Abstract
The Genomes OnLine Database (GOLD) (https://gold.jgi.doe.gov/) at the Department of Energy Joint Genome Institute (DOE-JGI) continues to maintain its role as one of the flagship genomic metadata repositories of the world. The ever-increasing number of projects and metadata are freely available to the user community world-wide. GOLD’s metadata is consumed by scientists and remains an important source for large-scale comparative genomics analysis initiatives. Encouraged by this active user engagement and growth, GOLD has continued to add new components and capabilities. The new features such as a public Application Programming Interface (API) and Ecosystem landing page as well as the growth of different entities in this current GOLD v.9 edition are described in detail in this manuscript.
INTRODUCTION
Genomes OnLine Database (GOLD) is a web-based resource that hosts a wealth of information from sequencing projects from all over the world. GOLD’s humble beginnings can be traced to 6 projects in an Excel spreadsheet on a personal computer back in 1997. The first published version of the database contained 20 complete genomes that were organized as a flat file (1). Today, 25 years later, GOLD has transformed into a relational database with web interface and Application Programming Interface (API) access to its curated metadata. Starting in 1999, we have published the developments and growth of the database periodically in several journals. The GOLD statistics page (https://gold.jgi.doe.gov/statistics) provides more detailed information on the growth of different types of projects over time.
A lot has changed in the sequencing world in these past 25 years. The explosive growth in the sheer volume of sequences has been astounding (2,3). To keep up with this increase in the number and diversity of genomes, the GOLD database has also evolved significantly (4–6). What has not changed is GOLD’s commitment to provide this information without any restrictions and by adhering to FAIR (Findable, Accessible, Interoperable, and Reusable) principles (7). GOLD continues to promote and comply with community-driven standards (8) including Minimum Information about any (x) Sequence (MIxS) standards (9), those for single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea (10) as well as standards for uncultivated viral genomes (MIUViG) (11). GOLD users have always been able to freely access the user-interface (UI), interact with its advanced search features, and download standardized metadata relevant to their field of research.
Genomic data sharing and comparative analyses advance all forms of research. When the data is accompanied by well-curated metadata, it leads to new discoveries and better insights. For example, the COVID-19 pandemic that has disrupted our lives in unimaginable ways over the last couple of years (12) has led to one of the most expansive efforts in viral genome sequencing across the world. And the fact that each new sequence has been accompanied with a curated set of metadata, such as the viral host, health condition, geographic location of isolation and more, has drastically enhanced the usability of the data. Along with the intricacies of identifying and tracking viral mutations, accurate recordkeeping of associated metadata has been instrumental in developing appropriate epidemiological responses. Unavailability, inaccuracies, and mismanagement of metadata can be detrimental and have far-reaching effects especially during public health emergencies like the COVID-19 pandemic (13,14). Thus, the importance of a large-scale, manually curated metadata management system such as GOLD cannot be overstated. Below we describe GOLD’s data management system and new updates in the last two years.
DATA MANAGEMENT
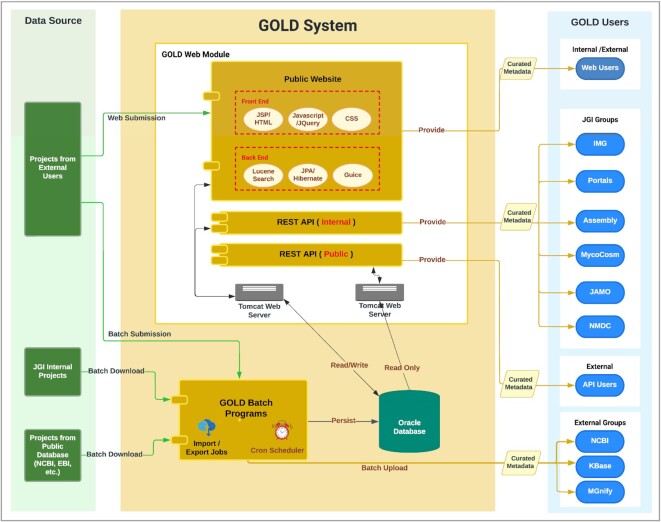
GOLD is comprised of various components that interoperate to enable the browsing and searching of curated microbiome metadata from around the world. It serves as a public facing metadata hub for Joint Genome Institute (JGI) by providing services to support processing, analysis, and publication of (meta)genomic data. GOLD’s metadata can also be downloaded in Excel format or accessed via a public API, to be discussed in more detail in later sections of this manuscript. Projects and their metadata get added to GOLD using a combination of automated and manual steps. They are imported through one of the following three routes: (Figure 1) (i) samples sequenced at JGI through one of its science programs; (ii) projects imported from public repositories such as GenBank (15) and SRA (2); and (iii) projects added manually by GOLD users in order to get their sequences annotated from the Integrated Microbial Genomes (IMG) data management system (16). Having a project defined in GOLD along with all the required metadata is a necessary step before any sequence can be submitted to IMG for annotation.
Figure 1.
Overview of GOLD system and processes. Application components of GOLD system consisting of Oracle database and front and backend components.
GOLD’s underlying codebase is written in a combination of Java, Python, Perl and Bash programming languages. The website and APIs run inside Apache Tomcat containers and use public web frameworks, such as Google Guice, Spring, and various Hibernate-related technologies. GOLD’s search and browsing capabilities are handled by the Apache Lucene search engine. GOLD’s internal components include a suite of Extract-Transform-Load (ETL) processes that ingest data from various sources. These processes can be broadly divided into three separate stages: (i) Extraction phase: Software pipelines process metadata from external repositories like NCBI and internal JGI sources. GOLD’s website also allows users to submit their own private data to GOLD for analysis in JGI’s systems such as IMG and MycoCosm (17), to name a few. (ii) Transformation phase: Since data originates from different sources, it tends to have its own unique terms and organization (i.e. a unique schema). Before such data can be imported into GOLD, these different schemas must be translated into a single unified schema. This is done through a set of automated and semi-automated pipelines, as well as by manual curation. (iii) Loading phase: In this phase, the unified data is loaded into GOLD’s Oracle database to be consumed by users.
CURRENT STATUS
GOLD data has increased significantly over the past years to keep up with the growth in genome sequencing initiatives worldwide. As of August 2022, there were 54 052 Studies in GOLD, representing an increase of 18% since the last release in September 2020. The number of Sequencing Projects (SP) and Analysis Projects (AP) has also gone up substantially. Currently, GOLD has 485 203 SPs, out of which 308 000 are isolate genome and transcriptome projects spread across bacteria (67%), eukaryotes (28%), virus (4.2%) and archaea (0.8%), followed by 149 642 metagenome and 27 560 metatranscriptome projects. The latest release also contains 368 875 APs in GOLD, representing over a 36% increase compared to the previous release. Approximately 42% of these APs have been submitted to IMG and have an IMG Taxon OID. Around 61.5% of the APs are for individual genomes while 38% of them are for metagenomes and metatranscriptomes, and the remaining 0.5% are combined assembly APs. There are 174 363 Biosamples in GOLD distributed across Environmental (43%), Host-associated (47%), and Engineered (9%) ecosystems. The number of GOLD Organisms has grown to 468 058, a 21% increase over the previous release. This can be largely attributed to the import of over 30 000 organisms with a rich set of metadata from Bacdive (18).
RESEARCH SUPPORTED BY GOLD METADATA
Metadata plays an important role in genomic data analysis by providing better correlations, interpretations, and insights into the analyzed data. Well curated metadata thus promotes large scale comparative genomic studies and new hypothesis testing, which otherwise would not have been possible. Here, we would like to highlight a select few publications that leveraged GOLD curated metadata. Edgar et al. (19) used virus host metadata from GOLD to characterize novel viruses and pinpoint their environmental reservoirs. Specifically, the authors studied genetic diversity of the Coronaviridae family to identify possible animal to human transmission routes. Vuong et al. (20) were analyzing the distribution of potential PHA-producing bacteria and archaea in various environments. For this goal, the authors used genome mining methods along with GOLD metadata—specifically, the taxonomic and ecological (ecosystem classification) ones. The metadata available in GOLD helped the researchers to find which classes of the PHA synthases (PhaC) had a diverse distribution. In another recent study, Yadav et al. (21) combined ecological distribution metadata from GOLD’s ecosystem classification to investigate metabolic and ecological markers of the UBA6911 Acidobacteria family.
NEW DEVELOPMENTS
In the past two years, the GOLD group has implemented several new features, some of which are described below.
Expanded download file
In order to facilitate easy access to GOLD’s growing list of projects and metadata, a dedicated ‘Downloads’ section was added to our home page. Users can download four separate files, each containing an export of different types of public GOLD data along with a preselected list of key metadata fields. These downloadable files are updated daily and have been popular among our users. In response to user requests, over the past couple of years, we have significantly expanded the list of available metadata fields that can be downloaded. Twenty-eight new metadata fields were added to the download file containing a public list of GOLD organisms; organism host name, sporulation, salinity, and motility were some of the metadata fields that were most requested. Similarly, genome publications associated with GOLD projects and data utilization status for JGI sequenced genomes were two of the additional fields that were added to the download file.
Downloadable search result upgrades
Fulfilling the goals of the future plans section of the previous GOLD publication (6), we have made two important changes to our search result downloads: (i) in GOLD v.9, users can download additional 10 000 rows of their search results for a total of 30 000 records. (ii) We have doubled the storage capacity, and users now have access to their search results for 4 weeks instead of 2 weeks in the earlier version.
Importing NCBI RefSeq viruses
Importing viral genomes from NCBI has been a long-standing challenge since NCBI virus projects and sequences do not always have the NCBI/GenBank accessions that are standard for isolate genomes. To include the growing number of viral sequences into GOLD and IMG, we designed a separate NCBI virus import process to circumvent the above challenges. As a result, we are now able to track all the viral genomes from NCBI’s Reference Sequence (RefSeq) (22) collection, and add them to the database on an ongoing basis.
API
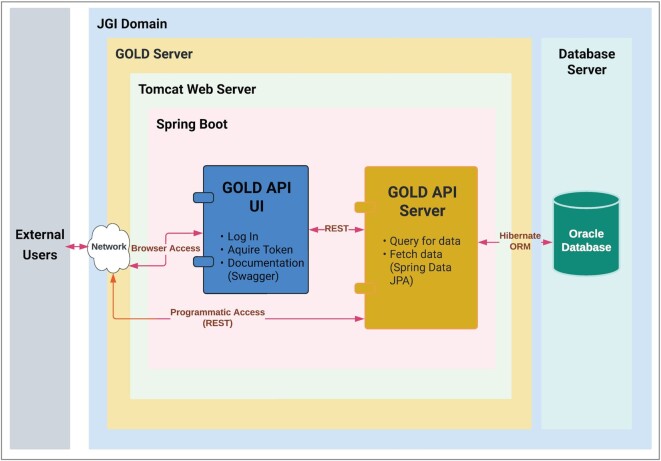
The GOLD API module was designed to provide users with a programmatic way of accessing GOLD’s metadata in a secure and reliable manner (Figure 2). Metadata can be retrieved in JavaScript Object Notation (JSON) format for all five of GOLD’s entities by referencing their associated GOLD IDs. The API also includes a website to handle user actions, such as signing in using JGI’s Single Sign On system, generating offline tokens to authorize programmatic access to the API’s metadata, as well as viewing the developer's manual and API documentation.
Figure 2.
Schematic representation of GOLD API implementation.
GOLD API uses Spring Boot 2.0 (https://spring.io/projects/spring-boot/#overview), a widely used framework for building RESTful web services. Its auto-configuration feature shortens code length and reduces boilerplate code, making web application development faster and easier. API’s Data Access Layer is implemented using Spring Data JPA on top of Hibernate ORM. Spring Data JPA provides enhanced support, such as dynamic query derivation from repository method names, reduced boilerplate code for CRUD operations, pagination, sorting, and auditing. API’s authentication layer leverages JGI’s Keycloak Single-Sign On system which acts as a centralized authentication server that provides and validates tokens for API users to access the API’s endpoints. Spring Boot Actuator is used to monitor the API and gathers metrics, traffic, and health information of the running application. API documentation is implemented by using Swagger (OpenAPI 3) technology (https://swagger.io/specification/). Not only does it provide detailed schema information for consuming the API’s data, but it also allows developers to interactively try out the API using a web browser.
MIxS Packages
Users entering a GOLD Biosample or Organism can either select the standard list of fields or choose from one of the following eight environmental packages: Soil, Water, Sediment, Plant, Microbial Mat, Hydrocarbon Core, Hydrocarbon FS, or Host-Associated package. The Host-Associated package was added in the most recent version of GOLD. This package has 48 new metadata fields including 2 new controlled vocabulary (CV) fields. All these environmental packages were updated to comply with the updated fields from the current MIxS v. 6.0.
Ecosystem Landing Page
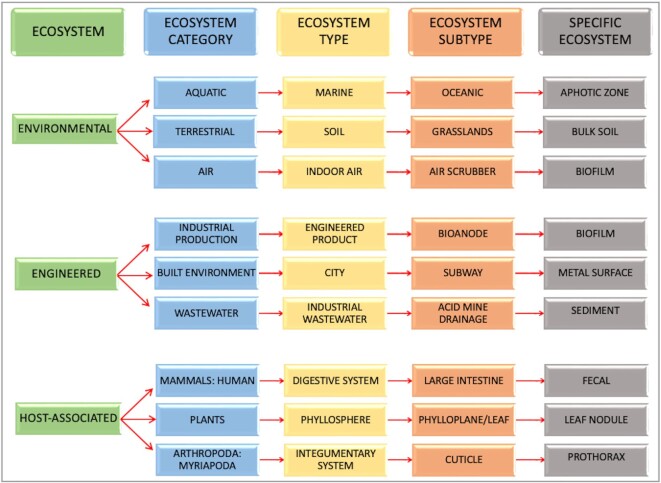
GOLD’s five-tiered Ecosystem Classification system was originally developed to systematically classify metagenome samples (23) in an ecosystem-diverse manner not available from any of the other metadata ontology systems. Accordingly, while it does bear some similarities to the Environmental Ontology (ENVO) (24) and Earth Microbiome Project Ontology (EMPO) (25) classification systems, it differs in several aspects, such as in simplicity and adaptability. GOLD’s ecosystem terms are not meant to be exhaustive and do not include all possible paths from a particular environment. Instead, the system contains a finite list of terms that cover environmental attributes from the samples that are entered. Periodically, the terms are reviewed and updated as more and more samples from novel environments are curated. New and existing users who are interested to know more about the Ecosystem Classification can access our new ecosystem landing page (https://gold.jgi.doe.gov/ecosystem_classification) to learn about the different ecosystem terms and explore distinct classification paths with examples. GOLD’s Ecosystem Classification is, however, the most diverse and inclusive habitat classification system to date, and remains unique in integrating environmental, host-associated, and engineered habitats in a single ontology. Accordingly, the top level consists of three broad Ecosystem terms: ‘Environmental’, ‘Engineered’ and ‘Host-Associated’. Each of them is further subcategorized into subsequent levels called Ecosystem Category, Ecosystem Type, Ecosystem Subtype and Specific Ecosystem to capture more details about the sample environment. For example, a sample isolated from a leaf nodule will have a GOLD Ecosystem Classification of Host-Associated: Plants: Phyllosphere: Phylloplane/Leaf: Leaf Nodule (Figure 3).
Figure 3.
A select list of GOLD’s 5-level ecosystem classification paths.
Type strains in GOLD
A bacterial or archaeal type strain is the strain used when a species is first reported, described, and officially named, following regulations of the International Code of Nomenclature of Prokaryotes (26). It serves as the cultured representative strain for its species and acts as an important taxonomic marker in the prokaryotic tree of life. By principles of nomenclature (27) as well as DNA:DNA hybridization and Average Nucleotide Identity (ANI) comparisons (28), no two type strains can be exactly similar to each other. As a result, studying the individual genomic sequences and associating them to their metabolism and phenotypes is very important. On its homepage, GOLD maintains a type strain tracker, which is reviewed and updated regularly. Users can click on individual type strain organisms and look at their respective metadata, access the list of type strains with IMG annotations, or view the projects imported from GenBank. As of August 2022, there are over 27,000 type strains in GOLD, including coidentical strains that have their respective type materials deposited in separate culture collections.
Updated help page
GOLD’s help page (https://gold.jgi.doe.gov/help) is frequently used by new and returning users to learn about different GOLD entities; it also teaches them how to enter one's own projects or to send a message to our technical team. The main landing page has five subsections: (a) GOLD Documentation, (b) Contact us with Feedback or Questions, (c) GOLD Terminology, (d) Frequently Asked Questions and (e) Trainings and Workshops. All these sections are routinely updated to add new information, based on user feedback. For example, the section on Trainings and Workshops is a completely new addition in the current release. It has links to a video tutorial that gives an overview of GOLD and provides step-by-step directions on how to enter different types of sequencing and analysis projects.
FUTURE DEVELOPMENT PLANS
The developments in microbial and microbiome research, both in terms of the volume of the data generated and the availability of novel computational and visualization tools, makes the need for curated metadata more relevant than ever. To support the needs of the research community, we plan to continue to curate and integrate projects from diverse environments and sources. This will include sourcing metadata and collaborating with the research community and other resources.
Expand access to metadata
We plan to expand the metadata fields in our download file and in our public API application, based on the user requests.
Collaborations
We closely work with two other DOE-funded projects such as National Microbiome Data Collaborative (NMDC) (29) and DOE Biology Knowledgebase (KBase) (30) in metadata curation, establishing metadata standards, and sample metadata exchange. We plan to further extend collaborative efforts in these areas of metadata curation, sharing, and enrichment.
‘How To’ short videos
As recommended by our Prokaryote Advisory Committee overseeing our program, we plan to develop short how-to videos to help our users in entering and updating metadata in GOLD. This will be in addition to the help pages and full-length help video we currently have available.
Environmental packages
As described above, we updated all our existing MIxS packages to version 6. We plan to include additional packages such as Built-environment, Human-associated, and Agricultural Microbiome packages in the near future.
Equivalent strains and metadata propagation
It is very common to have a strain deposited in multiple culture collections with distinct culture collection IDs. Type strain designation requires the strain be deposited in at least two culture collections. So, the proliferation of equivalent strains not only results in multiple taxonomies but also results in the variation or omission of metadata from one entry to the other. To address this, GOLD organizes equivalent strains into a single organism group and propagates metadata from one strain to another within that group. This is an ongoing process, and we continue to curate equivalent statins and enrich metadata through propagation.
Integration of Isolate and MAG taxonomy information form GTDB-tk
The Genome Taxonomy Database (GTDB) establishes a standardized microbial taxonomy based on genome phylogeny (31). Genomes used for phylogeny construction include those from GenBank as well as genomes of uncultured microorganisms obtained from metagenomes and single cells to ensure improved genomic representation of the microbial world. GOLD plans to develop stronger links with the GTDB, including the import and curation of new uncultured organisms proposed as type material.
SeqCode
As new community initiatives, such as SeqCode (32), become widely accepted, GOLD plans to adapt the proposed nomenclatural code for uncultivated prokaryotes with DNA sequences as type. This implies that as new uncultured organisms are sequenced and proposed as type strains, GOLD will expand its type strain catalog to include these new entries.
DATA AVAILABILITY
Genomes Online Database (GOLD) is freely available at the following URL: https://gold.jgi.doe.gov.
ACKNOWLEDGEMENTS
We would like to thank GOLD users and members of the microbiome research community for depositing metadata to GOLD. We are thankful to the JGI project management team, microbial genomics and metagenomics group, and our leadership team for their constant support and feedback. We thank members of the microbial genomics and metagenomics research and standards communities for their feedback and helpful discussions. Visualizations displayed in this manuscript have been created using MS-Office Suite, GNU Image Manipulation Program (GIMP) v 2.10, Adobe Acrobat Professional, and Lucid Charts.
Notes
Present address: Jagadish Chandrabose Sundaramurthi, Department of Genomic Medicine, The Jackson Laboratory, Farmington, CT 06032, USA.
Contributor Information
Supratim Mukherjee, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Dimitri Stamatis, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Cindy Tianqing Li, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Galina Ovchinnikova, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Jon Bertsch, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Jagadish Chandrabose Sundaramurthi, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Mahathi Kandimalla, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Paul A Nicolopoulos, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Alessandro Favognano, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
I-Min A Chen, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Nikos C Kyrpides, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
T B K Reddy, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
FUNDING
U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated [DE-AC02-05CH11231]; National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy; J.C.S. is supported by National Microbiome Data Collaborative (NMDC); Genomic Science Program in the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (BER) [DE-AC02-05CH11231 to L.B.N.L., 89233218CNA000001 to L.A.N.L., DE-AC05-00OR22725 to O.R.N.L., DE-AC05-76RL01830 to P.N.N.L.]. Funding for open access charge: U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated [DE-AC02-05CH11231].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kyrpides N.C. Genomes online database (GOLD 1.0): a monitor of complete and ongoing genome projects world-wide. Bioinformatics. 1999; 15:773–774. [DOI] [PubMed] [Google Scholar]
- 2. Katz K., Shutov O., Lapoint R., Kimelman M., Brister J.R., O’Sullivan C. The sequence read archive: a decade more of explosive growth. Nucleic Acids Res. 2022; 50:D387–D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arita M., Karsch-Mizrachi I., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021; 49:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Verezemska O., Isbandi M., Thomas A.D., Ali R., Sharma K., Kyrpides N.C.et al.. Genomes online database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017; 45:D446–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Chen I.-M.A., Kyrpides N.C., Reddy T.. Genomes online database (GOLD) v.7: updates and new features. Nucleic Acids Res. 2019; 47:D649–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Sundaramurthi J.C., Lee J., Kandimalla M., Chen I.-M.A., Kyrpides N.C., Reddy T.B.K.. Genomes online database (GOLD) v.8: overview and updates. Nucleic Acids Res. 2021; 49:D723–D733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E.et al.. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Field D., Sterk P., Kottmann R., De Smet J.W., Amaral-Zettler L., Cochrane G., Cole J.R., Davies N., Dawyndt P., Garrity G.M.et al.. Genomic standards consortium projects. Stand. Genomic Sci. 2014; 9:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yilmaz P., Kottmann R., Field D., Knight R., Cole J.R., Amaral-Zettler L., Gilbert J.A., Karsch-Mizrachi I., Johnston A., Cochrane G.et al.. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011; 29:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K., Schulz F., Jarett J., Rivers A.R., Eloe-Fadrosh E.A.et al.. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017; 35:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roux S., Adriaenssens E.M., Dutilh B.E., Koonin E.V., Kropinski A.M., Krupovic M., Kuhn J.H., Lavigne R., Brister J.R., Varsani A.et al.. Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol. 2019; 37:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du Toit A. Outbreak of a novel coronavirus. Nat. Rev. Microbiol. 2020; 18:123–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schriml L.M., Chuvochina M., Davies N., Eloe-Fadrosh E.A., Finn R.D., Hugenholtz P., Hunter C.I., Hurwitz B.L., Kyrpides N.C., Meyer F.et al.. COVID-19 pandemic reveals the peril of ignoring metadata standards. Scientific Data. 2020; 7:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gozashti L., Corbett-Detig R.. Shortcomings of SARS-CoV-2 genomic metadata. BMC Res. Notes. 2021; 14:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2022; 50:D161–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen I.-M.A., Chu K., Palaniappan K., Ratner A., Huang J., Huntemann M., Hajek P., Ritter S., Varghese N., Seshadri R.et al.. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 2021; 49:D751–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F.et al.. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014; 42:D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reimer L.C., Sardà Carbasse J., Koblitz J., Ebeling C., Podstawka A., Overmann J.. BacDive in 2022: the knowledge base for standardized bacterial and archaeal data. Nucleic Acids Res. 2022; 50:D741–D746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edgar R.C., Taylor J., Lin V., Altman T., Barbera P., Meleshko D., Lohr D., Novakovsky G., Buchfink B., Al-Shayeb B.et al.. Petabase-scale sequence alignment catalyses viral discovery. Nature. 2022; 602:142–147. [DOI] [PubMed] [Google Scholar]
- 20. Vuong P., Lim D.J., Murphy D.V., Wise M.J., Whiteley A.S., Kaur P.. Developing bioprospecting strategies for bioplastics through the large-scale mining of microbial genomes. Front. Microbiol. 2021; 12:697309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yadav A., Borrelli J.C., Elshahed M.S., Youssef N.H.. Genomic analysis of family UBA6911 (Group 18 acidobacteria) expands the metabolic capacities of the phylum and highlights adaptations to terrestrial habitats. Appl. Environ. Microbiol. 2021; 87:e0094721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W., O’Neill K.R., Haft D.H., DiCuccio M., Chetvernin V., Badretdin A., Coulouris G., Chitsaz F., Derbyshire M.K., Durkin A.S.et al.. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021; 49:D1020–D1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ivanova N., Tringe S.G., Liolios K., Liu W.-T., Morrison N., Hugenholtz P., Kyrpides N.C.. A call for standardized classification of metagenome projects. Environ. Microbiol. 2010; 12:1803–1805. [DOI] [PubMed] [Google Scholar]
- 24. Buttigieg P.L., Morrison N., Smith B., Mungall C.J., Lewis S.E.theENVO Consortium . The environment ontology: contextualising biological and biomedical entities. J. Biomed. Semantics. 2013; 4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson L.R., Sanders J.G., McDonald D., Amir A., Ladau J., Locey K.J., Prill R.J., Tripathi A., Gibbons S.M., Ackermann G.et al.. A communal catalogue reveals earth's multiscale microbial diversity. Nature. 2017; 551:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parker C.T., Tindall B.J., Garrity G.M.. International code of nomenclature of prokaryotes. Int. J. Syst. Evol. Microbiol. 2015; 69(1A):S1–S111. [DOI] [PubMed] [Google Scholar]
- 27. Wayne L.G., Brenner D.J., Colwell R.R., Grimont P.a.D., Kandler O., Krichevsky M.I., Moore L.H., Moore W.E.C., Murray R.G.E., Stackebrandt E.et al.. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987; 37:463–464. [Google Scholar]
- 28. Whitman W.B., Woyke T., Klenk H.-P., Zhou Y., Lilburn T.G., Beck B.J., De Vos P., Vandamme P., Eisen J.A., Garrity G.et al.. Genomic encyclopedia of bacterial and archaeal type strains, phase III: the genomes of soil and plant-associated and newly described type strains. Stand. Genomic Sci. 2015; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eloe-Fadrosh E.A., Ahmed F., Anubhav, Babinski M., Baumes J., Borkum M., Bramer L., Canon S., Christianson D.S., Corilo Y.E.et al.. The national microbiome data collaborative data portal: an integrated multi-omics microbiome data resource. Nucleic Acids Res. 2022; 50:D828–D836. [Google Scholar]
- 30. Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S.et al.. KBase: the united states department of energy systems biology knowledgebase. Nat. Biotechnol. 2018; 36:566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parks D.H., Chuvochina M., Rinke C., Mussig A.J., Chaumeil P.-A., Hugenholtz P.. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic. Acids. Res. 2022; 50:D785–D794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitman W.B., Chuvochina M., Hedlund B.P., Hugenholtz P., Konstantinidis K.T., Murray A.E., Palmer M., Parks D.H., Probst A.J., Reysenbach A.-L.et al.. Development of the seqcode: a proposed nomenclatural code for uncultivated prokaryotes with DNA sequences as type. Syst. Appl. Microbiol. 2022; 45:126305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomes Online Database (GOLD) is freely available at the following URL: https://gold.jgi.doe.gov.