Abstract
It is a challenge to efficiently integrate and present the tremendous amounts of single-cell data generated from multiple tissues of various species. Here, we create a new database named SPEED for single-cell pan-species atlas in the light of ecology and evolution for development and diseases (freely accessible at http://8.142.154.29 or http://speedatlas.net). SPEED is an online platform with 4 data modules, 7 function modules and 2 display modules. The ‘Pan’ module is applied for the interactive analysis of single cell sequencing datasets from 127 species, and the ‘Evo’, ‘Devo’, and ‘Diz’ modules provide comprehensive analysis of single-cell atlases on 18 evolution datasets, 28 development datasets, and 85 disease datasets. The ‘C2C’, ‘G2G’ and ‘S2S’ modules explore intercellular communications, genetic regulatory networks, and cross-species molecular evolution. The ‘sSearch’, ‘sMarker’, ‘sUp’, and ‘sDown’ modules allow users to retrieve specific data information, obtain common marker genes for cell types, freely upload, and download single-cell datasets, respectively. Two display modules (‘HOME’ and ‘HELP’) offer easier access to the SPEED database with informative statistics and detailed guidelines. All in all, SPEED is an integrated platform for single-cell RNA sequencing (scRNA-seq) and single-cell whole-genome sequencing (scWGS) datasets to assist the deep-mining and understanding of heterogeneity among cells, tissues, and species at multi-levels, angles, and orientations, as well as provide new insights into molecular mechanisms of biological development and pathogenesis.
INTRODUCTION
Heterogeneity is ubiquitous among cells, tissues, organs, and species. High cellular diversity exists in the mammalian lung (1,2). Early embryonic cells and pluripotent stem cells are differentiated into function-specific cells within tissues and organs by switching on/off diverse gene expression patterns during the development (3–5). The interactions between gene expression and stimuli play decisive roles in the maintenance of health status or disease development (6). Each cell type demonstrates different transcriptome changes in response to microbial and environmental insults (7,8). It is critical to compare gene expression profiles of cell types between normal and pathological tissues and clarify the biological significance and pathogenesis mechanisms.
Methods of cell classification by cell morphology or specific gene/protein expression panels characterize well-known cell types, while lacking the systematicity and correlation. The transcriptome or proteome at bulk cells and tissue fails to define the gene expression profile at single cell resolution. The scRNA-seq is an advanced high-throughput sequencing technology to capture the transcriptome of single cells on a large scale and to unbiasedly classify cell types in complex tissues and dissect the gene expression in individual cells (9). scRNA-seq provides opportunities to reliably identify novel or rare cell types (10), discriminate closely related cell populations (11), and define cell heterogeneity, differentiation, and development (12). With rapid development, scRNA-seq offers an unprecedented impetus to the initiation of an ambitious international consortium, the Human Cell Atlas (HCA) (13). In addition, the cellular atlas in multiple tissues or organs of other species has been accomplished by scRNA-seq in recent years, e.g. tissue cells from 11 non-model animals (14), circulating immune cells among 12 species (15), eight organs and tissues of mouse embryos at different stages (12), mice lung cells across aging stages (16,17), and pig cerebral cortex, hypothalamus (18), and lung (2). Single-cell transcriptomic data from pathologic tissues of patients and animal models of diseases is generated rapidly, e.g. cells from lower airways of asthmatic patients (19), fibrotic lungs of mice (20), and three brain regions of the Parkinson's disease model mouse (21).
Fast-increasing amounts of single-cell data on multiple tissues among different species are generated worldwide. The challenges are the deposition of raw data in scRNA-seq datasets and the difficulty of deep-mining the needed information for non-bioinformatics scientists. Therefore, effective integration and presentation of scRNA-seq datasets will facilitate and maximize the utilization by the research community. Raw data and expression matrix datasets from single-cell transcriptomes are submitted to several free academic websites such as Gene Expression Omnibus (22) and ArrayExpress (23). Several websites were established for collecting single-cell datasets across multiple species, such as Single Cell Portal with 11 species (https://singlecell.broadinstitute.org/single_cell), Single Cell Expression Atlas with 20 species (24), and UCSC Cell Browser with 15 species (25). Some portals were created specifically to collect scRNA-seq data for certain types of tissues or organs. LungMAP was established for illustrating the human and mouse lung molecular atlas (26), and ScdbLung for exploring single-cell data from the lung of human, mouse, rat, and pig (2). Of diseases-related scRNA-seq databases, CancerSEA provides distinct functional states of single-cell expression signatures for 25 cancer types (27), and CSEA-DB for 598 GWAS traits associated with the underlying cell types on basis of scRNA-seq (28). SC2disease is an accurate resource of gene expression profiles for 25 diseases (29). TISCH integrates scRNA-seq profiles from 76 tumor datasets (30). PBatlas was recently established as a versatile website to access the pig brain atlas (18). The CellMarker database records cell markers for human tissues and mouse tissues (31). VThunter presents the cell expression pattern of 107 virus receptors from 47 animal species (32).
Although some databases are focused on specific applications, there is still a practical need to construct databases with more species and with a special focus on the heterogeneities among cells, tissues, organs, and species. Thus, it is urgent to build a systematic, hierarchical, and comprehensive gene expression profiling database from public scRNA-seq data. To address those issues, we centralized multiple scRNA-seq datasets and established a comprehensive and freely accessible online database called SPEED.
DATA COLLECTION AND DATABASE CONTENT
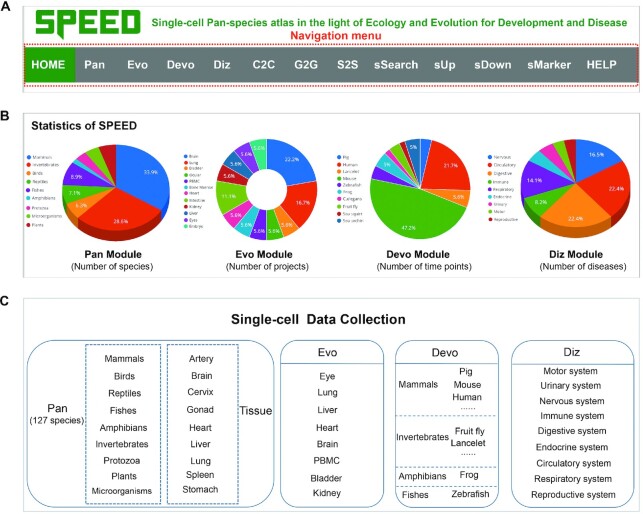
A total of 5,770,427 cells from 634 datasets across 127 species were collected into the SPEED database. The scRNA-seq datasets were downloaded from multiple sources, including NCBI/GEO (22), EMBL-EBI/SCEA (24), ArrayExpress (23), HCL (33), UCSC Cell Browser (25), MCA (34), and Single Cell Portal (https://singlecell.broadinstitute.org/single_cell), prioritizing studies with RDS files or gene expression matrix available. The navigation menu contains 13 modules, such as ‘HOME’, ‘Pan’, ‘Evo’, ‘Devo’, ‘Diz’, ‘C2C’, ‘G2G’, ‘S2S’, ‘sSearch’, ‘sUp’, ‘sDown’, ‘sMarker’, and ‘HELP’ (Figure 1A). All detailed information on scRNA-seq datasets of 122 species, is found on the ‘sDown’ module of SPEED, including data sources, timing, technologies, common and Latin names of species, sample tissues, treatments, and cell numbers. In addition, we collected the single-cell WGS (sc-WGS) datasets from 16 species in the ‘Pan’ module (35). The whole-genome sequencing (WGS) technology provides an in-depth description of single nucleotide polymorphisms (SNPs), small insertions, and deletions (INDELs).
Figure 1.
Overview of SPEED database. (A) Navigation menu of the SPEED website. (B) Statistics of Pan, Evo, Devo, and Diz modules in SPEED. (C) Single-cell data collection of the four data modules.
The scRNA-seq datasets were checked manually and processed using Seurat v4.1.1 (36). In brief, quality control was conducted for the downstream analysis by retaining cells with nFeature_RNA between 200 and 6000. The ‘NormalizeData’ function in Seurat v4.1.1 was then utilized to normalize gene expression matrix of the sparse single cell. The ‘FindVariableFeatures’ function was used to identify highly variable genes. The top 2000 highly variable genes were then dimensionally reduced using the principal component analysis (PCA). The top 30 principal components were picked for clustering. The visualization of single cell data sets was achieved using ShinyCell package (37). Cellular communication was analyzed in the ‘C2C’ module using CellChat (38). In the ‘G2G’ module, putative interactions between transcriptional factors (TFs) and downstream targets were predicted using GENIE3 (39) and GO enrichment was performed using clusterProfiler (40).
USER INTERFACE
The web application and search engine in SPEED are currently mounted on a high-performance Linux server. Users worldwide can freely access database for in-depth data visualization and custom analysis of 127 species and function modules for further personalized analysis. There are 13 modules in the navigation menu of SPEED (Figure 1A), which are grouped into three categories: display, data, and function. The display category includes ‘HOME’ and ‘HELP’ modules. The ‘HOME’ module introduces the statistics of datasets (Figure 1B). The ‘HELP’ module guides users to easily understand the SPEED user interface. A brief description and a frame-by-frame animation demonstration are provided for each module to help users to easily catch key points of the module usage.
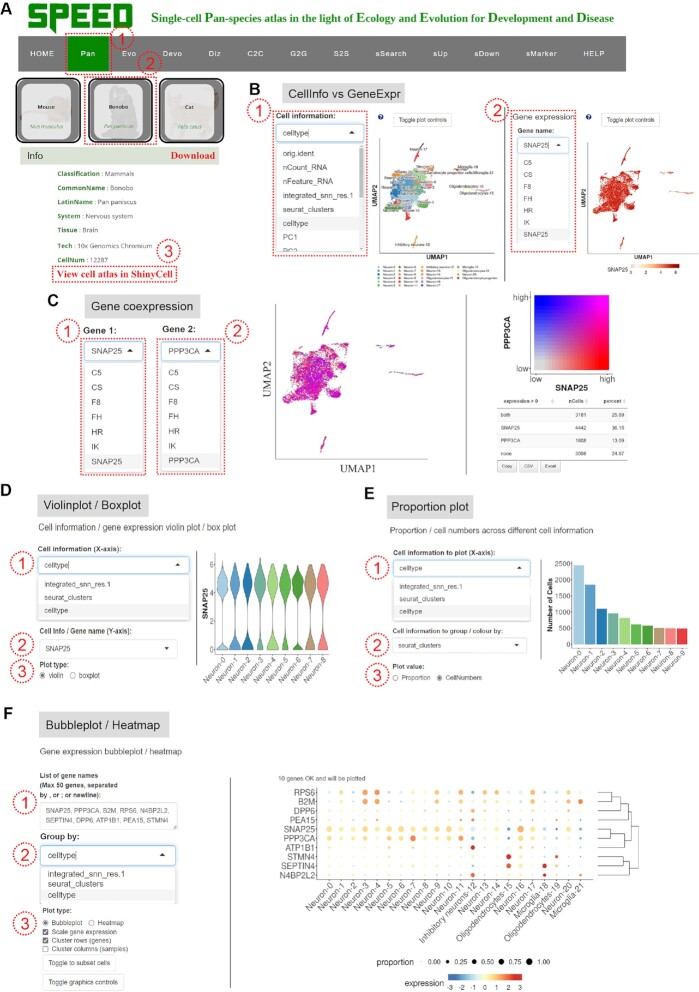
SPEED is designed mainly for the data categories: ‘Pan’, ‘Evo’, ‘Devo’, and ‘Diz’ modules in the navigation menu (Figure 1C). The ‘Pan’ module is one of the most core modules of SPEED, containing scRNA-seq datasets from 122 species and sc-WGS data for 16 mammalian species. On the ‘Pan’ page, the 122 species scRNA-seq datasets were grouped into 9 classes (i.e. mammals, birds, reptiles, fishes, amphibians, invertebrates, protozoa, plants, and microorganisms), according to different kingdoms and phyla (Figure 1C). The ‘Pan’ module allows the interactive exploration of each individual cell-type gene expression profile. If users wonder the cell-types-specific expression profile of a certain gene of interest in a certain tissue of a certain species, they can perform the following operations to obtain the desired information as shown in Figure 2A and B. When users move the cursor onto one species' image (e.g. ‘Bonobo’) (41), a sliding tab will pop up with the common species name in black font and its Latin name in green italics (Figure 2A). By clicking the sliding tab on the species image and the link in the column of ‘Rds’, users can browse the basic information of the species and the detailed information of single-cell atlas on a new tab page. Clicking the ‘View cell atlas in ShinyCell’ in red font guides users to a new page with seven tabs on the navigation menu (Figure 2A). In doing so, users may continue to visualize gene expression profiles and cell-type information for genes of interest. The ‘CellInfo vs GenExpr’ tab simultaneously visualizes cell information and gene expression side-by-side on low-dimensional representations. There are multiple options (e.g. ‘tSNE1’ for X-axis and ‘tSNE2’ for Y-axis) to choose from in the drop-down box under the X-axis and Y-axis of ‘Dimension Reduction’. Different dimensions of cell information are presented by switching the options (e.g. ‘celltype’) in the drop-down box under ‘Cell Information’ (Figure 2B, left). The cell-type gene expression profiles are visualized by choosing gene name of interest (e.g. ‘SNAP25’) in the drop-down box under ‘Gene Information’ (Figure 2B, right). The ‘CellInfo vs CellInfo’ and ‘GenExpr vs GenExpr’ tabs visualize two cell information and two gene expressions side-by-side on low-dimensional representations, respectively (Figure S1). Researchers can analyze the co-expression relationships between two genes on each cell-type of a certain tissue of interest, which may be realized through the following steps. The ‘Gene coexpression’ tab visualizes the cell-type co-expression of two genes (e.g. ‘SNAP25’ and ‘PPP3CA’) on low-dimensional representations when users choose or input two genes of interest in the boxes under ‘Gene Expression’ button (Figure 2C). The ‘Violinplot/Boxplot’ tab visualizes the gene expression or continuous cell information across groups of cells when users choose the option or input gene of interest in the boxes under ‘Cell information (X-axis)’ and ‘Cell Info/Gene name (Y-axis)’ (Figure 2D). The ‘Proportion plot’ tab shows the proportion and number of single cells (Figure 2E). The ‘Bubbleplot/Heatmap’ tab visualizes the gene expression patterns of multiple genes which are grouped by categorical cell information (e.g. celltype, RNA_snn_res.1, and seurat_clusters) (Figure 2F).
Figure 2.
Demonstration of Pan module. (A) Operation to access cell atlas in ShinyCell on the species profile page and download RDS files. (B) Illustration of viewing multi-classified cell and gene expression information in ShinyCell. (C) Illustration of viewing gene co-expression information. (D) Operation to view cell or gene information in Violinplot or Boxplot. (E) Operation to view the single cell composition.(F) Operation to view the gene expression pattern in Bubbleplot/Heatmap.
In addition to scRNA-seq datasets, sc-WGS datasets of 16 mammalian species are available in the tenth class on the ‘Pan’ module, which provide detailed information of SNPs and INDELs (35) (Figure S2A). By clicking the sliding tab on the species image (e.g. ‘King colobus’), a new page pops up and shows basic information of the dataset and two link buttons, ‘View SNP information’ and ‘View Indel information’ (Figure S2B). When users click the two links buttons, a new tab page appears with detailed information of the SNPs or INDELs from sc-WGS datasets (Figure S2C-D), including chromosomes, start and end sites, intergenic, intronic or exonic, sequencing methods, and others.
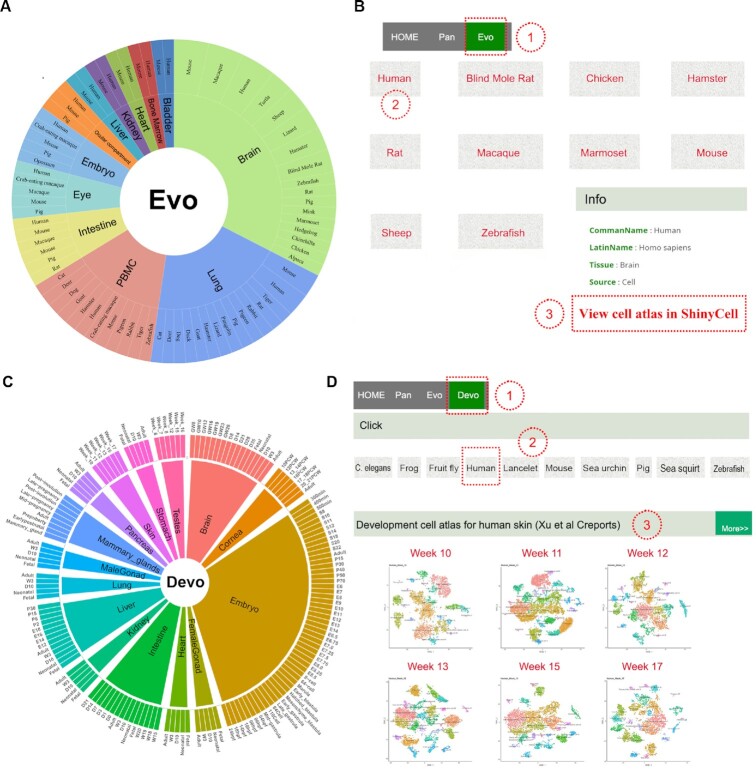
The ‘Evo’ module displays 18 datasets of multiple tissues from 28 species, including brain, lung, heart, bladder, ocular compartment, eye, bone marrow, intestine, kidney, liver, embryo, and peripheral blood mononuclear cell (PBMC) (Figure 3A). Cross-species comparisons of single-cell atlas of mammals, reptiles, and other species provide a reference evolutionary reservoir of developmental programs to explore potential networks among evolutionarily distant species. When users click the dataset picture (e.g. ‘Brain data sets (Geirsdottir et al Cell)’) on the ‘Evo’ module (Figure 3B), the current page switches to a new page to show all species pictures and their common names linked with the corresponding literature (42). By clicking the species picture (e.g. ‘Human’), users get more detailed information on the scRNA-seq atlas via the link button ‘View cell atlas in ShinyCell’ in a new tab page (Figure 3B).
Figure 3.
Overview of Evo and Devo modules. (A) Tissue types of each species in Evo module. Tissue types and species names are shown from the inner circle to the outer circle. (B) Operation to access the datasets in Evo module. (C) Distribution of tissue types in development time points in Devo module. (D) Operation to view the datasets in Devo module.
The developmental single-cell atlas is critical for understanding of stem cell biology. The ‘Devo’ module contains 28 single-cell datasets from 10 species (Figure 3C). On the ‘Devo’ page, the scRNA-seq dataset picture at each developmental stage (e.g. ‘Week 10’ under ‘Homo sapiens’) (43) (Figure 3D) was linked with a new tab page to show more details of the dataset by clicking the button ‘View cell atlas in ShinyCell’.
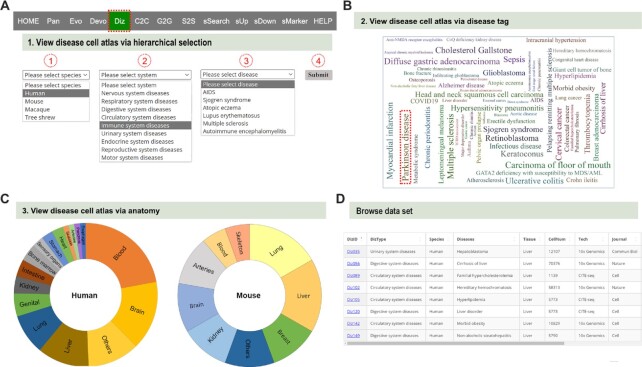
The ‘Diz’ module encompasses scRNA-seq datasets of 85 diseases (Figure 4A), including neurological, respiratory, digestive, cardiovascular, immunological, urinary, endocrinological, reproductive, and motor system diseases (Figure 4A). The disease-related scRNA-seq atlas (e.g. ‘Parkinson disease’) (44) can be viewed via the species-system-disease hierarchical select (Figure 4A), the disease tag (Figure 4B), or the tissue name (Figure 4C), to direct users to a new tab page with the basic information of the disease and dataset. By clicking the link button (e.g. ‘Diz035) (45) in the column of ‘DizlD’ (Figure 4D), users can obtain more detailed information on disease-related scRNA-seq atlas via the link button ‘View cell atlas in ShinyCell’ in a new tab page.
Figure 4.
Demonstration of Diz module. (A) Operation to search for the disease atlas by hierarchical selection. (B) Illustration of obtaining disease cell atlas by tissue name. (C) Illustration of viewing disease cell atlas by anatomical label. (D) Illustration of obtaining disease cell atlas in the summary table.
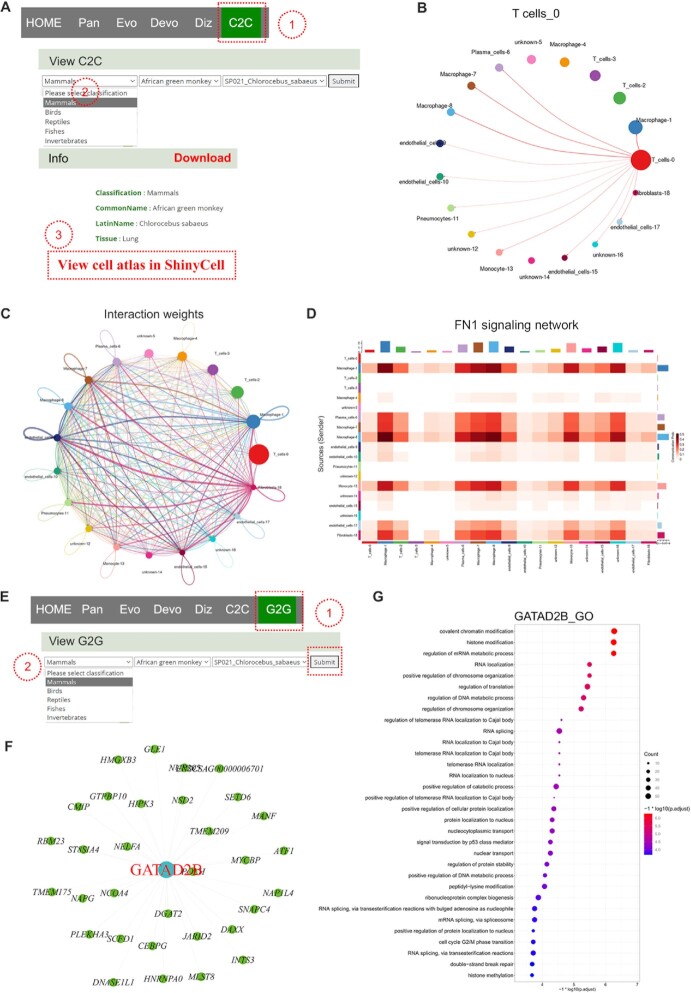
Seven function modules in the navigation menu include ‘C2C’, ‘G2G’, ‘S2S’, ‘sSearch’, ‘sUp’, ‘sDown’, and ‘sMarker’ (Figure 1A). The ‘C2C’ module provides potential cell interaction networks and signaling pathways between cell types. The potential intercellular communication mediated by ligand-receptor pairs analysis is performed using R package CellChat (38). The ligand-receptor pairs are assigned to different signaling pathways. On the ‘C2C’ page, when users choose one species (e.g. ‘African green monkey’) (46) under the ‘View C2C’ drop-down box via hierarchical selection and then click ‘Submit’ button (Figure 5A), a new tab page appears and presents the basic information of dataset, intercellular communication networks, and signaling pathways. The intercellular communications of one cell type with other cell types are shown in one image (Figure 5B), and all intercellular communications among cell types are merged in the last image (Figure 5C). The thickness of lines between cell types represents the strength of intercellular communication. Some signaling pathways are mainly enriched in intercellular communications between certain cell types. Each heatmap in Figure 5D shows one signaling pathway network among particular cell types. The table at the bottom shows the detailed information on every ligand-receptor interaction and related signaling pathway between any two cell types. Users can get customized cellular information by inputting the keywords in the ‘Search’ box.
Figure 5.
Demonstration of C2C and G2G modules. (A) Operation to search for the dataset in C2C module. (B) Intercellular communications between one cell type and other cell types. (C) Intercellular communications for all cell types. (D) Heatmap showing the FN1 signalling pathway network among cell types. (E) Operation to search for the dataset in G2G module. (F) Genetic regulatory networks (GRNs) of GATAD2B and its candidate target genes. (G) Gene Ontology (GO) term enrichment analysist of GATAD2B.
TFs and candidate target genes in each cell type are determined based on the scRNA-seq data. The ‘G2G’ module predicts the putative GRNs between TFs and candidate targets, to promote the understanding of the transcriptional regulation on the cell type level. Users view GRNs of each TF via hierarchical selection at the drop-down box of ‘View G2G’ (46) (Figure 5E). The TF name in red font is located in the hub of GRNs, around which candidate target genes are scattered (Figure 5F). The ‘View GO enrichment for each regulon’ catalogue performs GO (Gene Ontology) term enrichment analysis on candidate target genes of each regulon. The top GO terms for each regulon are shown under the ‘View GO enrichment for each regulon’ item (Figure 5G).
The ‘S2S’ module allows cross-species comparison of expression patterns of transcription factor encoding gene. Users query the cell-type expressions of one gene (Figure 6A) by selecting the tissue, choosing four species from four drop-down boxes, and selecting one gene and then clicking the ‘Plot’ button. Afterwards, a horizontal bar chart is generated to show the gene expression on cell types across species (14) (Figure 6A).
Figure 6.
Demonstration of S2S, sSearch, sUp, sDown, and sMarker modules. (A) Steps to view gene expression level for cell types of four species in S2S module. (B) Operation to search for the dataset in sSearch module. (C) Operation to upload the dataset to SPEED in sUp module. (D) Operation to download the dataset of four data modules in sDown module. (E) Operation to obtain marker genes of organs in sMarker module.
The ‘sSearch’ module provides an optional query mode combination for detailed information on scRNA-seq datasets. On the ‘sSearch’ page, users can query individually or in combination of multiple criteria (Figure 6B). After users click the ‘Apply’ button, the searched results are shown in the table below (Figure 6B). The RDS file of interest in the ‘Table’ could be downloaded via clicking the link in blue font of Rds (Figure 6B).
The ‘sUp’ module allows users to upload their own datasets to SPEED database. Users need to fill in email address, institution, species name, and tissue origin in the columns of ‘information collection’ before uploading the datasets (Figure 6C).
The ‘sDown’ module allows users to download all datasets in Rds file format. On the ‘sDown’ page, users choose one (e.g. ‘Pan’) of the ‘Pan’, ‘Evo’, ‘Devo’, and ‘Diz’ modules under the ‘Download RDS file for each module’, followed by the appearance of a key information table with details and download links for all Rds files in ‘Pan’ module (Figure 6D). Users can obtain the single-cell sequencing dataset of interest by clicking the ‘Download’ button.
Cell types in scRNA-seq data are identified by cell clustering with known marker genes. The ‘sMarker’ module gathers marker genes of different species in nine systems. On the ‘sMarker’ page, users query and download marker gene tables by clicking ‘Search cell markers by species’ or ‘Browse cell markers by tissue’. When users choose the classification/species/tissue/rds options in the drop-down box of ‘Search cell markers by species’ and press the ‘Submit’ button (Figure 6E), a new page appears with the species image and the basic information, as well as a link ‘View Marker Gene Table’ in red font. By clicking this link, users are directed to a new tab page with the table of marker genes. Users can search for any marker gene and download marker gene tables (Figure 6E). The entire datasets with marker gene tables are accessible via the link ‘View cell atlas in ShinyCell’. Alternatively, users can obtain the marker gene by choosing the tissue or organ image of interest under the ‘Browse cell markers by tissue’ item (Figure 6E). Once the tissue or organ image (e.g. ‘Brain’) is clicked, users will see a dataset table with the links of Rds files on a new page. Users can click the link to access the marker gene table.
SUMMARY AND FUTURE PERSPECTIVES
With rapid development of scRNA-seq technology, the number of scRNA-seq datasets is growing. The valuable data are systematically summarized and presented in a freely and publicly accessible database for global researchers to optimize the utility, explore scRNA-seq datasets, and facilitate scientific research, especially for scientists who lack bioinformatics experiences. Here, we collected publicly available single-cell sequencing datasets to create the freely accessible website SPEED, which enables researchers to easily interpret high-quality data resources. SPEED is a powerful tool to deeply mine and define the heterogeneity among cells, tissues, and species. We sorted relevant scRNA-seq datasets to establish the ‘Evo’, ‘Devo’, and ‘Diz’ modules for the convenience of researchers. Seven function modules were built in SPEED to conveniently perform personalized analysis and mining of these scRNA-seq datasets.
SPEED, with its dynamic integrations, will increase in value and importance as more large-scale scRNA-seq studies are performed and the amount of scRNA-seq data is growing exponentially. SPEED pays a special attention to multidimensional cell information from genomics, DNA methylation, chromatin accessibility sequencing, multi-omics (e.g. metabolome and proteome), and trans-omics at single-cell resolution. Data generated by spatial transcriptomics will be integrated into SPEED to interpret the original location of cells in the tissue. The feedbacks and suggestions from researchers will improve the user experience of SPEED.
Supplementary Material
Contributor Information
Yangfeng Chen, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
Xingliang Zhang, Department of Respiratory Diseases, Institute of Pediatrics, Shenzhen Children's Hospital, Shenzhen 518038, China; Department of Pediatrics, the Affiliated Hospital of Guangdong Medical University, Zhanjiang 524001, China.
Xi Peng, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yicheng Jin, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
Peiwen Ding, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
Jiedan Xiao, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
Changxiao Li, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
Fei Wang, Department of Biomedicine, Aarhus University, Aarhus 8000, Denmark.
Ashley Chang, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
Qizhen Yue, State Key Laboratory of Cell Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Mingyi Pu, Department of Medicine, Sun Yat-sen University, Shenzhen 518106, China.
Peixin Chen, Cam-Su Genomic Resource Center, Medical College of Soochow University, Suzhou 215123, China.
Jiayi Shen, Peninsula Cancer Research Center, School of Basic Medical Sciences, Binzhou Medical University, Yantai 264003, China.
Mengrou Li, Institutes of Biology and Medical Sciences (IBMS), Soochow University, Suzhou 215123, China.
Tengfei Jia, Institutes of Biology and Medical Sciences (IBMS), Soochow University, Suzhou 215123, China.
Haoyu Wang, College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Li Huang, The Future Laboratory, Tsinghua University, Beijing 100084, China.
Guoji Guo, Center for Stem Cell and Regenerative Medicine, Zhejiang University School of Medicine, Hangzhou 310058, China.
Wensheng Zhang, Cam-Su Genomic Resource Center, Medical College of Soochow University, Suzhou 215123, China; Peninsula Cancer Research Center, School of Basic Medical Sciences, Binzhou Medical University, Yantai 264003, China.
Hebin Liu, Institutes of Biology and Medical Sciences (IBMS), Soochow University, Suzhou 215123, China.
Xiangdong Wang, Zhongshan Hospital, Department of Pulmonary and Critical Care Medicine, Institute for Clinical Science, Shanghai Institute of Clinical Bioinformatics, Shanghai 200032, China.
Dongsheng Chen, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China; Suzhou Institute of Systems Medicine, Suzhou 215123, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The work was supported by Natural Science Foundation of Jiangsu Province (Grants No BK20220279), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-061), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2022-RC416-01), Gusu Innovation and Entrepreneurship Leading Talents Program (ZXL2022475) and the Shenzhen Science and Technology Program (JCYJ20210324125413036).
Conflict of interest statement. None declared.
REFERENCES
- 1. Tata P.R., Rajagopal J.. Plasticity in the lung: making and breaking cell identity. Development. 2017; 144:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang L., Zhu J., Wang H., Xia J., Liu P., Chen F., Jiang H., Miao Q., Wu W., Zhang L.et al.. A high-resolution cell atlas of the domestic pig lung and an online platform for exploring lung single-cell data. J Genet Genomics. 2021; 48:411–425. [DOI] [PubMed] [Google Scholar]
- 3. Chan M.M., Smith Z.D., Grosswendt S., Kretzmer H., Norman T.M., Adamson B., Jost M., Quinn J.J., Yang D., Jones M.G.et al.. Molecular recording of mammalian embryogenesis. Nature. 2019; 570:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen D., Jiang S., Ma X., Li F.. TFBSbank: a platform to dissect the big data of protein-DNA interaction in human and model species. Nucleic Acids Res. 2017; 45:D151–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma P., Liu X., Xu Z., Liu H., Ding X., Huang Z., Shi C., Liang L., Xu L., Li X.et al.. Joint profiling of gene expression and chromatin accessibility during amphioxus development at single-cell resolution. Cell Rep. 2022; 39:110979. [DOI] [PubMed] [Google Scholar]
- 6. Hollman A.L., Tchounwou P.B., Huang H.C.. The association between gene-environment interactions and diseases involving the human GST superfamily with SNP variants. Int. J. Environ. Res. Public Health. 2016; 13:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohen M., Giladi A., Gorki A.D., Solodkin D.G., Zada M., Hladik A., Miklosi A., Salame T.M., Halpern K.B., David E.et al.. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018; 175:1031–1044. [DOI] [PubMed] [Google Scholar]
- 8. Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S., Fernandez R., Akbarpour M., Chen C.I., Ren Z.et al.. Single-Cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019; 199:1517–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A.et al.. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009; 6:377–382. [DOI] [PubMed] [Google Scholar]
- 10. Farber D.L., Sims P.A.. Dissecting lung development and fibrosis at single-cell resolution. Genome Med. 2019; 11:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A.et al.. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015; 163:1663–1677. [DOI] [PubMed] [Google Scholar]
- 12. Dong J., Hu Y., Fan X., Wu X., Mao Y., Hu B., Guo H., Wen L., Tang F.. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 2018; 19:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M.et al.. The human cell atlas. Elife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen D., Sun J., Zhu J., Ding X., Lan T., Wang X., Wu W., Ou Z., Zhu L., Ding P.et al.. Single cell atlas for 11 non-model mammals, reptiles and birds. Nat. Commun. 2021; 12:7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Sun C., Wang F., Wang X., Zhu J., Luo L., Ding X., Zhang Y., Ding P., Wang H.et al.. Molecular mechanisms governing circulating immune cell heterogeneity across different species revealed by single-cell sequencing. Clin. Transl. Med. 2022; 12:e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Angelidis I., Simon L.M., Fernandez I.E., Strunz M., Mayr C.H., Greiffo F.R., Tsitsiridis G., Ansari M., Graf E., Strom T.M.et al.. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019; 10:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee J.H., Tammela T., Hofree M., Choi J., Marjanovic N.D., Han S., Canner D., Wu K., Paschini M., Bhang D.H.et al.. Anatomically and functionally distinct lung mesenchymal populations marked by lgr5 and lgr6. Cell. 2017; 170:1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu J., Chen F., Luo L., Wu W., Dai J., Zhong J., Lin X., Chai C., Ding P., Liang L.et al.. Single-cell atlas of domestic pig cerebral cortex and hypothalamus. Science Bulletin. 2021; 66:1448–1461. [DOI] [PubMed] [Google Scholar]
- 19. Vieira Braga F.A., Kar G., Berg M., Carpaij O.A., Polanski K., Simon L.M., Brouwer S., Gomes T., Hesse L., Jiang J.et al.. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019; 25:1153–1163. [DOI] [PubMed] [Google Scholar]
- 20. Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R.et al.. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019; 20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhong J., Tang G., Zhu J., Wu W., Li G., Lin X., Liang L., Chai C., Zeng Y., Wang F.et al.. Single-cell brain atlas of parkinson's disease mouse model. J Genet Genomics. 2021; 48:277–288. [DOI] [PubMed] [Google Scholar]
- 22. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Athar A., Fullgrabe A., George N., Iqbal H., Huerta L., Ali A., Snow C., Fonseca N.A., Petryszak R., Papatheodorou I.et al.. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res. 2019; 47:D711–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Papatheodorou I., Moreno P., Manning J., Fuentes A.M., George N., Fexova S., Fonseca N.A., Fullgrabe A., Green M., Huang N.et al.. Expression atlas update: from tissues to single cells. Nucleic Acids Res. 2020; 48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Speir M.L., Bhaduri A., Markov N.S., Moreno P., Nowakowski T.J., Papatheodorou I., Pollen A.A., Raney B.J., Seninge L., Kent W.J.et al.. UCSC cell browser: visualize your single-cell data. Bioinformatics. 2021; 37:4578–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ardini-Poleske M.E., Clark R.F., Ansong C., Carson J.P., Corley R.A., Deutsch G.H., Hagood J.S., Kaminski N., Mariani T.J., Potter S.S.et al.. LungMAP: the molecular atlas of lung development program. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017; 313:L733–L740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z.et al.. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; 47:D900–D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dai Y., Hu R., Manuel A.M., Liu A., Jia P., Zhao Z.. CSEA-DB: an omnibus for human complex trait and cell type associations. Nucleic Acids Res. 2021; 49:D862–D870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao T., Lyu S., Lu G., Juan L., Zeng X., Wei Z., Hao J., Peng J.. SC2disease: a manually curated database of single-cell transcriptome for human diseases. Nucleic Acids Res. 2021; 49:D1413–D1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun D., Wang J., Han Y., Dong X., Ge J., Zheng R., Shi X., Wang B., Li Z., Ren P.et al.. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021; 49:D1420–D1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M.et al.. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019; 47:D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen D., Tan C., Ding P., Luo L., Zhu J., Jiang X., Ou Z., Ding X., Lan T., Zhu Y.et al.. VThunter: a database for single-cell screening of virus target cells in the animal kingdom. Nucleic Acids Res. 2022; 50:D934–D942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W.et al.. Construction of a human cell landscape at single-cell level. Nature. 2020; 581:303–309. [DOI] [PubMed] [Google Scholar]
- 34. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F.et al.. Mapping the mouse cell atlas by microwell-seq. Cell. 2018; 172:1091–1107. [DOI] [PubMed] [Google Scholar]
- 35. Cagan A., Baez-Ortega A., Brzozowska N., Abascal F., Coorens T.H.H., Sanders M.A., Lawson A.R.J., Harvey L.M.R., Bhosle S., Jones D.et al.. Somatic mutation rates scale with lifespan across mammals. Nature. 2022; 604:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M.et al.. Integrated analysis of multimodal single-cell data. Cell. 2021; 184:3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ouyang J.F., Kamaraj U.S., Cao E.Y., Rackham O.J.L.. ShinyCell: Simple and sharable visualisation of single-cell gene expression data. Bioinformatics. 2021; 39:3374–3376. [DOI] [PubMed] [Google Scholar]
- 38. Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q.. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021; 12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huynh-Thu V.A., Irrthum A., Wehenkel L., Geurts P.. Inferring regulatory networks from expression data using tree-based methods. PLoS One. 2010; 5:e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L.et al.. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021; 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khrameeva E., Kurochkin I., Han, D., Guijarro P., Kanton S., Santel M., Qian Z., Rong S., Mazin P., Sabirov M.et al.. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res. 2020; 30:776–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geirsdottir L., David E., Keren-Shaul H., Weiner A., Bohlen S.C., Neuber J., Balic A., Giladi A., Sheban F., Dutertre C.-A.et al.. Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell. 2019; 179:1609–1622. [DOI] [PubMed] [Google Scholar]
- 43. Xu Y., Zhang J., Hu Y., Li X., Sun L., Peng Y., Sun Y., Liu B., Bian Z., Rong Z. Single-cell transcriptome analysis reveals the dynamics of human immune cells during early fetal skin development. Cell Reports. 2021; 36:109524. [DOI] [PubMed] [Google Scholar]
- 44. Kamath T., Abdulraouf A., Burris S.J., Langlieb J., Gazestani V., Nadaf N.M., Balderrama K., Vanderburg C., Macosko E.Z.. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat Neurosci. 2022; 25:588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bondoc A., Glaser K., Jin K., Lake C., Cairo S., Geller J., Tiao G., Aronow B.. Identification of distinct tumor cell populations and key genetic mechanisms through single cell sequencing in hepatoblastoma. Commun Biol. 2021; 4:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Pérez-Pérez L., Meade-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J.et al.. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med. 2021; 13:eabe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.