic activity of anti-e serum and cloning and sequencing of the structural gene. Infect Immun. 1991;59:3191–3198. doi: 10.1128/iai.59.9.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green B A, Vazquez M E, Zlotnick G W, Quigley-Reape G, Swarts J D, Green I, Cowell J L, Bluestone C D, Doyle W J. Evaluation of mixtures of purified Haemophilus influenzae outer membrane proteins in protection against challenge with nontypeable H. influenzae in the chinchilla otitis media model. Infect Immun. 1993;61:1950–1957. doi: 10.1128/iai.61.5.1950-1957.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove J, Busby S, Cole J. The role of the genes nrf EFG and ccmFH in cytochrome c biosynthesis in Escherichia coli. Mol Gen Genet. 1996;252:332–341. doi: 10.1007/BF02173779. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S D, Wu H C, Rick P D. A Salmonella typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J Bacteriol. 1997;179:4977–4984. doi: 10.1128/jb.179.16.4977-4984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heddleston K L, Gallagher J E, Rebers P A. Fowl cholera: immune response in turkeys. Avian Dis. 1970;14:626–635. [PubMed] [Google Scholar]
- 23.Heddleston K L, Rebers P A. Fowl cholera. Cross immunity induced in turkeys with formalin-killed in-vivo-propagated Pasteurella multocida. Avian Dis. 1972;16:578–586. [PubMed] [Google Scholar]
- 24.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heithoff D M, Conner C P, Mahan M J. Dissecting the biology of a pathogen during infection. Trends Microbiol. 1997;5:509–513. doi: 10.1016/S0966-842X(97)01153-0. [DOI] [PubMed] [Google Scholar]
- 26.Hunt L M, Ruffolo C G, Rajakumar K, Adler B. Physical and genetic map of the Pasteurella multocida A:1 chromosome. J Bacteriol. 1998;180:6054–6058. doi: 10.1128/jb.180.22.6054-6058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janson H, Carlén B, Cervin A, Forsgren A, Magnusdottir A B, Lindberg S, Runer T. Effects on the ciliated epithelium of protein D-producing and -nonproducing nontypeable Haemophilus influenzae in nasopharyngeal tissue cultures. J Infect Dis. 1999;180:737–746. doi: 10.1086/314921. [DOI] [PubMed] [Google Scholar]
- 28.Janson H, Melhus A, Hermansson A, Forsgren A. Protein D, the glycerophosphodiester phosphodiesterase from Haemophilus influenzae with affinity for human immunoglobulin D, influences virulence in a rat otitis model. Infect Immun. 1994;62:4848–4854. doi: 10.1128/iai.62.11.4848-4854.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 30.Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20922–20927. [PubMed] [Google Scholar]
- 31.Kashiwagi K, Watanabe R, Igarashi K. Involvement of ribonuclease III in the enhancement of expression of the speF-potE operon encoding inducible ornithine decarboxylase and polyamine transport protein. Biochem Biophys Res Commun. 1994;200:591–597. doi: 10.1006/bbrc.1994.1489. [DOI] [PubMed] [Google Scholar]
- 32.Le Gouill C, Parent J L, Rola-Pleszczynski M, Stankova J. Analysis of recombinant plasmids by a modified alkaline lysis method. Anal Biochem. 1994;219:164. doi: 10.1006/abio.1994.1250. [DOI] [PubMed] [Google Scholar]
- 33.Lin E E C. Dissimilatory pathways for sugars, polyols, and carbohydrates. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 244–284. [Google Scholar]
- 34.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 35.Mahan M J, Slauch J M, Hanna P C, Camilli A, Tobias J W, Waldor M K, Mekalanos J J. Selection for bacterial genes that are specifically induced in host tissues: the hunt for virulence factors. Infect Agents Dis. 1993;2:263–268. [PubMed] [Google Scholar]
- 36.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 37.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 38.Merrell D S, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 39.Missiakas D, Schwager F, Raina S. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 1995;14:3415–3424. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munson R S, Jr, Sasaki K. Protein D, a putative immunoglobulin D-binding protein produced by Haemophilus influenzae, is glycerophosphodiester phosphodiesterase. J Bacteriol. 1993;175:4569–4571. doi: 10.1128/jb.175.14.4569-4571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuhard J, Nygaard P. Purines and pyrimidines. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 445–473. [Google Scholar]
- 42.Page D M, Saunders N F, Ferguson S J. Disruption of the Pseudomonas aeruginosa dipZ gene, encoding a putative protein-disulfide reductase, leads to partial pleiotropic deficiency in c-type cytochrome biogenesis. Microbiology. 1997;143:3111–3122. doi: 10.1099/00221287-143-10-3111. [DOI] [PubMed] [Google Scholar]
- 43.Park Y K, Bearson B, Bang S H, Bang I S, Foster J W. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 44.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 47.Ruan R M, Akkoyunlu M, Grubb A, Forsgren A. Protein D of Haemophilus influenzae. A novel bacterial surface protein with affinity for human IgD. J Immunol. 1990;145:3379–3384. [PubMed] [Google Scholar]
- 48.Sasaki K, Munson R S., Jr Protein D of Haemophilus influenzae is not a universal immunoglobulin D-binding protein. Infect Immun. 1993;61:3026–3031. doi: 10.1128/iai.61.7.3026-3031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith M, Jessee J, Landers T, Jordan J. High efficiency bacterial electroporation: 1×1010E. coli transformants/μg. Focus. 1990;12:38–40. [Google Scholar]
- 50.Tabor C W, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 51.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wanner B L, Wilmes-Riesenberg M R. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J Bacteriol. 1992;174:2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss B, Wang L. De novo synthesis of thymidylate via deoxycytidine in dcd (dCTP deaminase) mutants of Escherichia coli. J Bacteriol. 1994;176:2194–2199. doi: 10.1128/jb.176.8.2194-2199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada M, Saier M H., Jr Physical and genetic characterization of the glucitol operon in Escherichia coli. J Bacteriol. 1987;169:2990–2994. doi: 10.1128/jb.169.7.2990-2994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]