Abstract
Metalloenzymes are attractive research targets in fields of chemistry, biology, and medicine. Given that metalloenzymes can manifest conservation of metal-coordination and ligand binding modes, the excavation and expansion of metalloenzyme-specific knowledge is of interest in bridging metalloenzyme-related fields. Building on our previous metalloenzyme-ligand association database, MeLAD, we have expanded the scope of metalloenzyme-specific knowledge and services, by forming a versatile platform, termed the Metalloenzyme Data Bank and Analysis (MeDBA). The MeDBA provides: (i) manual curation of metalloenzymes into different categories, that this M-I, M-II and M-III; (ii) comprehensive information on metalloenzyme activities, expression profiles, family and disease links; (iii) structural information on metalloenzymes, in particular metal binding modes; (iv) metalloenzyme substrates and bioactive molecules acting on metalloenzymes; (v) excavated metal-binding pharmacophores and (vi) analysis tools for structure/metal active site comparison and metalloenzyme profiling. The MeDBA is freely available at https://medba.ddtmlab.org.
INTRODUCTION
Metal ions are indispensable components of all living organisms. They play central roles in many biomedicinally important process, e.g. ferroptosis (1) and copper-induced cell death (2) amongst many others, and are functionally required adjuncts for a myriad of biological macromolecules. Metalloenzymes form an ubiquitous ensemble of enzymes that contain structurally and/or functionally important metal ions for bioactivity, structural, or signalling, or other reasons; more than one-third of all known enzymes are assigned as metalloenzymes. In a broader sense, nucleic acids with specific enzymatic functions (e.g. ribozymes and DNAzymes), which require one or more metal ions for activity, can also be categorized as metallo(en)zymes (3,4). Metalloenzymes have long been recognized as important in a variety of societally important and research fields. In synthetic biology, for example, functionally diverse artificial metalloenzymes have been developed, highlighting the potential to create new-to-nature biocatalytic transformations (5,6). Owing to their critical roles in various biological processes and diseases, including as recently reported in immunotherapy (7,8), metalloenzymes are important drug targets for therapeutic intervention. Although a number of new drugs or drug candidates for metalloenzymes have been established (9), most disease associations and therapeutic potentials for metalloenzymes remain largely unexploited, suggesting a vast space for drug discovery and development targeting metalloenzymes.
Metalloenzyme-specific knowledge, such as the metal ion(s) involved in catalysis, coordination chemistry, and ligand interactions, has had impact beyond biological catalysis. For instance, a deep understanding of how [FeFe]-hydrogenases interconvert protons, electrons and dihydrogen at their unique iron-based active site, has advanced the design of new catalytically active metalloenzymes and nucleic acids (4). Atomistic understanding of the metal-ligand interactions has assisted in the discovery of new metallodrugs and metalloenzyme inhibitors (10–12). Coupled with the fast-growing body of experimental data regarding metalloenzymes and ligands, the excavation and expansion of metalloenzyme-specific knowledge will thus be of great utility, not only in understanding and investigating metalloenzyme science (e.g. functions and ligand associations), but also in promoting and bridging metalloenzyme-related areas, including the design of non-protein metal using catalysts.
Previously, we established a metalloenzyme-ligand association database called MeLAD, with the aim of providing a general metalloenzyme-platform, in particular to assist metalloenzyme drug discovery (13). MeLAD provides centralized information exclusive to metalloenzyme-ligand associations, uniquely including excavated metal-binding pharmacophores (MBPs) and deduced metalloenzyme-ligand associations. Here, we reveal the integrated Metalloenzyme Data Bank and Analysis platform, termed MeDBA (https://medba.ddtmlab.org), which has a wider scope of metalloenzyme-specific knowledges and services. In MeDBA, metalloenzymes are divided into three general categories: M-I, M-II and M-III. M-I enzyme metal ions are directly involved in catalysis and are tightly bound at the active site, M-II enzyme metal ions are directly involved in catalysis, but are not tightly bound in active site, and M-III enzyme metal ions are important for enzyme activity, but are not present in active site. These metalloenzyme classifications will guide different objectives in metalloenzyme research, e.g. metal-binding drug discovery and de novo design of metalloenzymes. Comprehensive information on metalloenzymes, including names, catalytic reaction, and family, is provided by MeDBA. In particular, the overall profiles of metalloenzymes according to super-families and diseases are displayed, information which might help establish new research topics and open unexploited fields. Metalloenzyme structural information (e.g. metal sites and ligand interactions), together with substrates and bioactive molecules (e.g. PROTAC ligands, macrocycles, spiro compounds, drugs, and drug candidates) are comprehensively presented in an easy-to-use, cross-association manner. Importantly, MeDBA uniquely provides elaborately excavated MBPs for searching and servers for structure/metal active site comparisons and metalloenzyme profiling. New forms of webpage displays and interactive functions (e.g. structural comparisons, MBPs, and interaction display) are developed, which may be used as references for development of other database and web servers.
MATERIALS AND METHODS
Metalloenzyme identification
We began with data analysis of Protein Data Bank (PDB) structures (186,521 structures, available on 1 May 2022) using MeCOM, a tool for metal site detection and comparison (14). In MeCOM, the metal ions or metal-containing cofactors and corresponding chelating residues are defined as metal sites. Given a PDB structure, MeCOM detects an inferred metalloenzyme via the following steps: detecting all the metal sites in a structure; detecting whether any of the metal sites are adjacent to a potential active site pocket or a ligand within 6 Å; if yes, labelling the structure as ‘inferred metalloenzyme; recording the corresponding metal site contents including metal site composition, coordination mode, ligands, and pharmacophore files (for later ‘metal active site comparison’). To this end, 33 023 metal sites were extracted and 8790 proteins (with unique UniProt identifiers) were labelled as inferred metalloenzymes. Non-enzymatic proteins were then excluded according to UniProt information, leading to a list of 5836 inferred metalloenzymes with 25 160 corresponding metal sites.
We then manually curated all these inferred metalloenzymes and classified them into three general categories (M-I, M-II and M-III) by structure visualization and reference checking. Category M-I enzyme metal ions are directly involved in catalysis and are (apparently) tightly bound at the active site, M-II enzyme metal ions are directly involved in catalysis, but are not tightly bound at the active site, and M-III enzyme metal ions are important for enzyme activity (e.g. by stabilizing structural stability), but are not present at the active site. This process resulted in identification of 4722 metalloenzymes (3685 M-I metalloenzymes, 333 M-II metalloenzymes, and 704 M-III metalloenzymes) with corresponding 21 616 metal sites.
In addition to above described structure sources, we performed keyword searching in UniProt (15) and BRENDA (16) to identify potential metalloenzymes that were not successfully identified by MeCOM, or metalloenzymes which have no reported 3D structures, including uncharacterized metalloenzymes. In UniProt, keywords such as ‘metalloenzyme’ and ‘metallo*ase’, and metal binding information annotated with ‘catalytic’, were queried and filtered by organisms, which led to a list of 10,071 reviewed metalloenzymes and 54,301 unreviewed metalloenzymes. In BRENDA, the keywords (e.g. ‘require’, ‘depend’, ‘catalyze’, ‘metalloenzyme’ and ‘metalloprotease’) were queried specially for the enzymes with the ‘Metals/Ions’ records, which resulted in a list of 3,142 metalloenzymes. All these entries were manually reviewed and clustered in terms of catalytic metals, metal dependence, protein names, protein families and corresponding references. To this end, 46 927 metalloenzymes were retained.
According to all above-obtained metalloenzymes, we summarized a series of confidently assigned metalloenzyme families, such as metallo-β-lactamases, matrix metalloproteinases and 2-oxoglutarate-dependent oxygenases, which guided us to exploitation of metalloenzyme data from other free databases (15,17,18). Finally, we combined all the source data and removed duplicated entries (according to the UniProt identifier), then assigned metalloenzyme categories by referring to structural and family information. In total, 82,540 unique metalloenzymes were retained in MeDBA, consisting of 74 267 M-I metalloenzymes, 7338 M-II metalloenzymes and 935 M-III metalloenzymes.
Metalloenzyme information collection and process
To provide comprehensive information for above-obtained metalloenzymes, we first collected basic information, including protein name, gene name, organism, protein family, enzyme commission (EC) number, catalytic reaction, biofunction, and cofactor if present from UniProt. The families of metalloenzymes were collected from InterPro (19), which were used for the hierarchical family tree display. Metalloenzyme related disease data were collected from TTD (20) and ChEMBL (version 29) (21), and grouped by ICD-11 (22) and MeSH disease taxonomy, respectively. The distribution specificity data, such as tissue specificity, single cell type specificity, and brain specificity, were collected from ProteinAtlas (23). DNA/RNA binding metalloenzymes were identified by three sources: keywords (‘DNA-binding’ or ‘RNA-binding’) in UniProt, structural information from NPIDB (24), and RNA-binding protein annotations from RBP2GO (25). Intrinsically disordered metalloenzymes were annotated according to information from DisProt (26).
Metalloenzyme structure relevant information was then collected. Protein sequences were retrieved from UniProt (15), and protein domains were from InterPro (19). All experimental and predicted structures of metalloenzymes were collected from the PDB (27) and AlphaFoldDB (28), respectively. Notably, all metal site information, including metal site composition, coordination features, potential active pockets and nearby ligands, were generated using MeCOM (14); these metal sites were further grouped to metal site classes according to the metal ions and nature of the metal chelation.
Ligand identification and process
To sufficiently obtain metalloenzyme ligands, we considered different data sources. We first retrieved bioactive molecules for metalloenzymes from BindingDB (29) and ChEMBL (21). We then retrieved and curated drug/drug candidate information including mechanism of action, drug type, indication and maximum clinical phase, from TTD (20), ChEMBL (21) and recent references. Metalloenzyme-mediated reactions and corresponding substrates/products were collected from the Rhea database (30). Metalloenzyme-targeted PROTAC ligands were collected from PROTAC-DB (31). Notably, ligands from metalloprotein-ligand complex structures were collected separately. Finally, standard ‘InChI’ identifiers (32) were calculated for all the collected ligands, which guided removal of the duplicates and assignment of unique IDs (e.g. MeLig00000001) for all ligands. A total of 440 631 metalloenzyme-related ligands were included in MeDBA.
To provide more properties for browsing/searching ligands, we calculated the following properties: molecular weight, number of hydrogen bond acceptors, number of hydrogen bond donors, number of rotatable bonds, number of aromatic rings, topological polar surface area (TPSA), and ClogP by using RDKit tools. Notably, special ligands such as macrocycles (i.e. compounds containing a ring that consists of 12 or more atoms) and spiro compounds (i.e. containing at least one spiro atom) were analysed using RDKit. The SMILES string and MOL format files are available for all ligands.
Metalloenzyme-ligand association data collection and analysis
To better understand metalloenzyme-ligand associations, MeDBA provides 3D visualization of metalloenzymes complexed with ligands, proteins and DNAs/RNAs, and core metal binding pharmacophores (MBPs). The metalloenzyme–ligand complex structures were analysed by MeCOM (14) and related ligand information was extracted. The structures, UniProt ID and corresponding chains of metalloenzyme-binding proteins were collected from PDBe-KB (33). The metalloenzyme–DNA/RNA complexes were collected from NPIDB (24). Since MBPs are of great significance to drug discovery, we analysed all metalloenzyme-ligand complex structures in detail to obtain MBPs by the following steps: detecting the ligand metal-chelating atoms according to the rules of metal coordination as defined in AncPhore (34); labelling the metal-chelating atoms; (ii) splitting the ligand into fragments according to the bond cleavage rules as defined in LEADOPT (35); (iii) using the Floyd–Warshall algorithm (36) based shortest path algorithm to connect all the fragments if containing metal-chelating atoms, with the aim of defining an intact MBP for each metalloenzyme-ligand complex. Eventually, a total of 844 unique MBPs were obtained.
Metalloenzyme-specific analysis servers
To conveniently use the metalloenzyme data bank, we provided metalloenzyme-specific analysis servers. The 3D visualization of structure and interactions are provided using 3DMol (37), and the detailed views of protein-ligand interactions are supported by PLIP (38). The substructure search, conducted by OpenBabel (39), is designed for ligand and MBP searches. The structure and metal site comparison analyses employ MICAN (40) and MeCOM (14) for hind computations, respectively. Moreover, a ligand similarity server, implemented by OpenBabel (39), is provided for metalloenzyme profiling and for identification of new metalloenzyme-ligand associations.
Database implementation
MeDBA was developed using Python (version 3.9.12) and the Flask web framework (version 2.0.3), which runs on a Linux server (CentOS v7.04) with Nginx (version 1.20.1) and uWSGI (version 2.020) as the web servers, and the MySQL Community 8.0 as the database server. The user-friendly web interface is based on Bootstrap (version 4.61) and jQuery (version 3.60). Echarts (version 5.3) is exploited to provide intuitive statistical views of the data. Visualizations of the metalloenzyme-related 3D structures views and the chemical structures of ligands are supported by 3DMol (37) and RDKit, respectively. JSME is used to provide online molecular search service (41). API is used for data exchange, and asynchronous request and browser cache are utilized to increase data transmission efficiency and guarantee a better global service. The MeDBA website is HTTPS-enabled, which ensures that the data exchange between users and MeDBA servers is secured by an encrypted connection. MeDBA is freely accessible without registration and optimized for Google Chrome, Windows Edge, MacOS Safari, Opera and Firefox.
DATABASE CONTENT AND FEATURES
MeDBA overview
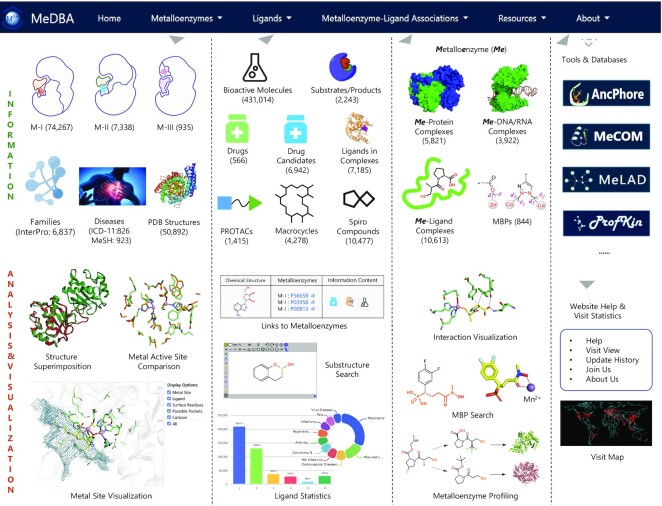
MeDBA aims to provide an integrated, distinctive, practicable, user-friendly platform for metalloenzyme-focused data and analysis. MeDBA contains three main sections: ‘Metalloenzymes’, ‘Ligands’ and ‘Metalloenzyme–Ligand Associations’ (Figure 1). The ‘Metalloenzymes’ section involves comprehensive metalloenzyme information, including metalloenzyme category, catalytic activity, expression, family, disease, structure, metal site, and related servers. The ‘Ligand’ section contains metalloenzyme substrates/products and bioactive molecules, in which special ligands (e.g. PROTAC ligands, macrocycles and spiro compounds), drugs and drug candidates are highlighted. The ‘Metalloenzyme–Ligand Associations’ section mainly involves detailed interactions of metalloenzyme–ligand, –protein and –DNA/RNA complexes, elaborately excavated MBPs, and related servers, facilitating to understand the association principles and derive new metalloenzyme-ligand associations. Related resources, help materials, and visit statistics are available. The current version of MeDBA contains data on 82 540 metalloenzymes (74 267 M-I metalloenzymes, 7338 M-II metalloenzymes and 935 M-III metalloenzymes), 440 631 ligands (431 014 bioactive molecules, 2243 substrates/products, 566 drugs, 6942 drug candidates and 1415 PROTACs), 10 613 metalloenzyme-ligand complexes, 21 616 metal sites and 844 MBPs.
Figure 1.
Overview of MeDBA resources. The numbers in parentheses refer to the number of entries in the current version.
Metalloenzymes
The Metalloenzyme menu gives direct access to the comprehensive metalloenzyme information and analysis modules. By using ‘Metalloenzyme Statistics’, users can quickly view basic information on metalloenzyme statistics (Supplementary Figure S1). On the ‘Metalloenzyme Search’ page, users can retrieve all metalloenzyme entries and corresponding information via nine kinds of annotations, including metalloenzyme name, gene name, metal ion, and metal site class. The retrieved results can be filtered by metalloenzyme category, enzyme class, and metal ion/metal-containing cofactor; they can be linked to the original data resources by clicking on the blue arrow or further accessed to the detail page for a specific metalloenzyme via UniProt ID (Supplementary Figure S2). The metalloenzyme detail page contains basic information (e.g. category, enzyme name, gene name, family, EC number, biofunction, cofactor, catalytic activity, expression, and disease), structural information (e.g. metalloenzyme sequence, InterPro domain, 3D structure, and metal site), and ligand information (e.g. chemical structure, source, association type, and association content) (Supplementary Figures S3 and S4).
In order to enable users to explore overall metalloenzyme families and their involvement in disease, metalloenzyme-family and metalloenzyme-disease associations are displayed in a hierarchical tree structure. On the ‘Metalloenzyme Families’ page, users can click or search a metalloenzyme family to obtain descriptions regarding the family and relevant metalloenzyme entries, along with corresponding categories and UniProt IDs. This tool is helpful for investigating biofunctions or catalytic mechanisms, including for uncharacterized metalloenzymes (Supplementary Figure S5). On the ‘Metalloenzyme Diseases’ page, metalloenzyme-related diseases are exhibited in ICD-11 or MeSH disease taxonomy, and can be filtered by keywords (Supplementary Figure S6). Users can obtain information regarding metalloenzymes related to a specific disease, a function that will be beneficial in drug discovery, in particular when targeting multiple metalloenzymes or in studies aimed at improving ligand selectivity.
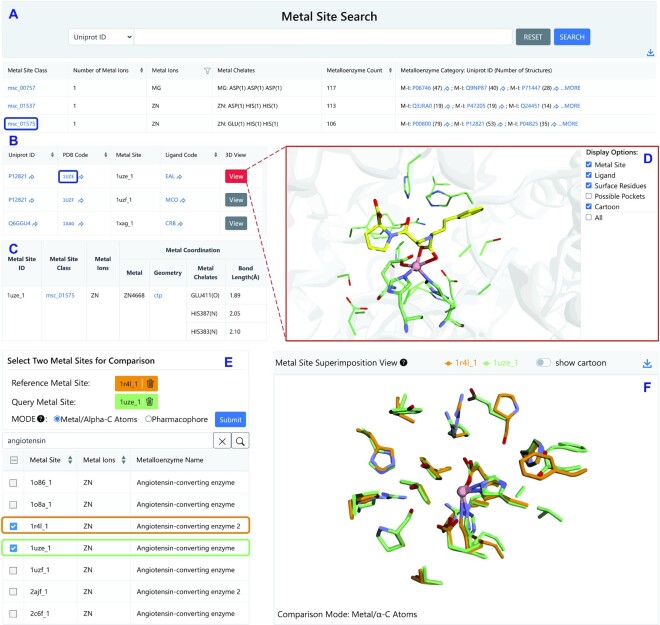
Given the unique characteristics of metalloenzymes, their structures are of special interest. As described above, the assigned metalloenzyme structures were analysed using MeCOM (14), and further manually curated into three categories: M-I, M-II and M-III. The metalloenzyme categories are useful to guide investigations on metal ion-involved interactions (e.g. ligand discovery). Currently, MeDBA contains 50 892 structures, covering 4205 M-I, 1342 M-II and 779 M-III metalloenzymes. The ‘Structure’ page supports searches by PDB code, metalloenzyme name, metal ion, metal-containing cofactor, and ligand code, and enables the user to obtain detailed structure information (e.g. 3D structure, Supplementary Figure S7) by clicking on the PDB code or details of metalloenzyme information by UniProt ID. All the metalloenzyme structures were analysed to generate separate metal binding sites, which were then grouped with the focus on metal ions and metal chelates. A total of 1738 metal site classes were obtained, which can be searched using the ‘Metal Site’ page (Figure 2A). Users can obtain a list of metalloenzymes for each metal site class, which groups the same or highly similar metal sites, and can access detailed structural information (Figure 2B–D). The metal site information will be useful both for metalloenzyme identification and de novo design.
Figure 2.
The metal site search, visualization, and comparison modules. (A) View of metal site classes, metal chelates, and relevant metalloenzymes in the ‘Metal Site’ page, which supports searching, internal and external linking. (B) View of related metalloenzymes when clicking the metal site class ‘msc_01575’. (C) View of detailed metal site information for PDB code 1UZE. (D) 3D visualization of angiotensin-converting enzyme's metal site. (E) Task panel of the ‘Metal Active Site Comparison’ module. (F) 3D visualization of the aligned metal sites for angiotensin-converting enzyme isoforms (reference metal site from PDB code 1R4L; query metal site from PDB code 1UZE).
To further exploit metalloenzyme structural information, we introduced two structural analysis modules (i.e. structure and metal active site comparison) in MeDBA. The ‘Structure Superimposition’ module enables users to compare metalloenzyme structures by superimpositions. By selecting two metalloenzyme structures and clicking the ‘Submit’ button, users can obtain calculated similarity scores and visualize the aligned metalloenzyme structures using the 3D graphical interface; users are also able to download the aligned structures for local analysis. By comparison, the ‘Metal Active Site Comparison’ module enables users to gain deeper insight into metal active site similarity by using MeCOM (14). Users can select two kinds of superimposition modes (i.e. ‘metal/alpha-C atoms’ and ‘pharmacophore’) for metal active site comparison (Figure 2E), and can obtain aligned metal active sites and the similarity score in a few seconds. Notably, this 3D graphical interface enables users to visualize the surface residues and binding pockets adjacent to the metal active site; moreover, the cartoon representations of the protein can be optionally switched off to focus on the aligned metal active sites (Figure 2F).
Special types of metalloenzymes, including DNA/RNA-binding and disordered metalloenzymes, are displayed individually in the Metalloenzyme section. The ‘DNA/RNA-binding metalloenzymes’ page contains the basic information (e.g. name, category, and UniProt ID), nucleic acid types (DNA, RNA, and Hybrid) and related experimental structures, and supports users to search, browse and download. The collected intrinsically disordered metalloenzymes are listed on the ‘Disordered Metalloenzymes’ page, where users can obtain basic information, including disordered sequences.
Ligands
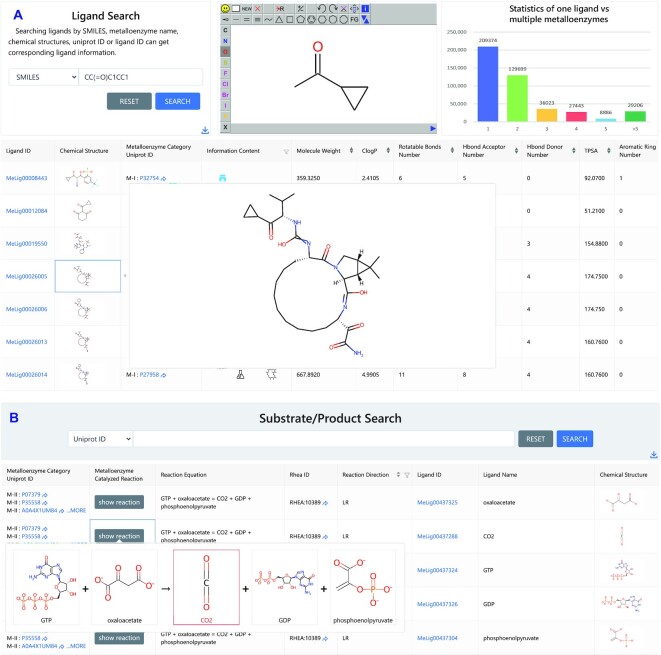
The Ligands section holds general information for bioactive molecules interacting with metalloenzymes and metalloenzyme substrates/products. The ‘Ligand Statistics’ page gives an overall view of ligand information (Supplementary Figures S8 and S9). Users can retrieve ligand information (e.g. chemical structures, information contents and basic properties) by querying a metalloenzyme name or UniProt ID on the ‘Ligand Search’ page. The Ligands section also supports chemical structure searches by inputting a SMILES or drawing a structure in the JSME applet window, an option which is valuable for users to quickly find ligands containing a given substructure or core scaffold (Figure 3A). The information contents (e.g. ‘Drug’ and ‘Macrocycle’) and ligand properties can be utilized as filter options to narrow search results. Through the ligand ID, users can access to the detailed ligand page, which gives basic properties, structure information and metalloenzyme association contents (Supplementary Figure S10).
Figure 3.
Ligand searching. (A) View of ligand structures, metalloenzymes, and the information content in the ‘Ligand Search’ module, exemplified by a substructure search with 1-cyclopropylethan-1-one (SMILES: CC(=O)C1CC1). (B) View of metalloenzyme-mediated reactions and related substrates and products, with an example of graphic showing the reaction equation for Rhea ID 10389.
On the ‘Bioactive Molecules’ page, users can obtain bioactivity data (‘IC50’, ‘EC50’, ‘Ki’ or ‘Kd’) and original references by searching a metalloenzyme name or UniProt ID (Supplementary Figure S11); the search results can be filtered by metalloenzyme category, assay type and activity source. The ‘Substrates/Products’ page contains metalloenzyme-mediated reactions and corresponding substrates/products, which users can search, view and download (Figure 3B). To improve the user experience, both graphical and textual reaction equations are provided. In addition, ligands of special interest including PROTACs, macrocycles, and spiro compounds, are provided in independent pages. For example, on the ‘PROTACs’ page, users can retrieve the PROTAC name, the relevant E3 ligase, bioassay data, source reference and related metalloenzymes by metalloenzyme name or UniProt ID. Identified macrocycles and spiro compounds are listed along with their chemical structures and bioactivity records, if available, on the ‘Macrocycles’ page and ‘Spiro Compounds’ page, respectively.
Data on drugs and drug candidates are important for metalloenzyme-targeted medicinal chemistry, and these are summarized on the ‘Approved Drugs’ and ‘Agents in Clinical Trials’ pages, respectively. Users can obtain information regarding drug name, chemical structure, metalloenzyme target category, metalloenzyme name, drug type, mechanism of action, clinical indication, and current clinical phase. The scope of drugs/drug candidates can be narrowed down by filtering metalloenzyme category, drug type and mechanism of action (Supplementary Figure S12). Notably, clicking a specific clinical indication will navigate to the corresponding ‘Metalloenzyme Diseases’ page for detailed information.
Metalloenzyme-ligand associations
This section of MeDBA involves structural bioinformation, MBP and metalloenzyme profiling modules, aiming to inform on the principles of metalloenzyme-ligand associations and to inspire or predict new metalloenzyme-ligand associations. Reported metalloenzyme-ligand complexes are listed on the ‘Metalloenzyme–Ligand Interactions’ page, which enables users to search by PDB code, metalloenzyme name, UniProt ID, ligand code and metal ions; interactions can be visualized in the graphical window by clicking the ‘3D view’ button (Supplementary Figure S13). The ‘Metalloenzyme–Protein Interactions’ and ‘Metalloenzyme–DNA/RNA Interactions’ pages are provided for users to analyse how metalloenzymes interact with proteins and nucleic acids, which may be useful for understanding roles of metalloenzymes in signal transduction beyond catalysis.
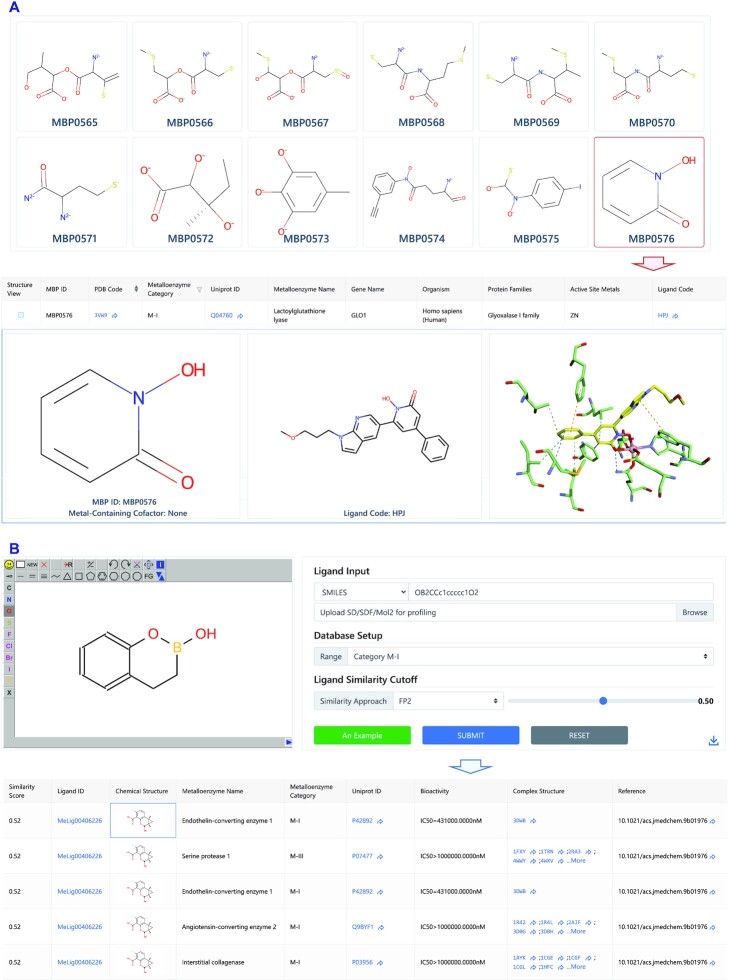
The ‘MBP Search’ module, in which a total of 844 excavated MBPs are displayed, not only enables links between different metalloenzymes to be made, but also provide clues to establish new metalloenzyme-ligand associations. Users can click a MBP chemical structure to obtain details of its metalloenzyme-ligand associations (ligand structure and interaction visualization, Figure 4A). Users can also search by substructure to identify related MBPs and metalloenzyme–ligand associations.
Figure 4.
Metal-binding pharmacophore (MBP) and metalloenzyme profiling modules. (A) View of MBPs listed in ‘MPB Search’ module and corresponding detailed information through clicking on the MBP0576 chemical structure. (B) View of task panel of the ‘Profiling by Ligand Similarity’ module, with an example of metalloenzyme profiling for the queried compound ‘3,4-dihydro-2H-benzo[e][1,2]oxaborinin-2-ol’, resulting in top-ranked similar ligands and their metalloenzyme associations.
The ‘Profiling by Ligand Similarity’ module provides a web server to predict potential metalloenzyme-ligand associations on the basis of ligand chemical similarity. Users can query a small molecule of interest by drawing its chemical structure, inputting SMILES or uploading a mol2/sdf format file, then setting up profiling parameters including database range (metalloenzyme category subsets or the entire database), similarity approach (‘FP2’, ‘FP3’, ‘FP4’ or ‘MACCS’), and similarity cutoff (Figure 4B). After submitting the job, users can obtain results within a few seconds, then can visualize ranked similar ligands and corresponding metalloenzymes (e.g. metalloenzyme categories, bioactivity records, complex structures, and references) (Figure 4B). Notably, the module is an efficient and convenient way to compare binding modes and MBP chemotypes.
SUMMARY AND FUTURE DIRECTIONS
MeDBA comprehensively integrates information regarding metalloenzymes, their ligands, metalloenzyme-ligand associations with metalloenzyme-specific computational analysis tools. It is the first database to provide general metalloenzyme categories based on approximate metal ion coordination strength and whether or not the metal ions are involved in catalysis. Importantly, MeDBA not only provides metalloenzyme bioactive molecules and substrates, but also provides experimentally determined metalloenzyme–ligand, –protein and –DNA/RNA complex structures as well as elaborately excavated metal-binding pharmacophores, information which will be important basis for deriving new metalloenzyme-related associations. The powerful yet simple to use analysis tools of MeDBA facilitate the retrieval, browsing, and querying of metalloenzymes and associated information, promoting information use and communication. Despite these features, we think there is still space for improvement and development of MeDBA, e.g. by expansion of MBPs, inclusion of metallo-ribozymes and DNAzymes, and improved analysis of metalloenzyme profiling. We intend MeDBA will serve as a valuable platform, coupled with advances in interface of chemistry, biology, and medicine, to foster research in the multifaceted field of metalloenzymes.
DATA AVAILABILITY
MeDBA, help materials and related tools/resources are freely available at https://medba.ddtmlab.org.
Supplementary Material
Contributor Information
Jun-Lin Yu, Key Laboratory of Drug Targeting and Drug Delivery System of Ministry of Education, Department of Medicinal Chemistry, West China School of Pharmacy, Sichuan University, Chengdu, Sichuan 610041, China.
Song Wu, National Clinical Research Center for Geriatrics and Department of Periodical Press, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, China.
Cong Zhou, Key Laboratory of Drug Targeting and Drug Delivery System of Ministry of Education, Department of Medicinal Chemistry, West China School of Pharmacy, Sichuan University, Chengdu, Sichuan 610041, China.
Qing-Qing Dai, Key Laboratory of Drug Targeting and Drug Delivery System of Ministry of Education, Department of Medicinal Chemistry, West China School of Pharmacy, Sichuan University, Chengdu, Sichuan 610041, China.
Christopher J Schofield, Department of Chemistry and the Ineos Oxford Institute for Antimicrobial Research, Chemistry Research Laboratory, Mansfield Road, University of Oxford, Oxford OX1 3TA, UK.
Guo-Bo Li, Key Laboratory of Drug Targeting and Drug Delivery System of Ministry of Education, Department of Medicinal Chemistry, West China School of Pharmacy, Sichuan University, Chengdu, Sichuan 610041, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [82122065, 82073698, 81874291 to G.B.L.], Sichuan Science and Technology Program [2022YFH0027 to G.B.L.]; 111 Project [B18035]; C.J.S. thank Cancer Research UK and the Biotechnological and Biological Research Council for support. Funding for open access charge: National Natural Science Foundation of China [82122065].
Conflict of interest statement. No conflicts are declared.
REFERENCES
- 1. Jiang X., Stockwell B.R., Conrad M.. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021; 22:266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kahlson M.A., Dixon S.J.. Copper-induced cell death. Science. 2022; 375:1231–1232. [DOI] [PubMed] [Google Scholar]
- 3. Sigel R.K.O., Pyle A.M.. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007; 107:97–113. [DOI] [PubMed] [Google Scholar]
- 4. Hemschemeier A., Happe T.. The plasticity of redox cofactors: from metalloenzymes to redox-active DNA. Nat. Rev. Chem. 2018; 2:231–243. [Google Scholar]
- 5. Wittwer M., Markel U., Schiffels J., Okuda J., Sauer D.F., Schwaneberg U.. Engineering and emerging applications of artificial metalloenzymes with whole cells. Nat. Catal. 2021; 4:814–827. [Google Scholar]
- 6. Vornholt T., Christoffel F., Pellizzoni M.M., Panke S., Ward T.R., Jeschek M.. Systematic engineering of artificial metalloenzymes for new-to-nature reactions. Sci. Adv. 2021; 7:eabe4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang T., Yin C., Fedorov A., Qiao L., Bao H., Beknazarov N., Wang S., Gautam A., Williams R.M., Crawford J.C.et al.. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature. 2022; 606:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barreira da Silva R., Leitao R.M., Pechuan-Jorge X., Werneke S., Oeh J., Javinal V., Wang Y., Phung W., Everett C., Nonomiya J.et al.. Loss of the intracellular enzyme QPCTL limits chemokine function and reshapes myeloid infiltration to augment tumor immunity. Nat. Immun. 2022; 23:568–580. [DOI] [PubMed] [Google Scholar]
- 9. Chen A.Y., Adamek R.N., Dick B.L., Credille C.V., Morrison C.N., Cohen S.M.. Targeting metalloenzymes for therapeutic intervention. Chem. Rev. 2019; 119:1323–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riccardi L., Genna V., De Vivo M.. Metal–ligand interactions in drug design. Nat. Rev. Chem. 2018; 2:100–112. [Google Scholar]
- 11. Yan Y.-H., Li G., Li G.-B.. Principles and current strategies targeting metallo-β-lactamase mediated antibacterial resistance. Med. Res. Rev. 2020; 40:1558–1592. [DOI] [PubMed] [Google Scholar]
- 12. Xiao Y.-C., Yu J.-L., Dai Q.-Q., Li G., Li G.-B.. Targeting metalloenzymes by boron-containing metal-binding pharmacophores. J. Med. Chem. 2021; 64:17706–17727. [DOI] [PubMed] [Google Scholar]
- 13. Li G., Su Y., Yan Y.-H., Peng J.-Y., Dai Q.-Q., Ning X.-L., Zhu C.-L., Fu C., McDonough M.A., Schofield C.J.et al.. MeLAD: an integrated resource for metalloenzyme-ligand associations. Bioinformatics. 2020; 36:904–909. [DOI] [PubMed] [Google Scholar]
- 14. Li G., Dai Q.-Q., Li G.-B.. MeCOM: a method for comparing three-dimensional metalloenzyme active sites. J. Chem. Inf. Model. 2022; 62:730–739. [DOI] [PubMed] [Google Scholar]
- 15. Consortium T.U. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang A., Jeske L., Ulbrich S., Hofmann J., Koblitz J., Schomburg I., Neumann-Schaal M., Jahn D., Schomburg D.. BRENDA, the ELIXIR core data resource in 2021: new developments and updates. Nucleic Acids Res. 2020; 49:D498–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naas T., Oueslati S., Bonnin R.A., Dabos M.L., Zavala A., Dortet L., Retailleau P., Iorga B.I.. Beta-lactamase database (BLDB) – structure and function. J. Enzy. Inh. Med. Chem. 2017; 32:917–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen Z., Yan Y.-H., Yang S., Zhu S., Yuan Y., Qiu Z., Jia H., Wang R., Li G.-B., Li H.. ProfKin: a comprehensive web server for structure-based kinase profiling. Eur. J. Med. Chem. 2021; 225:113772. [DOI] [PubMed] [Google Scholar]
- 19. Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S.et al.. The interpro protein families and domains database: 20 years on. Nucleic Acids Res. 2020; 49:D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F., Qiu Y., Chen Y.. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2021; 50:D1398–D1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños MaríaP., Mosquera JuanF., Mutowo P., Nowotka Met al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2018; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The L. icd-11. Lancet. 2019; 393:2275. [DOI] [PubMed] [Google Scholar]
- 23. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A.et al.. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 24. Zanegina O., Kirsanov D., Baulin E., Karyagina A., Alexeevski A., Spirin S.. An updated version of NPIDB includes new classifications of DNA–protein complexes and their families. Nucleic Acids Res. 2015; 44:D144–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caudron-Herger M., Jansen R.E., Wassmer E., Diederichs S.. RBP2GO: a comprehensive pan-species database on RNA-binding proteins, their interactions and functions. Nucleic Acids Res. 2020; 49:D425–D436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quaglia F., Mészáros B., Salladini E., Hatos A., Pancsa R., Chemes L.B., Pajkos M., Lazar T., Peña-Díaz S., Santos J.et al.. DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Res. 2021; 50:D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M.et al.. RCSB protein data bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2020; 49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A.et al.. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J.. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2015; 44:D1045–D1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bansal P., Morgat A., Axelsen K.B., Muthukrishnan V., Coudert E., Aimo L., Hyka-Nouspikel N., Gasteiger E., Kerhornou A., Neto T.B.et al.. Rhea, the reaction knowledgebase in 2022. Nucleic Acids Res. 2021; 50:D693–D700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weng G., Shen C., Cao D., Gao J., Dong X., He Q., Yang B., Li D., Wu J., Hou T.. PROTAC-DB: an online database of PROTACs. Nucleic Acids Res. 2020; 49:D1381–D1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heller S.R., McNaught A., Pletnev I., Stein S., Tchekhovskoi D. InChI, the IUPAC international chemical identifier. J. Cheminform. 2015; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. consortium P.-K. PDBe-KB: a community-driven resource for structural and functional annotations. Nucleic Acids Res. 2019; 48:D344–D353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dai Q., Yan Y., Ning X., Li G., Yu J., Deng J., Yang L., Li G.-B.. AncPhore: a versatile tool for anchor pharmacophore steered drug discovery with applications in discovery of new inhibitors targeting metallo-β-lactamases and indoleamine/tryptophan 2,3-dioxygenases. Acta Pharm. Sin. B. 2021; 11:1931–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li G.-B., Ji S., Yang L.-L., Zhang R.-J., Chen K., Zhong L., Ma S., Yang S.-Y.. LEADOPT: an automatic tool for structure-based lead optimization, and its application in structural optimizations of VEGFR2 and SYK inhibitors. Eur. J. Med. Chem. 2015; 93:523–538. [DOI] [PubMed] [Google Scholar]
- 36. Floyd R.W. Algorithm 97: Shortest path. Commun. ACM. 1962; 5:345. [Google Scholar]
- 37. Rego N., Koes D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2014; 31:1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adasme M.F., Linnemann K.L., Bolz S.N., Kaiser F., Salentin S., Haupt VJ., Schroeder M.. PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021; 49:W530–W534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R.. Open babel: an open chemical toolbox. J. Cheminform. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minami S., Sawada K., Chikenji G.. MICAN: a protein structure alignment algorithm that can handle Multiple-chains, inverse alignments, C(α) only models, alternative alignments, and non-sequential alignments. BMC Bioinform. 2013; 14:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bienfait B., Ertl P.. JSME: a free molecule editor in javascript. J. Cheminform. 2013; 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MeDBA, help materials and related tools/resources are freely available at https://medba.ddtmlab.org.