Abstract
It is 24 years since the IPD-IMGT/HLA Database, http://www.ebi.ac.uk/ipd/imgt/hla/, was first released, providing the HLA community with a searchable repository of highly curated HLA sequences. The database now contains over 35 000 alleles of the human Major Histocompatibility Complex (MHC) named by the WHO Nomenclature Committee for Factors of the HLA System. This complex contains the most polymorphic genes in the human genome and is now considered hyperpolymorphic. The IPD-IMGT/HLA Database provides a stable and user-friendly repository for this information. Uptake of Next Generation Sequencing technology in recent years has driven an increase in the number of alleles and the length of sequences submitted. As the size of the database has grown the traditional methods of accessing and presenting this data have been challenged, in response, we have developed a suite of tools providing an enhanced user experience to our traditional web-based users while creating new programmatic access for our bioinformatics user base. This suite of tools is powered by the IPD-API, an Application Programming Interface (API), providing scalable and flexible access to the database. The IPD-API provides a stable platform for our future development allowing us to meet the future challenges of the HLA field and needs of the community.
INTRODUCTION
The Immuno Polymorphism Database (IPD) provides a centralized repository for the polymorphic genes of the immune system. It is made up of four component databases with the IPD-IMGT/HLA Database being the largest (1–14). The IPD-IMGT/HLA Database contains the allelic sequences of the genes of the HLA system, located within the human Major Histocompatibility Complex (MHC). The HLA genes encode protein products that mediate human adaptive and innate immunity and influence the outcome of cell and organ transplantation. HLA molecules function by binding pathogen and cellular peptides and presenting them to cells of the immune system. The extended MHC contains over 420 genes and covers >7 Mb of the 6p21.3 region on the short arm of chromosome 6 (15). The core genes of interest are 41 highly polymorphic genes and pseudogenes. These genes include the most complex and polymorphic of the human genome, recently being described as hyperpolymorphic rather than simply polymorphic (16). The naming of new HLA genes and allele sequences is the responsibility of the WHO Nomenclature Committee for Factors of the HLA System (17–35). The IPD-IMGT/HLA Database provides the tools necessary for the curation and quality control of these sequences as well as the dissemination of this data. The dissemination of new allele names and sequences is of paramount importance in the clinical setting and occurs through quarterly releases of the IPD-IMGT/HLA Database. The first public release of the database was the 16 December 1998 (1) and included a total of 964 alleles. In this initial release only the coding sequences were included, with most alleles being represented by partial sequence, only having sequence covering the Antigen Recognition Domain (ARD)—exons 2 and 3 for HLA class I and exon 2 for HLA class II. These regions, involved in peptide binding, were prioritised in early HLA sequencing protocols as they were seen to encode the greatest levels of polymorphism and matching for these regions had been shown to impact on the successful outcome of transplantation (36). Since the first release, the database has been updated every 3 months for a total of 95 releases. During this time, the database has continually adapted to meet the demands of the HLA community including the addition of new genes and gene regions. Genomic sequences including introns were first added to the database for some HLA class I genes in October 2003 and have been included for more of the remaining genes as data has become available. The extension of these sequences has become more clinically relevant, as recent studies have shown that matching for intronic polymorphisms can improve clinical outcomes after Haematopoietic Cell Transplantation (HCT) (37,38).
DATA SOURCES
The IPD-IMGT/HLA Database is a global sequence repository with submitters of data from 47 countries and individuals accessing the database from 191 countries to date. Submissions to the database are curated, analysed and if they meet the strict requirements an official allele designation is assigned (35). This process includes both manual and automated components. Manual curation is central to the process to ensure all submissions meet the strict acceptance criteria and our curators work with submitters to gather missing information to allow for completion of as many submissions as possible. Automated processes have been developed to supplement, not replace, expert review. The IPD-IMGT/HLA Database is the official repository for the WHO Nomenclature Committee for factors of the HLA System (17–35) and submissions are made to the database prior to assignment of an official HLA allele designation by the committee. Once assigned the sequence will be incorporated into the subsequent quarterly release. Since the database was first released the IPD-IMGT/HLA Database curators have processed over 57 000 submissions. These submissions have come from a variety of sources, with the majority today submitted from HLA typing laboratories of large haematopoietic cell donor registries and commercial partners, performing routine HLA typing of potential donors for HCT. All submissions to the IPD-IMGT/HLA Database must also be submitted to one of the International Nucleotide Sequence Database Collaboration (INSDC) databases (39) including DNA DataBank of Japan (40), GenBank (41) and EMBL-ENA (42).
The rate of data submitted to the database continues to grow and has doubled recently from an average of 3000 submissions per year from 2013–2017 to 6000 a year from 2018–present, with the most submissions having been received in 2019. Despite reduced numbers due to the COVID-19 pandemic, more submissions were still received in 2020 than pre-2018. This increase is driven by the widespread uptake of Next Generation Sequencing (NGS) technology by HLA typing laboratories and donor registries. The IPD team have developed robust and high throughput data analysis pipelines to ensure that we are able to meet the increasing scale of this data without sacrificing any of the accuracy that has made it the gold standard repository for these clinically relevant sequences.
DATABASE GROWTH AND NEXT GENERATION SEQUENCING TECHNOLOGY
NGS technology has revolutionized genomic research. It has reduced the cost and effort of obtaining a DNA sequence to the point where it has now become routine in many clinical laboratories, which has broad implications for clinicians and researchers. NGS is a powerful tool in unpicking the complexity of HLA as it is now possible to routinely sequence the entirety of an HLA gene and identify novel variants in all exons and introns. This has led to the detection of many more novel HLA variants causing a surge in submissions to the IPD-IMGT/HLA Database. Since our previous publication in 2020 (14) an additional 11 246 alleles have been added to the database, an increase of 46.7% (Figure 1). The number of genes included in the database has also been expanded with HLA-DPA2, -DPB2, -DQA2 and -DQB2 added since 2020 (14). In addition, the database has seen longer and more complex submissions as NGS characterises the historically under-represented regions of the HLA genes outside the ARD, exons 2 and 3 for HLA class I and exon 2 for HLA class II. This has increased the number of coding variants outside exon 2 and 3 as well as the number of intronic variants identified. Based on models of variation in these exons alone the number of HLA variants in the human population has been predicted to be 2–3 million per locus for HLA class I (16). The number of new intronic variants suggest that this prediction is a vast underestimate.
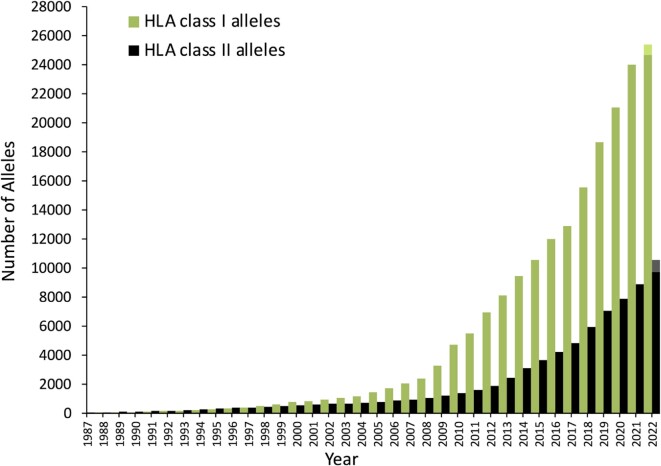
Figure 1.
Growth of the database in the number of alleles per year. Growth split into HLA class I (green) and HLA class II (black). The latest year (2022) is extrapolated for the end of the year in lighter shades.
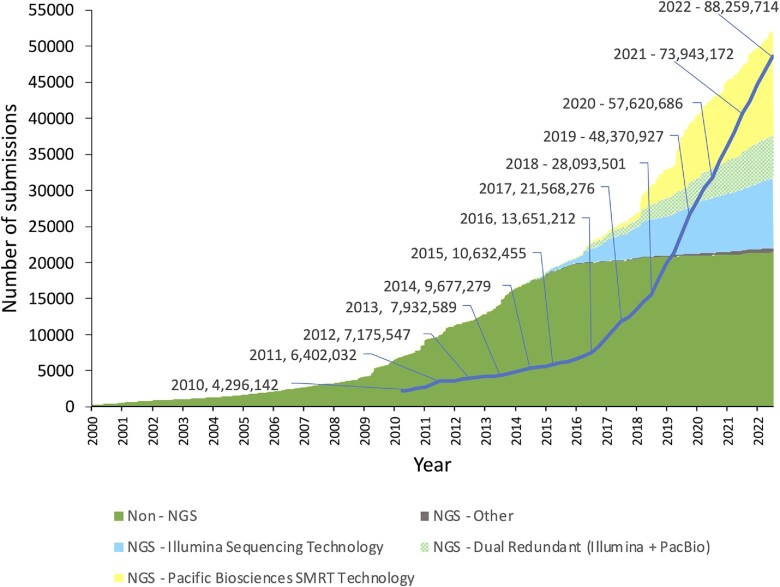
The rate of growth of the database has increased throughout its 24-year history and correlates with the dominant sequencing technology used at the time. Prior to 1998 the majority of new HLA variants were identified using serological methods generating a low-resolution HLA typing. After 1998 DNA based methods were developed using sets of oligonucleotide probes. Although this increased the resolution of HLA typing these probes had limited ability to detect novel variants. From 2010 the majority of sequences were identified using high resolution high-throughput Sequencing-Based Typing (SBT) using Sanger sequencing. This could identify the complete sequence of an HLA gene leading to an increase in new allele discovery, however SBT was cost and labour intensive. Since 2016 NGS technologies have become more widely adopted by the HLA community. This has provided a cost effective and routine solution to HLA typing leading to a significant increase in the number of alleles in the database. The latest release in July 2022 contains 35 077 allelic variants (Figure 1). NGS technology has also increased the length of sequences submitted to the database causing the rate of nucleotide growth to increase (Figure 2). The primary two NGS technologies used in submissions are Illumina—short read—and Pacific Biosciences (PacBio)—long read. Initially Illumina was the more dominant technology for sequencing HLA however the rate of PacBio submissions has increased dramatically from 2018 (Figure 2), being used on its own and in combination with Illumina in a ‘Dual Redundant’ strategy (43). The popularity of PacBio is again reflected in an increased rate of nucleotide growth from 2018. This is likely because long read sequencing is able to generate complete fully phased sequences for an entire HLA gene (44), particularly for HLA class II genes which are longer and have more complex intronic regions. As these longer sequences are submitted the rate of nucleotide growth increases. Since our previous publication in 2020 (14) the number of nucleotides in the database has almost doubled. The new full-length sequences include novel alleles as well as extensions of previously identified but partially sequenced alleles. Improving the coverage of the database both in terms of yet undiscovered alleles and completion of un-sequenced regions improves the quality of the data in the database and reduces the bioinformatics challenges of HLA typing. The number of partial sequences in the database continues to be a challenge, however the proportion of partial sequences has been decreasing for many years owing to the majority of newly identified alleles being sequenced for the full-length of the gene and targeted work by submitters to complete partial sequences in the database.
Figure 2.
Increasing use of NGS technologies in submissions to the IPD-IMGT/HLA Database. From 2000–2016 Sanger sequencing (green) was the dominant method in determining HLA sequences. Since 2016 there has been a sharp increase in NGS technologies starting with Illumina (light blue) and then PacBio (yellow). The increasing use of NGS technologies has resulted in an increased rate of nucleotide growth (dark blue line) in the database. Numbers of nucleotides in the database are given per year from 2010.
TOOLS AVAILABLE AT IPD-IMGT/HLA
The IPD-IMGT/HLA Database provides a diverse set of tools for the analysis of HLA sequences (Table 1.). These includes tools specifically developed for IPD as well as integration with existing tools provided by the European Molecular Biology Laboratory' s European Bioinformatics Institute (EMBL-EBI). Access to these tools is available via the IPD-IMGT/HLA website hosted by EBI at https://www.ebi.ac.uk/ipd/imgt/hla/.
Table 1.
Tools available from the IPD-IMGT/HLA Database
| Tool | Description | Input methods | Output |
|---|---|---|---|
| Alignment Tool | Pre-built alignments of sequences in the database | Select locus and feature (Exon 1, Exon 2, CDS …etc) then filter on specific alleles/groups and alignment formatting | Alignment matching the input rendered in HTML |
| Allele Query Tool | Search tool for retrieving information on alleles in the database | Web based form fill to build custom queries using drop downs menus and free text fields. Constructs a custom web address with the query. | Information on HLA alleles, with all queried fields, filtered on the parameters of the query. Results in HTML table or JSON |
| The same syntax is used to perform programmatic queries to the IPD API | |||
| Fields available to query include allele name, CWD status, confirmation status and more | |||
| Cell Query Tool | Search tool which allows complex queries on source cells material. | Web based form fill and IPD API in the same style as the Allele Query Tool. | Information on source cells, with all queried fields, filtered on the parameters of the query. Results in HTML table or JSON |
| Fields available to query include HLA Typing, homozygosity, ethnicity and more | |||
| HLA-DPB1-TCE Tool | Classification of HLA-DPB1 T-cell-epitope groups and matching status between patient and donor typing (53) | Web based form fill or IPD API. Provide HLA-DPB1 proteins for patient and donor. Multiple donors can be queried simultaneously | TCE Group and predicted immunogenicity of HLA-DPB1 proteins. Permissive status of DPB1 mismatching. Results in HTML table or JSON |
| HLA-B Leader Matching | Classification of HLA-B leader on presence of methionine or threonine at codon -21 and matching status between patient and donor typing (54) | Web based form fill or IPD API. Provide HLA-B proteins for patients and donors. Multiple donors can be queried simultaneously | HLA-B proteins with amino acid at -21 and patient donor B Leader matched status. Results in HTML table or JSON |
| KIR Ligand Calculator | Classification of KIR binding epitope in HLA-B and -C and matching status between patient and donor typing (55) | Web based form fill or IPD API. Provide HLA-B and -C proteins for patients and donors. Multiple donors can be queried simultaneously | HLA-B and -C proteins with KIR ligand motif and matching status. Results in HTML table or JSON |
Alignment Tool – This continues to be one of the most heavily used resources with ∼11 000 uses a month. It allows for custom filtering of pre-generated sequence alignments to their own specifications. These include protein, coding and full-length genomic alignments. Through collaboration with IPD-MHC (45) a new alignment tool has been developed and is currently in the beta testing stage. This new tool provides faster alignments as well as intuitive filtering options. While the sequence alignments produced with both tools are similar the mark up of some sequences, such as splice variants, may differ between the different versions of these tools.
Sequence Search Tools – Integration into the suite of search tools provided by the EMBL-EBI include EB-eye (46), FASTA and BLAST (47,48) search tools.
Downloads – Data from the current and previous releases is available for download in a variety of commonly used formats, such as FASTA, MSF and XML. The current release is available from an FTP directory hosted by EBI as well as a version-controlled git repository available on GitHub which also contains all previous releases as separate branches.
Matching Tools – The website provides access to a number of tools for improving donor selection using algorithms described in published work. Due to the high demand for use of these tools a set of matching Application Program Interfaces (APIs) have now been developed.
Allele Query Tool + Cell Query Tool – Access to detailed information on any HLA allele and source material. Due to the increased size and complexity of the database, as well as demand for more customizable and programmatic access, this tool has been replaced by the IPD API developed in collaboration with IPD-MHC (45).
IPD API
To meet the bioinformatic challenges given by the constant increase in volume and resolution, the IPD project has faced a recent reorganisation in its data presentation and the available tools. All static content of the IPD projects is now compiled at release time and available as HTML web pages—allowing a rapid and always available interrogation—while dynamic content is served by microservices, following the JAMstack (Javascript, API, Markdown) approach. JAMstack is an architectural approach that separates the content from the logic, isolating each function as an independent service. By decoupling the services needed to operate a site, each part can become scalable, easier to maintain and extend in functionality. This approach has not only secured the project for the years to come, future-proofing its architecture to the exponential increase in the amount of data and metadata collected but has also provided users with powerful tools for better consuming the available data. Information is organised via MongoDB (https://www.mongodb.org/) (Figure 3), a document-oriented database where data is stored as JavaScript Object Notation (JSON) data format, a flexible format that allows collecting a variable set of data formats, thus providing a flexible solution that can be easily modified or extended as new requirements emerge.
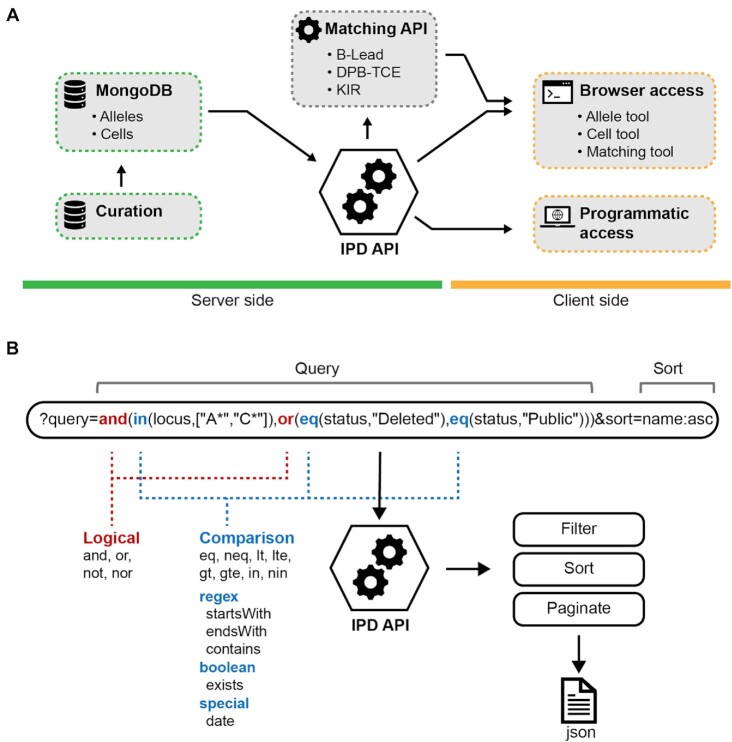
Figure 3.
The IPD API provides information both for allele and cell data. (A) Data is stored in a MongoDB database and is served through an API that can be accessed either programmatically or through a series of tools available on the official website. The Matching API functions in tandem with the IPD API providing a secondary entry point which includes specific logic to determine the matching criteria of patients and donors. (B) The API provides a powerful query schema that allows to virtually filter for any bit of information available on the database, providing a powerful tool for the study of genetic polymorphism in the immune system.
An application programming interface (API) provides access to the data and is both accessible via the conventional web interface and programmatic access (Figure 3.). The latter option allows expert users to programmatically access data and integrate the IPD project with custom pipelines. The IPD API features a query language that allows filtering the results by multiple criteria that can be concatenated with the most common logical operators; additional information is available at the IPD API help page (https://www.ebi.ac.uk/ipd/imgt/hla/about/help/api/). The flexibility of the implementation has allowed the IPD API to be integrated with all IPD databases, providing support for the Allele Query Tool, Cell Query Tool and the matching tools HLA-DPB1-T-Cell Epitope (TCE), HLA-B-Leader and Killer-cell Immunoglobulin-like Receptor (KIR) Ligand Calculator.
MATCHING APIS
Due to the widespread use of the HLA-DPB1-TCE, HLA-B Leader and KIR Ligand matching tools (Table 1.) for donor selection, as well as increased interest in programmatic access for bulk queries to these tools, we have developed a suite of matching APIs. Use of these APIs is available through web-based queries and programmatic access giving results in JSON format. These APIs function in tandem with the IPD API providing a secondary entry point which includes specific logic to determine the matching criteria of patients and donors. The Matching APIs currently available include:
HLA-DPB1-TCE Tool – Classification of HLA-DPB1 mismatches based on T-cell-epitope groups has been shown to predict whether an HLA-DPB1 mismatch is permissive and non-permissive after unrelated-donor Haematopoietic Cell Transplantation (HCT) (49–52). IPD-IMGT/HLA provides a tool to calculate the immunogenicity of HLA-DPB1 mismatches given prospective patient and donor typing. A second version of this tool has been developed using the more recently updated methodology for assessing the TCE group (53).
HLA-B Leader Matching – Polymorphism in the HLA-B leader sequence encodes either methionine (M) or threonine (T) at position –21 giving rise to TT, MT or MM genotypes. These genotypes have been shown to inform risk of Graft Versus Host Disease (GVHD) risk in HLA-B mismatched HCT. The risk of acute GVHD is higher when the patient has an M genotype and the leader sequence is mismatched (54). IPD-IMGT/HLA provides a tool to calculate the B leader mismatch given prospective patient and donor typing.
KIR Ligand Calculator – Research has shown that transplant strategies informed by KIR-ligand mismatch have resulted in less relapse, less GvHD and better overall survival in patients with Acute Myeloid Leukaemia (55). IPD-IMGT/HLA provides a tool to calculate the KIR Ligand mismatches given prospective patient and donor typing.
WEBSITE REDESIGN
The main access point to the database for most users is through our website hosted by EMBL-EBI at https://www.ebi.ac.uk/ipd/. The IPD-IMGT/HLA website was first released in 1998 (2) and was designed to provide a centralised resource and tools for accessing the database. Since then, the website has periodically been updated to improve the user experience as new web frameworks have evolved. As the size of the database has grown significantly in recent years this has challenged the performance of existing tools. We have recently refreshed our website seeking to improve the existing interface while leveraging recent developments in the tools available via the IPD API. The new website gives a more intuitive user interface and improved performance as well as allowing for the construction of more complex and customizable queries. This ensures that we continue to support our traditional users alongside the development of programmatic access for the emerging bioinformatics user base. Through collaboration between the development of the different IPD projects IPD-IMGT/HLA has seen increased functionality, user experience and ease of maintenance which allows us to better adapt to future challenges in the HLA field.
FUTURE DEVELOPMENTS
The development of the IPD API as well as matching APIs provide a robust platform for the future development of the IPD-IMGT/HLA Database. As the size of the database continues to grow and new findings in HLA research emerge IPD will continue to develop new tools for visualisation and access to this data while maintaining the high standards set in the presentation and quality of HLA sequences and nomenclature to the HLA community. These developments are in close collaboration with the development of other IPD projects. Recent tools developed by IPD-MHC include the IPD API, Primer Design Tool and in future a protein folding modelling tool. IPD-IMGT/HLA will make use of these tools building an integrated IPD ecosystem to better support the clinical and research HLA communities.
CONCLUSIONS
Recent developments in the HLA field including increases in the size and complexity of the database as well as published findings about the role of different HLA mismatching criteria in HCT outcomes have required the development of new approaches to access the IPD-IMGT/HLA Database. The work described here shows IPDs ability to adapt to changing environments and provides a framework for future development effectively future proofing the database through scalable solutions allowing the IPD-IMGT/HLA Database to fulfil its role as a centralised resource for researchers and clinicians in the HLA field. Through collaboration with IPD-MHC we will continue to develop resources to support the changing field of HLA and HLA bioinformatics.
DATA AVAILABILITY
The IPD-IMGT/HLA Database can be accessed at https://www.ebi.ac.uk/ipd/imgt/hla/. The IPD-IMGT/HLA Database provides an FTP site for the retrieval of sequences in a number of pre-formatted files. The sequences are provided as FASTA, PIR and MSF formats.
The FTP directory is available at the following address: ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/.
Version controlled access to current and previous versions of the database is available via a git repository hosted by GitHub at: https://www.github.com/ANHIG/IMGTHLA/.
For more information about the database or to subscribe to the IPD mailing list please contact hla@alleles.org.
ACKNOWLEDGEMENTS
The publication of the data online would not be possible without the ongoing support and collaboration of EMBL-EBI who provide the required infrastructure. In particular the Web Production and Infrastructure teams at EMBL-EBI. We would like to thank Prof. Parham, Departments of Structural Biology and Microbiology and Immunology at the Stanford University School of Medicine for continued advice and support. We would like to acknowledge the contributions of the developers of the IPD-MHC and IPD-NHKIR databases whose work has been instrumental to the latest developments of IPD-IMGT/HLA. Lastly, we would like to thank our submitters for contributing data to this project and our users for their ongoing support.
Contributor Information
Dominic J Barker, Anthony Nolan Research Institute, Royal Free Hospital, Pond Street, London, NW3 2QG, UK; UCL Cancer Institute, University College London (UCL), Royal Free Campus, Pond Street, London, NW3 2QG, UK.
Giuseppe Maccari, Data Science for Health (DaScH) Lab, Fondazione Toscana Life Sciences, Siena, Italy.
Xenia Georgiou, Anthony Nolan Research Institute, Royal Free Hospital, Pond Street, London, NW3 2QG, UK.
Michael A Cooper, Anthony Nolan Research Institute, Royal Free Hospital, Pond Street, London, NW3 2QG, UK.
Paul Flicek, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK.
James Robinson, Anthony Nolan Research Institute, Royal Free Hospital, Pond Street, London, NW3 2QG, UK; UCL Cancer Institute, University College London (UCL), Royal Free Campus, Pond Street, London, NW3 2QG, UK.
Steven G E Marsh, Anthony Nolan Research Institute, Royal Free Hospital, Pond Street, London, NW3 2QG, UK; UCL Cancer Institute, University College London (UCL), Royal Free Campus, Pond Street, London, NW3 2QG, UK.
FUNDING
The IPD-IMGT/HLA Database is funded by a grant from the National Marrow Donor Program/Be the Match #211392, with support from One Lambda (Thermo Fisher Scientific); DKMS; Histogenetics; American Society for Histocompatibility and Immunogenetics (ASHI); CareDX; European Federation for Immunogenetics (EFI); FujireBio; GenDX; Immucor; LabCorp; Omixon; Anthony Nolan; Asia-Pacific Histocompatibility and Immunogenetics Association (APHIA); BAG Diagnostics; Be The Match Foundation; National Marrow Donor Program (NMDP); Inno-Train Diagnostik GmBH; European Molecular Biology Laboratory. Funding for open access charge: Anthony Nolan Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Robinson J., Bodmer J.G., Malik K., Marsh S.G.E.. Development of the international immunogenetics HLA database. Hum. Immunol. 1998; 59:17. [Google Scholar]
- 2. Robinson J., Malik A., Parham P., Bodmer J.G., Marsh S.G.E.. IMGT/HLA - a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000; 55:280–287. [DOI] [PubMed] [Google Scholar]
- 3. Ruiz M., Giudecelli V., Ginestoux C., Stoehr P., Robinson J., Bodmer J., Marsh S.G.E., Bontrop R., Lemaitre M., Lefranc G.et al.. IMGT, the international immunogenetics database. Nucleic Acids Res. 2000; 28:219–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marsh S.G.E., Robinson J.. The IMGT/HLA sequence database. Rev. Immunogenet. 2000; 2:518–531. [PubMed] [Google Scholar]
- 5. Robinson J., Waller M.J., Parham P., Bodmer J.G., Marsh S.G.E.. IMGT/HLA - a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001; 29:210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robinson J., Waller M.J., Parham P., de Groot N., Bontrop R., Kennedy L.J., Stoehr P., Marsh S.G.E.. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003; 31:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robinson J., Waller M.J., Fail S.C., Marsh S.G.E.. The IMGT/HLA and IPD databases. Hum. Mutat. 2006; 27:1192–1199. [DOI] [PubMed] [Google Scholar]
- 8. Robinson J., Marsh S.G.E.. The IMGT/HLA database. Methods Mol. Biol. 2007; 409:43–60. [DOI] [PubMed] [Google Scholar]
- 9. Robinson J., Waller M.J., Fail S.C., McWilliam H., Lopez R., Parham P., Marsh S.G.E.. The IMGT/HLA database. Nucleic Acids Res. 2009; 37:D1013–D1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robinson J., Mistry K., McWilliam H., Lopez R., Parham P., Marsh S.G.E.. The IMGT/HLA database. Nucleic Acids Res. 2011; 39:D1171–D1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson J., Halliwell J.A., McWilliam H, Lopez R, Parham P., Marsh S.G.E.. The IMGT/HLA database. Nucleic Acids Res. 2013; 41:D1222–D1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robinson J., Halliwell J.A., Hayhurst J.H., Flicek P., Parham P., Marsh S.G.E.. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015; 43:D423–D431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson J., Soormally A.R., Hayhurst J.D., Marsh S.G.E.. The IPD-IMGT/HLA database - new developments in reporting HLA. Hum. Immunol. 2016; 77:233–237. [DOI] [PubMed] [Google Scholar]
- 14. Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E.. IPD-IMGT/HLA database. Nucleic Acids Res. 2020; 48:D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horton R., Wilming L., Rand V., Lovering R.C., Bruford E.A., Khodiyar V.K., Lush M.J., Povey S., Talbot C.C., Wright M.W.et al.. Gene map of the extended human MHC. Nat. Rev. Genet. 2004; 5:889–899. [DOI] [PubMed] [Google Scholar]
- 16. Robinson J., Guethlein L.A., Cereb N., Yang S.Y., Norman P.J., Marsh S.G.E., Parham P.. Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet. 2017; 13:e1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. WHO Nomenclature Committee Nomenclature for factors of the HL-a system. Bull. World Health Organ. 1968; 39:483–486. [PMC free article] [PubMed] [Google Scholar]
- 18. WHO Nomenclature Committee Terasaki P.I. WHO terminology report. Histocompatibility Testing, 1970. 1970; Munksgaard, Copenhagen: 49. [Google Scholar]
- 19. WHO Nomenclature Committee Nomenclature for factors of the HL-A system. Bull. World Health Organ. 1972; 47:659–662. [PMC free article] [PubMed] [Google Scholar]
- 20. WHO IUIS Terminology Committee Nomenclature for factofs of the HLA system. Bull. World Health Organ. 1975; 52:261–265. [PMC free article] [PubMed] [Google Scholar]
- 21. Albert E.D., Amos D.B., Bodmer W.F., Ceppellini R., Dausset J., Kissmeyer-Nielsen F., Mayr W., Payne R., Rood J.J., Terasaki P.I.et al.. Nomenclature for factors of the HLA system. Tissue Antigens. 1978; 1977; 11:81–86.77065 [Google Scholar]
- 22. WHO Nomenclature Committee Nomenclature for factors of the HLA system. Tissue Antigens. 1980; 16:113–117. [PubMed] [Google Scholar]
- 23. WHO Nomenclature Committee Nomenclature for factors of the HLA system. Tissue Antigens. 1984; 24:73–80. [PubMed] [Google Scholar]
- 24. WHO Nomenclature Commitee Nomenclature for factors of the HLA system. Tissue Antigens. 1988; 32:177–187. [DOI] [PubMed] [Google Scholar]
- 25. Bodmer J.G., Marsh S.G.E., Parham P., Erlich H.A., Albert E.D., Bodmer W.F., Dupont B., Mach B., Mayr W.R., Sasasuki T.et al.. Nomenclature for factors of the HLA system. Tissue Antigens. 1990; 1989; 35:1–8. [DOI] [PubMed] [Google Scholar]
- 26. Bodmer J.G., Marsh S.G.E., Albert E.D., Bodmer W.F., Dupont B., Erlich H.A., Mach B., Mayr W.R., Parham P., Sasasuki T.et al.. Nomenclature for factors of the HLA system. Tissue Antigens. 1991; 1990; 37:97–104. [DOI] [PubMed] [Google Scholar]
- 27. Bodmer J.G., Marsh S.G.E., Albert E.D., Bodmer W.F., Dupont B., Erlich H.A., Mach B., Mayr W.R., Parham P., Sasasuki T.et al.. Nomenclature for factors of the HLA system. Hum. Immunol. 1992; 1991; 34:4–18. [DOI] [PubMed] [Google Scholar]
- 28. Bodmer J.G., Marsh S.G.E., Albert E.D., Bodmer W.F., Dupont B., Erlich H.A., Mach B., Mayr W.R., Parham P., Sasasuki T.et al.. Nomenclature for factors of the HLA system, 1994. Tissue Antigens. 1994; 44:1–18. [DOI] [PubMed] [Google Scholar]
- 29. Bodmer J.G., Marsh S.G.E., Albert E.D., Bodmer W.F., Bontrop R.E., Charron D., Dupont B., Erlich H.A., Mach B., Mayr W.R.et al.. Nomenclature for factors of the HLA system, 1995. Tissue Antigens. 1995; 46:1–18. [DOI] [PubMed] [Google Scholar]
- 30. Bodmer J.G., Marsh S.G.E., Albert E.D., Bodmer W.F., Bontrop R.E., Charron D., Dupont B., Erlich H.A., Fauchet R., Mach B.et al.. Nomenclature for factors of the HLA system. Tissue Antigens. 1997; 1996; 49:297–321. [DOI] [PubMed] [Google Scholar]
- 31. Bodmer J.G., Marsh S.G.E., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Hansen J.A., Mach B., Mayr W.R.et al.. Nomenclature for factors of the HLA system. Tissue Antigens. 1999; 1998; 53:407–446. [DOI] [PubMed] [Google Scholar]
- 32. Marsh S.G.E., Bodmer J.G., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Hansen J.A., Mach B., Mayr W.R.et al.. Nomenclature for factors of the HLA system. Tissue Antigens. 2001; 2000; 57:236–283. [DOI] [PubMed] [Google Scholar]
- 33. Marsh S.G.E., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Geraghty D.E., Hansen J.A., Mach B., Mayr W.R.et al.. Nomenclature for factors of the HLA system, 2002. Tissue Antigens. 2002; 60:407–464. [DOI] [PubMed] [Google Scholar]
- 34. Marsh S.G.E., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Geraghty D.E., Hansen J.A., Hurley C.K., Mach B.et al.. Nomenclature for factors of the HLA system, 2004. Tissue Antigens. 2005; 65:301–369. [DOI] [PubMed] [Google Scholar]
- 35. Marsh S.G.E., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Fernandez-Vina M., Geraghty D.E., Holdsworth R., Hurley C.K.et al.. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010; 75:291–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersdorf E.W., Hansen J.A., Martin P.J., Woolfrey A., Malkki M., Gooley T., Storer B., Mickelson E., Smith A., Anasetti C.. Major-Histocompatibility-Complex class i alleles and antigens in hematopoietic-cell transplantation. N. Engl. J. Med. 2001; 345:1794–1800. [DOI] [PubMed] [Google Scholar]
- 37. Mayor N.P., Hayhurst J.D., Turner T.R., Szydlo R.M., Shaw B.E., Bultitude W.P., Sayno J.R., Tavarozzi F., Latham K., Anthias C.et al.. Recipients receiving better HLA-Matched hematopoietic cell transplantation grafts, uncovered by a novel HLA typing method, have superior survival: a retrospective study. Biol. Blood Marrow Transplant. 2019; 25:443–450. [DOI] [PubMed] [Google Scholar]
- 38. Mayor N.P., Wang T., Lee S.J., Kuxhausen M., Vierra-Green C., Barker D.J., Auletta J., Bhatt V.R., Gadalla S.M., Gragert L.et al.. Impact of previously unrecognized HLA mismatches using ultrahigh resolution typing in unrelated donor hematopoietic cell transplantation. J. Clin. Oncol. 2021; 39:2397–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karsch-Mizrachi I., Nakamura Y., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2012; 40:D33–D37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kodama Y., Mashima J., Kaminuma E., Gojobori T., Ogasawara O., Takagi T., Okubo K., Nakamura Y.. The DNA data bank of japan launches a new resource, the DDBJ omics archive of functional genomics experiments. Nucleic Acids Res. 2012; 40:D38–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2013; 41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amid C., Birney E., Bower L., Cerdeno-Tarraga A., Cheng Y., Cleland I., Faruque N., Gibson R., Goodgame N., Hunter C.et al.. Major submissions tool developments at the european nucleotide archive. Nucleic Acids Res. 2012; 40:D43–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Albrecht V., Zweiniger C., Surendranath V., Lang K., Schofl G., Dahl A., Winkler S., Lange V., Bohme I., Schmidt A.H.. Dual redundant sequencing strategy: Full-length gene characterisation of 1056 novel and confirmatory HLA alleles. HLA. 2017; 90:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayor N.P., Robinson J., McWhinnie A.J.M., Ranade S., Eng K., Midwinter W., Bultitude W.P., Chin C.S., Bowman B., Marks P.et al.. HLA typing for the next generation. PLoS One. 2015; 10:e0127153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maccari G., Robinson J., Ballingall K., Guethlein L.A., Grimholt U., Kaufman J., Ho C.S., De Groot N.G., Flicek P., Bontrop R.E.et al.. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res. 2017; 45:D860–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valentin F., Squizzato S., Goujon M., McWilliam H., Paern J., Lopez R.. Fast and efficient searching of biological data resources–using EB-eye. Brief. Bioinf. 2010; 11:375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Labarga A., Valentin F., Anderson M., Lopez R.. Web services at the european bioinformatics institute. Nucleic Acids Res. 2007; 35:W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D.et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fleischhauer K., Shaw B.E., Gooley T., Malkki M., Bardy P., Bignon J.D., Dubois V., Horowitz M.M., Madrigal J.A., Morishima Y.et al.. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012; 13:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crocchiolo R., Zino E., Vago L., Oneto R., Bruno B., Pollichieni S., Sacchi N., Sormani M.P., Marcon J., Lamparelli T.et al.. Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009; 114:1437–1444. [DOI] [PubMed] [Google Scholar]
- 51. Zino E., Vago L., Di Terlizzi S., Mazzi B., Zito L., Sironi E., Rossini S., Bonini C., Ciceri F., Roncarolo M.G.et al.. Frequency and targeted detection of HLA-DPB1 t cell epitope disparities relevant in unrelated hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2007; 13:1031–1040. [DOI] [PubMed] [Google Scholar]
- 52. Zino E., Frumento G., Marktel S., Sormani M.P., Ficara F., Di Terlizzi S., Parodi A.M., Sergeant R., Martinetti M., Bontadini A.et al.. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004; 103:1417–1424. [DOI] [PubMed] [Google Scholar]
- 53. Crivello P., Zito L., Sizzano F., Zino E., Maiers M., Mulder A., Toffalori C., Naldini L., Ciceri F., Vago L.et al.. The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2015; 21:233–241. [DOI] [PubMed] [Google Scholar]
- 54. Petersdorf E.W., Stevenson P., Bengtsson M., De Santis D., Dubois V., Gooley T., Horowitz M., Hsu K., Madrigal J.A., Malkki M.et al.. HLA-B leader and survivorship after HLA-mismatched unrelated donor transplantation. Blood. 2020; 136:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ruggeri L., Capanni M., Casucci M., Volpi I., Tosti A., Perruccio K., Urbani E., Negrin R.S., Martelli M.F., Velardi A.. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999; 91:333–339. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The IPD-IMGT/HLA Database can be accessed at https://www.ebi.ac.uk/ipd/imgt/hla/. The IPD-IMGT/HLA Database provides an FTP site for the retrieval of sequences in a number of pre-formatted files. The sequences are provided as FASTA, PIR and MSF formats.
The FTP directory is available at the following address: ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/.
Version controlled access to current and previous versions of the database is available via a git repository hosted by GitHub at: https://www.github.com/ANHIG/IMGTHLA/.
For more information about the database or to subscribe to the IPD mailing list please contact hla@alleles.org.