Abstract
The human leukocyte antigen (HLA) system, or the human version of the major histocompatibility complex (MHC), is known for its extreme polymorphic nature and high heterogeneity. Taking advantage of whole-genome and whole-exome sequencing data, we developed PGG.MHC to provide a platform to explore the diversity of the MHC in Asia as well as in global populations. PGG.MHC currently archives high-resolution HLA alleles of 53 254 samples representing 190 populations spanning 66 countries. PGG.MHC provides: (i) high-quality allele frequencies for eight classical HLA loci (HLA-A, -B, -C, -DQA1, -DQB1, -DRB1, -DPA1 and -DPB1); (ii) visualization of population prevalence of HLA alleles on global, regional, and country-wide levels; (iii) haplotype structure of 134 populations; (iv) two online analysis tools including ‘HLA imputation’ for inferring HLA alleles from SNP genotyping data and ‘HLA association’ to perform case/control studies for HLA-related phenotypes and (v) East Asian–specific reference panels for HLA imputation. Equipped with high-quality frequency data and user-friendly computer tools, we expect that the PGG.MHC database can advance the understanding and facilitate applications of MHC genomic diversity in both evolutionary and medical studies. The PGG.MHC database is freely accessible via https://pog.fudan.edu.cn/pggmhc or https://www.pggmhc.org/pggmhc.
INTRODUCTION
The major histocompatibility complex (MHC) region, located at chromosome position 6p21 of the human genome, harbors multiple class I and class II human leukocyte antigen (HLA) genes, which are co-dominantly expressed to present intracellular and extracellular antigens, respectively. The HLA genes are associated with various autoimmune diseases (1,2), infectious diseases (3), cancers (4) and psychiatric diseases (5). HLA genes also play a vital role in transplantation, for certain encoded HLA molecules can function as antigens which can incite immune response and lead to rejection in transplantation (6). The HLA genes are highly polymorphic, and there are currently 34 422 identified alleles according to the HLA sequence database IPD-IMGT/HLA (Release 3.49 July 2022) (7), allowing fine-tuning of the immune system. The HLA genes display a high degree of diversity among populations and geographic regions. The diversity of HLA alleles can explain the genetic difference in some diseases among populations. For example, previous studies have shown that the HLA-B*27 carrier rate correlates with the population prevalence of ankylosing spondylitis, a type of arthritis of the human spine (8). Moreover, the disease-associated HLA alleles can vary among populations of different ancestry; for instance, pemphigus vulgaris is associated with HLA-DRB1*04:02 in Ashkenazi Jews, while in non-Jewish patients with European or Asian ethnic origins, the disease is associated with HLA-DRB1*14:01/04 and HLA-DQB1*05:03 (9). HLA haplotype denotes a certain combination of HLA alleles on a chromosome, which is very important in matching unrelated donors and recipients in hematopoietic stem cell transplantation (HSCT). A study of German donors used the observed regional differences in HLA-A, -B and -DR alleles and haplotype frequencies to optimize HSCT donor recruitment strategies (10). Therefore, it is essential to investigate the diversity of HLA genes and dissect their haplotype structures within and between populations.
To enable researchers to estimate the allele frequencies of HLA genes and other immune-related genes among populations, the allele frequency net database (AFND) has been constructed to serve as a repository for allele frequency data (11). In the AFND’s latest update, a gold-standard dataset classification criterion was introduced, including allele frequencies summing up to 1 [±1.5%], sample size larger than 50, and 2-field resolution HLA allele. However, there is a lack of unified integration and HLA typing process in the AFND database, which can affect the comparability of allele frequencies between different datasets. Moreover, a limited number of loci have been genotyped in most AFND datasets. For instance, there are fewer than 10 datasets that have seven loci genotyped among golden datasets (12). In addition, to the best of our knowledge, there are no computer tools available in AFND or any other MHC databases for functional analysis of the MHC region.
To address these issues, we developed a database towards understanding the Population Genomics and Genetics (PGG) of MHC, namely PGG.MHC, to explore the diversity of the MHC region in global populations. Eight classical HLA loci (HLA-A, -B, -C, -DQA1, -DQB1, -DRB1, -DPA1 and -DPB1) were genotyped for each sample. By the definition of AFND, 65 datasets in PGG.MHC can be categorized as golden datasets, providing users with high-quality frequency data. For HLA alleles, we offer convenient queries of HLA allele frequencies and visualization of allele frequencies at global, regional, and country-wide levels. For HLA haplotypes, we offer haplotype frequency estimation and haplotype structure visualization of 134 populations.
In addition, we also implemented two computer tools with online interfaces, namely, HLA imputation and HLA association. The HLA imputation analysis tool imputes HLA genotypes for low-density single-nucleotide polymorphism (SNP) genotyping data using three public reference panels, i.e. 1000G_REF, Pan-Asian panel, and Korean panel, as well as two newly constructed East Asian–specific reference panels, i.e. HuaBiao_REF and PGG_REF. The HLA association analysis tool was developed for phenotype associations based on HLA genotypes with a case/control design, where users can customize control data in the PGG.MHC database.
MATERIALS AND METHODS
Data collection
PGG.MHC currently archives 2181 HLA alleles of 53 254 samples representing 190 global populations spanning 66 countries, of which 6054 are whole-genome sequencing (WGS) data, 5002 are whole-exome sequencing (WES) data, and 42 198 are high-density genome-wide genotyping data. The data were collected from the 1000 Genomes Project (KGP) (13), the Human Genome Diversity Project (14), and the Simons Genomic Diversity Project (15), as well as from the HuaBiao project (16), which provides samples from a variety of regional Han Chinese populations, and the PGG project (17,18), which covers samples from diverse populations in East Asia. Taken together, data from the HuaBiao project and the PGG project constitute most of the PGG.MHC data source (>50 000), which captures the intricate regional genetic diversity in East Asian ethnic groups. Detailed information about data sources of PGG.MHC is provided in Supplementary Table S1.
HLA typing
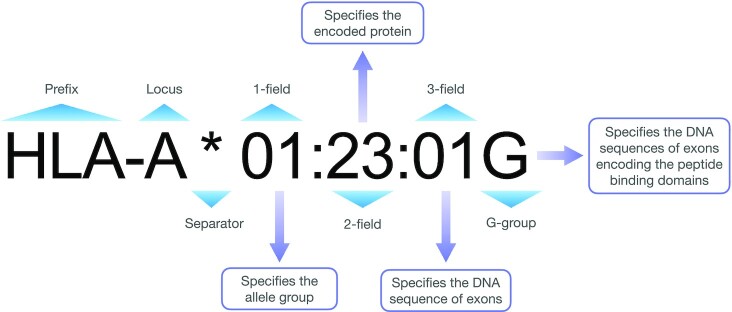
We performed HLA typing for three class I HLA genes (HLA-A, -B and -C) and five class II HLA genes (HLA-DRB1, -DQA1, -DQB1, -DPA1 and -DPB1). These genes were chosen for their important roles in antigen presentation and the fact that they are highly polymorphic (19). We applied corresponding HLA typing techniques for WGS, WES and genotyping data to determine the HLA alleles for each sample. In addition, for WES and WGS samples with high sequencing coverage, we obtained G-group resolution and 3-field resolution HLA alleles when applicable. Details about HLA nomenclature can be found in Figure 1. The complete workflow of HLA typing and genotype integration is described below:
Figure 1.
A schematic diagram for HLA nomenclature.
Direct typing for WGS and WES data
We performed HLA typing using various methods. For WGS data, we adopted kourami (20), HLA*LA (21), HLA-HD (22) and OptiType (23) to perform HLA typing. OptiType could only provide HLA alleles at the 2-field resolution, while HLA*LA and kourami could provide HLA alleles at the G-group resolution. For uniformity of HLA typing resolution, we mapped G-group resolution alleles to 2-field alleles based on each G-group allele's nomenclature. For WES data, we performed typing using HLA-HD, OptiType, and POLYSOLVER (24) on class I genes and HLA-HD, PHLAT (25), and HLA-Genotyper (https://pypi.org/project/hla-genotyper) on class II genes. Next, we integrated the typing results based on different methods for each HLA gene by designating the one with the highest frequency of occurrence as the final HLA genotype. In addition, we used HLA*LA to perform 3-field HLA typing for WGS data and POLYSOLVER for WES data.
Imputation-based HLA typing for genotyping data
Unlike WGS and WES data on which HLA typing can be directly carried out, genotyping data need to be imputed with sufficient SNPs to perform HLA typing. Hence, we adopted HLA-TAPAS (26) for imputation-based HLA typing of genotyping data. In particular, we first preprocessed the data by extracting SNPs of the HLA region using BCFtools (27) and converting the genome coordinates to GRCh38 using CrossMap (28). Second, we annotated the SNPs with dbSNP version 151 (29) and filtered out variants with missing call rates exceeding 0.1 or minor allele frequency below 0.001. Finally, we performed HLA imputation using HLA-TAPAS and summarized the 2-field HLA typing results.
We verified the accuracy of our HLA typing workflow using samples from the KGP. WGS, WES, and genotyping data were acquired for these samples. Benchmark HLA types were obtained from (30). We demonstrated the HLA typing results for different data types and the benchmark HLA types in Supplementary Table S2. It turned out that the overall typing accuracy for WGS/WES/genotyping data are 100%, 100%, 100% at the 1-field resolution, and 98.75%, 92.5%, 98.75% at the 2-field resolution, respectively.
Haplotype structure building
We merged samples of the same population from different data sources to build a haplotype structure. For each population with a sample size larger than 20, haplotype frequencies were estimated between adjacent loci and based on eight HLA loci's 2-field resolution HLA alleles, using an expectation–maximization (EM) algorithm implemented in Hapl-o-Mat (31).
Database implementation
We used HTTPS protocol to process, deliver, and render web resources and to ensure the secure transmission of private user data. The functionality of PGG.MHC has been tested in mainstream web browsers, including Chrome, Safari, IE, Opera and Firefox. We adopted a state-of-the-art architecture (Supplementary Figure S1) to curate data storage and database management. For front-end development, we built the web user interfaces using a progressive JavaScript framework called Vue.js (https://vuejs.org), and data visualization was powered by echarts.js (https://echarts.apache.org). For back-end development, the HTTP web server was created using Node.js (https://nodejs.org), a JavaScript runtime environment. For data storage, the structured data were stored in MySQL (https://www.mysql.com), and the unstructured document data, such as haplotype structures, were stored in MongoDB (https://www.mongodb.com). We used Nginx (https://nginx.org) as the reverse proxy server.
DATABASE CONTENT
Overview
The database has three major functional modules. First, it has easy-to-use interfaces for querying allele frequency and affiliated populations (Figure 2). We also integrated external links to PGG.Population (32) database, containing detailed population genetic structure, links to AFND databases, containing the allele reports, and links to IPG-IMGT/HLA database, containing the sequences of HLA alleles. Second, there is an interactive visualization of allele population prevalence and haplotype structure (Figure 3). The query interfaces and the visualization functions combined can illuminate the genetic structure of populations. Third, there are online analysis functions for genotype imputation and phenotype association (Figure 3). HLA imputation can infer HLA types for low-density SNP genotyping data uploaded by users using five reference panels compiled in our server. Considering the potential bias resulting from the heterogeneity in genetic background between case and control samples when performing association studies, we implemented an online interface for HLA associations where users can choose PGG.MHC populations that share a similar genetic background as their own controls.
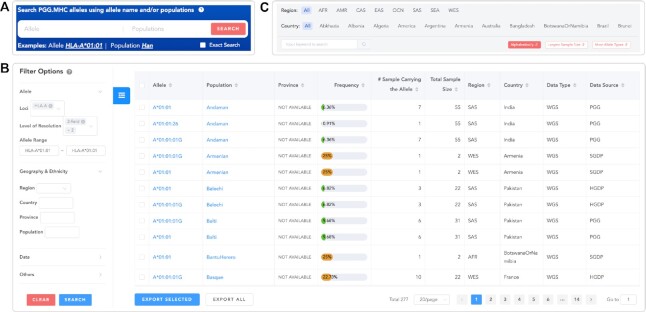
Figure 2.
Interfaces for data query in PGG.MHC. (A) The query interface on the Home page. (B) Filter options and an example of query results. (C) The population query interface on the Population List page.
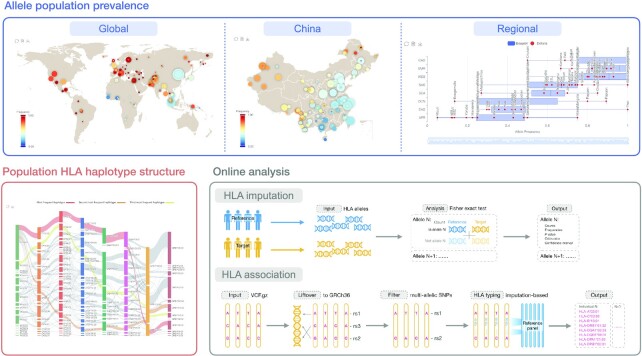
Figure 3.
Sitemap of PGG.MHC. The database consists of three modules: (i) visualization of allele frequencies at global, country-wide, and regional levels; (ii) estimation of haplotype frequency and visualization of population haplotype structure; (iii) HLA imputation and HLA association online analysis tools.
Database query
HLA allele frequency query
PGG.MHC provides powerful query interfaces for searching and browsing HLA allele frequencies by offering an intuitive search box on the home page (Figure 2A) and an extensive query interface on the HLA alleles page (Figure 2B). In the search box on the home page, users can build their queries by providing allele names or populations to search for PGG.MHC allele frequency entries. A click on the search button redirects the web page to the HLA alleles page, demonstrating the matched entries, where users can compare the differences in population frequencies of a specific allele or investigate the variety of HLA alleles in populations of interest. Users can click on the allele name to access the web page for allele details. In the sidebar on the HLA alleles page, users can fine-tune their query by four groups of filter options: (i) designating HLA loci, allele resolution, or allele range; (ii) selecting geographic locations or ethnic groups; (iii) limiting the source and type of data; and (iv) restricting the range of allele frequencies. Moreover, this interface allows users to sort query results by allele, population, frequency, and sample size by clicking the triangles on the right, and it enables users to export the desired entries as an Excel file.
Population query
For users who are interested in HLA alleles of particular populations, we also offer a population query interface on a web page with the population list (Figure 2C), where they can select a region and a country (the web UI will show a complete list of countries when the mouse hovers on country items), or enter a keyword to query populations, and sort populations by different criteria. The population list below displays the population profile cards according to the query. Clicking on the card body redirects the web page to the corresponding population details page.
Population prevalence visualization of HLA alleles
A key point in studying the polymorphisms of HLA alleles in populations is investigating the differences in allele frequencies among populations. PGG.MHC offers population prevalence visualization interfaces of HLA alleles at global, country-wide (China), and regional levels. At global and country-wide levels, we depict population prevalence using a scatter plot on a map, where each scatter denotes a set of samples from the same ethnic group and the same data source, with its color mapping the allele frequency and size mapping the sample size, facilitating users to develop an intuitive sense about population prevalence. At the regional level, we used box plots to summarize allele frequency variation in different geographical regions.
To illustrate the potential application of population prevalence visualization in genetic and clinical studies, we picked the allele HLA-DPA1*01:03 as a showcase and attached the screenshots of its population prevalence visualizations in Figure 3. This allele was previously reported in a genetic study to be associated with an inflammatory rheumatic disease called ankylosing spondylitis (AS) (P = 0.027) (2). In the global population prevalence map, we can see a pattern of allele frequency distribution, in which the allele is more common in Caucasian populations like European and American populations but less common in Japanese and African populations. This pattern fits the observations of previous genetic studies of AS (8), suggesting that the population prevalence visualization has potential value for genetic diversity studies of diseases.
We are aware that the AFND database also visualizes the allele frequency by scatter plot on a global map. However, we improved and expanded the visualization in the following aspects: (i) for the functionality of the interface, we implemented a frequency bar in global and China maps with which users can toggle the upper and lower bounds to selectively display datasets limited to a specific frequency range; (ii) for a different perspective of allele frequency data, we not only offer population prevalence visualization of global and regional perspectives but also zoom-in to the most populous country in the world to depict the HLA allele frequencies in China and (iii) for the manner of display, unlike in AFND, where the allele frequencies in different regions are displayed in separate bar plots, we display the regional frequencies in a single box plot so that users can compare the frequency difference between regions and evaluate the variety of frequencies in a single region.
HLA haplotype structures
We estimated haplotype frequencies based on eight HLA loci at the 2-field resolution and displayed the results in the Sankey diagram and table views. The Sankey diagram view offers an intuitive understanding of the population haplotype structure, where we highlight the top three most frequent HLA haplotypes. Users can export the table view data as an Excel file to compare haplotype frequencies between populations and investigate the population diversity of HLA haplotypes.
The haplotype structure shown in Figure 3 belongs to the Japanese population in our database. The most frequent haplotype we observe (HLA-A*24:02–HLA-C*12:02–HLA-B*52:01–HLA-DRB1*15:02–HLA-DQB1*06:01–HLA-DPA1*02:01–HLA-DPB1*09:01) is in concordance with the previous literature (33), largely validating our estimation of the haplotype frequency.
To the best of our knowledge, long-range haplotype frequency information based on eight loci is only available for the three populations in the AFND database (until 15 July 2022). The PGG.MHC, however, integrates the haplotype structure of 134 populations, including 33 regional subpopulations in China. The population haplotype frequencies we offer are fundamental to disease association studies, population genetics studies, and HLA-matching for HSCT.
ONLINE ANALYSIS
HLA imputation
HLA imputation is an approach that infers HLA alleles for SNP genotyping data using a pre-constructed HLA reference panel, which can subsequently contribute to association studies and disease fine-mapping. PGG.MHC implements an imputation pipeline involving SNP2HLA (34) and offers users an online interface to perform imputation tasks with their data. As for reference panels, we not only include three public reference panels: 1000G_REF (34), Pan-Asian (35) and KOR_REF (36), but also offer two newly constructed reference panels, i.e. HuaBiao_REF and PGG_REF. HuaBiao_REF is a population-specific reference panel based on 5002 WES samples, which we built to facilitate HLA imputation for Han Chinese data. PGG_REF is based on 1836 WGS samples from diverse populations in Asia containing 13 350 SNPs in the HLA region, which can improve the imputation accuracy and typing resolution. Detailed information about reference panels can be found in Supplementary Table S3. The results containing the imputed genotype files in VCF format and the imputed 2-field resolution HLA alleles in TXT format will be sent to users via email. To facilitate the comprehension of this pipeline, users can download example input and output files or utilize the automatic filling button to have a try before operating their data.
HLA association
HLA association is a method for performing case/control studies and investigating the HLA alleles associated with phenotypes or diseases. We implemented an association pipeline in the PGG.MHC server involving PyHLA (37) and offered a user-friendly online interface where users can upload HLA genotypes of samples or choose PGG.MHC samples to perform association. The HLA association interface allows users to choose PGG.MHC populations with a similar genetic structure to their target populations as controls can help to eliminate the influences of the heterogeneous genetic structure of case and control data on the statistical analysis. Moreover, the large sample size of PGG.MHC population can improve the power of association studies.
FUTURE DIRECTIONS
Currently, PGG.MHC archives HLA alleles of 53 254 samples to capture global population diversity, especially in Asia. After HLA typing and genotype integration of the samples, we calculated the allele frequencies and estimated the haplotype frequencies of populations. We provided powerful query and interactive visualization interfaces for allele frequencies and populations. Moreover, we offered two online computer tools for genotype imputation and phenotype association of user-upload data. We expect PGG.MHC to serve as a repository for high-quality HLA allele and haplotype frequency data and facilitate the study of human genetics, vaccine and immunotherapy development, and other related fields.
In the future, we will make an effort to archive as many samples as possible and offer HLA alleles of higher resolution. For the functionality of the database, we plan to develop an advanced haplotype frequency query interface where users can directly input HLA haplotypes or alleles to search. We also have a plan to improve the online analysis functions by visualizing the results to facilitate data interpretation.
DATA AVAILABILITY
The complete workflow of HLA typing and HLA imputation for genotyping data can be found on the group website (https://pog.fudan.edu.cn/#/software) and GitHub repository (https://github.com/Shuhua-Group/PGG.MHC). The use of the data by this work is approved by the Ministry of Science and Technology of the People's Republic of China (No. 2022BAT2237). For validation of the HLA typing workflow, we download WGS (https://www.internationalgenome.org/data-portal/data-collection/30x-grch38), WES (https://www.internationalgenome.org/data-portal/data-collection/phase-3), and genotyping (https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/hd_genotype_chip) samples from the KGP data portal. The benchmark HLA alleles for these samples were downloaded from https://doi.org/10.1371/journal.pone.0206512.s010.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all participants of the Han100K Initiative. We thank the HumPOG IT team for database deployment and technical support. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Contributor Information
Xiaohan Zhao, State Key Laboratory of Genetic Engineering, Center for Evolutionary Biology, Collaborative Innovation Center for Genetics and Development, School of Life Sciences, FudanUniversity, Shanghai 200438, China; Human Phenome Institute, Zhangjiang Fudan International Innovation Center, and Ministry of Education Key Laboratory of Contemporary Anthropology, FudanUniversity, Shanghai 201203, China.
Sen Ma, Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy ofSciences, Shanghai 200031, China.
Baonan Wang, State Key Laboratory of Genetic Engineering, Center for Evolutionary Biology, Collaborative Innovation Center for Genetics and Development, School of Life Sciences, FudanUniversity, Shanghai 200438, China; Human Phenome Institute, Zhangjiang Fudan International Innovation Center, and Ministry of Education Key Laboratory of Contemporary Anthropology, FudanUniversity, Shanghai 201203, China.
Xuetong Jiang, State Key Laboratory of Genetic Engineering, Center for Evolutionary Biology, Collaborative Innovation Center for Genetics and Development, School of Life Sciences, FudanUniversity, Shanghai 200438, China.
Shuhua Xu, State Key Laboratory of Genetic Engineering, Center for Evolutionary Biology, Collaborative Innovation Center for Genetics and Development, School of Life Sciences, FudanUniversity, Shanghai 200438, China; Human Phenome Institute, Zhangjiang Fudan International Innovation Center, and Ministry of Education Key Laboratory of Contemporary Anthropology, FudanUniversity, Shanghai 201203, China; Department of Liver Surgery and Transplantation Liver Cancer Institute, Zhongshan Hospital, FudanUniversity, Shanghai 200032, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Basic Science Center Program [32288101]; The National Natural Science Foundation of China (NSFC) [32030020, 31961130380]; The Strategic Priority Research Program [XDPB17, XDB38000000] of the Chinese Academy of Sciences (CAS); UK Royal Society-Newton Advanced Fellowship [NAF\R1\191094]; Shanghai Municipal Science and Technology Major Project [2017SHZDZX01]. Funding for open access charge: NSFC [31961130380, RS NAF\R1\191094].
Conflict of interest statement. None declared.
REFERENCES
- 1. Apps R., Qi Y., Carlson J.M., Chen H., Gao X., Thomas R., Yuki Y., Del Prete G.Q., Goulder P., Brumme Z.L.et al.. Influence of HLA-C expression level on HIV control. Science. 2013; 340:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Díaz-Peña R., Castro-Santos P., Aransay A.M., Brüges-Armas J., Pimentel-Santos F.M., López-Larrea C.. Genetic study confirms association of HLA-DPA1∗01:03 subtype with ankylosing spondylitis in HLA-B27-positive populations. Hum. Immunol. 2013; 74:764–767. [DOI] [PubMed] [Google Scholar]
- 3. Migliorini F., Torsiello E., Spiezia F., Oliva F., Tingart M., Maffulli N.. Association between HLA genotypes and COVID-19 susceptibility, severity and progression: a comprehensive review of the literature. Eur. J. Med. Res. 2021; 26:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zeestraten E.C.M., Reimers M.S., Saadatmand S., Dekker J.W.T., Liefers G.J., van den Elsen P.J., van de Velde C.J.H., Kuppen P.J.K.. Combined analysis of HLA class i, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br. J. Cancer. 2014; 110:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.-H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H.et al.. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014; 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montgomery R.A., Tatapudi V.S., Leffell M.S., Zachary A.A.. HLA in transplantation. Nat. Rev. Nephrol. 2018; 14:558–570. [DOI] [PubMed] [Google Scholar]
- 7. Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E.. IPD-IMGT/HLA database. Nucleic Acids Res. 2019; 48:D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu X., Wang G., Zhang L., Xu H.. Genetics of ankylosing spondylitis-focusing on the ethnic difference between east asia and europe. Front Genet. 2021; 12:671682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bystryn J.-C., Rudolph J.L.. Pemphigus. Lancet North Am. Ed. 2005; 366:61–73. [DOI] [PubMed] [Google Scholar]
- 10. Schmidt A.H., Solloch U.V., Baier D., Stahr A., Wassmuth R., Ehninger G., Rutt C.. Regional differences in HLA antigen and haplotype frequency distributions in germany and their relevance to the optimization of hematopoietic stem cell donor recruitment. Tissue Antigens. 2010; 76:362–379. [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez-Galarza F.F., McCabe A., Santos E.J.M.d., Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A.et al.. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020; 48:D783–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santos E.J.M.d., McCabe A., Gonzalez-Galarza F.F., Jones A.R., Middleton D.. Allele frequencies net database: improvements for storage of individual genotypes and analysis of existing data. Hum. Immunol. 2016; 77:238–248. [DOI] [PubMed] [Google Scholar]
- 13. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L.et al.. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008; 319:1100–1104. [DOI] [PubMed] [Google Scholar]
- 15. Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A.et al.. The simons genome diversity project: 300 genomes from 142 diverse populations. Nature. 2016; 538:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hao M., Pu W., Li Y., Wen S., Sun C., Ma Y., Zheng H., Chen X., Tan J., Zhang G.et al.. The huabiao project: whole-exome sequencing of 5000 han chinese individuals. J. Genet. Genomics. 2021; 48:1032–1035. [DOI] [PubMed] [Google Scholar]
- 17. Gao Y., Zhang C., Yuan L., Ling Y., Wang X., Liu C., Pan Y., Zhang X., Ma X., Wang Y.et al.. PGG.Han: the han chinese genome database and analysis platform. Nucleic Acids Res. 2020; 48:D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang C., Gao Y., Ning Z., Lu Y., Zhang X., Liu J., Xie B., Xue Z., Wang X., Yuan K.et al.. PGG.SNV: understanding the evolutionary and medical implications of human single nucleotide variations in diverse populations. Genome Biol. 2019; 20:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanchez-Mazas A. A review of HLA allele and SNP associations with highly prevalent infectious diseases in human populations. Swiss Med. Wkly. 2020; 150:w20214. [DOI] [PubMed] [Google Scholar]
- 20. Lee H., Kingsford C.. Kourami: graph-guided assembly for novel human leukocyte antigen allele discovery. Genome Biol. 2018; 19:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dilthey A.T., Mentzer A.J., Carapito R., Cutland C., Cereb N., Madhi S.A., Rhie A., Koren S., Bahram S., McVean G.. HLA* LA—HLA typing from linearly projected graph alignments. Bioinformatics. 2019; 35:4394–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawaguchi S., Higasa K., Shimizu M., Yamada R., Matsuda F.. HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 2017; 38:788–797. [DOI] [PubMed] [Google Scholar]
- 23. Szolek A., Schubert B., Mohr C., Sturm M., Feldhahn M., Kohlbacher O.. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014; 30:3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shukla S.A., Rooney M.S., Rajasagi M., Tiao G., Dixon P.M., Lawrence M.S., Stevens J., Lane W.J., Dellagatta J.L., Steelman S.. Comprehensive analysis of cancer-associated somatic mutations in class i HLA genes. Nat. Biotechnol. 2015; 33:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bai Y., Ni M., Cooper B., Wei Y., Fury W.. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics. 2014; 15:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo Y., Kanai M., Choi W., Li X., Sakaue S., Yamamoto K., Ogawa K., Gutierrez-Arcelus M., Gregersen P.K., Stuart P.E.. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet. 2021; 53:1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M.et al.. Twelve years of SAMtools and BCFtools. Gigascience. 2021; 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao H., Sun Z., Wang J., Huang H., Kocher J.-P., Wang L.. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014; 30:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abi-Rached L., Gouret P., Yeh J.-H., Di Cristofaro J., Pontarotti P., Picard C., Paganini J.. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLoS One. 2018; 13:e0206512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schäfer C., Schmidt A.H., Sauter J.. Hapl-o-Mat: open-source software for HLA haplotype frequency estimation from ambiguous and heterogeneous data. BMC Bioinf. 2017; 18:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C., Gao Y., Liu J., Xue Z., Lu Y., Deng L., Tian L., Feng Q., Xu S.. PGG.Population: a database for understanding the genomic diversity and genetic ancestry of human populations. Nucleic Acids Res. 2018; 46:D984–D993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okada Y., Momozawa Y., Ashikawa K., Kanai M., Matsuda K., Kamatani Y., Takahashi A., Kubo M.. Construction of a population-specific HLA imputation reference panel and its application to graves' disease risk in Japanese. Nat. Genet. 2015; 47:798–802. [DOI] [PubMed] [Google Scholar]
- 34. Jia X., Han B., Onengut-Gumuscu S., Chen W.M., Concannon P.J., Rich S.S., Raychaudhuri S., de Bakker P.I.. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013; 8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pillai N.E., Okada Y., Saw W.Y., Ong R.T., Wang X., Tantoso E., Xu W., Peterson T.A., Bielawny T., Ali M.et al.. Predicting HLA alleles from high-resolution SNP data in three southeast Asian populations. Hum. Mol. Genet. 2014; 23:4443–4451. [DOI] [PubMed] [Google Scholar]
- 36. Kim K., Bang S.Y., Lee H.S., Bae S.C.. Construction and application of a Korean reference panel for imputing classical alleles and amino acids of human leukocyte antigen genes. PLoS One. 2014; 9:e112546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan Y., Song Y.Q.. PyHLA: tests for the association between HLA alleles and diseases. BMC Bioinf. 2017; 18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete workflow of HLA typing and HLA imputation for genotyping data can be found on the group website (https://pog.fudan.edu.cn/#/software) and GitHub repository (https://github.com/Shuhua-Group/PGG.MHC). The use of the data by this work is approved by the Ministry of Science and Technology of the People's Republic of China (No. 2022BAT2237). For validation of the HLA typing workflow, we download WGS (https://www.internationalgenome.org/data-portal/data-collection/30x-grch38), WES (https://www.internationalgenome.org/data-portal/data-collection/phase-3), and genotyping (https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/hd_genotype_chip) samples from the KGP data portal. The benchmark HLA alleles for these samples were downloaded from https://doi.org/10.1371/journal.pone.0206512.s010.