Abstract
Numerous studies have shown that RNA plays an important role in the occurrence and development of diseases, and RNA-disease associations are not limited to noncoding RNAs in mammals but also exist for protein-coding RNAs. Furthermore, RNA-associated diseases are found across species including plants and nonmammals. To better analyze diseases at the RNA level and facilitate researchers in exploring the pathogenic mechanism of diseases, we decided to update and change MNDR v3.0 to RNADisease v4.0, a repository for RNA-disease association (http://www.rnadisease.org/ or http://www.rna-society.org/mndr/). Compared to the previous version, new features include: (i) expanded data sources and categories of species, RNA types, and diseases; (ii) the addition of a comprehensive analysis of RNAs from thousands of high-throughput sequencing data of cancer samples and normal samples; (iii) the addition of an RNA-disease enrichment tool and (iv) the addition of four RNA-disease prediction tools. In summary, RNADisease v4.0 provides a comprehensive and concise data resource of RNA-disease associations which contains a total of 3 428 058 RNA-disease entries covering 18 RNA types, 117 species and 4090 diseases to meet the needs of biological research and lay the foundation for future therapeutic applications of diseases.
INTRODUCTION
Continuously emerging evidence shows that RNA dysregulation or dysfunctions are important causes of disease development (1–4), and they also play a crucial part in disease-targeted therapy and prevention of viral infection (5,6). Therefore, research on RNA-disease has always attracted attention (7–9). However, there are currently a few databases of the association between miRNAs, lncRNAs, circRNAs and other noncoding RNAs with diseases in humans, mice and other mammals. Nonetheless, with the rapid advance in high-throughput sequencing technologies, many types of RNAs have been found to play a significant role in diseases (10,11), such as tRNA, which is considered to be a diagnostic marker for Alzheimer's disease, breast cancer, and other diseases or is directly related to disease prognosis (12,13). On the other hand, RNA affects plant traits and diseases; similar to ginseng rusty root symptoms, ncRNAs were found to play a regulatory role in diseased tissues (14,15).
Moreover, The Cancer Genome Atlas (TCGA) (16), International Cancer Genome Consortium (ICGC) (17) and Therapeutically Applicable Research to Generate Effective Treatments (TARGET) have generated a large amount of deep sequencing data and are widely used by researchers because of its large number of samples, rich clinical information, and reliable data. Analysis based on sequencing data from these databases can help to understand the function of RNA in cancers. Although mRNA in the TCGA is generally studied, ncRNAs in these three databases also deserve to be analyzed. Therefore, there is a great need to analyze and summarize these data in these databases to help explore the dynamic expression, clinical significance, and function of RNA in the physiological and pathological conditions of various cancer types.
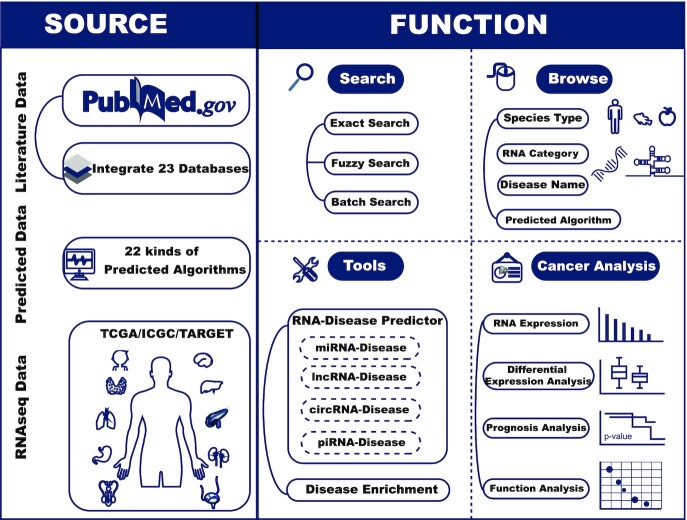
Here, RNADisease v4.0 updated from MNDR v3.0 (18) was produced based on demand. RNADisease v4.0 integrates manual curation of numerous literature, other experimentally validated databases, prediction algorithms, and RNA sequencing data of 44 different cancer types under one common framework (Figure 1). Moreover, it also provides two types of tools: RNA-disease prediction tool and disease enrichment tool. Overall, RNADisease v4.0 has integrated >3 400 000 entries, over a threefold increment in data, and an increase to 4090 diseases across 117 species. It provides a comprehensive and easily accessible data resource to assist researchers in better understanding disease mechanisms.
Figure 1.
Overview of the RNADisease v4.0 repository.
MATERIALS AND METHODS
Data collection and organization
RNADisease v4.0 generally contains three types of data: experimentally validated data, computationally predicted data, and RNA sequencing data about 44 types of cancers. Regarding the first type of data, we paid more attention to the regulation and pathogenesis of RNA in diseases, and we also focused on the complementary binding of mRNA and ncRNA to play a role in disease. Accordingly, we reviewed >40 000 published studies and acquired over 180 000 experimental RNA-disease associations. Furthermore, we integrated 23 other related experimentally validated databases (19–41) and finally obtained nearly 350 000 literature validated entries (Supplemental Table S1). Then, we used 22 different computationally predicted algorithms or web servers including CD-LNLP, DeepDCR, PreCDA, DincRNA, GMCLDA, HDncRNA, LDAI-ISPS, LDAP, LRLSLDA-LNCSIM, RWRlnD, TAM, BRMDA, MCLPMDA, MDHGI, MDNNMTF, miRPD, PBMDA, MMGCN, SPM, TDRC, iPiDi-PUL (29,42–60) to predict the RNA-disease data of four different types of RNAs (Supplemental Table S2). For the RNA sequencing data, we integrated RNA sequence data from TCGA, ICGC and TARGET, covering 44 cancer types, to explore the expression profiles, functions, and prognosis of miRNAs, lncRNAs, and mRNAs in cancers.
To standardize the data and increase the reference value, we linked the data from different sources to the authoritative reference database to annotate the data in RNADisease v4.0 in detail. mRNA, lncRNA, tRNA, rRNA, snoRNA and snRNA symbols were mapped to NCBI Gene (61) and Ensembl (62), while miRNA, circRNA, and piRNA symbols were mapped to miRBase (63), circBank (64) or circBase (65) and piRBase (66), respectively. The disease terms were mapped to the Disease Ontology (67), MeSH vocabularies and KEGG DISEASE. In extension, RNA-related subcellular localization from RNALocate v2.0 (68), RNA interactions from RNAInter v4.0 (69), and drug-related information were obtained from five databases: ncDR (38), NoncoRNA (22), NRDTD (70), RNAInter v4.0 (69) and KEGG DISEASE. Furthermore, we mapped the drug annotations to PubChem Compound.
Cancer analysis
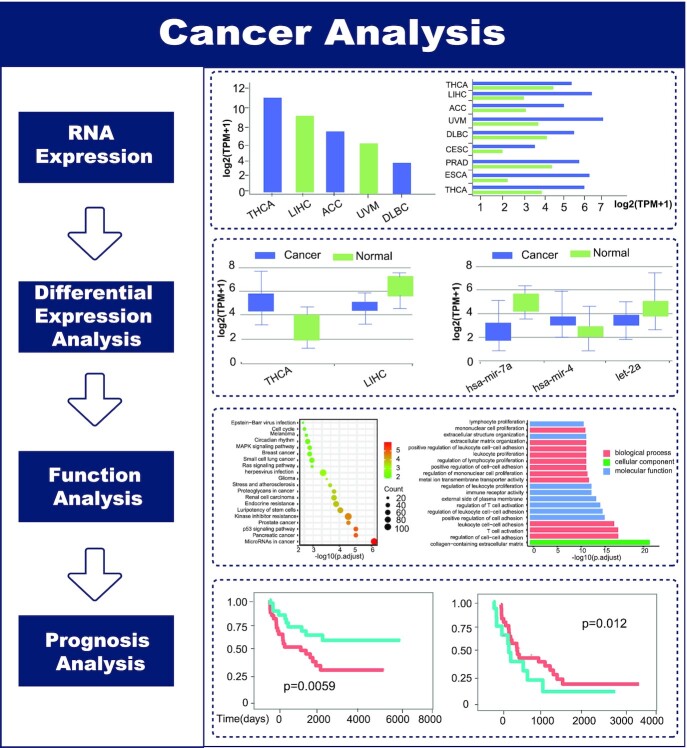
For RNA sequencing data, we downloaded the RNA read count value from TCGA, ICGC and TARGET. With gene annotation (Release 36 from GENCODE) and miRNA annotation (Release 20 from miRbase), mRNA, lncRNA and miRNA expression profiles were extracted. Then, the RNA expression levels were normalized by transcripts per million (TPM), and the average of the normalized RNA expression values in all disease samples of one cancer was taken as the RNA expression value in cancer. DEseq2 (71), edgeR (72) and Wilcoxon rank sum test were used to perform differential expression analysis between cancer and normal samples (normal sample > 2). Set padj (False Discovery Rate) value < 0.05 to obtain differentially expressed RNAs, and screen out unique, differentially expressed RNAs in one cancer. In the following, we used the differentially expressed mRNAs and the target genes of differentially expressed miRNAs predicted by miRWalk (73) to perform functional annotation and show some of the most significantly related pathways or functions, usually the top 20. Finally, we obtained clinical data from the TCGA, ICGC and TARGET performed survival analysis on differentially expressed RNAs or top 200 RNAs with the highest expression levels in a cancer without normal control using the survival R package and set a P-value <0.05 as the threshold to filter which RNAs were meaningful for patient survival (Figure 2).
Figure 2.
Introduction and usage of the Cancer Analysis page.
Disease enrichment
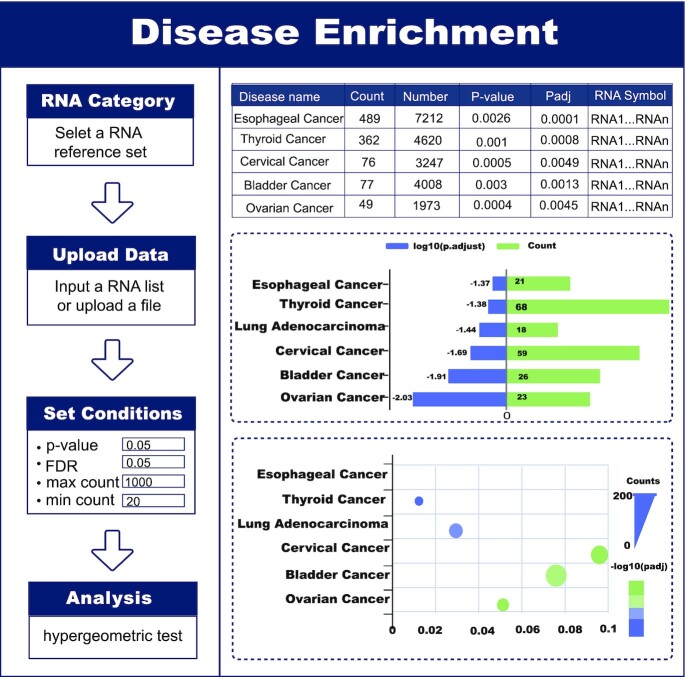
In particular, RNADisease v4.0 offers a disease enrichment tool based on all or one type of RNA in the repository as a reference set to infer RNA function from the RNA-disease perspective. This tool supports users in inputting an RNA symbol or RNA ID list, setting a series of conditions, and then using a hypergeometric test to calculate the significance p-value based on all experimentally validated RNA-disease in RNADisease. The enrichment significance P-value is calculated as:
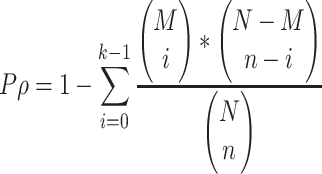 |
(1) |
where N represents the number of all experimentally validated RNAs or a certain type of experimentally validated RNAs in RNADisease v4.0, n is the number of all RNAs input by the user, M represents the total number of all RNAs or a type of RNAs contained in a certain disease, and k represents the number of intersections between the RNA list of interest to the user and all RNAs or a type of RNAs contained in a certain disease in RNADisease v4.0 (M∩n). Users can adjust the number of RNAs required to be enriched and set thresholds for P-value and false discovery rates (FDR) to control the number of returned results (Figure 3).
Figure 3.
Introduction and usage of the Disease Enrichment tool.
Prediction tools
RNADisease v4.0 provides a prediction tool for four RNA types based on different predicted algorithms on the website: MDNNMTF (Module-based Dynamic Neighborhood Non-negative Matrix Tri-Factorization) was used for miRNA-disease prediction, while GMCLDA (Geometric Matrix Completion lncRNA-Disease Association) was used for lncRNA-disease prediction, CD-LNLP based on linear neighborhood label propagation method to do circRNA-disease prediction, and iPiDi-PUL could be used to calculate the associations between piRNAs and diseases. MDNNMTF, GMCLDA, CD-LNLP are all based on RNA similarity matrix, disease similarity matrix and RNA-disease association matrix for RNA-disease prediction. Accordingly, we reconstructed three RNA-disease association matrices of different RNA types based on the experimentally validated data in RNADisease, and based on this matrix, we used the corresponding predicted algorithm to provide users with predicted results. On ‘prediction tool’ page, users can make predictions in the form RNA sequences, screening different types of RNAs and corresponding predicted algorithms, and finally choosing the number of results to display.
RESULTS
RNADisease statistics
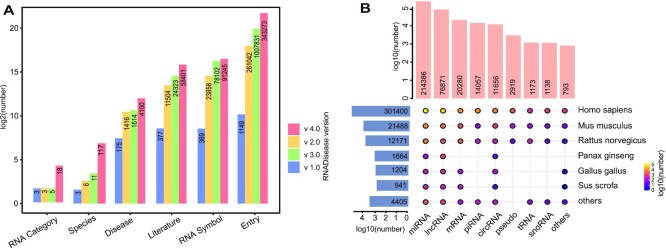
Overall, RNADisease v4.0 contains a total of 343 273 experimentally validated entries, of which 204 296 entries were collected by us (marked by * in RNADisease), and 183 911 entries came from other databases. Moreover, RNADisease v4.0 features a wider coverage, the experimentally validated data in the database is extracted from 58 401 pieces of literature, covering 117 species, 18 RNA types (such as snRNA, tRNA, mtRNA, scRNA, mRNA, YRNA), and 4090 diseases (Figure 4A, B). Concerning predicted data, RNADisease v4.0 includes 2 323 376 miRNA-associated, 345 725 lncRNA-associated, 48 779 piRNA-associated and 362 454 circRNA-associated entries for Homo sapiens, as well as 2434 and 28 predicted lncRNA-disease associations for Mus musculus and Rattus norvegicus, respectively. Through analysis, we obtained a total of 38 094 differentially expressed RNAs from the RNA sequencing data, including 20 005 mRNAs, 16 701 lncRNAs, and 1388 miRNAs. Among these differentially expressed RNAs, 22 333 RNAs were significant for patient survival, and these RNAs contained 15 897 mRNAs, 5876 lncRNAs and 560 miRNAs (Supplementary Table S3).
Figure 4.
Statistics of RNADisease v4.0. (A) The features and development across the RNADisease versions. Different colors represent versions of RNADisease over the decade. (B) The distribution of experimentally validated RNA-disease associations of 18 types of RNA in 117 species. Orange bars represent the number of RNA-disease entries in each RNA type, blue bars stand for the number of RNA-disease entries in each species, The color of the dots in the bubble plot represents the number of RNA-disease entries in a specific species-RNA type.
Database usage
RNADisease v4.0 builds a user-friendly platform to meet the needs of all kinds of research. It provides not only basic RNA, disease, reference-related information, and official annotation, but also the classification of strong and weak entry evidence, experimental methods, expression in diseases and RNA-disease score obtained by the scoring method of RNA-disease entries in MNDR v2.0 launched in 2018, which can be viewed on the ‘detail’ page. In addition, users can access data in three ways: (i) a quick search based on the RNA symbol, RNA ID or disease name on the ‘Home’ page; (ii) an ‘Exact Search’, ‘Fuzzy Search’ and ‘Batch search’ to retrieve data on the ‘Search’ page; and (iii) browse data according to RNA type, disease name, predicted algorithm, or species on the ‘Browse’ page.
To illustrate the RNA sequencing data of different cancers, the ‘Cancer Analysis’ page provides two methods to search the analysis results based on three differentially expressed analysis. The expression of RNA in cancer, differentially expressed or specific differentially expressed RNA in cancer, and the functional analysis and survival analysis of differentially expressed RNA can be obtained by searching cancer name or RNA symbol/ID and setting Method of Differential Expression Analysis. Moreover, RNADisease v4.0 provides two types of tools, one is disease enrichment tool, on ‘Disease Enrichment’ page, it first returns the RNA symbols matched in RNADisease v4.0 according to the RNA list entered by the user and then uses a hypergeometric test to obtain the diseases significantly related to the input RNA list and visualize the top 20 significant diseases. The other is the prediction tool, On the ‘Prediction Tool’ page, the user first selects the RNA type and corresponding predicted algorithm, then inputs the RNA sequence with fasta format to be predicted, and finally chooses the number of the predicted results to display.
CONCLUSION AND FUTURE PERSPECTIVES
We present an RNA-disease resource, which recruits more than three million RNA-disease association entries, over 3-fold of the old version, and the experimental data includes nearly 350 000 and contains 18 RNA types and 4090 diseases. Furthermore, the species coverage increased from 11 to 117 species. Moreover, RNADisease v4.0 adds a comprehensive analysis of RNAs from thousands of high-throughput sequencing data of cancer samples and normal samples, miRNA/lncRNA/circRNA/mRNA/piRNA-disease enrichment tools, and four different RNA types of RNA-disease prediction tool.
In the face of a massive and growing amount of RNA-disease data, data collection and integration have become a major challenge. While we continue to expand the amount of data, we will develop related natural language processing algorithms to assist data collection and integration as well as improving the scoring system to provide a more reliable scoring mode for RNA-disease. In addition, we will strive to design more friendly functions from the perspective of user-experience and more efficient search methods to cope with complex types of user input. In the future, we will insist on continuously updating and maintaining RNADisease, as in the past ten years, making RNADisease v4.0 the most comprehensive RNA-disease resource and a more comprehensive RNA-disease analysis platform to satisfy different requirements.
DATA AVAILABILITY
All our data is stored in RNADisease: http://www.rnadisease.org or http://www.rna-society.org/mndr/.
Supplementary Material
Contributor Information
Jia Chen, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Jiahao Lin, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Yongfei Hu, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Meijun Ye, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Linhui Yao, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Le Wu, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Wenhai Zhang, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Meiyi Wang, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Tingting Deng, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Feng Guo, School of Medicine, Tsinghua University, Beijing 100084, China.
Yan Huang, Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Bofeng Zhu, Guangzhou Key Laboratory of Forensic Multi-Omics for Precision Identification, School of Forensic Medicine, Southern Medical University, Guangzhou 510515, China.
Dong Wang, Department of Bioinformatics, Guangdong Province Key Laboratory of Molecular Tumor Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China; Dermatology Hospital, Southern Medical University, Guangzhou 510091, China; Department of Bioinformatics, Fujian Key Laboratory of Medical Bioinformatics, School of Medical Technology and Engineering, Fujian Medical University, Fuzhou 350122, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Project of China [2021YFC2500300, 2019YFA0801800]; National Natural Science Foundation of China [82070109, 62002153, 81930055]; Guangdong Basic and Applied Basic Research Foundation [2022A1515011253, 2019A1515010784]; Medical Scientific Research Foundation of Guangdong Province, China [A2022058]. Funding for open access charge: National Key Research and Development Project of China [2021YFC2500300, 2019YFA0801800].
Conflict of interest statement. None declared.
REFERENCES
- 1. Toden S., Zumwalt T.J., Goel A.. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta Rev. Cancer. 2021; 1875:188491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rogoyski O.M., Pueyo J.I., Couso J.P., Newbury S.F.. Functions of long non-coding RNAs in human disease and their conservation in drosophila development. Biochem. Soc. Trans. 2017; 45:895–904. [DOI] [PubMed] [Google Scholar]
- 3. Harries L.W. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 2012; 40:902–906. [DOI] [PubMed] [Google Scholar]
- 4. Liu T.Y., Zhang Y.C., Lin Y.Q., Hu Y.F., Zhang Y., Wang D., Wang Y., Ning L.. Exploration of invasive mechanisms via global ncRNA-associated virus-host crosstalk. Genomics. 2020; 112:1643–1650. [DOI] [PubMed] [Google Scholar]
- 5. Cheng J., Lin Y., Xu L., Chen K., Li Q., Xu K., Ning L., Kang J., Cui T., Huang Y.et al.. ViRBase v3.0: a virus and host ncRNA-associated interaction repository with increased coverage and annotation. Nucleic Acids Res. 2022; 50:D928–D933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu H., Fu H., Cui T., Ning L., Shao H., Guo Y., Ke Y., Zheng J., Lin H., Wu X.et al.. RNAPhaSep: a resource of RNAs undergoing phase separation. Nucleic Acids Res. 2022; 50:D340–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chow M.Y.T., Qiu Y., Lam J.K.W.. Inhaled RNA therapy: from promise to reality. Trends Pharmacol. Sci. 2020; 41:715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wild E.J., Tabrizi S.J.. Therapies targeting DNA and RNA in huntington's disease. Lancet Neurol. 2017; 16:837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang Y., Wang J., Zhao Y., Wang H., Liu T., Li Y., Cui T., Li W., Feng Y., Luo J.et al.. cncRNAdb: a manually curated resource of experimentally supported RNAs with both protein-coding and noncoding function. Nucleic Acids Res. 2021; 49:D65–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu M., Dai L., Wan L., Zhang S., Peng H.. Dynamic increase of red cell distribution width predicts increased risk of 30-Day readmission in patients with acute exacerbation of chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2021; 16:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu W., Lee I., Spratt H., Fang X., Bao X.. tRNA-Derived Fragments in alzheimer's disease: implications for new disease biomarkers and neuropathological mechanisms. J. Alzheimers Dis. 2021; 79:793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Y., Yang X., Jiang G., Zhang H., Ge L., Chen F., Li J., Liu H., Wang H.. 5'-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis. Genome Med. 2021; 13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu L., Li J., Gong Y., Wu Q., Tan S., Sun D., Xu X., Zuo Y., Zhao Y., Wei Y.Q.et al.. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer. 2019; 18:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bian X., Yu P., Dong L., Zhao Y., Yang H., Han Y., Zhang L.. Regulatory role of non-coding RNA in ginseng rusty root symptom tissue. Sci. Rep. 2021; 11:9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang M., Wu B., Chen C., Lu S.. Identification of mRNA-like non-coding RNAs and validation of a mighty one named MAR in panax ginseng. J. Integr. Plant Biol. 2015; 57:256–270. [DOI] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Research Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International Cancer Genome Consortium Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabe R.R., Bhan M.K., Calvo F., Eerola I.et al.. International network of cancer genome projects. Nature. 2010; 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ning L., Cui T., Zheng B., Wang N., Luo J., Yang B., Du M., Cheng J., Dou Y., Wang D. MNDR v3.0: mammal ncRNA-disease repository with increased coverage and annotation. Nucleic Acids Res. 2021; 49:D160–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou B., Ji B., Liu K., Hu G., Wang F., Chen Q., Yu R., Huang P., Ren J., Guo C.et al.. EVLncRNAs 2.0: an updated database of manually curated functional long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2021; 49:D86–D91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao H., Shi J., Zhang Y., Xie A., Yu L., Zhang C., Lei J., Xu H., Leng Z., Li T.et al.. LncTarD: a manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 2020; 48:D118–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019; 47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L., Wu P., Wang Z., Meng X., Zha C., Li Z., Qi T., Zhang Y., Han B., Li S.et al.. NoncoRNA: a database of experimentally supported non-coding RNAs and drug targets in cancer. J. Hematol. Oncol. 2020; 13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao Y., Shang S., Guo S., Li X., Zhou H., Liu H., Sun Y., Wang J., Wang P., Zhi H.et al.. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021; 49:D1251–D1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., Cao Y., Zhang H., Wang T., Tian Q., Lu X., Lu X., Kong X., Liu Z., Wang N.et al.. NSDNA: a manually curated database of experimentally supported ncRNAs associated with nervous system diseases. Nucleic Acids Res. 2017; 45:D902–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma L., Cao J., Liu L., Du Q., Li Z., Zou D., Bajic V.B., Zhang Z.. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019; 47:D128–D134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan C., Lei X., Tie J., Zhang Y., Wu F., Pan Y.. CircR2Disease v2.0: an updated web server for experimentally validated circRNA-disease associations and its application. Genomics Proteomics Bioinformatics. 2021; 2021: 10.1016/j.gpb.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao D., Zhang L., Zheng M., Sun X., Lu Y., Liu P.. Circ2Disease: a manually curated database of experimentally validated circRNAs in human disease. Sci. Rep. 2018; 8:11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang W., Zeng B., Yang M., Yang H., Wang J., Deng Y., Zhang H., Yao G., Wu S., Li W.. ncRNAVar: a manually curated database for identification of noncoding RNA variants associated with human diseases. J. Mol. Biol. 2021; 433:166727. [DOI] [PubMed] [Google Scholar]
- 29. Wang W.J., Wang Y.M., Hu Y., Lin Q., Chen R., Liu H., Cao W.Z., Zhu H.F., Tong C., Li L.et al.. HDncRNA: a comprehensive database of non-coding RNAs associated with heart diseases. Database (Oxford). 2018; 2018:bay067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao Y., Li X., Shang S., Guo S., Wang P., Sun D., Gan J., Sun J., Zhang Y., Wang J.et al.. LincSNP 3.0: an updated database for linking functional variants to human long non-coding RNAs, circular RNAs and their regulatory elements. Nucleic Acids Res. 2021; 49:D1244–D1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Muhammad A., Waheed R., Khan N.A., Jiang H., Song X.. piRDisease v1.0: a manually curated database for piRNA associated diseases. Database (Oxford). 2019; 2019:baz052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z., Shi J., Gao Y., Cui C., Zhang S., Li J., Zhou Y., Cui Q.. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019; 47:D1013–D1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang W., Yao G., Wang J., Yang M., Wang J., Zhang H., Li W.. ncRPheno: a comprehensive database platform for identification and validation of disease related noncoding RNAs. RNA Biol. 2020; 17:943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruepp A., Kowarsch A., Theis F.. PhenomiR: microRNAs in human diseases and biological processes. Methods Mol. Biol. 2012; 822:249–260. [DOI] [PubMed] [Google Scholar]
- 35. Wang D., Gu J., Wang T., Ding Z.. OncomiRDB: a database for the experimentally verified oncogenic and tumor-suppressive microRNAs. Bioinformatics. 2014; 30:2237–2238. [DOI] [PubMed] [Google Scholar]
- 36. Yue M., Zhou D., Zhi H., Wang P., Zhang Y., Gao Y., Guo M., Li X., Wang Y., Zhang Y.et al.. MSDD: a manually curated database of experimentally supported associations among miRNAs, SNPs and human diseases. Nucleic Acids Res. 2018; 46:D181–D185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie B., Ding Q., Han H., Wu D. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013; 29:638–644. [DOI] [PubMed] [Google Scholar]
- 38. Dai E., Yang F., Wang J., Zhou X., Song Q., An W., Wang L., Jiang W.. ncDR: a comprehensive resource of non-coding RNAs involved in drug resistance. Bioinformatics. 2017; 33:4010–4011. [DOI] [PubMed] [Google Scholar]
- 39. Cheng W.C., Chung I.F., Huang T.S., Chang S.T., Sun H.J., Tsai C.F., Liang M.L., Wong T.T., Wang H.W.. YM500: a small RNA sequencing (smRNA-seq) database for microRNA research. Nucleic Acids Res. 2013; 41:D285–D294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao Z., Wang K., Wu F., Wang W., Zhang K., Hu H., Liu Y., Jiang T.. circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death. Dis. 2018; 9:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Z., Wu L., Wang A., Tang W., Zhao Y., Zhao H., Teschendorff A.E.. dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res. 2017; 45:D812–D818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng L., Hu Y., Sun J., Zhou M., Jiang Q.. DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics. 2018; 34:1953–1956. [DOI] [PubMed] [Google Scholar]
- 43. Lu C., Yang M., Li M., Li Y., Wu F.X., Wang J.. Predicting human lncRNA-Disease associations based on geometric matrix completion. IEEE J. Biomed. Health Inform. 2020; 24:2420–2429. [DOI] [PubMed] [Google Scholar]
- 44. Sun J., Shi H., Wang Z., Zhang C., Liu L., Wang L., He W., Hao D., Liu S., Zhou M.. Inferring novel lncRNA-disease associations based on a random walk model of a lncRNA functional similarity network. Mol. Biosyst. 2014; 10:2074–2081. [DOI] [PubMed] [Google Scholar]
- 45. Zhu R., Wang Y., Liu J.X., Dai L.Y.. IPCARF: improving lncRNA-disease association prediction using incremental principal component analysis feature selection and a random forest classifier. BMC Bioinf. 2021; 22:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y., Nie C., Zang T., Wang Y.. Predicting circRNA-Disease associations based on circRNA expression similarity and functional similarity. Front. Genet. 2019; 10:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peng W., Du J., Dai W., Lan W.. Predicting miRNA-Disease association based on modularity preserving heterogeneous network embedding. Front. Cell Dev. Biol. 2021; 9:603758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu S.P., Liang C., Xiao Q., Li G.H., Ding P.J., Luo J.W.. MCLPMDA: a novel method for miRNA-disease association prediction based on matrix completion and label propagation. J. Cell. Mol. Med. 2019; 23:1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei H., Xu Y., Liu B.. iPiDi-PUL: identifying Piwi-interacting RNA-disease associations based on positive unlabeled learning. Brief. Bioinform. 2021; 22:bbaa058. [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y., Chen M., Li A., Cheng X., Jin H., Liu Y.. LDAI-ISPS: lncrna-Disease associations inference based on integrated space projection scores. Int. J. Mol. Sci. 2020; 21:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lan W., Li M., Zhao K., Liu J., Wu F.X., Pan Y., Wang J.. LDAP: a web server for lncRNA-disease association prediction. Bioinformatics. 2017; 33:458–460. [DOI] [PubMed] [Google Scholar]
- 52. Li J., Han X., Wan Y., Zhang S., Zhao Y., Fan R., Cui Q., Zhou Y.. TAM 2.0: tool for MicroRNA set analysis. Nucleic Acids Res. 2018; 46:W180–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang W., Yu C.L., Wang X.C., Liu F.. Predicting circRNA-disease associations through linear neighborhood label propagation method. IEEE Access. 2019; 7:83474–83483. [Google Scholar]
- 54. Zeng X., Zhong Y., Lin W., Zou Q.. Predicting disease-associated circular RNAs using deep forests combined with positive-unlabeled learning methods. Brief. Bioinform. 2020; 21:1425–1436. [DOI] [PubMed] [Google Scholar]
- 55. Zhu C.C., Wang C.C., Zhao Y., Zuo M., Chen X.. Identification of miRNA-disease associations via multiple information integration with bayesian ranking. Brief Bioinform. 2021; 22:bbab302. [DOI] [PubMed] [Google Scholar]
- 56. Chen X., Yin J., Qu J., Huang L.. MDHGI: matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Comput. Biol. 2018; 14:e1006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zeng X., Liu L., Lu L., Zou Q.. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics. 2018; 34:2425–2432. [DOI] [PubMed] [Google Scholar]
- 58. Tang X., Luo J., Shen C., Lai Z.. Multi-view multichannel attention graph convolutional network for miRNA-disease association prediction. Brief Bioinform. 2021; 22:bbab174. [DOI] [PubMed] [Google Scholar]
- 59. Mork S., Pletscher-Frankild S., Palleja Caro A., Gorodkin J., Jensen L.J.. Protein-driven inference of miRNA-disease associations. Bioinformatics. 2014; 30:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ding P., Luo J., Xiao Q., Chen X.. A path-based measurement for human miRNA functional similarities using miRNA-disease associations. Sci. Rep. 2016; 6:32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R.et al.. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015; 43:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J.et al.. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu M., Wang Q., Shen J., Yang B.B., Ding X.. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019; 16:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Glazar P., Papavasileiou P., Rajewsky N.. circBase: a database for circular RNAs. RNA. 2014; 20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang J., Zhang P., Lu Y., Li Y., Zheng Y., Kan Y., Chen R., He S.. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019; 47:D175–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schriml L.M., Munro J.B., Schor M., Olley D., McCracken C., Felix V., Baron J.A., Jackson R., Bello S.M., Bearer C.et al.. The human disease ontology 2022 update. Nucleic Acids Res. 2022; 50:D1255–D1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cui T., Dou Y., Tan P., Ni Z., Liu T., Wang D., Huang Y., Cai K., Zhao X., Xu D.et al.. RNALocate v2.0: an updated resource for RNA subcellular localization with increased coverage and annotation. Nucleic Acids Res. 2022; 50:D333–D339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kang J., Tang Q., He J., Li L., Yang N., Yu S., Wang M., Zhang Y., Lin J., Cui T.et al.. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 2022; 50:D326–D332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen X., Sun Y.Z., Zhang D.H., Li J.Q., Yan G.Y., An J.Y., You Z.H.. NRDTD: a database for clinically or experimentally supported non-coding RNAs and drug targets associations. Database (Oxford). 2017; 2017:bax057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sticht C., De La Torre C., Parveen A., Gretz N.. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018; 13:e0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All our data is stored in RNADisease: http://www.rnadisease.org or http://www.rna-society.org/mndr/.