Abstract
snoDB is an interactive database of human small nucleolar RNAs (snoRNAs) that includes up-to-date information on snoRNA features, genomic location, conservation, host gene, snoRNA–RNA targets and snoRNA abundance and provides links to other resources. In the second edition of this database (snoDB 2.0), we added an entirely new section on ribosomal RNA (rRNA) chemical modifications guided by snoRNAs with easy navigation between the different rRNA versions used in the literature and experimentally measured levels of modification. We also included new layers of information, including snoRNA motifs, secondary structure prediction, snoRNA–protein interactions, copy annotations and low structure bias expression data in a wide panel of tissues and cell lines to bolster functional probing of snoRNA biology. Version 2.0 features updated identifiers, more links to external resources and duplicate entry resolution. As a result, snoDB 2.0, which is freely available at https://bioinfo-scottgroup.med.usherbrooke.ca/snoDB/, represents a one-stop shop for snoRNA features, rRNA modification targets, functional impact and potential regulators.
Graphical Abstract
Graphical Abstract.
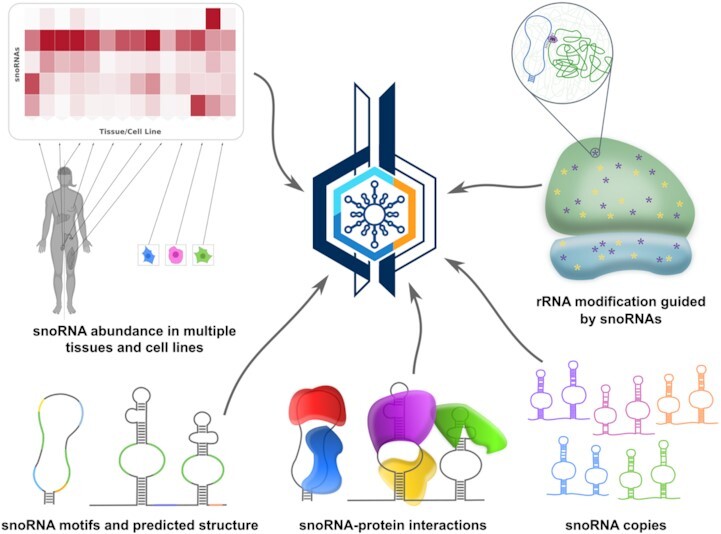
snoDB provides extensive characterization of human small nucleolar RNAs (snoRNAs). Version 2.0 features added sequence motifs and secondary structure, protein binders, snoRNA copies, abundance levels across diverse samples and snoRNA-guided ribosomal RNA modifications.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) are small non-coding RNAs ranging in size from 60 to 300 nucleotides and encoded in all eukaryotic genomes annotated so far. Based on their characteristic RNA motifs and protein-binding partners, they are classified into two groups, the box C/D snoRNAs and the box H/ACA snoRNAs, best characterized for their role in guiding, respectively, the 2′-O-ribose methylation and the pseudouridylation of ribosomal RNAs (rRNAs) and small nuclear RNAs (snRNAs). However, in the last two decades, a handful of snoRNAs have been shown to be involved in the regulation of a multitude of other cellular processes such as alternative splicing (1–3), 3′ end processing (4), exosome recruitment (5) and chromatin remodelling (6) through either RNA or protein interactions. Moreover, a plethora of studies found that snoRNAs are dysregulated in many cancer types and other diseases (7–11).
The study of snoRNAs and their complex family relationships involves several technical challenges. These include the very stable structure of snoRNAs and, more specific to mammals, their high copy numbers in genomes and their genomic location in introns of longer genes referred to as their host genes (12–14). These challenges are compounded by other factors related to our limited knowledge of these small RNAs and the few reliable datasets detecting them and their targets accurately. For example, the functions of chemical modification in rRNAs guided by snoRNAs, while being extensively studied, are not yet fully understood [e.g. (15–20)]. Indeed, for a long time, it was accepted that these chemical modifications are present on all rRNA molecules to maintain their conserved structures. However, with the emergence of advanced techniques to detect the level of chemical modification in rRNAs, such as RiboMethSeq (21) or HydraPsiSeq (22), it was recently shown that some positions are only partially modified and that the level of modification could vary in different cell lines, conditions or in certain pathologies (15,17,20,22–24). Furthermore, although there is currently a massive amount of publicly available RNA-seq datasets, few provide a reliable and realistic quantification for snoRNAs. TGIRT-seq, a sequencing method using a thermostable reverse transcriptase, is one such method that performs well for highly structured RNAs such as transfer RNAs and snoRNAs (12,25,26).
For the study of human snoRNAs, four dedicated databases are currently publicly available. snoRNABase (27), available since 2005, provides information about snoRNA sequences, rRNA and snRNA targets as well as their host genes, but is no longer regularly updated and is lacking many human snoRNAs present in other resources. snoRNA Atlas provides information about chromosomal location, conservation, average abundance and targets (28), while snOPY is focused on snoRNA orthology relationships across eukaryotes (29). As a complementary resource, we developed in 2018 snoDB (30), an interactive specialized human snoRNA database offering up-to-date information obtained from general and snoRNA-specific resources as well as our low structure bias TGIRT-seq datasets. We now provide snoDB 2.0, a second version with updated identifiers and updated links to other resources, correction for duplicate entries and most importantly diverse new data presented in an interactive format. snoDB now includes snoRNA–protein interaction data, information on the different copies of each snoRNA, snoRNA motifs and domains, secondary structure prediction, TGIRT-seq abundance data for 9 tissues and 5 cell lines (for a total of 37 samples) and a brand new section on rRNA modifications guided by snoRNAs.
DATABASE CONTENT
To ensure that the information is as up to date as possible, we obtained data from the latest versions of all the resources that were originally considered for the first version of snoDB. One of the problems we faced with the original version was the redundancy of some entries (Supplementary Figure S1). Indeed, some overlapping snoRNAs having as few as one nucleotide difference in 5', 3' or both were present as two or more distinct entries in snoDB (as well as in other resources), with their own sets of IDs and missing information. To solve this problem in version 2.0, we merged overlapping entries, ensuring that one genomic location was associated with only one snoRNA. Because we had no way of knowing the true beginning and end of a snoRNA, we prioritized information (e.g. genomic location, gene name, etc.) from some resources over others. We arbitrarily prioritized, in the following order, Ensembl (V104) (31), RefSeq (V109) (32), snoRNA Atlas (July 2021) (28) and snOPY (July 2021) (29) annotations. Ensembl itself has duplicate entries for some snoRNAs in its annotation file. In these cases, we chose snoRNAs starting with ‘SNOR’ first; otherwise, we prioritized the longest of the entries. In all cases, we used a concatenation of the IDs (e.g. ENSG00000281910;ENSG00000206952) to help the user to easily identify their snoRNAs of interest. We also included links (all obtained in July 2021) from HGNC (33), NCBI (34), snoRNABase (27), RNAcentral (35) and Rfam (36). When possible, we used the Infernal package to add missing Rfam IDs as recommended by Rfam (36). snoDB 2.0 now contains 2123 non-redundant entries.
Along with all the features that were originally included in the first version of snoDB (30), we now have additional evolutionary conservation information, sequence annotations, a predicted secondary structure, protein interaction data and copy information for all snoRNAs in snoDB as well as the level of abundance of snoRNAs and of their host gene. snoDB now presents a conservation score that was extracted from the UCSC Genome Browser, PhastCons track for 100 vertebrates (37,38) for each snoRNA. In addition, each entry now also features a secondary structure predicted using RNAfold and RNAplot from the Vienna package (39) in which the predicted snoRNA sequence motifs and guide regions can be highlighted. snoDB already contained information on RNA interacting partners of snoRNAs, to which we have added a section on snoRNA–protein interactions, extracted from eCLIP datasets of 150 RNA-binding proteins from the Encyclopedia of DNA Elements (ENCODE) Consortium (40). We also added a section on family members (based on Rfam classification), which is important when analysing snoRNAs by PCR, for example, since such snoRNAs often have very similar sequences. Finally, the abundance of snoRNAs and their host genes was obtained from 37 datasets that use the low structure bias TGIRT-seq approach (12,22,23) (GEO entries GSE126797, GSE157846, GSE99065 and GSE209924, and SRA entries SRX1426160 and SRX1426193). For all these newly added characteristics, more details are available in the About/Help section of the database.
To improve snoRNA quantification from RNA-seq datasets, we create a simple tool called snoRupdate. Indeed, to avoid biases due to the lack of some snoRNAs from current annotation files (e.g. gene transfer format), it is important to complete these annotations before performing RNA-seq analysis. For this purpose, snoRupdate, accessible from the main page or directly from GitHub (https://github.com/scottgroup/snoRupdate), is designed to add all missing snoRNAs from snoDB to the provided annotation file.
Finally, in this second version of the database, we added a completely new section on chemical modifications of rRNAs, which includes snoRNA–rRNA interaction sites, widely used rRNA sequences with conversion between them and modification levels for known modified rRNA positions. The rRNA modification sites were taken from snoRNABase (27), snoRNA Atlas (28), Krogh et al. (41) and Kehr et al. (42). Modification site annotations are dependent on rRNA sequences, and several versions of rRNAs are used in the literature (Supplementary Table S1). Lastly, rRNA modification levels were extracted from four studies using RiboMethSeq (17,23), HydraPsiSeq (22) or SILNAS (24).
WEB INTERFACE
Main page
On the main page, we made changes taking care to keep the interface similar enough to allow a smooth transition for users of the previous version. The navigation bar, on the top right panel, now includes a change log page, which allows to track changes made in future versions and to have access to old versions, a new rRNA chemical modification section (discussed further below), a link to snoRupdate (described above) and finally a sequence search tool section embedding a widget designed by RNAcentral (35) that enables the comparison of an input sequence against a large collection of non-coding RNAs.
The bottom of the page presents the main snoRNA interactive table, the columns of which can be controlled using the ‘Column visibility’ button. In version 2.0, we generated unique snoDB identifiers and use them for navigation through the website. The table can be easily filtered using the multiple search fields (see the tutorial section for advanced usage). As in the previous version, all the information in the main table, or only a selection, can be exported in different formats (xlsx, tsv, bed and now in fasta). Individual rows can also be selected to export only a part of the visible data.
snoRNA abundance visualization page
snoDB now provides abundance data from a considerably larger number of samples (9 tissues and 5 cell lines consisting in total of 37 datasets). We thus dedicated an independent page to the visualization of abundance, accessible through a button at the top right of the table of the main page, employing the Clustergrammer-JS and Clustergrammer-PY libraries (43), to provide an interactive heatmap. From the main table, the abundance profiles for all entries, or only a subpart (filtered and/or selected), can be observed simultaneously. The abundance data of all 37 samples can also be easily downloaded from this page.
Individual snoRNA pages
Aside from the main page, snoDB also has an individual page dedicated to each snoRNA. In this detailed view, all the information contained in the table from the main page on a particular snoRNA is recorded. In this updated version, it is possible to visualize important motifs (boxes and guide regions) of a snoRNA, and its predicted secondary structure by toggling two buttons in the first section. Furthermore, since snoRNA copies can often be a problem, as they are not so easy to find and are thus often overlooked, we included in this page a section listing all copies of a snoRNA. These copies can also be further investigated by clicking on ‘Inspect copies in main table’. When redirected, the information for the different entries can be exported in different formats and the level of abundance of all the snoRNA copies can be viewed in a heatmap. Next, in addition to snoRNA–RNA interactions, we also added a section dedicated to snoRNA–protein interactions. This section lists the RNA-binding proteins reported to bind to the snoRNA, the cell line in which the experiment was conducted, a P-value for the interaction and a score [obtained from ENCODE (40)]. Finally, instead of using a table for the level of abundance of the snoRNA and its host gene, they are now available as interactive box plots.
rRNA chemical modifications
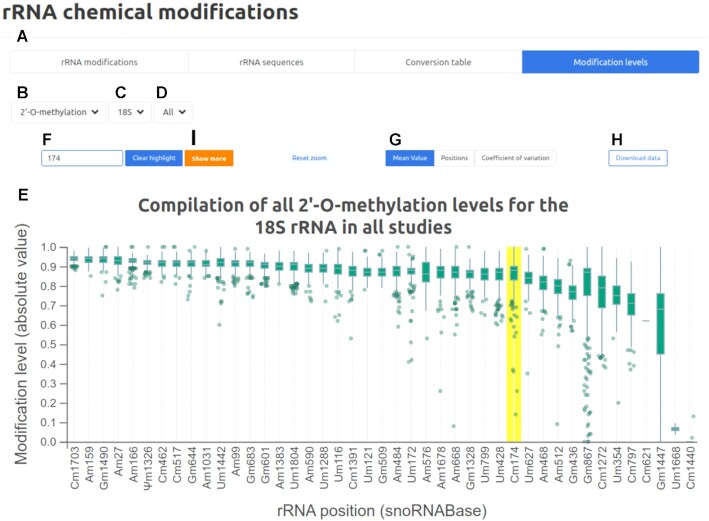
One of the significant additions of the snoDB 2.0 new release is the rRNA modification section, which contains four subsections accessible via the different top tabs: rRNA modifications, rRNA sequences, conversion table and modification levels (Figure 1A). The rRNA modification subsection includes an interactive table of all listed snoRNA–rRNA interactions. The status of the modification (i.e. whether the modification site has been validated by one or more techniques) is also mentioned to avoid confusion due to some predicted sites not being modified. The whole, or only the desired part, can be copied or exported in csv or xlsx formats. Next, the rRNA sequence tab enables visualization of the most widely used rRNA sequences. Validated modified positions are annotated and color-coded [2′-O-ribose methylation (Nm), pseudouridylation (Ψ) or both], and clicking on any of these modified sites returns snoRNAs predicted to modify this position, as well as a clickable link to the detailed view for the snoRNAs. The third subsection is a conversion table between different versions of rRNAs used in different articles and references. Any position can be searched, and the table contains the position in each different rRNA reference sequence as well as the nucleotide at this specific position. The whole table or a part can also be copied or exported in csv or xlsx formats. The last tab refers to a section where one can visualize the level of modification observed by different techniques and independent groups, in different tissues and cell lines (Figure 1), with the provision that rigorous comparisons cannot be made between the datasets because different methods were employed. By selecting the modification type (2′-O-ribose methylation or pseudouridylation; Figure 1B), the rRNA molecule (18S, 28S or 5.8S; Figure 1C) and the study (all the studies or only a single one; Figure 1D), the user sees a box plot displaying a compilation of all the modification levels for each position (Figure 1E). From there, it is possible to search a position with the top left input field (or by clicking on the chart; Figure 1F), reorder the rRNA positions according to position, coefficient of variation or mean value (default sorting) (Figure 1G) and change the zoom of the chart. The entire dataset used to generate the box plot can be exported using the ‘Download data’ button (Figure 1H). When a position is highlighted in the chart, an orange button appears (Figure 1I), which provides the user with a more detailed view of the level of modification, in a dot plot representation, of a single site across different previously selected cell lines and studies (Supplementary Figure S2).
Figure 1.
Screenshot of the ‘Modification levels’ tab, one of the four subsections of the new rRNA chemical modification section. (A) Four tabs corresponding to each of the new subsections: rRNA modifications, rRNA sequences, conversion table and modification levels. (B–D) Modification type (2′-O-ribose methylation or pseudouridylation), rRNA molecule (18S, 28S or 5.8S) and study (all the studies or only a single one) to choose from. (E) Box plot displaying a compilation of the levels of all modified positions. (F) Search field used to look for a specific modified position. (G) Buttons to reorder the box plot according to mean value, position or coefficient of variation. (H) ‘Download data’ button to download the raw data used to generate the chart. (I) ‘Show more’ button used to get a more detailed view of the level of modification of a single position (see Supplementary Figure S2).
CONCLUSION AND FUTURE PLANS
snoDB aims to provide the community with an efficient way to navigate through the most up-to-date snoRNA data. This second version of snoDB has kept its user-friendly interface with its main overview table and individual snoRNA pages, but on top of the consolidation and addition of missing snoRNAs, many more characteristics of snoRNAs are now available, including sequence motifs and structures, copy identification, considerably more abundance data and snoRNA–protein interactions. We also added a completely new section on rRNA modifications. Our future plans, in addition to periodic updates of the current information contained in the database, include providing the same level of information on snoRNAs from other species.
DATA AVAILABILITY
snoDB 2.0 is freely available at https://bioinfo-scottgroup.med.usherbrooke.ca/snoDB/.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to members of their groups for useful discussions, to Leandro Fequino for technical support and to Yuri Motorin and Virginie Marchand for providing their ribosome modification datasets. M.S.S. and S.A.E. are members of the RNA group and the Centre de Recherche du Centre Hospitalier Universitaire de Sherbrooke (CRCHUS). V.M. is an Institut National de la Santé et de la Recherche Médicale (Inserm) fellow and F.C. is a Centre National de Recherche Scientifique (CNRS) fellow.
Contributor Information
Danny Bergeron, Département de biochimie et génomique fonctionnelle, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
Hermes Paraqindes, Inserm U1052, CNRS UMR5286 Centre de Recherche en Cancérologie de Lyon, F-69000 Lyon, France; Centre Léon Bérard, F-69008 Lyon, France; Université de Lyon 1, F-69000 Lyon, France.
Étienne Fafard-Couture, Département de biochimie et génomique fonctionnelle, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
Gabrielle Deschamps-Francoeur, Département de biochimie et génomique fonctionnelle, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
Laurence Faucher-Giguère, Département de microbiologie et d’infectiologie, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
Philia Bouchard-Bourelle, Département de biochimie et génomique fonctionnelle, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
Sherif Abou Elela, Département de microbiologie et d’infectiologie, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
Frédéric Catez, Inserm U1052, CNRS UMR5286 Centre de Recherche en Cancérologie de Lyon, F-69000 Lyon, France; Centre Léon Bérard, F-69008 Lyon, France; Université de Lyon 1, F-69000 Lyon, France; Institut Convergence PLAsCAN, F-69373 Lyon, France.
Virginie Marcel, Inserm U1052, CNRS UMR5286 Centre de Recherche en Cancérologie de Lyon, F-69000 Lyon, France; Centre Léon Bérard, F-69008 Lyon, France; Université de Lyon 1, F-69000 Lyon, France; Institut Convergence PLAsCAN, F-69373 Lyon, France.
Michelle S Scott, Département de biochimie et génomique fonctionnelle, Faculté de médecine et des sciences de la santé, Université de Sherbrooke, Sherbrooke, Québec J1E 4K8, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research [PJT-153171 to M.S.S. and S.A.E.]; Fonds de Recherche du Québec—Nature et Technologies [2022-PR-298214 to M.S.S. and S.A.E.; to G.D.-F.]; Fonds de Recherche du Québec—Santé [296343 to M.S.S.]; Fondation ARC pour la Recherche sur le Cancer [20161204686 MARACAS.v0 to V.M. and F.C.]; SIRIC program [INCa-DGOS-Inserm 12563 LyRICAN to V.M. and F.C.]; Labex program [DEVCAN2UMAN to V.M. and F.C.]; Synergie Lyon Cancer Foundation [to V.M. and F.C.]; Agence Nationale de la Recherche [PLAsCAN—ANR-17-CONV-0002 and ActiMeth—ANR-19-CE12-0004 to V.M. and F.C.]; Cancéropôle Lyon Auvergne Rhône-Alpes—AAP international 2021 [MARACAS.v3.0 to V.M.]; Institut National du Cancer [PLBio 2019-138 MARACAS.v1 to V.M.]; Inserm-ITMO Cancer [non-coding RNA, REMOTE to V.M.]; Natural Sciences and Engineering Research Council of Canada [to D.B. and E.F.-C.]; La Ligue Nationale Contre le Cancer [to H.P.]; Fonds de Recherche du Québec—Santé [to L.F.-G.]. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cavaillé J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Hüttenhofer A.. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. U.S.A. 2000; 97:14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falaleeva M., Pages A., Matuszek Z., Hidmi S., Agranat-Tamir L., Korotkov K., Nevo Y., Eyras E., Sperling R., Stamm S.. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl Acad. Sci. U.S.A. 2016; 113:E1625–E1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott M.S., Ono M., Yamada K., Endo A., Barton G.J., Lamond A.I.. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 2012; 40:3676–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang C., Shi J., Guo Y., Huang W., Huang S., Ming S., Wu X., Zhang R., Ding J., Zhao W.et al.. A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017; 45:8647–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhong F., Zhou N., Wu K., Guo Y., Tan W., Zhang H., Zhang X., Geng G., Pan T., Luo H.et al.. A snoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015; 43:10474–10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He X., Chen X., Zhang X., Duan X., Pan T., Hu Q., Zhang Y., Zhong F., Liu J., Zhang H.et al.. An lnc RNA (GAS5)/snoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015; 43:3712–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang J., Wen J., Huang Z., Chen X.-P., Zhang B.-X., Chu L.. Small nucleolar RNAs: insight into their function in cancer. Front. Oncol. 2019; 9:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Werf J., Chin C.V., Fleming N.I.. SnoRNA in cancer progression, metastasis and immunotherapy response. Biology (Basel). 2021; 10:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deogharia M., Majumder M.. Guide snoRNAs: drivers or passengers in human disease?. Biology (Basel). 2018; 8:E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang Z.-H., Du Y.-P., Wen J.-T., Lu B.-F., Zhao Y.. snoRNAs: functions and mechanisms in biological processes, and roles in tumor pathophysiology. Cell Death Discov. 8:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faucher-Giguère L., Roy A., Deschamps-Francoeur G., Couture S., Nottingham R.M., Lambowitz A.M., Scott M.S., Abou Elela S.. High-grade ovarian cancer associated H/ACA snoRNAs promote cancer cell proliferation and survival. NAR Cancer. 2022; 4:zcab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boivin V., Deschamps-Francoeur G., Couture S., Nottingham R.M., Bouchard-Bourelle P., Lambowitz A.M., Scott M.S., Abou-Elela S.. Simultaneous sequencing of coding and noncoding RNA reveals a human transcriptome dominated by a small number of highly expressed noncoding genes. RNA. 2018; 24:950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fafard-Couture É., Bergeron D., Couture S., Abou-Elela S., Scott M.S.. Annotation of snoRNA abundance across human tissues reveals complex snoRNA–host gene relationships. Genome Biol. 2021; 22:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bergeron D., Laforest C., Carpentier S., Calvé A., Fafard-Couture É., Deschamps-Francoeur G., Scott M.S.. SnoRNA copy regulation affects family size, genomic location and family abundance levels. BMC Genomics. 2021; 22:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S.E., Garcia M., Laforêts F., Marcel V., Baudin-Baillieu A.et al.. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl Acad. Sci. U.S.A. 2017; 114:12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sloan K.E., Warda A.S., Sharma S., Entian K.-D., Lafontaine D.L.J., Bohnsack M.T.. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017; 14:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Motorin Y., Quinternet M., Rhalloussi W., Marchand V.. Constitutive and variable 2′-O-methylation (Nm) in human ribosomal RNA. RNA Biol. 2021; 18:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monaco P.L., Marcel V., Diaz J.-J., Catez F.. 2′-O-Methylation of ribosomal RNA: towards an epitranscriptomic control of translation?. Biomolecules. 2018; 8:E106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Penzo M., Montanaro L.. Turning uridines around: role of rRNA pseudouridylation in ribosome biogenesis and ribosomal function. Biomolecules. 2018; 8:E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaafar M., Paraqindes H., Gabut M., Diaz J.-J., Marcel V., Durand S.. 2′O-Ribose methylation of ribosomal RNAs: natural diversity in living organisms, biological processes, and diseases. Cells. 2021; 10:1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marchand V., Blanloeil-Oillo F., Helm M., Motorin Y.. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 2016; 44:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchand V., Pichot F., Neybecker P., Ayadi L., Bourguignon-Igel V., Wacheul L., Lafontaine D.L.J., Pinzano A., Helm M., Motorin Y.. HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 2020; 48:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marcel V., Kielbassa J., Marchand V., Natchiar K.S., Paraqindes H., Nguyen Van Long F., Ayadi L., Bourguignon-Igel V., Lo Monaco P., Monchiet D.et al.. Ribosomal RNA 2′O-methylation as a novel layer of inter-tumour heterogeneity in breast cancer. NAR Cancer. 2020; 2:zcaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N.et al.. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018; 46:9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nottingham R.M., Wu D.C., Qin Y., Yao J., Hunicke-Smith S., Lambowitz A.M.. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA. 2016; 22:597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin Y., Yao J., Wu D.C., Nottingham R.M., Mohr S., Hunicke-Smith S., Lambowitz A.M.. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016; 22:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lestrade L., Weber M.J.. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006; 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., Gruber A.R.. An updated human snoRNAome. Nucleic Acids Res. 2016; 44:5068–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoshihama M., Nakao A., Kenmochi N.. snOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes. 2013; 6:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bouchard-Bourelle P., Desjardins-Henri C., Mathurin-St-Pierre D., Deschamps-Francoeur G., Fafard-Couture É., Garant J.-M., Elela S.A., Scott M.S.. snoDB: an interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res. 2020; 48:D220–D225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J.et al.. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gale R.P., Hochhaus A., Cross N.C.P., Harrison C.J.. HGNC nomenclature for fusion genes. Leukemia. 2021; 35:3039. [DOI] [PubMed] [Google Scholar]
- 34. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., Mcveigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. RNAcentral Consortium RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021; 49:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., Griffiths-Jones S., Toffano-Nioche C., Gautheret D., Weinberg Z.et al.. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021; 49:D192–D200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S.et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K.et al.. The Encyclopedia of DNA Elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krogh N., Jansson M.D., Häfner S.J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A.H., Nielsen H.. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016; 44:7884–7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kehr S., Bartschat S., Tafer H., Stadler P.F., Hertel J.. Matching of soulmates: coevolution of snoRNAs and their targets. Mol. Biol. Evol. 2014; 31:455–467. [DOI] [PubMed] [Google Scholar]
- 43. Fernandez N.F., Gundersen G.W., Rahman A., Grimes M.L., Rikova K., Hornbeck P., Ma’ayan A.. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data. 2017; 4:170151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
snoDB 2.0 is freely available at https://bioinfo-scottgroup.med.usherbrooke.ca/snoDB/.