Abstract
The National Center for Biotechnology Information (NCBI) provides online information resources for biology, including the GenBank® nucleic acid sequence database and the PubMed® database of citations and abstracts published in life science journals. NCBI provides search and retrieval operations for most of these data from 35 distinct databases. The E-utilities serve as the programming interface for most of these databases. New resources include the Comparative Genome Resource (CGR) and the BLAST ClusteredNR database. Resources receiving significant updates in the past year include PubMed, PMC, Bookshelf, IgBLAST, GDV, RefSeq, NCBI Virus, GenBank type assemblies, iCn3D, ClinVar, GTR, dbGaP, ALFA, ClinicalTrials.gov, Pathogen Detection, antimicrobial resistance resources, and PubChem. These resources can be accessed through the NCBI home page at https://www.ncbi.nlm.nih.gov.
INTRODUCTION
NCBI overview
The National Center for Biotechnology Information (NCBI), a center within the National Library of Medicine at the National Institutes of Health, was created in 1988 to develop information systems for molecular biology (1). In this article we provide a brief overview of the NCBI collection of databases, followed by a summary of resources that we significantly updated in the past year. We provide more complete discussions of NCBI resources on the home pages of individual databases, on the NCBI Learn page (https://www.ncbi.nlm.nih.gov/learn/), and in the NCBI Handbook (https://www.ncbi.nlm.nih.gov/books/NBK143764/).
NCBI databases
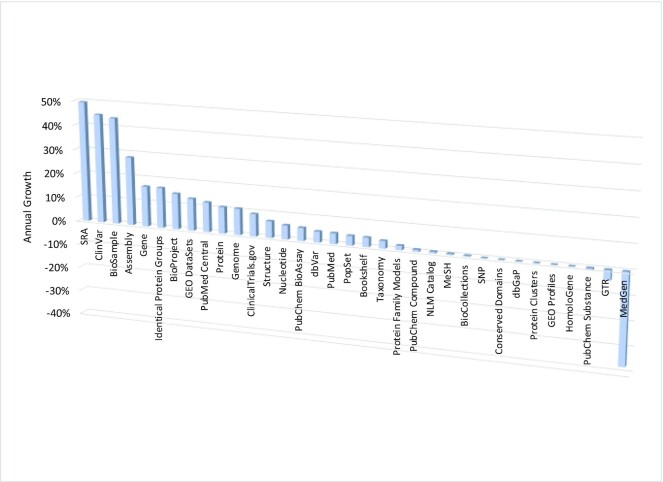
NCBI maintains a diverse set of 35 databases that together contain 3.8 billion records (Table 1 and Figure 1), most of which are available through the Entrez retrieval system (2) at https://www.ncbi.nlm.nih.gov/search/. Each database supports text searching using simple Boolean queries, downloading of data in various formats, and linking records between databases based on asserted relationships. Records retrieved in Entrez can be displayed in many formats and downloaded singly or in batches. An Application Programming Interface for Entrez functions (the E-utilities) is available, and detailed documentation is provided at https://eutils.ncbi.nlm.nih.gov/.
Table 1.
NCBI databases (as of 12 August 2022)
| Database | Records | Description |
|---|---|---|
| Literature | ||
| PubMed | 34 477 874 | scientific and medical abstracts/citations |
| PubMed Central | 8 226 092 | full-text journal articles |
| NLM Catalog | 1 640 320 | index of NLM collections |
| Bookshelf | 926 456 | books and reports |
| MeSH | 349 801 | ontology used for PubMed indexing |
| Genomes | ||
| Nucleotide | 503 629 990 | DNA and RNA sequences from GenBank and RefSeq |
| BioSample | 28 001 796 | descriptions of biological source materials |
| SRA | 23 813 19 | high-throughput DNA/RNA sequence read archive |
| Taxonomy | 2 571 112 | taxonomic classification and nomenclature catalog |
| Assembly | 1 388 980 | genome assembly information |
| BioProject | 614 936 | biological projects providing data to NCBI |
| Genome | 71 826 | genome sequencing projects by organism |
| BioCollections | 8 492 | museum, herbaria, and biorepository collections |
| Genes | ||
| GEO Profiles | 128 414 055 | gene expression and molecular abundance profiles |
| Gene | 39 247 239 | collected information about gene loci |
| GEO DataSets | 5 415 327 | functional genomics studies |
| PopSet | 381 462 | sequence sets from phylogenetic/population studies |
| HomoloGene | 141 268 | homologous gene sets for selected organisms |
| Clinical | ||
| dbSNP | 1 076 992 604 | short genetic variations |
| dbVar | 7 435 613 | genome structural variation studies |
| ClinVar | 1 550 791 | human variations of clinical significance |
| ClinicalTrials.gov | 424 545 | registry of clinical studies |
| MedGen | 210 691 | medical genetics literature and links |
| GTR | 74 358 | genetic testing registry |
| dbGaP | 1 405 | genotype/phenotype interaction studies |
| Proteins | ||
| Protein | 1 073 575 272 | protein sequences from GenBank and RefSeq |
| Identical Protein Groups | 522 090 692 | protein sequences grouped by identity |
| Protein Clusters | 1 137 329 | sequence similarity-based protein clusters |
| Structure | 194 274 | experimentally-determined biomolecular structures |
| Protein Family Models | 182 436 | conserved domain architectures, HMMs, and BlastRules |
| Conserved Domains | 62 852 | conserved protein domains |
| Chemicals | ||
| PubChem Substance | 282 038 443 | deposited substance and chemical information |
| PubChem Compound | 111 567 444 | chemical information with structures, information, and links |
| PubChem BioAssay | 1 466 011 | bioactivity screening studies |
| BioSystems | 983 968 | molecular pathways with links to genes, proteins and chemicals |
Figure 1.
Annual growth rates of the number of records in each NCBI database as of 12 August 2022.
Data sources and collaborations
NCBI receives data from three sources: direct submissions from researchers, national and international collaborations or agreements with data providers and research consortia, and internal curation efforts. For example, NCBI manages the GenBank database (3) and participates with the EMBL-EBI European Nucleotide Archive (ENA) (4) and the DNA Data Bank of Japan (DDBJ) (5) as a partner in the International Nucleotide Sequence Database Collaboration (INSDC) (6). Details about direct submission processes are available from the NCBI Submit page (https://www.ncbi.nlm.nih.gov/home/submit.shtml) and from the resource home pages (e.g. the GenBank page, https://www.ncbi.nlm.nih.gov/genbank/). More information about the various collaborations, agreements, and curation efforts are also available through the home pages of the individual resources.
RECENT DEVELOPMENTS
Literature updates
PubMed
PubMed provides free online access to citations and abstracts for biomedical literature and facilitates searching across the MEDLINE, PubMed Central and Bookshelf literature resources. In the past year, PubMed added over 1.3 million citations, growing the database to >34.4 million total citations.
We are continuing to develop new features and update existing offerings in PubMed to consistently improve the user experience. For example, we added additional page navigation options to the web interface displaying PubMed search results. These options include one-click navigation to the first, previous, next, and last page of results, and these buttons are conveniently placed both above and below the list of records retrieved. We added more checks and validations to processes that prepare data for single citation matching (7) that enables the tool to be more reliable and stable. Work continues to ensure that Best Match (8) continues to return the most relevant and useful articles for each query. Part of this effort was a recent codeathon where teams investigated PubMed search behavior so that the search engine treats each article fairly and equitably (https://www.ncbi.nlm.nih.gov/pubs/techbull/jf22/brief/jf22_ncbi_codeathon.html).
As of May 2022, NLM transitioned to fully automated MeSH indexing of MEDLINE citations in PubMed both to provide users with timely access to MeSH indexed metadata and to enable MeSH indexing to keep pace with the rapidly expanding volume of published biomedical literature (https://www.ncbi.nlm.nih.gov/pubs/techbull/nd21/nd21_medline_2022.html). New MEDLINE citations are now typically indexed with MeSH terms within 48 h of appearing in the PubMed database. NLM staff continue to be involved in the refinement of automated indexing algorithms and they also provide quality assurance for automated indexing.
PubMed Central (PMC)
PMC is NCBI’s free full-text archive of biomedical and life sciences journal literature. In 2022, the PMC archive surpassed 8 million publicly available full-text journal articles, author manuscripts, and preprints. Additionally, over the last year, PMC’s Public Health Emergency COVID-19 Initiative (https://www.ncbi.nlm.nih.gov/pmc/about/covid-19/) continued to make coronavirus-related articles accessible in PMC in formats and under license terms that facilitate text mining and secondary analysis. We have added >280 000 articles to PMC because of this collaboration with the publishing community. PMC has also continued to pilot the curation and ingest of NIH-supported preprints reporting COVID-19 research (https://www.ncbi.nlm.nih.gov/pmc/about/nihpreprints/). Through the first 2 years of the pilot (July 2020–July 2022), we added 3400 preprint records to PMC, accelerating and expanding discovery of NIH research relating to the ongoing public health emergency.
PMC launched an updated website in March 2022, marking the first step in an ongoing modernization effort to ensure the long-term sustainability of the PMC infrastructure. This was the first major update to the website since 2012. For PMC website visitors, the changes were primarily to the look and feel of the site, including a redesigned and reorganized homepage, easier to navigate documentation, a revamped user guide, and a streamlined article display. All pages on the PMC website are now responsive to device screen sizes and are more mobile friendly. We will continue to release new features on an ongoing basis guided by feedback from website visitors, usability testing, and user research.
Bookshelf
The NCBI Bookshelf provides free online access to full-text books and documents in the life sciences, healthcare, and medicine. In the past year, Bookshelf added over 1000 books, growing the repository to over 10 600 total books from over 150 content providers. Significant new peer-reviewed collections added in 2022 were in the subjects of toxicology, diabetes, nutrition, health disparities, and public health.
Bookshelf is in the process of releasing a new content management system (CMS) to support its submission and conversion workflows. Integrated with PMC architecture, it streamlines and further automates Bookshelf's submission pipeline from point of submission to public access on the Bookshelf website. The initial version permits the conversion and ingest of both entire submitted books and individual chapters added and updated at different times by a team of users. For large integrated resources in which individual chapters are added and updated independently, editors can also intuitively manage the order and display of their content and update the bibliographic metadata and funding information pertaining to the resource. The CMS automatically reports validation and other data integrity and style errors to users so they can fix these issues themselves, allowing them both to preserve and ensure the quality of their content according to archival standards. Users are also provided previews of their content as it will display on the Bookshelf website for further quality assurance, which is of value when creating new content.
Genome updates
NCBI Virus
The NCBI Virus resource facilitates easy access to viral sequence data and normalized metadata. More than 7.3 million unique sequence samples have been submitted to the Sequence Read Archive (SRA) and GenBank during the COVID-19 pandemic. NCBI has developed an analysis pipeline that consistently calculates nucleotide and protein variations across these sequence samples and captures the genetic variations in each sample in a Variant Call Format (VCF) file (https://www.ncbi.nlm.nih.gov/sra/docs/sars-cov-2-variant-calling/). VCF files are available through the COVID-19 Genome Sequence Dataset on Amazon Web Services (AWS) Registry of Open Data (RODA) Open Data Portal (https://registry.opendata.aws/ncbi-covid-19/) and the COVID-19 Genome Sequence Dataset on the Google Cloud Platform (GCP) Public Dataset Program (https://console.cloud.google.com/marketplace/product/national-library-of-medicine/ncbi-covid-data). The underlying nucleotide and protein variation data, information on coverage and read depth, sequence sample lineage assignment, and descriptive metadata are available from AWS Athena (https://www.ncbi.nlm.nih.gov/sra/docs/sra-athena/) and GCP BigQuery (https://www.ncbi.nlm.nih.gov/sra/docs/sra-bigquery/).
We have added several features to the NCBI Virus Resource (https://www.ncbi.nlm.nih.gov/labs/virus/) to better support search, retrieval, and analysis of SARS-CoV-2 sequence data. These include a current classification of GenBank sequences into Pangolin lineages (9), links between GenBank records and underlying SRA or BioSample samples, sample isolate names, and identification of sequences that were collected for the purpose of baseline surveillance. A new SARS-CoV-2 Variants Overview dashboard (https://www.ncbi.nlm.nih.gov/activ) has also been added to support the NIH Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) Tracking Resistance and Coronavirus Evolution (TRACE) initiative (https://www.nih.gov/research-training/medical-research-initiatives/activ/tracking-resistance-coronavirus-evolution-trace). This dashboard leverages data from the NCBI SARS-CoV-2 variation analysis pipeline and includes information lineage defining mutations, links to experimental data associated with specific lineages in the COVID-19 Open Data Portal (10), and visualizations of the geographic and temporal distribution of lineages.
Comparative Genome Resource
The NIH Comparative Genomics Resource (CGR) is a multi-year National Library of Medicine (NLM) project being developed at NCBI to maximize the impact of eukaryotic research organisms and their genomic data resources to biomedical research. The CGR project (https://www.ncbi.nlm.nih.gov/comparative-genomics-resource/) will facilitate reliable comparative genomics analyses for all eukaryotic organisms in collaboration with the genomics community. Consistently annotated and uncontaminated genomes are the inputs to reliable comparative analyses. NCBI is developing publicly accessible, cloud-ready tools for contamination screening and annotation to support the creation and deposition of these data in GenBank. The project's efforts will also enrich genome-associated content held at NCBI with community-supplied content and facilitate access to, analyses of, and downloads of genome and genome associated data through a streamlined online experience, as well as command line tools and public APIs.
In 2022, we released several CGR-associated developments. A beta version of a new foreign contamination screening (FCS) tool (https://github.com/ncbi/fcs) that detects adaptors and cross-species contamination in assembled genomes is now available for download. NCBI is rescreening genomes in GenBank and RefSeq with this tool to identify previously undetected contamination. To date, we have removed 131 Mb of contamination from 64 genomes in both databases. A recent set of CGR-associated content enhancements added new content to Gene records. We added publication references programmatically sourced from the Alliance of Genome Resources (https://www.alliancegenome.org/) to Gene records for organisms including Caenorhabditis elegans, Saccharomyces cerevisiae, and Drosophila melanogaster. We also added descriptive information to Gene records for the above three organisms as well as for Homo sapiens, Mus musculus, Rattus norvegicus, and Danio rerio. In the past year, we added nearly 10 000 curated and/or published protein architectures to SPARCLE (11). Our focus was to provide coverage for proteins annotated on assemblies from taxa identified as ‘small eukaryotes’ such as nematodes, fungi, and protists, but most of the added architectures provide significant coverage of taxa beyond that scope and extending to other metazoans. These annotations, as well as others providing information relevant to protein function, are now available in Conserved Domain Search (CD-Search) results and through the Protein Families database.
Additional CGR releases included a new visualization tool, the Comparative Genome Viewer (CGV) (https://ncbi.nlm.nih.gov/genome/cgv/), that allows users to compare two genomes based on assembly-assembly alignments provided by NCBI. In support of CGR, NCBI Datasets (https://www.ncbi.nlm.nih.gov/datasets/), a resource providing web, command line, and programmatic access to data from multiple NCBI databases, introduced new genome pages that facilitate both browsing and downloading of packages for genome and associated metadata. These packages include sections containing BioSample data, gene annotation information, publications, and Benchmarking Universal Single-Copy Orthologs (BUSCO) scores (12). These pages also provide the corresponding commands to access the data via the Datasets command line tool and API, links to corresponding tables with filterable lists of genomes and genes, and relevant analysis tools such as BLAST and GDV.
BLAST clusteredNR
The protein BLAST web interface now offers the clusteredNR database derived from clustering the standard protein nr database at 90% identity and 90% length (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch). Searches using this database are faster and allow users to see more taxonomically diverse results. The summary of clusters displays the common ancestor for the sequences in each cluster, the number of cluster members and organisms, as well as the protein title for the representative sequence of each cluster. The results interface for clusteredNR also supports multiple ways to explore the contents of an individual cluster that includes a COBALT (13) multiple alignment, a tree display and taxonomic information.
IgBLAST
We have updated IgBLAST (14) by adding the ability to annotate the constant (C) gene region for human and mouse immunoglobulin (Ig) sequences in IgBLAST results. This is particularly helpful for users who want to identify the Ig isotypes while also analyzing VDJ rearrangements. We also improved the ability of IgBLAST to correctly identify rearranged J genes in cases where unrearranged J genes are included in a query sequence. For example, a rearranged genomic VDJ sequence containing a rearranged IGHJ1 gene may still have other IGHJ genes in unrearranged germline configurations downstream. Previously, in some cases IgBLAST may have erroneously reported such downstream J genes as rearranged.
Genome data viewer (GDV)
NCBI’s Genome Data Viewer (https://www.ncbi.nlm.nih.gov/genome/gdv/) supports visualization and analysis of annotated eukaryotic assemblies. Researchers can examine NCBI-provided gene annotation, variation, and RNA-seq analysis as data tracks in the browser, or stream or upload their own BLAST searches, mapped analyses, or annotations of genes or features. Population variation data from the European Variation Archive (EVA) database are shown as tracks in the browser where available. Significantly, recent improvements allow users to add study data from the SRA, GEO and dbGaP databases directly to the GDV view (https://ncbiinsights.ncbi.nlm.nih.gov/2021/10/21/geo-sra-dbgap-tracks-genome-data-viewer/).
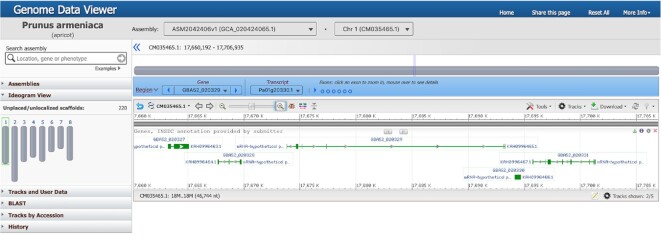
Genome assemblies from over 1500 animals, plants, fungi, and protist species are currently available in GDV. To better support the growing number of eukaryotic genomes, we are now including selected complete, high-quality assemblies with annotations provided by GenBank submitters to our catalog of viewable genomes. As shown in Figure 2, researchers can view submitter-provided annotations as tracks in GDV for these genomes and upload or stream their own custom data as well. Over 1400 NCBI-annotated (RefSeq) and over 500 submitter-annotated assemblies are available in GDV as of this writing.
Figure 2.
GDV visualization of a region of Chromosome 1 from the Prunus armeniaca genome assembly ASM2042406v1 showing a track of gene annotations provided by the GenBank submitter.
RefSeq
The NCBI RefSeq collection now includes 258 143 prokaryote and 1433 eukaryote genomes as of 23 August 2022, representing yearly growth of 15% and 11%, respectively. Genomes from 898 species are now annotated with NCBI’s Eukaryotic Genome Annotation Pipeline, including all vertebrates and most other multicellular eukaryotes, with most models completely based on RNA-seq and/or protein alignment evidence. The RefSeq annotation of the human genome prominently incorporates the Matched Annotation from the NCBI and EMBL-EBI (MANE) (15) dataset. This collaborative project aims to converge on human gene and transcript annotation between RefSeq and Ensembl/GENCODE to define a genome-wide set of representative protein-coding transcripts, called MANE Select, that serve as universal standards for clinical variant reporting. The latest annotation of the human GRCh38.p14 reference genome, Annotation Release 110, includes MANE Select transcripts for over 99% of protein-coding genes and a set of high-value transcripts referred to as MANE Plus Clinical for those loci where the MANE Select alone is not sufficient to report all currently known pathogenic variants. We encourage adoption of MANE transcripts to increase the consistency of clinical reporting, streamline clinical interpretation, and facilitate the comparison and exchange of data between resources.
The human and mouse RefSeq genome annotations also include over 17 000 non-genic RefSeq Functional Element (RefSeqFE) features, such as enhancers, silencers or recombination regions, that have been derived from published experimental studies (16). RefSeqFE data have multiple uses for basic functional discovery and bioinformatics studies and may be particularly useful for clinically relevant genetic variant interpretation. They also have additional uses as known positive controls in various epigenomic studies and as reference standards for functional interactions.
Lastly, RefSeq features a complete annotation of the human T2T-CHM13v2.0 assembly produced by the Telomere-to-Telomere (T2T) Consortium (17), the first gap-less assembly available for the human genome. The RefSeq annotation of T2T-CHM13v2.0 includes both projection of curated genes, transcripts, and RefSeqFEs, and de novo predictions of novel protein-coding and non-coding genes as well as pseudogenes. The GRCh38.p14 and T2T-CHM13v2.0 annotations are fully supported through NCBI Gene, BLAST, GDV, and complete datasets are available by FTP and through NCBI Datasets.
Type assemblies
NCBI collects and curates prokaryotic type strains and their genomes, referred to as ‘type assemblies.’ Type assemblies act as unambiguous references for taxonomic names and play an important role in comparative genomics, for example when computing average nucleotide identity (ANI), and especially when verifying and reclassifying a taxon (18). NCBI evaluates all GenBank assemblies, including type assemblies, for potential anomalies such as contamination, misassembly, and taxonomic misidentification. We have identified 173 such anomalous type assemblies and excluded them from being considered as a type in NCBI resources. In addition, NCBI applies additional criteria to identify potentially problematic type assemblies, and these are considered as types but are not used for making changes such as reclassifying or modifying existing taxonomies of other assemblies. A type assembly is considered potentially problematic if it is either significantly different from other type assemblies or the majority of non-type assemblies from its own species, or if it is identical to type assemblies from a different species. We have found 1123 assemblies to be potentially problematic. Using type assemblies as references, we verified the taxonomy of over 1.1 million GenBank genomes and the taxonomy of over 7000 new submissions before accepting them in GenBank. In addition, we reclassified over 1800 existing genomes in GenBank. The public NCBI Genomes FTP site (https://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS) contains detailed reports such as a full list of all prokaryotic type assemblies with their ANI validation status. Each file is accompanied by a detailed README file describing its contents.
Protein updates
iCn3D
During the past year, we have updated the 3D structure viewer iCn3D (19,20) frequently to accommodate new features. iCn3D can be used to conveniently examine more than 200 million 3D structures predicted by AlphaFold, as found in the AlphaFoldDB (21). iCn3D provides a richly annotated view of these coordinate sets and provides functionality to align and superimpose them with experimentally derived 3D structures from the Protein Data Bank (PDB). Annotations include the location of conserved domains, functional sites, SNPs (22) and variation tracked by ClinVar (23) for human proteins, post translational modifications (PTMs) available from UniProt (https://www.uniprot.org/help/post-translational_modification), 3D domains, disulfide bonds, and more. 3D neighbors for an AlphaFold structure can be found using VAST search (24) or Foldseek (25), and multiple PDB or AlphaFold structures can be aligned directly in iCn3D based on 3D structure or sequence similarities. iCn3D also supports basic Virtual Reality (VR) and Augmented Reality (AR) views. The VR view requires a VR headset, and the AR view requires an Android phone. We have enhanced the analysis of molecular interactions by showing both the common and differing interactions in the comparative analysis of 3D structures. We have added scripting support using Node.js and Python for automated analysis of many structures. The script ‘annotation.js’ (https://github.com/ncbi/icn3d/tree/master/icn3dnode) retrieves annotations from iCn3D, and Python scripts (https://github.com/ncbi/icn3d/tree/master/icn3dpython) support the retrieval of any data accessible in the user interface of iCn3D.
Clinical updates
ClinVar
ClinVar (23) is NCBI’s archive of genetic variants and interpretations of their significance for human health. Over the past year, ClinVar added to the database 392 000 new variants processed from 582 000 new submitted records. We improved ClinVar's pre-submission validation by adding validation for citations, gene symbols, and some special cases for the clinical significance of the variant. We also enhanced ClinVar's submission API that now allows the deletion of records, the submission of pharmacogenomic variants, and a test environment. These improvements have helped the ClinVar team maintain a median turnaround time for submissions of six days.
We added several features to ClinVar to make finding data easier and more intuitive. Search in ClinVar now supports querying for a genomic location using UCSC-style chromosome coordinates, e.g. chr1:11,102,837–11,267,747. Searches that use chromosome coordinates show the results in both the traditional table format and a new genome view that gives a better visual context for the results. Searches for gene symbols show the results in a table format and a new lollipop diagram for the gene that makes it easy to visualize the distribution of variants across the gene. Additionally, the Variation pages were updated to make it easier to find the details of each submitted record. The table of ‘Submitted interpretations and evidence’ continues to show high-level information for each SCV record, but each row in the table can be expanded in place to see additional details provided by the submitter.
We added other features to help users assess the interpretations of variants found in ClinVar. Based on a recommendation from ClinGen (https://clinicalgenome.org/site/assets/files/4531/clingenrisk_terminology_recomendations-final-02_18_20.pdf), we added new terms to describe the interpretation of variants that have low penetrance or are risk alleles. To accommodate these new terms, we updated the algorithms that calculate an overall clinical significance and conflicts in interpretations in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/docs/clinsig/#clinsig_agg). To help web users better evaluate variants with conflicts in the interpretation, we added new filters to the search results page so that the user can focus on certain subsets of conflicts, such as a pathogenic or likely pathogenic (P/LP) variant versus a variant of uncertain significance (VUS).
Genetic Testing Registry
The Genetic Testing Registry (GTR, https://www.ncbi.nlm.nih.gov/gtr/) is NCBI’s online resource for clinical and research genetic tests (26). GTR was founded in 2012 with the goal of providing transparent genetic testing information and advancing public health and research into the genetic basis of health and disease. In 2020, GTR expanded its scope to include molecular and serologic tests for microbes. As of July 2022, GTR includes 74 350 tests by 516 laboratories, covering 22 511 conditions and 18 738 genes. Of these, 102 tests are for microbes and 81 for SARS-CoV-2 molecular and antigen tests. GTR contains a variety of genetic tests including those for somatic phenotypes, Mendelian disorders, and pharmacogenetic responses. Tests can be single gene or panels, and GTR worked to improve its submission pipeline to be able to process large tests that include exomes. Currently, the largest test in GTR interrogates 4672 genes and 5133 conditions. Each test in GTR is orderable, and descriptive information for each test is intended to help health care providers find the most appropriate test for their patients. To aid in standardizing test data, GTR provides list of values for most data fields or validation that ensures compliance with standards.
In 2022, GTR developed a submission API to enable laboratories to submit new tests, update existing tests, and delete tests they no longer offer. The API will help laboratories with informatics capacity to maintain test data in GTR automatically. Fulgent Genetics is the laboratory with the largest number of tests in GTR, having 19 321 tests or ∼26% of all GTR tests. The GTR API aims to encourage similar labs to fully register their test catalog. We developed GTR’s API with input from current submitters, and it validates content including genes and variants, conditions, and CPT codes while also providing a test environment.
dbGaP
The Database of Genotypes and Phenotypes (dbGaP) provides unprecedented access to very large genetic and phenotypic datasets funded by the NIH and other agencies. Scientists from the global research community may access all public data and apply for controlled access to data for thousands of studies. The information contained in dbGaP includes individual-level molecular and phenotype data, analysis results, medical images, general information about the study, and documents that contextualize phenotypic variables, such as research protocols and questionnaires. To facilitate interoperability and exchange of dbGaP data between disparate systems, we developed the dbGaP FHIR API for data exchange using the FHIR standard (https://www.hl7.org/fhir/). Users can explore the mappings between other dbGaP types and FHIR resources at https://dbgap-api.ncbi.nlm.nih.gov/fhir-mapping/interactive. All dbGaP research studies' metadata are available as open-access data (https://dbgap-api.ncbi.nlm.nih.gov/fhir/x1/ResearchStudy). In addition, the API will soon host over 1 billion individual level values and combinations of observation values across demographic, clinical, and exposure variables. A test dataset is available for preview (https://dbgap-api.ncbi.nlm.nih.gov/fhir/x1/Patient).
ALFA
The current release (Release 2) of the ALFA project (https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/) provides allele frequencies for 200 000 subjects from dbGaP studies. We will provide additional studies and subjects with updates in future releases. We computed the subject's ancestry for 12 ALFA reported populations using the GRAF-pop feature of GRAF (https://www.ncbi.nlm.nih.gov/snp/docs/gsr/data_inclusion/#Sample). GRAF-pop infers subject ancestry from genotypes, estimates population structure, and uses the results to validate self-reported populations in studies. The GRAF-pop feature has been upgraded and is now provided as a separate software package called GrafPop, independent of GRAF. The enhancements included using ten times more SNP markers (100 000) for ancestry inference, support for VCF file format input, and a variety of other features. GrafPop and source codes are available for download at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/Software.cgi.
ClinicalTrials.gov
Launched in 2000, ClinicalTrials.gov (https://clinicaltrials.gov) is an online database of information provided by sponsors or investigators for ∼430 000 clinical research studies conducted around the world, including summary results for nearly 56 000 studies. Since October 2019, NLM has been engaging stakeholders and using feedback to modernize ClinicalTrials.gov to deliver an improved user experience on an updated platform that will accommodate growth and enhance efficiency (https://www.nlm.nih.gov/od/bor/clinicaltrialswg/NLM_BOR_CTG_WG_Modernization_Update_Report.pdf). In December 2021 (at the beginning of Year 3 of the modernization effort) (https://nlmdirector.nlm.nih.gov/2021/12/08/clinicaltrials-gov-modernization-effort-beta-releases-now-available/), NLM released the first beta version of the ClinicalTrials.gov website that introduced users to a new technology platform and allowed us to evaluate its real-world performance. Three subsequent releases throughout 2022 built upon this foundational beta framework by enhancing features in response to user feedback. The new design includes improved record navigation, content in plain language, updated search functionality, and download functions while allowing users to expand and collapse content sections. In January 2022, NLM released the initial version of the beta Protocol Registration and Results System (PRS), the data entry and management system for ClinicalTrials.gov. Two additional releases followed in 2022. These beta sites function in parallel to their legacy counterparts. This allows for usability research and iterative improvements to the beta sites without disrupting current operations and the existing user experience.
Pathogen detection
The NCBI Pathogen Detection Project (https://www.ncbi.nlm.nih.gov/pathogens/) helps public health scientists investigate disease outbreaks by integrating pathogen genomic sequences obtained from cultured bacterial isolates and quickly clustering and identifying related sequences (27). It has been used successfully to help uncover an international outbreak due to contaminated mushrooms (28) and has been shown to contribute significantly to reducing illness and the burden of disease in the US for foodborne pathogens (29). As of 11 August 2022, over 1 162 000 pathogen isolates covering 52 bacterial taxa and one emerging fungal pathogen, Candida auris, are actively being analyzed. The analysis results are available in the Isolates Browser daily (https://www.ncbi.nlm.nih.gov/pathogens/isolates).
Antimicrobial resistance
The Pathogen Detection team has continued to release updated resources for antimicrobial resistance (AMR) (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/). We recently described the additions of virulence and stress response genes to AMRFinderPlus (30). In 2017, NCBI described the transfer of the beta lactamase allele registry previously hosted at the Lahey Clinic to NCBI (31). In 2021 an international group of beta-lactamase experts developed a consensus protocol for naming naturally occurring beta-lactamase genes at a meeting organized by the American Society for Microbiology, during which NCBI was defined as the curator of record for beta-lactamase nomenclature (32). Our curation efforts and browser interfaces are described elsewhere (33).
The AMR database release on 8 August 2022 included 7374 total proteins (6333 AMR proteins, 235 stress response proteins and 806 virulence proteins) as well as 991 point mutations. We have added over 2500 publication references. Two new web interfaces are now available: the Pathogen Detection Reference HMM Catalog, a portal to the curated database of reference hidden Markov models (HMMs) used by AMRFinderPlus (https://www.ncbi.nlm.nih.gov/pathogens/hmm/), and the Reference Gene Hierarchy that displays the hierarchy of genes, families and upstream nodes used to organize genes and HMMs (https://www.ncbi.nlm.nih.gov/pathogens/genehierarchy/). We added a download feature to the Reference Gene Catalog that allows users to download reference protein and nucleotide sequences in FASTA format.
We analyze all bacterial isolates in the Pathogen Detection Isolates Browser with AMRFinderPlus (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/), and the three categories of genes (AMR, stress, and virulence) are available in the Isolates Browser. Currently over 1 137 000 isolates have at least one identified AMR gene, over 964 000 have at least one identified stress response gene, and over 648 000 have at least one identified virulence gene. An antibiogram template for capturing antibiotic susceptibility data is available and is tied to BioSample submissions (https://www.ncbi.nlm.nih.gov/pathogens/submit-data/#ast), and the S/I/R calls area is available in the Isolates Browser for over 19 000 isolates. For the subset of isolates that have genome assemblies available in GenBank and that have genes identified by AMRFinderPlus, a tabular viewer called the Microbial Browser for Genetic and Genomic Elements (MicroBIGG-E) is available (27,33). It has been used, for example, to evaluate the health risk of AMR genes (34). MicroBIGG-E is available both through a web interface and through Google Cloud Platform (https://www.ncbi.nlm.nih.gov/pathogens/docs/microbigge_gcp). This allows bulk access to the tabular data that cannot be accessed from the web interface due to web download limits.
Chemical updates
Thanks to data integration with more than 60 new sources over the past year, PubChem (35,36) now provides chemical information for 112 million compounds collected from over 860 data sources. Among the newly added data to PubChem are annotations about drug products (from the U.S. Food and Drug Administration (FDA)’s National Drug Code (NDC) Directory) and FDA-licensed biological products (from the FDA Purple Book). Information on drugs for HIV/AIDS and related opportunistic infections have also been integrated from the drug database available at the HIV Clinical Info website (https://clinicalinfo.hiv.gov). In addition, occupational health information on hazardous chemicals and associated diseases from Haz-Map (https://haz-map.com/) has been added to PubChem.
We released the PubChem Cell Line and Taxonomy pages (37) that present all data available in PubChem for a given cell line and organism, respectively. The Cell Line and Taxonomy pages contain annotations about cell lines and organisms collected from authoritative data sources, and this helps users understand data from bioassays performed against a particular cell line or organism. In addition, PubChem now supports programmatic access to target-specific bioactivity data and relevant annotations (e.g. for a given protein, gene, pathway, cell line, or organism), through PUG-REST (38) and PUG-View (39), which are PubChem's representation state transfer (REST)-like interfaces.
FOR FURTHER INFORMATION
The resources described here include documentation, other explanatory materials, and references to collaborators and data sources on their respective web sites. The NCBI Help Manual and the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/) describe the principal NCBI resources in detail. The NCBI Learn page (www.ncbi.nlm.nih.gov/learn/) provides links to documentation, tutorials, webinars, courses, and upcoming conference exhibits. A variety of video tutorials are available on the NLM YouTube channel that can be accessed through links in the standard NCBI page footer. User-support staff are available to answer questions at info@ncbi.nlm.nih.gov, and users can view support articles at https://support.nlm.nih.gov. Updates on NCBI resources and database enhancements are described on the NCBI Insights blog (https://ncbiinsights.ncbi.nlm.nih.gov/), NCBI social media sites (FaceBook, Twitter, and LinkedIn), and the several mailing lists and RSS feeds that provide updates on services and databases. Links to these resources are in the NCBI page footer and on NCBI Insights.
DATA AVAILABILITY
The data and resources discussed in this paper are publicly available at https://www.ncbi.nlm.nih.gov.
ACKNOWLEDGEMENTS
The authors would like to thank all the NCBI staff who through their dedicated efforts continue to allow NCBI to provide our full collection of services to the community. This work was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health.
Contributor Information
Eric W Sayers, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Evan E Bolton, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
J Rodney Brister, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kathi Canese, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jessica Chan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Donald C Comeau, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Catherine M Farrell, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Michael Feldgarden, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Anna M Fine, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kathryn Funk, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Eneida Hatcher, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Sivakumar Kannan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Christopher Kelly, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Sunghwan Kim, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
William Klimke, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Melissa J Landrum, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Stacy Lathrop, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Zhiyong Lu, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Thomas L Madden, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Adriana Malheiro, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Aron Marchler-Bauer, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Terence D Murphy, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Lon Phan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Shashikant Pujar, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Sanjida H Rangwala, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Valerie A Schneider, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Tony Tse, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jiyao Wang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jian Ye, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Barton W Trawick, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kim D Pruitt, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Stephen T Sherry, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
FUNDING
Funding for open access charge: National Center for Biotechnology Information of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A.. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996; 266:141–162. [DOI] [PubMed] [Google Scholar]
- 3. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2022; 50:D161–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cummins C., Ahamed A., Aslam R., Burgin J., Devraj R., Edbali O., Gupta D., Harrison P.W., Haseeb M., Holt S.et al.. The european nucleotide archive in 2021. Nucleic Acids Res. 2022; 50:D106–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okido T., Kodama Y., Mashima J., Kosuge T., Fujisawa T., Ogasawara O.. DNA data bank of japan (DDBJ) update report 2021. Nucleic Acids Res. 2022; 50:D102–D105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arita M., Karsch-Mizrachi I., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021; 49:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yeganova L., Comeau D.C., Kim W., Wilbur W.J., Lu Z.. SingleCite: towards an improved single citation search in pubmed. Proceedings of the BioNLP 2018 Workshop. 2018; Melbourne, Australia: Association for Computational Linguistics; 151–155. [Google Scholar]
- 8. Fiorini N., Canese K., Starchenko G., Kireev E., Kim W., Miller V., Osipov M., Kholodov M., Ismagilov R., Mohan S.et al.. Best match: new relevance search for pubmed. PLoS Biol. 2018; 16:e2005343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B.et al.. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021; 7:veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brimacombe K.R., Zhao T., Eastman R.T., Hu X., Wang K., Backus M., Baljinnyam B., Chen C.Z., Chen L., Eicher T.et al.. An opendata portal to share COVID-19 drug repurposing data in real time. 2020; bioRxiv doi:05 June 2020, pre-print: not peer-reviewed 10.1101/2020.06.04.135046. [DOI]
- 11. Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S.et al.. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020; 48:D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015; 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- 13. Papadopoulos J.S., Agarwala R.. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007; 23:1073–1079. [DOI] [PubMed] [Google Scholar]
- 14. Ye J., Ma N., Madden T.L., Ostell J.M.. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013; 41:W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morales J., Pujar S., Loveland J.E., Astashyn A., Bennett R., Berry A., Cox E., Davidson C., Ermolaeva O., Farrell C.M.et al.. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature. 2022; 604:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farrell C.M., Goldfarb T., Rangwala S.H., Astashyn A., Ermolaeva O.D., Hem V., Katz K.S., Kodali V.K., Ludwig F., Wallin C.L.et al.. RefSeq functional elements as experimentally assayed nongenic reference standards and functional interactions in human and mouse. Genome Res. 2022; 32:175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A.et al.. The complete sequence of a human genome. Science. 2022; 376:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ciufo S., Kannan S., Sharma S., Badretdin A., Clark K., Turner S., Brover S., Schoch C.L., Kimchi A., DiCuccio M.. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018; 68:2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Youkharibache P., Zhang D., Lanczycki C.J., Geer R.C., Madej T., Phan L., Ward M., Lu S., Marchler G.H.et al.. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics. 2020; 36:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J., Youkharibache P., Marchler-Bauer A., Lanczycki C., Zhang D., Lu S., Madej T., Marchler G.H., Cheng T., Chong L.C.et al.. iCn3D: from web-based 3D viewer to structural analysis tool in batch mode. Front. Mol. Biosci. 2022; 9:831740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with alphafold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W.et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gibrat J.F., Madej T., Bryant S.H.. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996; 6:377–385. [DOI] [PubMed] [Google Scholar]
- 25. van Kempen M., Kim S.S., Tumescheit C., Mirdita M., Gilchrist L.M., Söding J., Steinegger M.. Foldseek: fast and accurate protein structure search. 2022; bioRxiv doi:09 February 2022, preprint: not peer reviewed 10.1101/2022.02.07.479398. [DOI] [PMC free article] [PubMed]
- 26. Rubinstein W.S., Maglott D.R., Lee J.M., Kattman B.L., Malheiro A.J., Ovetsky M., Hem V., Gorelenkov V., Song G., Wallin C.et al.. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013; 41:D925–D935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sayers E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2020; 48:D9–D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pettengill J.B., Markell A., Conrad A., Carleton H.A., Beal J., Rand H., Musser S., Brown E.W., Allard M.W., Huffman J.et al.. A multinational listeriosis outbreak and the importance of sharing genomic data. Lancet Microbe. 2020; 1:e233–e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown B., Allard M., Bazaco M.C., Blankenship J., Minor T.. An economic evaluation of the whole genome sequencing source tracking program in the u.S. PLoS One. 2021; 16:e0258262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feldgarden M., Brover V., Gonzalez-Escalona N., Frye J.G., Haendiges J., Haft D.H., Hoffmann M., Pettengill J.B., Prasad A.B., Tillman G.E.et al.. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021; 11:12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resource Coordinators, N. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017; 45:D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bradford P.A., Bonomo R.A., Bush K., Carattoli A., Feldgarden M., Haft D.H., Ishii Y., Jacoby G.A., Klimke W., Palzkill T.et al.. Consensus on beta-Lactamase nomenclature. Antimicrob. Agents Chemother. 2022; 66:e0033322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feldgarden M., Brover V., Fedorov B., Haft D.H., Prasad A.B., Klimke W.. Curation of the AMRFinderPlus databases: applications, functionality and impact. Microb. Genom. 2022; 8:mgen000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang A.N., Gaston J.M., Dai C.L., Zhao S., Poyet M., Groussin M., Yin X., Li L.G., van Loosdrecht M.C.M., Topp E.et al.. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021; 12:4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim S. Getting the most out of pubchem for virtual screening. Expert Opin. Drug Discov. 2016; 11:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim S., Cheng T., He S., Thiessen P.A., Li Q., Gindulyte A., Bolton E.E.. PubChem protein, gene, pathway, and taxonomy data collections: bridging biology and chemistry through target-centric views of pubchem data. J. Mol. Biol. 2022; 434:167514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim S., Thiessen P.A., Cheng T., Yu B., Bolton E.E.. An update on PUG-REST: RESTful interface for programmatic access to pubchem. Nucleic Acids Res. 2018; 46:W563–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim S., Thiessen P.A., Cheng T., Zhang J., Gindulyte A., Bolton E.E.. PUG-View: programmatic access to chemical annotations integrated in pubchem. J. Cheminform. 2019; 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and resources discussed in this paper are publicly available at https://www.ncbi.nlm.nih.gov.