Abstract
The National Genomics Data Center (NGDC), part of the China National Center for Bioinformation (CNCB), provides a family of database resources to support global academic and industrial communities. With the explosive accumulation of multi-omics data generated at an unprecedented rate, CNCB-NGDC constantly expands and updates core database resources by big data archive, integrative analysis and value-added curation. In the past year, efforts have been devoted to integrating multiple omics data, synthesizing the growing knowledge, developing new resources and upgrading a set of major resources. Particularly, several database resources are newly developed for infectious diseases and microbiology (MPoxVR, KGCoV, ProPan), cancer-trait association (ASCancer Atlas, TWAS Atlas, Brain Catalog, CCAS) as well as tropical plants (TCOD). Importantly, given the global health threat caused by monkeypox virus and SARS-CoV-2, CNCB-NGDC has newly constructed the monkeypox virus resource, along with frequent updates of SARS-CoV-2 genome sequences, variants as well as haplotypes. All the resources and services are publicly accessible at https://ngdc.cncb.ac.cn.
INTRODUCTION
The National Genomics Data Center (NGDC) is affiliated to Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (CAS) & China National Center for Bioinformation (CNCB). Since its foundation in 2019, CNCB-NGDC has been constructed in collaborations with additional two CAS institutions, viz., Institute of Biophysics and Shanghai Institute of Nutrition and Health, as well as by joint efforts with partners (https://ngdc.cncb.ac.cn/partners). Over the last decades, an increasing number of large-scale high-throughput sequencing projects have been conducted globally, advancing the understanding of the genetic basis of diseases, genetic epidemiology and public health (1–3) For example, UK Biobank collects a rich variety of genome-wide genotype data and enables population-based cohort studies on genetic and epidemiological associations for a broad range of health-related traits (1). Such large-scale cohort studies have uncovered novel biomarkers and drug targets, which have greatly contributed to disease molecular diagnosis and precision medicine. Meanwhile, single-cell sequencing technologies have been rapidly developed and widely adopted to elucidate genomic (4), transcriptomic (5), epigenomic (6) and proteomic (7) heterogeneities in cellular populations and to disentangle complex mechanisms of diseases at single-cell resolution (8). As a result, immense amount of multi-omics data has been generated at an ever-increasing rate and scale. Therefore, synthesizing and sharing such massive quantities of data and knowledge is increasingly important for a wide range of research activities worldwide.
In the past year, CNCB-NGDC has made continuous efforts in developing new resources and updating relevant resources, accordingly providing open access to a family of resources for advancing life and health sciences globally (9–18) Particularly, in the context of monkeypox outbreak and COVID-19 pandemic, considerable efforts have been devoted to integrating, analyzing and updating the virus genome sequences, variants, and haplotypes (19–21). Importantly, several core database resources have been recommended by major publishers, greatly accelerating the efficient deposition and open sharing of biomedical data. Meanwhile, in addition to data sharing of SARS-CoV-2 genomes with NCBI, CNCB-NGDC is building close collaborations with INSDC (22) by mirroring the metadata and sequence data from NCBI SRA (23). Here, we provide a brief overview of new developments and recent updates in CNCB-NGDC and describe its core resources and services (Figure 1). All these resources and services are publicly available in the home page of CNCB-NGDC (https://ngdc.cncb.ac.cn).
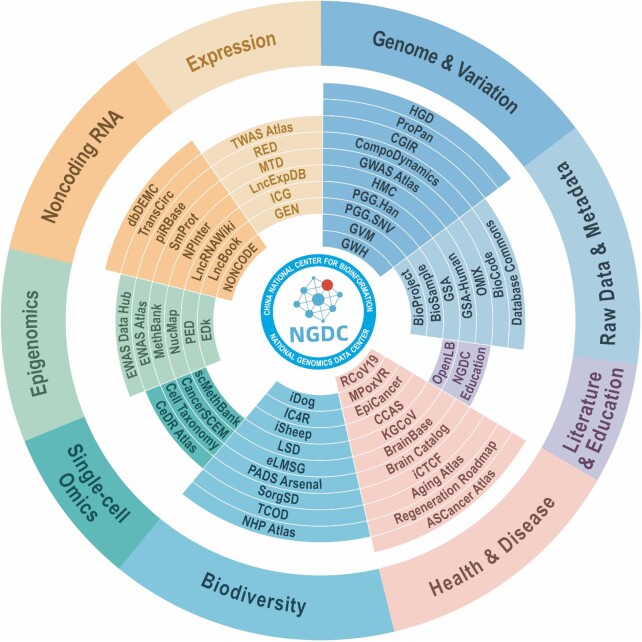
Figure 1.
Core database resources of CNCB-NGDC classified by database categories. These database resources are publicly available and searchable through the home page of CNCB-NGDC at https://ngdc.cncb.ac.cn. A full list of data resources is shown at https://ngdc.cncb.ac.cn/databases.
NEW DEVELOPMENTS
Health and disease
MPoxVR
The Monkeypox Virus Resource (MPoxVR; https://ngdc.cncb.ac.cn/gwh/poxvirus) is a one-stop portal that integrates monkeypox virus genome sequences, variants, publications and online tools (21). MPoxVR collects all public genome sequences and metadata for the Poxviridae family from GenBank (24), accompanying with daily update through in-house automatic pipelines. Of note, MPoxVR performs systematic analysis to obtain a dynamic landscape of genomic variations from a global perspective, providing all identified variants and detailed statistics for each virus isolate and attributing functional annotation and population frequency to each variant. It also equips with online tools for sequence alignment, genome annotation and variant annotation. In addition, it provides a full list of relevant publications, including published articles from PubMed as well as preprints from bioRxiv and medRxiv. Furthermore, MPoxVR offers data submission services to accept raw sequences and assembled genomes in cooperation with the Genome Sequence Archive (25) and Genome Warehouse (26) of CNCB-NGDC. Given the spread of recent outbreak caused by monkeypox virus, MPoxVR serves as a valuable resource for the global research community.
KGCoV
The Knowledge Graph of SARS-CoV-2 (KGCoV; https://www.biosino.org/kgcov) (27) is an online database centered on virus genomic information and epidemiological data, which is helpful to identify relevant knowledge and devise epidemic prevention and control policies in collaboration with disease control personnel. To help analyze the spread and evolution of the virus, KGCoV collates a wide range of data covering viral genomes, sequence variations, and locations in temporal and spatial distribution from GISAID. Thus, it collects 445 470 genomic records and 2 571 621 epidemiological records from Wikipedia and research papers. As a result, a total of 11 412 genome-case pairs are generated for the surveillance of virus transmission and reconstruction of infection paths. In conclusion, KGCoV conducts standardized processing on viral genomic and epidemiological data and produces a new genomic epidemiology knowledge graph to show the genomic mutation sites, epidemiological information and their connections in SARS-CoV-2.
ASCancer Atlas
ASCancer Atlas (https://ngdc.cncb.ac.cn/ascancer) is a comprehensive knowledgebase designed to provide a complete landscape of carcinogenic alternative splicing (AS) in human cancers (28). The current version of ASCancer Atlas houses about 2 million computationally putative splicing events identified from large-scale cancer transcriptome datasets. Different from existing databases of AS in cancers, it has unique features as follows: (i) high-confidence collection of 2,006 experimentally validated cancer-associated splicing events; (ii) complete splicing regulatory network and (iii) a suite of multi-dimensional online splicing analysis tools. In summary, ASCancer Atlas provides a repository of oncogenic AS to help researchers study a full spectrum of splicing disorders in human cancers.
Brain catalog
Brain Catalog (https://ngdc.cncb.ac.cn/braincatalog) presents a resource for a variety of disorders, diseases, and risk factors that are broadly related to the dysfunctions of the brain (29). Specifically, we collected more than 500 GWAS summary statistics datasets for psychiatric disorders, neurodevelopmental disorders, cognitive disorders, substance use disorders, behavioral habits, psychosocial and personality traits, and neurodegenerative diseases. Brain Catalog estimates the SNP-based heritability, the partitioning heritability based on functional annotations, and genetic correlations among traits. Augmented by a list of comprehensive annotation datasets including 58 QTL datasets spanning 6 types of QTLs, Brain Catalog hosts inferring results from multiple methods for the candidate causal variants, causal genes, and functional tissues and cell types for each trait. Finally, Brain Catalog presents inferred risk factors that are likely causal to each trait. In conclusion, Brain Catalog serves as a valuable resource to delineate the genetic components of brain-related traits.
CCAS
CCAS (https://ngdc.cncb.ac.cn/ccas) is a one-stop and comprehensive annotation system for individual cancer genome at multi-omics level (30). CCAS integrates 20 widely recognized resources in the field to support data annotation for 10 categories of cancers covering 395 subtypes. Data from each resource are curated and standardized by using multiple ontology frameworks. The inputs of CCAS include abnormalities on single nucleotide variant/insertion or deletion, expression, copy number variation, and methylation level. Consensus outputs are arranged in a tabular form and visualized in figures. Expanded panels with additional information are used for conciseness, and most figures are interactive to show additional information. Moreover, CCAS offers multi-dimensional annotation information, including mutation signature pattern, gene set enrichment analysis, pathways, and clinical trial related information. In summary, CCAS is designed to help users intuitively understand the molecular mechanisms of tumors and discover key functional genes.
Genome and variation
HGD
The Homologous Gene Database (HGD; https://ngdc.cncb.ac.cn/hgd) integrates multi-species and multi-omics data and provides one-stop public data services for browsing, retrieval, comparison and downloading (31). By integrating several existing homologous resources that vary in terms of inferring method and homology relationship, HGD is able to efficiently eliminate the difficulties for researchers in choosing and mapping homology results from one species to another. Besides, by offering various gene function annotations, HGD makes it convenient to conduct comprehensive homologous functional studies on large-scale genome sequences. Currently, HGD houses a total of 112 383 644 homologous pairs for 37 species, including 19 animals, 16 plants and 2 microorganisms. Specifically, 10 of the 37 species are model organisms. Meanwhile, HGD integrates various annotations including 16 909 homologs with traits, 276 670 homologs with variants, 398 573 homologs with expression profiles and 536 852 homologs with gene ontology annotations, which can help users gain a deeper understanding of homologous gene function.
ProPan
ProPan (https://ngdc.cncb.ac.cn/propan) is a public database for comprehensively profiling prokaryotic pan-genome dynamics (32). In the current version, it covers 51 882 high-quality strain genomes and provides a total of 1504 pangenomes, with 23 in archaea and 1481 bacteria. ProPan offers multi-dimensional insights into species, such as pan-genome dynamics characteristics, multiple gene functional annotation, functional protein association networks, pathway map association, resistance gene prediction for 126 substances (antimicrobial drug, biocide, and metal), and evaluation of 31 metabolic cycle processes (e.g. organic carbon oxidation, nitrite oxidation, and sulfur oxidation). Collectively, ProPan bears great utility for studying prokaryotic pan-genome dynamics, species classification and identification, pan-genome metabolism and further beyond.
Biodiversity
TCOD
The Tropical Crops Omics Database (TCOD; https://ngdc.cncb.ac.cn/tcod) is a comprehensive omics data resource for tropical crops. By integrating diverse data from five economic tropical crops, namely, cassava, rubber tree, sugarcane, mango and pineapple, TCOD houses 1253 samples with whole-genome raw sequencing data, 14 chromosome-level genome assemblies, 565 185 genes with functional annotations, 111 934 324 unique variants, 10 433 germplasm items and 23 279 publications. In addition, TCOD embeds BLAST for finding homologous genes across multiple species as well as a genome browser for visualizing the distribution of SNPs and indels on the genome. Taken together, TCOD functions as a multi-omics data platform for tropical crops and thereby provides data services for researchers to conduct selective breeding and trait improvement research.
Expression
TWAS Atlas
TWAS Atlas (https://ngdc.cncb.ac.cn/twas) is a curated knowledgebase of transcriptome-wide association studies (TWAS) (33). Based on manual curation of TWAS related publications and integration of external relevant datasets, the current implementation of TWAS Atlas contains a curated collection of 401 266 gene-trait associations, which are derived from a total of 200 publications and encompass 22 247 genes and 257 traits that are classified into diseases, phenotypic abnormalities, measurements and others. Most importantly, an interactive knowledge graph covering all catalogued gene-trait associations is constructed, which can be used to add remarkable SNP-gene associations to achieve the integration and visualization of SNP-gene-trait associations. Collectively, TWAS Atlas provides comprehensive and visualized regulatory relationships, enabling researchers to better understand the genetic mechanisms of various phenotypes and complex diseases.
RECENT UPDATES
Raw data and metadata
BioProject and BioSample
BioProject (https://ngdc.cncb.ac.cn/bioproject) and BioSample (https://ngdc.cncb.ac.cn/biosample) are two public repositories of biological research projects and samples, respectively. They collect descriptive metadata on biological projects and samples investigated in experiments and provide centralized accesses to all public projects and samples as well as cross links to their related data resources. Till September 2022, there are a total of 7906 biological projects and 783 267 biological samples submitted by 4312 users from 1027 organizations (Figure 2A), clearly showing a rapid increase by comparison with 4514 projects and 482 577 samples in August 2021. In addition, this year, BioProject and BioSample have mirrored the data of INSDC (International Nucleotide Sequence Database Collaboration) by downloading and integrating all metadata of 596 052 projects and 27 977 897 samples from NCBI.
Figure 2.
Statistics of data submissions to CNCB-NGDC. (A) Data statistics of BioProject and BioSample. (B) Data statistics of Experiments and Runs in GSA. (C) Timeline of data growth in GSA. (D) Statistics of genome assemblies in GWH. All statistics are frequently updated and publicly available at https://ngdc.cncb.ac.cn/bioproject, https://ngdc.cncb.ac.cn/biosample, https://ngdc.cncb.ac.cn/gsa and https://ngdc.cncb.ac.cn/gwh.
GSA, GSA-Human and OMIX
The Genome Sequence Archive (GSA; https://ngdc.cncb.ac.cn/gsa) (25,34) is a public data repository for raw sequence reads, which accepts worldwide data submissions, performs data curation and quality control for all submitted data, and provides free open data for sharing services for sharing all publicly available data. GSA for Human (GSA-Human; https://ngdc.cncb.ac.cn/gsa-human) (25) is a data archive specialized for human genetic related omics data with controlled-access and security services. As of September 2022, GSA and GSA-Human have together collected 654 635 experiments and 773 032 runs and archived a total of 16.3 PB data, showing a rapid increase of data volume by comparison with the previous release last September (∼10 PB) (Figure 2B and 2C). Similar to BioProject and BioSample, GSA has also mirrored the INSDC’s data by collecting and integrating the relevant metadata and raw data from NCBI SRA, covering 20 488 321 experiments, 21 963 869 runs and 962 TB of sequence files. The Open Archive for Miscellaneous Data database (OMIX; https://ngdc.cncb.ac.cn/omix), as a member of the GSA family, accepts miscellaneous data with different types as well as supplementary information and materials with various formats. OMIX has archived 952 submissions with 20.30 TB, demonstrating its dramatic growth in contrast to 269 submissions and 13.3 TB last September.
Database commons
Database Commons (https://ngdc.cncb.ac.cn/databasecommons) is a global catalog of biological databases. It provides easy access and retrieval to a full collection of worldwide biological databases, assesses the database impact by factoring both citation and age, and delivers a series of useful statistics and trends to investigate their status and impact on biomedical research. Since its inception in 2015, it has been expanded frequently to incorporate more databases and enriched gradually with a series of user-friendly functionalities. Currently, with the efforts of more than 50 curators, it catalogues a total of 5825 databases, involving 8929 publications and 1976 institutions throughout the world. Notably, Database Commons has been recommended by Cell Press and Bioinformatics Advances to provide registry services for biological data repositories. In addition, Database Commons, in collaboration with Nucleic Acids Research (NAR), provides registration services for databases published in the NAR Database Issue.
Genome and variation
Genome warehouse
The Genome Warehouse (GWH; https://ngdc.cncb.ac.cn/gwh) is a public resource for archiving assembled genome sequences and their detailed metadata (26). Compared to 20 606 assemblies last August, GWH hosts a total of 24 781 assemblies for 1792 species as of September 2022 (Figure 2D). Among them, 12 887 assemblies are publicly released and reported in 206 journal articles, by comparison with 9886 assemblies and 97 articles in August 2021. All released sequences have passed strict quality control, and are searchable and freely accessible in GWH. Of interest, 25 protist genome assemblies mainly from the Protist 10 000 Genomes Project (P10K) (35) are deposited in the current release of GWH. Particularly, GWH has received and released telomere-to-telomere gap-free assemblies for organisms such as Arabidopsis thaliana (36). In support of MPoxVR (21), GWH has integrated NCBI genome and protein sequences of all viruses in the Poxviridae family (https://ngdc.cncb.ac.cn/gwh/browse/virus/poxviridae), with the aim to enable researchers to conduct comparative analysis on poxviruses.
GVM and GWAS atlas
The Genome Variation Map (GVM; https://ngdc.cncb.ac.cn/gvm) (37,38) and GWAS Atlas (https://ngdc.cncb.ac.cn/gwas) (39,40) are two public variation-related resources. GVM is a public repository of genome variations, including single nucleotide polymorphisms (SNP) and small insertions and deletions (indel), features data collection for a wide range of species and accepts data submissions from all over the world. GWAS Atlas (39,40) is a curated resource of genome-wide variant-trait associations in plants and animals. Till August 2022, GVM has received 244 data submissions involving 165 243 samples from 37 species and contained a total of ∼1055 million variants, encompassing 330 projects and 65 862 samples and covering 18 animals, 26 plants and 3 viruses. GWAS Atlas integrates 233 599 associations across ten cultivated plants and five domesticated animals that were manually curated from 3072 studies in 771 publications. As a result, a total of 36 874 genes and 1395 traits are annotated based on a set of ontologies. To prioritize the most important loci for functional follow-up studies, a total of 4492 unique lead SNPs for 407 traits and 361 unique experiment-validated causal variants for 131 traits are newly provided. To facilitate comparative analysis across species, GWAS Atlas unifies trait vocabularies and defines new ontology terms for 1056 traits, resulting in a total of 1172 Plant Phenotype/Trait Ontology (PPTO) and 431 Animal Phenotype/Trait Ontology (APTO) terms. Together, GVM provides high-density reference variations and GWAS Atlas integrates high-quality curated GWAS associations for plants and animals, which serve as valuable resources for genomic variation research of important traits.
Health and disease
RCoV19
The 2019 Novel Coronavirus Resource (RCoV19; https://ngdc.cncb.ac.cn/ncov) (41,42) provides a series of functional modules on SARS-CoV-2 genome sequences, genomic mutations, variant monitoring, online data analysis toolkits and literatures. As of September 2022, a total of 13 345 020 SARS-CoV-2 sequences and meta data are integrated, and 188 100 genomic mutations are identified based on those complete and high-quality genome sequences. To meet the need for near real-time mutation surveillance and early-warning of high-risk variants, RCoV19 offers the genomic prevalence of lineages globally and nationally, and allows mutation prevalence comparison across lineages. It also provides potential high-risk variants predicted weekly by a machine learning model based on several important haplotype network features, and estimates the percentage of high-risk lineages among all sequences since its emergence. More importantly, RCoV19 provides curated knowledge of host susceptibility to SARS-CoV-2 and mutation effects on transmission and pathogenicity, which are of great usefulness for origin tracing and transmission preventing.
Expression
Gene expression nebulas
Gene Expression Nebulas (GEN; https://ngdc.cncb.ac.cn/gen) is a data portal integrating transcriptomic profiles at both bulk and single-cell levels in various conditions across multiple species (43). In the current release, 146 gene expression profiling datasets related to SARS-CoV-2 infection (101 bulk and 45 scRNA-seq) derived from 140 original high-throughput sequencing projects are systematically incorporated. In comparison to the previous release (August 2021), total number of incorporated datasets increases from 323 to 469, covering 54 448 samples and 18 966 983 cells of 34 species, involving 21 animals, 10 plants, 2 protists and 1 fungus. In addition to the enrichment of data volume, GEN has also been significantly upgraded by providing an easy-to-use one-stop offline RNA-seq data analysis pipeline, named GENToolkit, which aims to facilitate the standardization of expression profiling analysis of both bulk RNA-seq and scRNA-seq data across various technical and biological conditions.
Internal control genes
The database of Internal Control Genes (ICG; https://ngdc.cncb.ac.cn/icg) is a well-established knowledgebase of experiment-validated internal control genes and their respective applicable scenarios for RT-qPCR normalization across a wide variety of species (44). In the current version, ICG houses a total of 2514 high-quality verified internal control genes from 509 species (188 animals, 264 plants, 28 fungi and 29 bacteria), associating with 2725 corresponding applicable scenarios. Particularly, a new module ‘Health & Disease portal’ is set up to facilitate the application of RT-qPCR in precision medicine research, which currently supports 27 cancer types, 15 diseases and 17 human molecular biological models. In addition to mRNAs, effective normalization strategies for diverse types of non-coding RNAs are also integrated. Moreover, to improve the flexibility and functionality, ICG is implemented based on MySQL/Java, greatly facilitating structured management, access, utilization and knowledge enrichment.
Epigenomics
EWAS open platform
The EWAS Open Platform (https://ngdc.cncb.ac.cn/ewas) (45) is a one-stop resource for epigenome-wide association studies (EWAS). It is made up of three parts: EWAS Data Hub (46) for data collection and standardized normalization, EWAS Atlas (47) for knowledge extraction and curation, and EWAS Toolkit for downstream analysis and visualization. The current version of EWAS Open Platform is updated by adding 17 820 samples and 25 526 associations. Among them, all DNA methylation array data was normalized with batch effect removal using GMQN (48), a reference-based method for correcting batch effects as well as probe bias in the Human Methylation BeadChip. At present, EWAS Open Platform houses 133 672 DNA methylation array data, including 1099 tissues/cell types and 612 diseases, as well as 642 544 high-quality EWAS associations manually curated from 1586 studies in 991 publications, covering 717 traits and 3497 cohorts. In addition, taking advantage of the high-quality knowledge and data, EWAS Open Platform provides a number of reference DNA methylation profiles and offers online services for enrichment, annotation, and network visualization.
MethBank
The Methylation Bank (MethBank; https://ngdc.cncb.ac.cn/methbank) (49–51) is a comprehensive database of whole-genome DNA methylation across a variety of species. In the current release of MethBank, significant improvements and updates have been made in data volume, downstream data mining for differential methylation and web interfaces. Specifically, the updated MethBank features: (i) an increase in single-base resolution methylomes, from 855 last August to 1449 across 23 species and 236 tissues/cell lines in different biological scenarios including development, cancer and physiology; (ii) computational identification of differentially methylated regions related to 887 different biological groups and characterization of enriched biological pathways to expand the methylation traits/biomarkers resource; (iii) a new knowledge module that consists of a curation network for 266 associations of biological contexts and featured differentially methylated genes; (iv) an increasing amount of microarray data with up to 111 tissue/cell type specific samples and (v) significant improvements in visualization together with the completely redesigned web interfaces.
Noncoding RNA
LncBook (https://ngdc.cncb.ac.cn/lncbook) is a comprehensive database of human long non-coding RNAs (lncRNAs) as well as their annotations (52,53). The updated release of LncBook integrates more lncRNA genes, characterizes their molecular signatures in more biological contexts, and incorporates more annotations by including new omics features. First, it incorporates 119 722 new transcripts and 9632 new genes, updates gene structure of 21 305 lncRNAs, and provides 323 950 high-quality lncRNA transcripts and 95 243 genes. Second, it enriches the expression and methylation annotations with more biological contexts, highlights disease/trait-associated variants, and predicts lncRNA-miRNA binding sites. Third, it integrates new omics features of lncRNA genes including sequence conservation across 40 vertebrates, small protein expression, and interaction with proteins. LncRNAWiki (https://ngdc.cncb.ac.cn/lncrnawiki), a knowledgebase of human lncRNAs, incorporates comprehensive annotations of functional lncRNAs based on a standardized curation model and provides user-friendly web interfaces to facilitate data curation, retrieval and visualization (54,55). This year, based on manual curation of 535 publications, we have expanded LncRNAWiki by adding 97 experiment-validated human lncRNAs, updating 191 existing lncRNAs and integrating 4761 newly-curated associations.
Single-cell omics
Cell taxonomy
Cell Taxonomy (https://ngdc.cncb.ac.cn/celltaxonomy) is a comprehensive and curated repository of cell types and associated cell markers encompassing a wide range of species, tissues and conditions (56). Combined with literature curation and data integration, up to September 2022, Cell Taxonomy houses 3143 cell types and 26 613 associated cell markers in 257 conditions and 387 tissues across 34 species based on 4299 publications and scRNA-seq profiles of ∼3.5 million cells. It presents a significant increase in comparison to the last version in September 2021 (containing 2650 cell types and 25 087 cell markers in 157 conditions, 296 tissues, 21 species supported by 3402 publications and 1.9 million scRNA-seq profiles). Collectively, Cell Taxonomy represents a fundamentally useful reference to systematically and accurately characterize cell types and thus lays an important foundation for deeply understanding and exploring cellular biology in diverse species.
CONCLUDING REMARKS
With the explosive growth of multi-omics data, CNCB-NGDC keeps putting efforts to provide a suite of newly developed and updated database resources, with the aim to accept data submissions and provide value-added annotations and curated knowledge for the global research community. Ongoing efforts include, but not limited to, automation of data submission, curation, integration and analysis procedures, infrastructure upgrades for big data storage and transmission, and development of new tools and pipelines in aid of big data analysis. As one of major global centers, CNCB-NGDC will continue to expand and provide a family of data resources and services to support knowledge discovery for a wide range of research activities in life and health sciences.
DATA AVAILABILITY
All resources and services are publicly available in the home page of CNCB-NGDC (https://ngdc.cncb.ac.cn).
ACKNOWLEDGEMENTS
We thank our users for submitting data, sending suggestions, reporting bugs and getting involved in community curation. CNCB-NGDC is indebted to its funders, including the Ministry of Science & Technology and the Ministry of Finance of the People's Republic of China as well as Chinese Academy of Sciences. We would like to express our sincere thanks to the late Professor Weimin Zhu, a scientific advisor of our center, who had provided valuable advice for the center development since its inception in 2016.
APPENDIX
Corresponding author: Yongbiao Xue1,2,3,*
Co-corresponding authors: Yiming Bao1,2,3,*, Zhang Zhang1,2,3,*, Wenming Zhao1,2,3,*, Jingfa Xiao1,2,3,*, Shunmin He3,4,*, Guoqing Zhang3,5,*, Yixue Li3,5,*, Guoping Zhao3,5,6,7,*, Runsheng Chen4,8,*
CNCB-NGDC MEMBERS (Arranged by project role and then by contribution except for Team Leader (TL), as indicated)
MPoxVR: Yingke Ma1,2,#, Meili Chen1,2,#, Cuiping Li1,2,#, Shuai Jiang1,2, Dong Zou1,2, Zheng Gong1,2, Xuetong Zhao1,2,3, Yanqing Wang1,2, Junwei Zhu1,2, Zhang Zhang1,2,3, Wenming Zhao1,2,3, Yongbiao Xue1,2,3, Yiming Bao1,2,3,*(TL), Shuhui Song1,2,3,# (TL)
KGCoV: Guoqing Zhang5,#, Yunchao Ling5, Yiwei Wang5, Jiaxin Yang5, Xinhao Zhuang5
HGD: Guangya Duan1,2,3,#, Gangao Wu1,2,3,#, Xiaoning Chen1,2,3, Dongmei Tian1,2, Zhaohua Li1,2,3, Yanling Sun1,2, Zhenglin Du1,2, Lili Hao1,2, Shuhui Song1,2,3, Yuan Gao1,2,3, Jingfa Xiao1,2,3, Zhang Zhang1,2,3, Yiming Bao1,2,3, Bixia Tang1,2,#, Wenming Zhao1,2,3,*
ProPan: Yadong Zhang1,2,#, Hao Zhang1,2,3,#, Zaichao Zhang9, Qiheng Qian1,2,3, Zhewen Zhang1,2,#, Jingfa Xiao1,2,3,*
TCOD: Hailong Kang1,2,3,#, Tianhao Huang1,2,3,#, Xiaoning Chen1,2,3,#, Zhiqiang Xia10, Xincheng Zhou11, Jinquan Chao12, Bixia Tang1,2, Zhonghuang Wang1,2,3, Junwei Zhu1,2, Zhenglin Du1,2, Sisi Zhang1,2, Jingfa Xiao1,2,3, Weimin Tian12, Wenquan Wang10,#, Wenming Zhao1,2,3,*
ASCancer Atlas: Song Wu1,2,3,#, Yue Huang2,3,13,#, Mochen Zhang1,2,3, Zheng Gong1,2,3, Guoliang Wang1,2,3, Xinchang Zheng1,2, Wenting Zong1,2,3, Wei Zhao1,2,3, Peiqi Xing2,13, Rujiao Li1,2,3,# (TL), Zhaoqi Liu2,3,13,# (TL), Yiming Bao1,2,3,* (TL)
TWAS Atlas: Mingming Lu1,2,3,#, Yadong Zhang1,2,#, Fengchun Yang14,#, Jialin Mai 1,2,3,#, Qianwen Gao1,2,3,15, Xiaowei Xu14, Hongyu Kang14, Li Hou14, Yunfei Shang1,2,3, Qiheng Qain1,2,3, Jie Liu16, Meiye Jiang1,2,3, Hao Zhang1,2,3, Congfan Bu1,2, Jinyue Wang17, Zhewen Zhang1,2, Zaichao Zhang9, Jingyao Zeng1,2,#, Jiao Li14,#, Jingfa Xiao1,2,3,*
Brain Catalog: Siyu Pan2,3,13,#, Hongen Kang2,3,13,#, Xinxuan Liu2,13,18,#, Shiqi Lin2,3,13, Na Yuan2,13, Zhang Zhang1,2,3, Yiming Bao1,2,3, Peilin Jia2,13,#
CCAS: Xinchang Zheng1,2,#, Wenting Zong1,2,3,#, Zhaohua Li1,2,3,#, Yanling Sun1,2,#, Yingke Ma1,2, Zhuang Xiong1,2,3, Song Wu1,2,3, Fei Yang1,2,3, Wei Zhao1,2,3, Congfan Bu1,2, Zhenglin Du1,2, Jingfa Xiao1,2,3,*, Yiming Bao1,2,3,*
BioProject & BioSample & GSA & BIG Submission: Xu Chen1,2,#, Tingting Chen1,2,#, Sisi Zhang1,2,#, Yanling Sun1,2,#, Caixia Yu1,2, Bixia Tang1,2, Junwei Zhu1,2, Lili Dong1,2, Shuang Zhai1,2, Yubin Sun1,2, Qiancheng Chen1,2, Xiaoyu Yang1,2, Xin Zhang1,2, Zhengqi Sang1,2, Yonggang Wang1,2, Yilin Zhao1,2, Huanxin Chen1,2, Li Lan1,2, Yanqing Wang1,2,# (TL), Wenming Zhao1,2,3,* (TL)
OMIX: Anke Wang1,2,#, Caixia Yu1,2,#, Yanqing Wang1,2, Sisi Zhang1,2,# (TL)
GWH: Yingke Ma1,2,#, Yaokai Jia1,2,#, Xuetong Zhao1,2, Meili Chen1,2,# (TL)
GVM: Cuiping Li1,2,#, Dongmei Tian1,2,#, Bixia Tang1,2,#, Yitong Pan1,2,3, Lili Dong1,2, Xiaonan Liu1,2,3, Shuhui Song1,2,3,# (TL)
GWAS Atlas: Xiaonan Liu1,2,3,#, Dongmei Tian1,2,#, Cuiping Li1,2,#, Bixia Tang1,2, Zhonghuang Wang1,2,3, Rongqin Zhang1,2,3, Yitong Pan1,2,3, Yi Wang1,2,3, Dong Zou1,2, Shuhui Song1,2,3,# (TL)
RCoV19: Cuiping Li1,2,#, Dong Zou1,2,#, Lina Ma1,2,3,#, Zheng Gong1,2,3,#, Junwei Zhu1,2, Xufei Teng1,2,3, Lun Li1,2, Na Li1,2, Ying Cui1,2,3, Guangya Duan1,2,3, Mochen Zhang1,2,3, Tong Jin1,2,3, Hailong Kang1,2,3, Zhonghuang Wang1,2,3, Gangao Wu1,2,3, Tianhao Huang1,2,3, Wei Zhao1,2,3, Enhui Jin1,2,3, Tao Zhang1,2,3, Zhang Zhang1,2,3, Wenming Zhao1,2,3, Yongbiao Xue1,2,3, Yiming Bao1,2,3,* (TL), Shuhui Song1,2,3,# (TL)
GEN: Tianyi Xu1,2,#, Dong Zou1,2,#, Ming Chen1,2,3,#, Guangyi Niu1,2,3,#, Rong Pan1,2,3, Tongtong Zhu1,2,3, Yuan Chu1,2,3, Lili Hao1,2,# (TL)
ICG: Jian Sang1,2,3,#, Rong Pan1,2,3,#, Dong Zou1,2,#, Yuanpu Zhang19, Zhennan Wang20, Ming Chen1,2,3, Yuansheng Zhang1,2,3, Tianyi Xu1,2, Qiliang Yao21, Tongtong Zhu1,2,3, Guangyi Niu1,2,3, Lili Hao1,2,# (TL)
EWAS Open Platform: Zhuang Xiong1,2,3,#, Fei Yang1,2,3,#, Guoliang Wang1,2,3,#, Rujiao Li1,2,3,# (TL)
MethBank: Wenting Zong1,2,3,#, Mochen Zhang1,2,3,#, Dong Zou1,2,#, Wei Zhao1,2,3,#, Guoliang Wang1,2,3, Fei Yang1,2, Song Wu1,2,3, Xinran Zhang1,2,3, Xutong Guo1,2,3, Yingke Ma1,2, Zhuang Xiong1,2,3, Rujiao Li1,2,3,# (TL)
LncBook: Zhao Li1,2,3,#, Lin Liu1,2,#, Changrui Feng1,2,3,#, Yuxin Qin1,2,3, Jingfa Xiao1,2,3, Lina Ma1,2,3,# (TL)
LncRNAWiki: Wei Jing1,2,3,#, Sicheng Luo1,2,22,#, Zhao Li1,2,3, Lina Ma1,2,3,# (TL)
Cell Taxonomy: Shuai Jiang1,2,#, Qiheng Qian1,2,3,#, Tongtong Zhu1,2,3,#, Wenting Zong1,2,3, Yunfei Shang1,2,3, Tong Jin1,2,3, Yuansheng Zhang1,2,3, Ming Chen1,2,3, Zishan Wu1,2,3, Yuan Chu1,2,3, Rongqin Zhang1,2,3, Sicheng Luo1,2,3, Wei Jing1,2,3, Dong Zou1,2, Yiming Bao1,2,3, Jingfa Xiao1,2,3,* (TL), Zhang Zhang1,2,3,* (TL)
Database Commons: Dong Zou1,2,#, Lin Liu1,2,#, Yuxin Qin1,2,3, Sicheng Luo1,2,22, Wei Jing1,2,3, Qianpeng Li1,2,3, Pei Liu40, Yongqing Sun40, Lina Ma1,2,3,# (TL)
Writing Group: Shuai Jiang1,2, Zhuojing Fan1,2, Wenming Zhao1,2,3,*, Jingfa Xiao1,2,3,*, Yiming Bao1,2,3,*, Zhang Zhang1,2,3,*
CNCB-NGDC PARTNERS (Listed in alphabetical order by database names)
AnimalTFDB: Wen-Kang Shen23, An-Yuan Guo23
BBCancer: Zhixiang Zuo24, Jian Ren24
CancerSEA: Xinxin Zhang25, Yun Xiao25, Xia Li25
CellMarker: Xinxin Zhang25, Yun Xiao25, Xia Li25
CGDB: Dan Liu23, Chi Zhang23, Yu Xue23
CGGA: Zheng Zhao26, Tao Jiang26
circAtlas: Wanying Wu27, Fangqing Zhao27
CirFunBase: Xianwen Meng28, Ming Chen28
CPLM: Yujie Gou23, Miaomiao Chen23, Yu Xue23
dbPSP & THANATOS: Di Peng23, Yu Xue23
DEG & DoriC: Hao Luo29,30,31, Feng Gao29,30,31
DrLLPS: Wanshan Ning23, Yu Xue23
eLMSG: Wan Liu5, Yunchao Ling5, Ruifang Cao5, Guoqing Zhang5
EPSD & WERAM: Yuxiang Wei23, Yu Xue23
EVAtlas: Chun-Jie Liu23, An-Yuan Guo23
EVmiRNA: Gui-Yan Xie23, An-Yuan Guo23
GenTree: Hao Yuan3,20, Tianhan Su3,20, Yong E. Zhang3,20,32
GTDB: Chenfen Zhou5, Pengyu Wang5, Guoqing Zhang5
HCL: Yincong Zhou28, Ming Chen28, Guoji Guo33
hTFtarget: Qiong Zhang23, An-Yuan Guo23
iEKPD: Shanshan Fu23, Xiaodan Tan23, Yu Xue23
iPCD: Dachao Tang23, Yu Xue23
iUUCD: Weizhi Zhang23, Yu Xue23
LeukemiaDB: Mei Luo23, An-Yuan Guo23
lnCAR: Yubin Xie24, Jian Ren24
lncRNASNP2: Ya-Ru Miao23, An-Yuan Guo23
MCA: Yincong Zhou28, Ming Chen28, Guoji Guo33
MiCroKiTS: Xinhe Huang23, Zihao Feng23, Yu Xue23
miRNASNP: Chun-Jie Liu23, An-Yuan Guo23
msRepDB: Xingyu Liao34,35, Xin Gao34, Jianxin Wang35
PEA: Guiyan Xie23, An-Yuan Guo23
PceRBase: Chunhui Yuan28, Ming Chen28
PlantRegMap: Dechang Yang36, Feng Tian36, Ge Gao36
PncStres: Wenyi Wu28, Ming Chen28
PTMD: Cheng Han23, Yu Xue23, Qinghua Cui37,38
RhesusBase: Chunfu Xiao39, Chuan-Yun Li39
RMVar: XiaoTong Luo24, Jian Ren24
SEECancer: Xinxin Zhang25, Yun Xiao25, Xia Li25
SEGreg: Qing Tang23, An-Yuan Guo23
ZCURVE_CoVdb: Hao Luo29,30,31, Feng Gao29,30,31
*To whom correspondence should be addressed: Yongbiao Xue (ybxue@big.ac.cn).
Correspondence may also be addressed to Yiming Bao (baoym@big.ac.cn), Zhang Zhang (zhangzhang@big.ac.cn), Wenming Zhao (zhaowm@big.ac.cn), Jingfa Xiao (xiaojingfa@big.ac.cn), Shunmin He (heshunmin@ibp.ac.cn), Guoqing Zhang (gqzhang@picb.ac.cn), Yixue Li (yxli@sibs.ac.cn), Guoping Zhao (gpzhao@sibs.ac.cn) and Runsheng Chen (crs@ibp.ac.cn).
# The authors wish it to be known that, in their opinion, these authors should be regarded as Joint First Authors.
1 National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
2 China National Center for Bioinformation, Beijing 100101, China
3 University of Chinese Academy of Sciences, Beijing 100049, China
4 National Genomics Data Center & Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China
5 National Genomics Data Center & Bio-Med Big Data Center, Key Laboratory of Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Xuhui, Shanghai 200031, China
6 CAS-Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, 300 Fenglin Road, Xuhui, Shanghai 200032, China
7 Center for Quantitative Synthetic Biology, Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
8 Guangdong Geneway Decoding Bio-Tech Co. Ltd, Foshan, 528316, China
9 Department of Biology, The University of Western Ontario, London, Ontario N6A 5B7, Canada
10 College of Tropical Crops, Hainan University, Haikou 570228, China
11 Institute of Tropical Biosciences and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou 571101, China
12 Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou 571101, China
13 CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
14 Institute of Medical Information, Chinese Academy of Medical Sciences/Peking Union Medical College, Beijing 100020, China
15 Current address: Beijing Novogene Bioinformatics Technology Co.,Ltd, Beijing 100000, China
16 North China University of Science and Technology Affiliated Hospital, Tangshan 063000, China
17 Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China
18 School of Future Technology, University of Chinese Academy of Sciences, Beijing 100049, China
19 College of Computer Science Technology, Inner Mongolia Normal University, Hohhot 010010, China
20 Key Laboratory of Zoological Systematics and Evolution and State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China
21 Department of Civil and Environmental Engineering, The Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong, China
22 Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China
23 MOE Key Laboratory of Molecular Biophysics, Hubei Bioinformatics and Molecular Imaging Key Laboratory, Center for Artificial Intelligence Biology, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, Hubei, China
24 State Key Laboratory of Oncology in South China, Cancer Center, Collaborative Innovation Center for Cancer Medicine, School of Life Sciences, Sun Yat-sen University, Guangzhou 510060, China
25 College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China
26 Beijing Neurosurgical Institute, Capital Medical University, Beijing 100070, China
27 Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing 100101, China
28 Department of Bioinformatics, College of Life Sciences; The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China
29 Department of Physics, School of Science, Tianjin University, Tianjin 300072, China
30 Frontiers Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin 300072, China
31 SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300072, China
32 CAS Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
33 Center for Stem Cell and Regenerative Medicine, Zhejiang University School of Medicine, Hangzhou, China
34 Computational Bioscience Research Center (CBRC), Computer, Electrical and Mathematical Sciences and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal 23955, Saudi Arabia
35 Hunan Provincial Key Lab on Bioinformatics, School of Computer Science and Engineering, Central South University, Changsha 410083, China
36 State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Biomedical Pioneering Innovative Center (BIOPIC) & Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), Peking University, Beijing 100871, China
37 Department of Biomedical Informatics, School of Basic Medical Sciences, MOE Key Lab of Cardiovascular Sciences, Center for Noncoding RNA Medicine, Peking University, Beijing 100190, China
38 Center of Bioinformatics, Key Laboratory for NeuroInformation of Ministry of Education, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, Sichuan 610054, China
39 Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Institute of Molecular Medicine, College of Future Technology, Peking University, Beijing, China
40 Huazhong University of Science and Technology, Wuhan, Hubei 430074, China
Present address: Jian Sang, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Contributor Information
CNCB-NGDC Members and Partners:
Yongbiao Xue, Yiming Bao, Zhang Zhang, Wenming Zhao, Jingfa Xiao, Shunmin He, Guoqing Zhang, Yixue Li, Guoping Zhao, Runsheng Chen, Yingke Ma, Meili Chen, Cuiping Li, Shuai Jiang, Dong Zou, Zheng Gong, Xuetong Zhao, Yanqing Wang, Junwei Zhu, Zhang Zhang, Wenming Zhao, Yongbiao Xue, Yiming Bao, Shuhui Song, Guoqing Zhang, Yunchao Ling, Yiwei Wang, Jiaxin Yang, Xinhao Zhuang, Guangya Duan, Gangao Wu, Xiaoning Chen, Dongmei Tian, Zhaohua Li, Yanling Sun, Zhenglin Du, Lili Hao, Shuhui Song, Yuan Gao, Jingfa Xiao, Zhang Zhang, Yiming Bao, Bixia Tang, Wenming Zhao, Yadong Zhang, Hao Zhang, Zaichao Zhang, Qiheng Qian, Zhewen Zhang, Jingfa Xiao, Hailong Kang, Tianhao Huang, Xiaoning Chen, Zhiqiang Xia, Xincheng Zhou, Jinquan Chao, Bixia Tang, Zhonghuang Wang, Junwei Zhu, Zhenglin Du, Sisi Zhang, Jingfa Xiao, Weimin Tian, Wenquan Wang, Wenming Zhao, Song Wu, Yue Huang, Mochen Zhang, Zheng Gong, Guoliang Wang, Xinchang Zheng, Wenting Zong, Wei Zhao, Peiqi Xing, Rujiao Li, Zhaoqi Liu, Yiming Bao, Mingming Lu, Yadong Zhang, Fengchun Yang, Jialin Mai, Qianwen Gao, Xiaowei Xu, Hongyu Kang, Li Hou, Yunfei Shang, Qiheng Qain, Jie Liu, Meiye Jiang, Hao Zhang, Congfan Bu, Jinyue Wang, Zhewen Zhang, Zaichao Zhang, Jingyao Zeng, Jiao Li, Jingfa Xiao, Siyu Pan, Hongen Kang, Xinxuan Liu, Shiqi Lin, Na Yuan, Zhang Zhang, Yiming Bao, Peilin Jia, Xinchang Zheng, Wenting Zong, Zhaohua Li, Yanling Sun, Yingke Ma, Zhuang Xiong, Song Wu, Fei Yang, Wei Zhao, Congfan Bu, Zhenglin Du, Jingfa Xiao, Yiming Bao, Xu Chen, Tingting Chen, Sisi Zhang, Yanling Sun, Caixia Yu, Bixia Tang, Junwei Zhu, Lili Dong, Shuang Zhai, Yubin Sun, Qiancheng Chen, Xiaoyu Yang, Xin Zhang, Zhengqi Sang, Yonggang Wang, Yilin Zhao, Huanxin Chen, Li Lan, Yanqing Wang, Wenming Zhao, Anke Wang, Caixia Yu, Yanqing Wang, Sisi Zhang, Yingke Ma, Yaokai Jia, Xuetong Zhao, Meili Chen, Cuiping Li, Dongmei Tian, Bixia Tang, Yitong Pan, Lili Dong, Xiaonan Liu, Shuhui Song, Xiaonan Liu, Dongmei Tian, Cuiping Li, Bixia Tang, Zhonghuang Wang, Rongqin Zhang, Yitong Pan, Yi Wang, Dong Zou, Shuhui Song, Cuiping Li, Dong Zou, Lina Ma, Zheng Gong, Junwei Zhu, Xufei Teng, Lun Li, Na Li, Ying Cui, Guangya Duan, Mochen Zhang, Tong Jin, Hailong Kang, Zhonghuang Wang, Gangao Wu, Tianhao Huang, Wei Zhao, Enhui Jin, Tao Zhang, Zhang Zhang, Wenming Zhao, Yongbiao Xue, Yiming Bao, Shuhui Song, Tianyi Xu, Dong Zou, Ming Chen, Guangyi Niu, Rong Pan, Tongtong Zhu, Yuan Chu, Lili Hao, Jian Sang, Rong Pan, Dong Zou, Yuanpu Zhang, Zhennan Wang, Ming Chen, Yuansheng Zhang, Tianyi Xu, Qiliang Yao, Tongtong Zhu, Guangyi Niu, Lili Hao, Zhuang Xiong, Fei Yang, Guoliang Wang, Rujiao Li, Wenting Zong, Mochen Zhang, Dong Zou, Wei Zhao, Guoliang Wang, Fei Yang, Song Wu, Xinran Zhang, Xutong Guo, Yingke Ma, Zhuang Xiong, Rujiao Li, Zhao Li, Lin Liu, Changrui Feng, Yuxin Qin, Jingfa Xiao, Lina Ma, Wei Jing, Sicheng Luo, Zhao Li, Lina Ma, Shuai Jiang, Qiheng Qian, Tongtong Zhu, Wenting Zong, Yunfei Shang, Tong Jin, Yuansheng Zhang, Ming Chen, Zishan Wu, Yuan Chu, Rongqin Zhang, Sicheng Luo, Wei Jing, Dong Zou, Yiming Bao, Jingfa Xiao, Zhang Zhang, Dong Zou, Lin Liu, Yuxin Qin, Sicheng Luo, Wei Jing, Qianpeng Li, Pei Liu, Yongqing Sun, Lina Ma, Shuai Jiang, Zhuojing Fan, Wenming Zhao, Jingfa Xiao, Yiming Bao, Zhang Zhang, Wen-Kang Shen, An-Yuan Guo, Zhixiang Zuo, Jian Ren, Xinxin Zhang, Yun Xiao, Xia Li, Xinxin Zhang, Yun Xiao, Xia Li, Dan Liu, Chi Zhang, Yu Xue, Zheng Zhao, Tao Jiang, Wanying Wu, Fangqing Zhao, Xianwen Meng, Ming Chen, Yujie Gou, Miaomiao Chen, Yu Xue, Di Peng, Yu Xue, Hao Luo, Feng Gao, Wanshan Ning, Yu Xue, Wan Liu, Yunchao Ling, Ruifang Cao, Guoqing Zhang, Yuxiang Wei, Yu Xue, Chun-Jie Liu, An-Yuan Guo, Gui-Yan Xie, An-Yuan Guo, Hao Yuan, Tianhan Su, Yong E Zhang, Chenfen Zhou, Pengyu Wang, Guoqing Zhang, Yincong Zhou, Ming Chen, Guoji Guo, Qiong Zhang, An-Yuan Guo, Shanshan Fu, Xiaodan Tan, Yu Xue, Dachao Tang, Yu Xue, Weizhi Zhang, Yu Xue, Mei Luo, An-Yuan Guo, Yubin Xie, Jian Ren, Ya-Ru Miao, An-Yuan Guo, Yincong Zhou, Ming Chen, Guoji Guo, Xinhe Huang, Zihao Feng, Yu Xue, Chun-Jie Liu, An-Yuan Guo, Xingyu Liao, Xin Gao, Jianxin Wang, Guiyan Xie, An-Yuan Guo, Chunhui Yuan, Ming Chen, Dechang Yang, Feng Tian, Ge Gao, Wenyi Wu, Ming Chen, Cheng Han, Yu Xue, Qinghua Cui, Chunfu Xiao, Chuan-Yun Li, XiaoTong Luo, Jian Ren, Xinxin Zhang, Yun Xiao, Xia Li, Qing Tang, An-Yuan Guo, Hao Luo, Feng Gao, Yongbiao Xue, Yiming Bao, Zhang Zhang, Wenming Zhao, Jingfa Xiao, Shunmin He, Guoqing Zhang, Yixue Li, Guoping Zhao, and Runsheng Chen
FUNDING
National Key Research & Development Program of China [2021YFF0703704, 2021YFF0703703, 2020YFA0907001]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38030200, XDA19050302, XDA24040201, XDB38030100, XDB38030400, XDA12030100, XDB38040300, XDB38030202, XDA16021403, XDB38000000, XDB38030000, XDB38010400, XDB38010401]; National Key Research & Development Program of China [2021YFF0703700, 2021YFF0703702, 2021YFF0704500, 2021YFC2301502, 2021YFC0863300, 2019YFA0801801, 2018YFA0801405, 2018YFD1000505, 2018YFC2000100, 2018YFC1406902, 2018YFC0910400, 2018YFC0310602, 2018YFA0903700, 2018YFA0900704, 2018YFA0900700]; National Natural Science Foundation of China [31970565, 31871328, 31871294, 31970647, 31801104, 32000475, 1470330, 31961130380, 31822030, 31801113, 31801154, 91940303, 91940306, 31871281, 31970634, 31930021, 32025009, 31970633, 32100520, 32170669, 32100506, 32100511, 62002388, 82161148009, 32270718, 32030021]; International Partnership Program of the Chinese Academy of Sciences [153D31KYSB20170121]; Genomics Data Center Construction of Chinese Academy of Sciences [WX145XQ07-04]; Fundamental Research Funds for the Central Universities [2019kfyRCPY043]; UK Royal Society-Newton Advanced Fellowship [NAF\R1\191094]; Key Research Program of Frontier Sciences of the Chinese Academy of Sciences [QYZDJ-SSW-SYS009]; Key Technology Talent Program of the Chinese Academy of Sciences; the 100 Talent Program of the Chinese Academy of Sciences; K.C. Wong Education Foundation; the Youth Innovation Promotion Association of the Chinese Academy of Sciences [2019104, 2018134, 2017141, 2021038, 2022098]; Special Project on Precision Medicine under the National Key R&D Program [SQ2017YFSF090210]; China Postdoctoral Science Foundation [2019M652623, 2018M632830, 2021M693109]; Open Biodiversity and Health Big Data Program of IUBS; Professional Association of the Alliance of International Science Organizations [ANSO-PA-2020-07]; Funds for Basic Resources Investigation Research of the Ministry of Science and Technology [2018FY10080002]; Special Project on National Science and Technology Basic Resources Investigation [2019FY100102]; CAS Pioneer 100-Talent program; Key Research Program of the Chinese Academy of Sciences [KFZD-SW-219-5]; Zhang jiang special project of national innovation demonstration zone [ZJ2018-ZD-013]; Science and Technology Service Network Initiative of Chinese Academy of Sciences; Hunan Provincial Science and technology Program [2018wk4001], 111 Project [B18059], King Abdullah University of Science and Technology (KAUST) Office of Sponsored Research (OSR) [FCC/1/1976-18-01, FCC/1/1976-23-01, FCC/1/1976-25-01, FCC/1/1976-26-01, REI/1/0018-01-01, REI/1/4216-01-01, REI/1/4437-01-01, REI/1/4473-01-01, URF/1/4352-01-01, URF/1/4379-01-01, REI/1/4742-01-01, URF/1/4098-01-01]; Biological Resources Programme, Chinese Academy of Sciences [KFJ-BRP-017-79]; Specialized Research Assistant Program of the Chinese Academy of Sciences [202044]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J.et al.. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018; 562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The GTEx Consortium The Human Cell Atlas. Elife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen L., Fan R., Tang F.. Advanced Single-cell omics technologies and informatics tools for genomics, proteomics, and bioinformatics analysis. Genomics Proteomics Bioinformatics. 2021; 19:343–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tabula Sapiens Consortium Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P.et al.. The tabula sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022; 376:eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sinha S., Satpathy A.T., Zhou W., Ji H., Stratton J.A., Jaffer A., Bahlis N., Morrissy S., Biernaskie J.A.. Profiling chromatin accessibility at single-cell resolution. Genomics Proteomics Bioinformatics. 2021; 19:172–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balog J.A., Honti V., Kurucz E., Kari B., Puskas L.G., Ando I., Szebeni G.J.. Immunoprofiling of drosophila hemocytes by Single-cell mass cytometry. Genomics Proteomics Bioinformatics. 2021; 19:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng L., Qin S., Si W., Wang A., Xing B., Gao R., Ren X., Wang L., Wu X., Zhang J.et al.. Pan-cancer single-cell landscape of tumor-infiltrating t cells. Science. 2021; 374:abe6474. [DOI] [PubMed] [Google Scholar]
- 9. CNCB-NGDC Members and Partners Database resources of the national genomics data center, china national center for bioinformation in 2022. Nucleic Acids Res. 2022; 50:D27–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. CNCB-NGDC Members and Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021; 49:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. National Genomics Data Center Members and Partners Database Resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020; 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. BIG Data Center Members Database Resources of the BIG Data Center in 2019. Nucleic Acids Res. 2019; 47:D8–D14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. BIG Data Center Members Database Resources of the BIG Data Center in 2018. Nucleic Acids Res. 2018; 46:D14–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. BIG Data Center Members The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang S., Du Q., Feng C., Ma L., Zhang Z.. CompoDynamics: a comprehensive database for characterizing sequence composition dynamics. Nucleic Acids Res. 2022; 50:D962–D969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Kang H., Xu T., Hao L., Bao Y., Jia P.. CeDR Atlas: a knowledgebase of cellular drug response. Nucleic Acids Res. 2022; 50:D1164–D1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao J., Zhang Y., Tan S., Yang Q., Wang H., Xia X., Luo J., Guo H., Zhang Z., Li Z.. LSD4.0: an improved database for comparative studies of leaf senescence. Mol Horticulture. 2022; 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hua Z., Tian D., Jiang C., Song S., Chen Z., Zhao Y., Jin Y., Huang L., Zhang Z., Yuan Y.. Towards comprehensive integration and curation of chloroplast genomes. Plant Biotechnol. J. 2022; 20:2239–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao W.M., Song S.H., Chen M.L., Zou D., Ma L.N., Ma Y.K., Li R.J., Hao L.L., Li C.P., Tian D.M.et al.. The 2019 novel coronavirus resource. Yi Chuan = Hereditas /Zhongguo Yi Chuan Xue Hui Bian ji. 2020; 42:212–221. [DOI] [PubMed] [Google Scholar]
- 20. Song S., Ma L., Zou D., Tian D., Li C., Zhu J., Chen M., Wang A., Ma Y., Li M.et al.. The Global Landscape of SARS-CoV-2 Genomes, Variants, and Haplotypes in 2019nCoVR. Genomics Proteomics Bioinformatics. 2020; 18:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Y., Chen M., Bao Y., Song S., MPoxVR Team.. MPoxVR – a comprehensive genomic resource for monkeypox virus variants surveillance. Innovation. 2022; 3:100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arita M., Karsch-Mizrachi I., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021; 49:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. International Nucleotide Sequence Database Collaboration Leinonen R., Sugawara H., Shumway M.. The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2022; 50:D161–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y.et al.. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021; 19:578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., Xu X., Hao L., Li Z., Gong Z.et al.. Genome warehouse: a public repository housing Genome-scale data. Genomics Proteomics Bioinformatics. 2021; 19:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y., Yang J., Zhuang X., Ling Y., Cao R., Xu Q., Wang P., Xu P., Zhang G.. Linking genomic and epidemiologic information to advance the study of COVID-19. Sci Data. 2022; 9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu S., Huang Y., Zhang M., Gong Z., Wang G., Zheng X., Zong W., Zhao W., Xing P., Li R.et al.. ASCancer atlas: a comprehensive knowledgebase of alternative splicing in human cancers. Nucleic Acids Res. 2023; 10.1093/nar/gkac955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan S., Kang H., Liu X., Lin S., Yuan N., Zhang Z., Bao Y., Jia P.. Brain catalog: a comprehensive resource for the genetic landscape of brain-related traits. Nucleic Acids Res. 2023; 10.1093/nar/gkac895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng X., Zong W., Li Z., Ma Y., Sun Y., Xiong Z., Wu S., Yang F., Zhao W., Bu C.et al.. CCAS: one-stop and comprehensive annotation system for individual cancer genome at multi-omics level. Front. Genet. 2022; 13:956781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duan G., Wu G., Chen X., Tian D., Li Z., Sun Y., Du Z., Hao L., Song S., Gao Y.et al.. HGD: an integrated homologous gene database across multiple species. Nucleic Acids Res. 2023; 10.1093/nar/gkac970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Zhang H., Zhang Z., Qian Q., Zhang Z., Xiao J.. ProPan: a comprehensive database for profiling prokaryotic pan-genome dynamics. Nucleic Acids Res. 2023; 10.1093/nar/gkac832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu M., Zhang Y., Yang F., Mai J., Gao Q., Xu X., Kang H., Hou L., Shang Y., Qian Q.et al.. TWAS atlas: a curated knowledgebase of transcriptome-wide association studies. Nucleic Acids Res. 2023; 10.1093/nar/gkac821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q.et al.. GSA: genome sequence archive. Genomics Proteomics Bioinformatics. 2017; 15:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miao W., Song L., Ba S., Zhang L., Guan G., Zhang Z., Ning K.. Protist 10,000 genomes project. Innovation (Camb). 2020; 1:100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang B., Yang X., Jia Y., Xu Y., Jia P., Dang N., Wang S., Xu T., Zhao X., Gao S.et al.. High-quality Arabidopsis thaliana genome assembly with nanopore and hifi long reads. Genomics Proteomics Bioinformatics. 2021; 20:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song S., Tian D., Li C., Tang B., Dong L., Xiao J., Bao Y., Zhao W., He H., Zhang Z.. Genome variation map: a data repository of genome variations in BIG data center. Nucleic Acids Res. 2018; 46:D944–D949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li C., Tian D., Tang B., Liu X., Teng X., Zhao W., Zhang Z., Song S.. Genome variation map: a worldwide collection of genome variations across multiple species. Nucleic Acids Res. 2021; 49:D1186–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian D., Wang P., Tang B., Teng X., Li C., Liu X., Zou D., Song S., Zhang Z.. GWAS atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020; 48:D927–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X., Tian D., Li C., Tang B., Wang Z., Zhang R., Pan Y., Wang Y., You D., Zhang Z.et al.. GWAS atlas: an updated knowledgebase integrating more curated associations in plants and animals. Nucleic Acids Res. 2023; 10.1093/nar/gkac924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gong Z., Zhu J.W., Li C.P., Jiang S., Ma L.N., Tang B.X., Zou D., Chen M.L., Sun Y.B., Song S.H.et al.. An online coronavirus analysis platform from the national genomics data center. Zool Res. 2020; 41:705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan J., Zou D., Li C., Zhang Z., Song S., Wang X.. SR4R: an integrative SNP resource for genomic breeding and population research in rice. Genomics Proteomics Bioinformatics. 2020; 18:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Zou D., Zhu T., Xu T., Chen M., Niu G., Zong W., Pan R., Jing W., Sang J.et al.. Gene expression nebulas (GEN): a comprehensive data portal integrating transcriptomic profiles across multiple species at both bulk and single-cell levels. Nucleic Acids Res. 2022; 50:D1016–D1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sang J., Wang Z., Li M., Cao J., Niu G., Xia L., Zou D., Wang F., Xu X., Han X.et al.. ICG: a wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 2018; 46:D121–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiong Z., Yang F., Li M., Ma Y., Zhao W., Wang G., Li Z., Zheng X., Zou D., Zong W.et al.. EWAS open platform: integrated data, knowledge and toolkit for epigenome-wide association study. Nucleic Acids Res. 2022; 50:D1004–D1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiong Z., Li M., Yang F., Ma Y., Sang J., Li R., Li Z., Zhang Z., Bao Y.. EWAS data hub: a resource of DNA methylation array data and metadata. Nucleic Acids Res. 2020; 48:D890–D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li M., Zou D., Li Z., Gao R., Sang J., Zhang Y., Li R., Xia L., Zhang T., Niu G.et al.. EWAS atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019; 47:D983–D988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiong Z., Li M., Ma Y., Li R., Bao Y.. GMQN: a reference-based method for correcting batch effects and probe bias in humanmethylation beadchip. Front. Genet. 2021; 12:810985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li R., Liang F., Li M., Zou D., Sun S., Zhao Y., Zhao W., Bao Y., Xiao J., Zhang Z.. MethBank 3.0: a database of DNA methylomes across a variety of species. Nucleic Acids Res. 2018; 46:D288–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zou D., Sun S., Li R., Liu J., Zhang J., Zhang Z.. MethBank: a database integrating next-generation sequencing single-base-resolution DNA methylation programming data. Nucleic Acids Res. 2015; 43:D54–D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang M., Zong W., Zou D., Wang G., Zhao W., Yang F., Wu S., Zhang X., Guo X., Ma Y.et al.. MethBank 4.0: anupdated database of DNA methylation across a variety of species. Nucleic Acids Res. 2023; 10.1093/nar/gkac969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ma L., Cao J., Liu L., Du Q., Li Z., Zou D., Bajic V.B., Zhang Z.. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019; 47:D128–D134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Z., Liu L., Feng C., Qin Y., Xiao J., Zhang Z., Ma L.. LncBook 2.0: integrating human long non-coding RNAs with multi-omics annotations. Nucleic Acids Res. 2023; 10.1093/nar/gkac999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma L., Li A., Zou D., Xu X., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu L., Li Z., Liu C., Zou D., Li Q., Feng C., Jing W., Luo S., Zhang Z., Ma L.. LncRNAWiki 2.0: a knowledgebase of human long non-coding RNAs with enhanced curation model and database system. Nucleic Acids Res. 2022; 50:D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang S., Qian Q., Zhu T., Zong W., Shang Y., Jin T., Zhang Y., Chen M., Wu Z., Chu Y.et al.. Cell taxonomy: a curated repository of cell types with multifaceted characterization. Nucleic Acids Res. 2023; 10.1093/nar/gkac816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All resources and services are publicly available in the home page of CNCB-NGDC (https://ngdc.cncb.ac.cn).