Abstract
GWAS Atlas (https://ngdc.cncb.ac.cn/gwas/) is a manually curated resource of genome-wide genotype-to-phenotype associations for a wide range of species. Here, we present an updated implementation of GWAS Atlas by curating and incorporating more high-quality associations, with significant improvements and advances over the previous version. Specifically, the current release of GWAS Atlas incorporates a total of 278,109 curated genotype-to-phenotype associations for 1,444 different traits across 15 species (10 plants and 5 animals) from 830 publications and 3,432 studies. A collection of 6,084 lead SNPs of 439 traits and 486 experiment-validated causal variants of 157 traits are newly added. Moreover, 1,056 trait ontology terms are newly defined, resulting in 1,172 and 431 terms for Plant Phenotype and Trait Ontology and Animal Phenotype and Trait Ontology, respectively. Additionally, it is equipped with four online analysis tools and a submission platform, allowing users to perform data analysis and data submission. Collectively, as a core resource in the National Genomics Data Center, GWAS Atlas provides valuable genotype-to-phenotype associations for a diversity of species and thus plays an important role in agronomic trait study and molecular breeding.
INTRODUCTION
Genome-wide Association Study (GWAS) has revealed large numbers of genetic variants that are associated with many complex traits in human (1), plants (2–4) and animals (5), and several resources have been developed to provide publicly available GWAS associations (6–11). GWAS Atlas (2), an important genotype-to-phenotype (G2P) association knowledgebase officially released in 2020 in the National Genomics Data Center, part of the China National Center for Bioinformation (12–14), has been rapidly developed to accommodate hundreds of thousands manually curated high-quality G2Ps in both crops and domesticated animals. With the rapid advances in high-throughput sequencing technology and its broad application in population genomics (15–17), over the past several years, increasing GWAS studies have been conducted globally in various species, resulting in more G2P associations to be curated and incorporated in GWAS Atlas. Meanwhile, among these G2P-associated variants, identifying lead SNPs that are significantly independent from neighboring SNPs and determining causal variants that can directly affect phenotypes are of crucial significance for better characterizing the genetic architecture of human diseases (18) and importantly agronomical traits (19–21). Moreover, evidence has accumulated that morphological/physiological/biochemical traits can be under convergent selection, leading to similar traits in different species (22–24). Thus, it is highly needed for GWAS Atlas to incorporate more G2P associations from literatures, prioritize lead SNPs and causal variants from G2P-associated variants, and establish a comprehensive collection of standardized trait vocabularies and descriptions, particularly for uncharacterized traits in crops and animals. Here, we present an updated release of GWAS Atlas with significant updates and enhancements, which incorporates a curated collection of 278,109 G2P associations for 15 species (10 plants and 5 animals), 6,084 lead SNPs and 486 experimental validated causal variants. Moreover, in the updated release, we newly define 1,056 trait ontology terms, provide four online analysis tools and a submission functionality for GWAS studies or summaries, and develop more friendly web user interfaces.
MATERIALS AND METHODS
To provide high-quality G2Ps, an improved curation model was built (Supplementary Figure S1; https://ngdc.cncb.ac.cn/gwas/documentation) to guide curators to extract G2P associations and causal variants. Specifically, publications were retrieved in PubMed using keywords of ‘species name’ and ‘GWAS’. After manual curation, publications containing significant GWAS associations with necessary description on biological traits were incorporated in GWAS Atlas. The curated information was further grouped into four aspects, including general information (genomic location, trait, P-value, etc.), population information (population, sample size, condition, etc.), allele function (e.g. trait impact, allele effect) and publication (PMID, title, journal, etc.).
When multiple associations for any trait were obtained, linkage disequilibrium (LD)-based clumping analysis was performed to identify lead SNPs based on P-values using PLINK v.1.9 (25). Specifically, associated SNPs were filtered when their statistical significance level (P-value) and pairwise correlation coefficient (R2) are not satisfied with the standard threshold (P-value < 5 × 10−8, R2 < 0.6 within a 1000 kb window). Then, lead SNPs were identified if the R2 of retaining SNPs was smaller than 0.1 (26–28). To investigate the association at the gene level, GWAS summary data analysis was performed using Multi-marker Analysis of GenoMic Annotation (MAGMA) (29). That is, all associated SNPs were further aggregated to the corresponding genes, and P-values and sample sizes of any SNP within a gene were combined to test the joint association of all markers in the gene with phenotypes. To this end, the imputed population-relevant references of ten species were downloaded from Plant-ImputeDB (30) and Animal-ImputeDB (31), and the lead SNP analysis was performed. A detailed pipeline was summarized in Supplementary Figure S2.
All curated biological trait entities were mapped to several bio-ontologies, including Plant Trait Ontology (PTO) (32), Plant Phenotype and Trait Ontology (PPTO), Animal Trait Ontology for Livestock (ATOL) (33) and Animal Phenotype and Trait Ontology (APTO). Among these bio-ontologies, the PTO and ATOL are two widely used ontologies for plant and livestock trait ontology mapping. While the PPTO and APTO are additionally established by using semantic mapping to integrate more newly curated traits that cannot be mapped to any known terms in PTO and ATOL.
DATA GROWTH AND DATA MODULES
Over the past two years, GWAS Atlas has been significantly updated regarding data volume and data modules (Table 1). The number of G2P associations is growing rapidly from 75,467 in 9 species in September 2020 to 278,109 in 15 species in September 2022. In the current version of GWAS Atlas, a total of six species, 576 publications, 2,440 studies, 202,642 associations, 97,468 variants, 24,083 genes and 830 traits are additionally curated and included. More importantly, 6,084 lead SNPs and 486 experimental validated causal variants are newly identified and integrated. To better present G2P associations, all relevant entities and metadata in GWAS Atlas are organized into eight modules in terms of species, association, causal variant, trait, variant, gene, study, publication (Table 2), and all traits are organized into the ‘Ontologies’ module. In addition, a submission platform and four online analysis toolkits are provided for GWAS data submission, lead SNP analysis, gene-trait association search, Manhattan plot and quantile-quantile plot.
Table 1.
Statistics of two versions of GWAS Atlas
| GWAS Atlas (2023) | GWAS Atlas (2020) | |
|---|---|---|
| GWAS associations | ||
| Species | 15 | 9 |
| Publications | 830 | 254 |
| Studies | 3,432 | 992 |
| Associations | 278,109 | 75,467 |
| Variants | 145,534 | 48,066 |
| Genes | 55,036 | 30,953 |
| Traits | 1,444 | 614 |
| Lead and causal variants | ||
| Lead SNPs | 6,084 | NA |
| Causal variants | 486 | NA |
| Toolkits | ||
| LeadSNPFinder | Available | NA |
| GeneFinder | Available | NA |
| MHPlotter | Available | NA |
| QQPlotter | Available | NA |
| Submission | Available | NA |
Table 2.
Detailed statistics of curated items in 15 species in GWAS Atlas
| Species | # Publications | # Studies | # Traits | # Associations | # Causal variants | # Variants | # Genes | |
|---|---|---|---|---|---|---|---|---|
| Bread wheat | 94 | 762 | 186 | 20,452 | 57 | 9,938 | 6,464 | |
| Cassava | 7 | 27 | 46 | 260 | - | 201 | 259 | |
| Cotton | 4 | 23 | 24 | 21,955 | - | 6,115 | 991 | |
| Plant | Japanese apricot | 2 | 3 | 10 | 1,865 | - | 1,556 | 625 |
| Maize | 153 | 485 | 314 | 38,127 | - | 30,245 | 9,280 | |
| Oilseed rape | 37 | 129 | 74 | 3,137 | - | 2,440 | 2,135 | |
| Rice | 219 | 1,184 | 461 | 163,479 | 333 | 74,455 | 22,412 | |
| Rye | 2 | 12 | 2 | 2,084 | - | 392 | 23 | |
| Sorghum | 39 | 176 | 151 | 8,829 | - | 6,805 | 5,490 | |
| Soybean | 73 | 324 | 145 | 8,950 | 96 | 6,148 | 5,123 | |
| Cattle | 19 | 36 | 50 | 1,651 | - | 1,481 | 256 | |
| Chicken | 64 | 103 | 130 | 3,782 | - | 2,701 | 1,096 | |
| Animal | Goat | 15 | 19 | 51 | 971 | - | 857 | 59 |
| Pig | 67 | 87 | 146 | 1,767 | - | 1,494 | 509 | |
| Sheep | 35 | 62 | 70 | 800 | - | 706 | 314 |
Note: Species in bold are newly included.
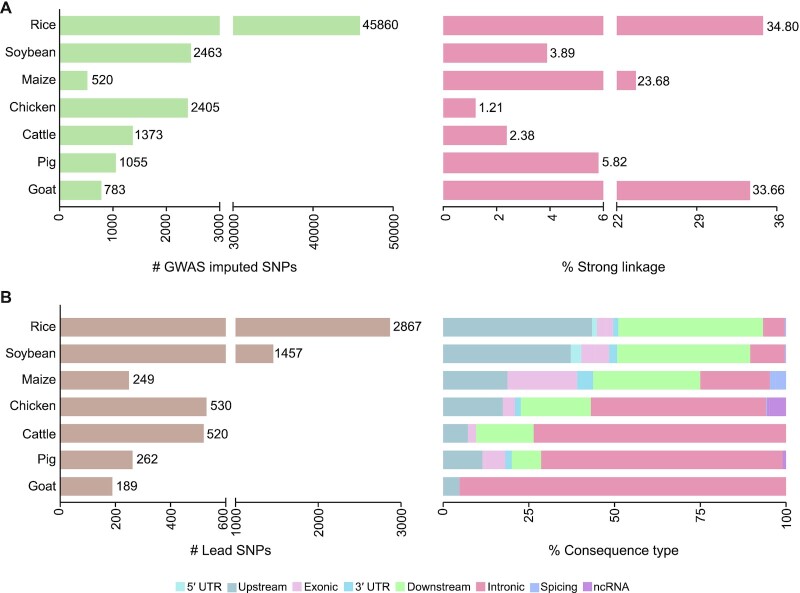
The GWAS associations provide statistical evidence that a region is likely to harbour a causal variant, but it is still needed to discriminate functional variants from variants in LD. Based on those species with the imputed population-relevant references, we perform LD analysis and reveal that 1.21–34.80% of the associated variant pairs in different species show strong linkage (Figure 1A). This suggests that a few of associated variants are highly correlated with neighboring SNPs, thereby serving as surrogates. Then, we perform lead SNP analysis to select significantly independent SNPs, resulting in 6,084 lead SNPs. By functional annotation, we find that these lead SNPs frequently occur in intronic region, upstream and downstream regions of genes, while only a small number of lead SNPs cause nonsynonymous substitutions (Figure 1B). To facilitate users to prioritize potential causal variants for each phenotype, lead SNPs are labelled specifically in the ‘Associations’ module.
Figure 1.
Data statistics and characterization of variants. (A) The number of GWAS variants and the proportion of strong LD pairs (R2 ≥ 0.6) in seven species (three plants and four animals) with imputed references. (B) The number and corresponding consequence type of lead SNPs in seven species.
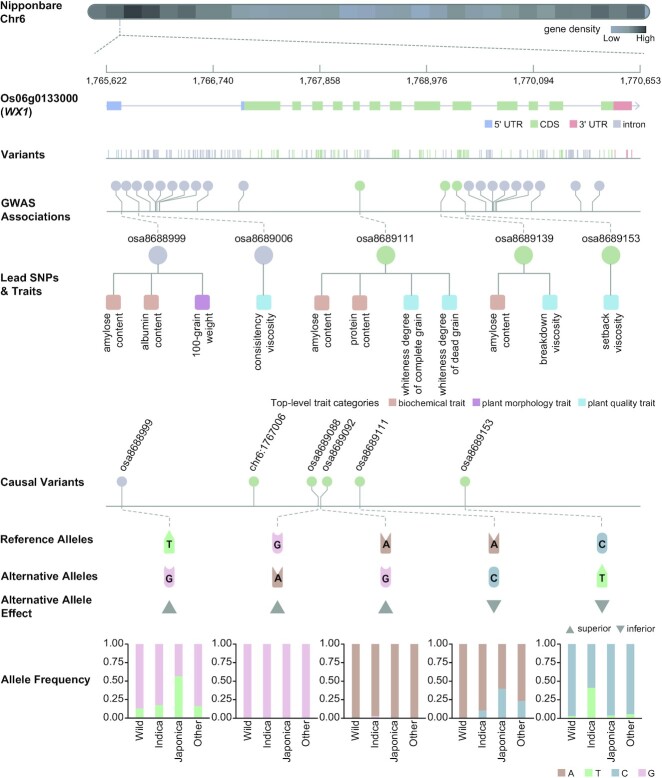
Lead SNPs identified by computational methods are potential causal variants, yet requiring further experimental validation. Up to now, a lot of known causal variants have been experimentally identified and applied in plant breeding (34–36). Based on literature curation from 358 publications, therefore, we obtain a comprehensive collection of 486 causal variants corresponding to 265 genes in rice, soybean, and bread wheat. Among these genes, 57 genes have two or more causal variants, and these variants may cause incremental changes in targeted traits. For instance, WX1 (Os06g0133000), a multiple-allele gene in rice, has been reported to be involved in regulating amylose content, which is a crucial physicochemical property responsible for the eating and cooking quality of rice grain (37) (Figure 2). A total of 22 (9.61%) SNPs recorded in the Genome Variation Map (GVM) (38,39) has been identified to be associated with more than ten traits by GWAS analysis, including amylose content, consistency viscosity, etc. Among these 22 SNPs, there are five lead SNPs and six causal variants, and strikingly, the three lead SNPs (osa8688999, osa8689111 and osa8689153) are causal variants. Additionally, the alternative alleles of osa8689153 and osa8689111 have been validated as inferior alleles causing the increase of amylose content (37,40). In contrast, the alternative alleles of the other four variants (osa8688999, osa8689092, osa8689088 and chr6:1767006) are superior alleles resulting in decreased amylose content (37,41). Moreover, users could learn more about the allele and genotype frequency distribution in different populations in the ‘Causal variants’ module. Taking osa8688999 as an example, the G allele is less frequent in japonica (44%) when compared with indica (82%) or wild (87%), suggesting that users could select a donor line whose genotype is GG at osa8688999 if they want to improve varieties with low amylose content. Together, the integrated causal variants are of great usefulness for better understanding the genetic architecture of complex traits and for aiding precise designing and breeding.
Figure 2.
Variants in WX1 gene. Variants in WX1 are organized as polymorphic variants, GWAS associated variants, lead SNPs identified by computational methods, and causal variants curated from publication.
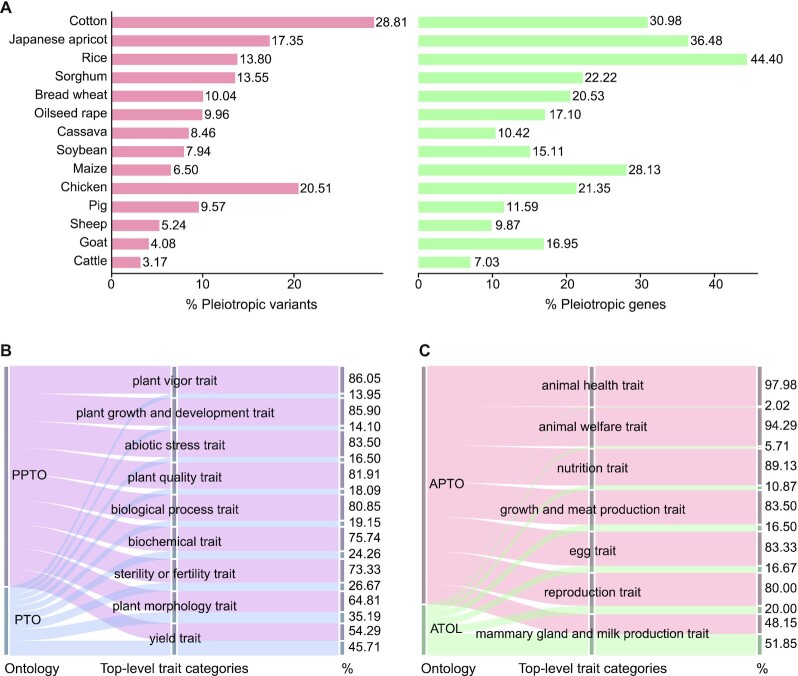
If a variant or gene is detected to be associated with two or more traits, it will be regarded as pleiotropic (26,42). Based on all curated G2P associations, we find a total of 17,755 (12.23%) pleiotropic variants and 17,161 (31.19%) pleiotropic genes over 14 species. These species differ in the percentage of pleiotropic variants and genes, varying from 3.17% in cattle to 28.81% in cotton and from 7.03% in cattle to 44.40% in rice, respectively (Figure 3A). Among these pleiotropic variants, 309 (1.74%) are also identified as lead SNPs. Taking WX1 as an example, it is detected to be significantly associated with 18 agronomical traits (including amylose content, 100-grain weight, protein content, etc.). As mentioned above, three lead variants in WX1, viz., osa8688999, osa8689111 and osa8689139 are also pleiotropic according to GWAS Atlas (Supplementary Figure S3). Specially, osa8688999 is associated with one plant morphology trait (100-grain weight) and two biochemical traits (amylose content and albumin content), osa8689111 is associated with two biochemical traits (amylose content and protein content) and two plant quality traits (whiteness degree of complete grain and whiteness degree of dead grain), and osa8689139 is associated with one biochemical trait (amylose content) and one plant quality trait (breakdown viscosity) (43–48). Together, variants/genes with pleiotropy are especially salient in the context of identifying molecular targets for agricultural breeding, so that GWAS Atlas provides users with easy access to variants and genes of interest in the ‘Variants’ and ‘Genes’ modules.
Figure 3.
Data statistics of pleiotropic variants and genes and distribution of trait terms in several reference ontologies. (A) The proportion of pleiotropic variants and genes in 14 species (9 plants and 5 animals). (B) The proportion of PPTO and PTO ontology terms in each top-level trait category. (C) The proportion of APTO and ATOL ontology terms in each top-level trait category.
The current implementation of GWAS Atlas is also equipped with more enhanced PPTO and APTO. In contrast to the previous version, PPTO and APTO are enriched by a total of 1,056 traits as well as their controlled vocabularies and descriptions, which cannot map to any known trait terms in PTO and ATOL. There are totally 16 broad and upper-level trait categories in the PPTO and APTO, with 65.85–98.02% trait terms newly defined (Figure 3B and C), which are curated from published papers, books, and Biomedical Ontology Repository (BioPortal) (49). To sum up, all curated G2P associations to a particular trait could be easily accessed in the ‘Ontology’ or ‘Trait’ module.
ONLINE ANALYSIS TOOLKITS
GWAS Atlas provides four commonly used tools, including LeadSNPFinder, GeneFinder, MHPlotter, and QQPlotter. LeadSNPFinder is developed to accept user-provided GWAS summary data and identify significantly independent lead SNPs by calculating pairwise LD (R2) for all G2P associations according to the species-specific reference panel. GeneFinder is another interactive tool developed to obtain a list of genes that are statistically associated with one or multiple phenotypes of interest. Besides, MHPlotter and QQPlotter are developed to visualize GWAS results with Manhattan plot and quantile-quantile (QQ) plot. MHPlotter allows users to visualize user-uploaded statistical associations and highlighted SNPs in distinct regions. Also, QQPlotter allows users to evaluate the GWAS result by sorting the observed P-values for each SNP and plotting against the expected P-values from the null hypothesis.
GWAS DATA SUBMISSION PLATFORM
The ‘Submit’ module offers GWAS studies or summary data submission services. For convenience, users just need to provide PMID and publication title, so that the G2P association information will be curated and incorporated into GWAS Atlas by expert curators. While for those unpublished data, users are allowed to be a curator to upload GWAS meta and summary data based on our provided template. Detailed instructions for data submission are available at https://ngdc.cncb.ac.cn/gwas/documentation.
DISCUSSION AND FUTURE DIRECTIONS
GWAS Atlas, as an important knowledgebase for G2P in plants and animals, features massive high-quality statistical associations by manual curation. So far, the current release of GWAS Atlas includes 278,109 G2P associations of 1,444 traits across 15 species. Along with the development and application of GWAS, many biological and computational post-GWAS approaches are used to perform fine-mapping and gene prioritization (50,51). In this update, we identify lead SNPs and gene-trait associations by statistical methods, define new ontology terms for uncharacterized traits, and curate experimentally validated causal variants and genes from publications. To enable users to easily manoeuvre the association knowledge, the updated GWAS Atlas equips with friendly web interfaces, powerful online analysis tools, and GWAS data submission functionality. Thus, future directions of GWAS Atlas include continuous curation of G2Ps and causal variants from a broader range of species and comprehensive integration of multi-omics data to identify more potential causal variants and genes for various traits. We also plan to develop semantic mapping algorithms to summarize and reorganize hierarchical structure of ontology terms to help users to achieve precision annotation for pleiotropic variants, loci and genes. At last, we call for worldwide collaborations to participate in community curation to build GWAS Atlas into a valuable resource covering more comprehensive associations across multiple species.
DATA AVAILABILITY
GWAS Atlas is freely available online at https://ngdc.cncb.ac.cn/gwas/ and does not require user to register.
Supplementary Material
ACKNOWLEDGEMENTS
We thank a number of users for reporting bugs and providing suggestions.
Contributor Information
Xiaonan Liu, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dongmei Tian, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Cuiping Li, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Bixia Tang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Zhonghuang Wang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Rongqin Zhang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yitong Pan, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformatics, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yi Wang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dong Zou, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Zhang Zhang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Shuhui Song, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China; University of Chinese Academy of Sciences, Beijing 100049, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDA24040201 to S.S., XDA19050302 to Z.Z.]; National Key Research & Development Program of China [2021YFF0703703 to S.S.]; National Natural Science Foundation of China [32000475 to D.T., 31871328 to Z.Z., 32030021 to Z.Z.]; Youth Innovation Promotion Association of the Chinese Academy of Sciences [Y2021038 to S.S.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J.. 10 Years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 2017; 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tian D., Wang P., Tang B., Teng X., Li C., Liu X., Zou D., Song S., Zhang Z.. GWAS atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020; 48:D927–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu S., Li C., Wang H., Wang S., Yang S., Liu X., Yan J., Li B., Beatty M., Zastrow-Hayes G.et al.. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 2020; 21:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song S., Tian D., Zhang Z., Hu S., Yu J.. Rice genomics: over the past two decades and into the future. Genomics Proteomics Bioinformatics. 2018; 16:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma A., Lee J.S., Dang C.G., Sudrajad P., Kim H.C., Yeon S.H., Kang H.S., Lee S.H.. Stories and challenges of genome wide association studies in livestock - A Review. Asian-Australas. J. Anim. Sci. 2015; 28:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E.et al.. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J.et al.. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L.et al.. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014; 42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beck T., Shorter T., Brookes A.J.. GWAS central: a comprehensive resource for the discovery and comparison of genotype and phenotype data from genome-wide association studies. Nucleic Acids Res. 2020; 48:D933–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li M.J., Wang P., Liu X., Lim E.L., Wang Z., Yeager M., Wong M.P., Sham P.C., Chanock S.J., Wang J.. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012; 40:D1047–D1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z.et al.. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CNCB-NGDC Members and Partners Database resources of the national genomics data center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022; 50:D27–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. CNCB-NGDC Members and Partners Database resources of the national genomics data center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021; 49:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. National Genomics Data Center Members and Partners Database resources of the national genomics data center in 2020. Nucleic Acids Res. 2020; 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. The IC4R Project Consortium Information commons for rice (IC4R). Nucleic Acids Res. 2016; 44:D1172–D1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sang J., Zou D., Wang Z., Wang F., Zhang Y., Xia L., Li Z., Ma L., Li M., Xu B.et al.. IC4R-2.0: rice genome reannotation using massive RNA-seq data. Genomics Proteomics Bioinformatics. 2020; 18:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan J., Zou D., Li C., Zhang Z., Song S., Wang X.. SR4R: an integrative SNP resource for genomic breeding and population research in rice. Genomics Proteomics Bioinformatics. 2020; 18:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abell N.S., DeGorter M.K., Gloudemans M.J., Greenwald E., Smith K.S., He Z., Montgomery S.B.. Multiple causal variants underlie genetic associations in humans. Science. 2022; 375:1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miao C., Yang J., Schnable J.C.. Optimising the identification of causal variants across varying genetic architectures in crops. Plant Biotechnol. J. 2019; 17:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hafliger I.M., Spengeler M., Seefried F.R., Drogemuller C.. Four novel candidate causal variants for deficient homozygous haplotypes in holstein cattle. Sci. Rep. 2022; 12:5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang C., Yan J., Jiang S., Li X., Min H., Wang X., Hao D.. Resequencing 250 soybean accessions: new insights into genes associated with agronomic traits and genetic networks. Genomics Proteomics Bioinformatics. 2022; 20:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen W., Chen L., Zhang X., Yang N., Guo J., Wang M., Ji S., Zhao X., Yin P., Cai L.et al.. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science. 2022; 375:eabg7985. [DOI] [PubMed] [Google Scholar]
- 23. Di Vittori V., Gioia T., Rodriguez M., Bellucci E., Bitocchi E., Nanni L., Attene G., Rau D., Papa R.. Convergent evolution of the seed shattering trait. Genes (Basel). 2019; 10:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Y., Wu Q., Ma S., Ma T., Shan L., Wang X., Nie Y., Ning Z., Yan L., Xiu Y.et al.. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Slifer S.H. PLINK: key functions for data analysis. Curr. Protoc. Hum. Genet. 2018; 97:e59. [DOI] [PubMed] [Google Scholar]
- 26. Watanabe K., Stringer S., Frei O., Umicevic Mirkov M., de Leeuw C., Polderman T.J.C., van der Sluis S., Andreassen O.A., Neale B.M., Posthuma D.. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019; 51:1339–1348. [DOI] [PubMed] [Google Scholar]
- 27. Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017; 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiel D.P., Kemp J.P., Rivadeneira F., Westendorf J.J., Karasik D., Duncan E.L., Imai Y., Muller R., Flannick J., Bonewald L.et al.. The musculoskeletal knowledge portal: making omics data useful to the broader scientific community. J. Bone Miner. Res. 2020; 35:1626–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015; 11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao Y., Yang Z., Yang W., Yang Y., Gong J., Yang Q.Y., Niu X.. Plant-ImputeDB: an integrated multiple plant reference panel database for genotype imputation. Nucleic Acids Res. 2021; 49:D1480–D1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang W., Yang Y., Zhao C., Yang K., Wang D., Yang J., Niu X., Gong J.. Animal-ImputeDB: a comprehensive database with multiple animal reference panels for genotype imputation. Nucleic Acids Res. 2020; 48:D659–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cooper L., Meier A., Laporte M.A., Elser J.L., Mungall C., Sinn B.T., Cavaliere D., Carbon S., Dunn N.A., Smith B.et al.. The planteome database: an integrated resource for reference ontologies, plant genomics and phenomics. Nucleic Acids Res. 2018; 46:D1168–D1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hughes L.M., Bao J., Hu Z.L., Honavar V., Reecy J.M.. Animal trait ontology: the importance and usefulness of a unified trait vocabulary for animal species. J. Anim. Sci. 2008; 86:1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiong Q., Ma B., Lu X., Huang Y.H., He S.J., Yang C., Yin C.C., Zhao H., Zhou Y., Zhang W.K.et al.. Ethylene-Inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell. 2017; 29:1053–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma J., Yang S., Wang D., Tang K., Feng X.X., Feng X.Z.. Genetic mapping of a light-dependent lesion mimic mutant reveals the function of coproporphyrinogen III oxidase homolog in soybean. Front. Plant Sci. 2020; 11:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z.G., Lv G.D., Li B., Wang J.J., Zhao Y., Kong F.M., Guo Y., Li S.S.. Isolation and characterization of the tasnrk2.10 gene and its association with agronomic traits in wheat (Triticum aestivum l.). PLoS One. 2017; 12:e0174425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang C., Zhu J., Chen S., Fan X., Li Q., Lu Y., Wang M., Yu H., Yi C., Tang S.et al.. Wx(lv), the ancestral allele of rice waxy gene. Mol Plant. 2019; 12:1157–1166. [DOI] [PubMed] [Google Scholar]
- 38. Li C., Tian D., Tang B., Liu X., Teng X., Zhao W., Zhang Z., Song S.. Genome variation map: a worldwide collection of genome variations across multiple species. Nucleic Acids Res. 2021; 49:D1186–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song S., Tian D., Li C., Tang B., Dong L., Xiao J., Bao Y., Zhao W., He H., Zhang Z.. Genome variation map: a data repository of genome variations in BIG data center. Nucleic Acids Res. 2018; 46:D944–D949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Isshiki M., Morino K., Nakajima M., Okagaki R.J., Wessler S.R., Izawa T., Shimamoto K.. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5' splice site of the first intron. Plant J. 1998; 15:133–138. [DOI] [PubMed] [Google Scholar]
- 41. Anacleto R., Badoni S., Parween S., Butardo V.M., Misra G., Cuevas R.P., Kuhlmann M., Trinidad T.P., Mallillin A.C., Acuin Cet al.. Integrating a genome-wide association study with a large-scale transcriptome analysis to predict genetic regions influencing the glycaemic index and texture in rice. Plant Biotechnol. J. 2019; 17:1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Solovieff N., Cotsapas C., Lee P.H., Purcell S.M., Smoller J.W.. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 2013; 14:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen P., Shen Z., Ming L., Li Y., Dan W., Lou G., Peng B., Wu B., Li Y., Zhao D.et al.. Genetic basis of variation in rice seed storage protein (Albumin, globulin, prolamin, and glutelin) content revealed by genome-wide association analysis. Front. Plant Sci. 2018; 9:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong H., Liu S., Zhao G., Zhang C., Peng Z., Wang Z., Yang J., Li Y.. Genetic diversity relationship between grain quality and appearance in rice. Front. Plant Sci. 2021; 12:708996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H., Xu X., Vieira F.G., Xiao Y., Li Z., Wang J., Nielsen R., Chu C.. The power of inbreeding: NGS-Based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant. 2016; 9:975–985. [DOI] [PubMed] [Google Scholar]
- 46. Zhou H., Xia D., Zhao D., Li Y., Li P., Wu B., Gao G., Zhang Q., Wang G., Xiao J.et al.. The origin of Wx(la) provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 2021; 63:878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X., Chen Z., Zhang G., Lu H., Qin P., Qi M., Yu Y., Jiao B., Zhao X., Gao Q.et al.. Analysis of genetic architecture and favorable allele usage of agronomic traits in a large collection of chinese rice accessions. Sci. China Life Sci. 2020; 63:1688–1702. [DOI] [PubMed] [Google Scholar]
- 48. Cruz M., Arbelaez J.D., Loaiza K., Cuasquer J., Rosas J., Graterol E.. Genetic and phenotypic characterization of rice grain quality traits to define research strategies for improving rice milling, appearance, and cooking qualities in latin america and the caribbean. Plant Genome. 2021; 14:e20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Whetzel P.L., Noy N.F., Shah N.H., Alexander P.R., Nyulas C., Tudorache T., Musen M.A.. BioPortal: enhanced functionality via new web services from the national center for biomedical ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011; 39:W541–W545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Broekema R.V., Bakker O.B., Jonkers I.H.. A practical view of fine-mapping and gene prioritization in the post-genome-wide association era. Open Biol. 2020; 10:190221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schaid D.J., Chen W., Larson N.B.. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018; 19:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS Atlas is freely available online at https://ngdc.cncb.ac.cn/gwas/ and does not require user to register.