Abstract
SulfAtlas (https://sulfatlas.sb-roscoff.fr/) is a knowledge-based resource dedicated to a sequence-based classification of sulfatases. Currently four sulfatase families exist (S1–S4) and the largest family (S1, formylglycine-dependent sulfatases) is divided into subfamilies by a phylogenetic approach, each subfamily corresponding to either a single characterized specificity (or few specificities in some cases) or to unknown substrates. Sequences are linked to their biochemical and structural information according to an expert scrutiny of the available literature. Database browsing was initially made possible both through a keyword search engine and a specific sequence similarity (BLAST) server. In this article, we will briefly summarize the experimental progresses in the sulfatase field in the last 6 years. To improve and speed up the (sub)family assignment of sulfatases in (meta)genomic data, we have developed a new, freely-accessible search engine using Hidden Markov model (HMM) for each (sub)family. This new tool (SulfAtlas HMM) is also a key part of the internal pipeline used to regularly update the database. SulfAtlas resource has indeed significantly grown since its creation in 2016, from 4550 sequences to 162 430 sequences in August 2022.
INTRODUCTION
Sulfation is a crucial modification found in nearly all prokaryotic and eukaryotic organisms which drastically changes the physicochemical and biological properties of compounds. Sulfated biomolecules are highly diverse in structure and function and they include small compounds (e.g. steroid in humans and other vertebrates, secondary metabolites in plants), proteins (i.e. tyrosine sulfation), lipids (e.g. sphingolipids), polyphenols (e.g. sulfated polyphenols in red and brown algae) and carbohydrates (e.g. glycosaminoglycans and mucins in animals, sulfated polysaccharides in marine algae and seagrasses, sulfated exopolysaccharides in some bacteria) (1). Sulfatases are key enzymes in sulfate metabolism, catalyzing the removal of sulfate groups according to hydrolytic or oxidative mechanisms (sulfuric ester hydrolases EC 3.1.6.-, sulfamidases EC 3.10.1.- and dioxygenase EC 1.14.11.-) (2,3). Before 2016, the vast majority of characterized sulfatases were studied in human and animals in the context of severe metabolic disorders (2). A few sulfatases were also characterized in bacteria with diverse substrate specificities (1). However, this limited number of characterized sulfatases was far from reflecting the huge diversity of sulfated biomolecules. With the explosion of genomic data, the gap between new sequences and characterized enzymes was increasing and annotation of sulfatase sequences was prone to errors. Inspired by expert-curated databases such as the Carbohydrate-Active enZymes database (CAZY, http://www.cazy.org/) (4), we thus decided in 2016 to create the SulfAtlas database. SulfAtlas proposes a classification system of sulfatases based on sequence homology, allowing a better prediction of substrate specificity (1).
Currently sulfatases are divided into four protein families based on sequence homology: formylglycine-dependent sulfatases (S1 family) (2); alkylsulfodioxygenases (S2 family), represented by the alkylsufatase AtsK from Pseudomonas putida S-313 (3); the alkylsulfohydrolases (S3 family), represented by the alkylsulfatase SdsA1 from Pseudomonas aeruginosa PAO1 (5); and the arylsulfohydrolases (S4 family), represented by the arylsulfatase AtsA from Pseudoalteromonas carrageenovora 9T (6). These four types of sulfatases also strongly differ in their catalytic mechanisms. The S1 sulfatases contain a peculiar catalytic residue, the Cα-formylglycine (FGly) which is post-translationally generated from a conserved cysteine or serine (7,8). The S2 sulfatases are also fairly unusual enzymes, using an oxidative mechanism involving iron and alpha-ketoglutarate cofactors to desulfate akyl substrates (3). Despite their extreme sequence divergence (<15% identity), the S3 and S4 sulfatases adopt the same fold and belong to the metallo-β-lactamase superfamily with a conserved cation-binding catalytic machinery (9). Extensive details on the folds and catalytic machineries of the different types of sufatases have been reviewed in the SulfAtlas founding article (1).
The S1 family encompasses the vast majority of sulfatases and is the most diverse in term of substrate specificities. For this reason, we have divided the S1 family into subfamilies, based on phylogenetic analyses. In 2016, we had thus defined 73 S1 subfamilies, each subfamily corresponding to either a known specificity or to uncharacterized substrates (1). An additional S1 subfamily named S1_NC (for non-classified) contains S1 sequences which cannot be reliably assigned to a phylogenetic subfamily. These sequences are thus considered orphans until close homologues are found.
In SulfAtlas, each family or subfamily page provides (sub)family descriptors (known enzymatic activities, catalytic residues and available 3D structures) and a table with all the UniProt accession numbers of sulfatases belonging to this (sub)family with the protein or locus name, the organism taxon and the EC and PDB numbers when they exist. All these fields are linked to corresponding databases (UniProt (10), ExplorEnz (11), the NCBI Taxonomy database (12) and the Protein Data Bank (13)). Different tools are provided to explore SulfAtlas: a search engine using keywords and a sequence similarity BLAST-based server (BLASTP and BLASTX, (14)) to query single or multiple sequences. Selected sulfatase sequences can be exported in FASTA format (15). (Sub)family tables can be also downloaded as excel, CSV or PDF format. SulfAtlas content is regularly updated, both in terms of sequences and of experimental knowledge based on day-to-day inspection of new sulfatase characterizations in the literature. In the present article we outline the evolution of the field and the changes implemented in SulfAtlas since 2016. Notably we have developed new tools: (i) a search engine using Hidden Markov model (HMM) for each (sub)family allowing the query of single sequences or complete proteomes and (ii) an internal pipeline for the semi-automatic update of sequence content in order to cope with the ever-increasing flow of genomic sequences.
State of the art of characterized sulfatases
In 2016, SulfAtlas contained 4,550 sequences (version 1.0, July 2013 dataset; S1: 4,061; S2: 104; S3: 370; S4: 15, Table 1). Forty nine characterized sulfatases were initially known, although they only represented 16 distinct substrate specificities (1). Only one sulfatase activity was known in the families S2 (alkylsulfatase), S3 (alkylsulfatase) and S4 (arylsulfatase) and all the other characterized enzymes were found in the S1 family (from S1_1 to S1_12 and in S1_19). All of these S1 subfamilies were monospecific at the time of the SulfAtlas creation, with the exception of the subfamilies S1_7 [iduronate 2-sulfatase (16), endo-4S-kappa-carrageenan sulfatase (17,18)], S1_11 [Mucin-desulfating sulfatase (19), heparan sulfate 6-O-sulfatase (20)] and S1_19 [endo-4S-iota-carrageenan sulfatase and endo-4S-kappa-carrageenan sulfatase (18,21)]. Although several substrate specificities are found in the S1_11 and S1_19 subfamilies, to this date, the regioselectivities are identical within each subfamily (d-glucose-6-sulfatase and d-galactose-4-sulfatase, respectively).
Table 1.
Growth of the SulfAtlas database in the past 9 years. The SulfAtlas database was created in December 2016 (1) based on sequences available in July 2013
| Subfamilies | Sequences | Characterized sequences | Unique activities | Sequences with structures | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Family | 2013 | 2022 | 2013 | 2022 | 2013 | 2022 | 2013 | 2022 | 2013 | 2022 |
| S1 | 73 | 110 | 4,061 | 148,634 | 43 | 77 | 13 | 32 | 8 | 40 |
| S2 | - | - | 105 | 4,280 | 2 | 2 | 1 | 1 | 2 | 6 |
| S3 | - | - | 370 | 9,155 | 3 | 3 | 1 | 1 | 3 | 4 |
| S4 | - | - | 15 | 271 | 1 | 1 | 1 | 1 | 0 | 0 |
In the last six years, 19 new sulfatase activities have been discovered (Supplementary Table S1), indicating an increase of the discovery rate. Interestingly, almost all these studies have adopted the SulfAtlas classification and use it as a guide to select subfamilies with unknown substrate specificity. Without exception, the new sulfatase activities have been S1 family sulfatases originating from bacteria and specific for complex carbohydrates (polysaccharides or proteoglycans). This is a significant change in trend, since most sulfatases were previously studied in human and animals (1,2). Moreover, all the studied microorganisms were isolated from either human gut microbiota or marine environments. These trend changes are clearly related to the explosion of microbial genomic data in these two types of habitats rich in sulfated carbohydrate sources. The acceleration in sulfatase characterization is also mainly due to several remarkable large studies which have analyzed entire Polysaccharide Utilization Loci (PUL) (22) or catabolic pathways. Notably, new sulfatases have been discovered in the catabolism of heparan sulfate/heparin (23), dermatan sulfate (24), colonic mucins (25,26), red algal carrageenans (18,27) and agars (28) and green algal ulvans (29). As initially predicted (1), most new sulfatase activities have been found in previously uncharacterized S1 subfamilies [Supplementary Table S1: S1_13, S1_15, S1_16, S1_17, S1_20, S1_22, S1_25, S1_27, S1_46 and S1_81 (a new S1 subfamily, see below)]. A few new activities have been also identified in subfamilies already containing characterized sulfatases: [Colonic mucin] endo-d-galactose-3-sulfate 3-sulfatase (S1_4) and [Colonic mucin] d-galactose-6-sulfate 6-sulfatase (S1_4) (25); [Ulvan] endo-xylose-2-sulfate 2-sulfatase (S1_7) and [Ulvan] exo-xylose-2-sulfate 2-sulfatase (S1_8) (29); [Porphyran] exo-l-galactose-6-sulfate 6-O-sulfatase (S1_11) (28). This phenomenon is also observed in several newly characterized S1 subfamilies (S1_15, S1_17, S1_25 and S1_27, Supplementary Table S1). As previously observed in the subfamilies S1_11 and S1_19 (1), in most of these polyspecific subfamilies there is a common regioselectivity or at least a similar spatial position of the sulfate group which is removed in different substrates: (S1_8) [heparin] 2-N-sulfaminidase (30) and [Ulvan] xylose-2-O-sulfatases (29); (S1_11) [heparin, chondroitin or mucins] N-acetylglucosamine 6-O-sulfatases (31) and [porphyran] l-galactose-6-sulfate 6-O-sulfatases (28); (S1_17) [carrageenan] endo-3,6-anhydro-d-galactose-2-sulfate 2-O-sulfatase (18) and [sulfated fucans] exo-Fucose-2-sulfate 2-O-sulfatase (32); (S1_25) [sulfated fucans] exo-fucose-3-sulfate 3-O-sulfatase (33) or [ulvan] exo-l-rhamnose-3-sulfate 3-O-sulfatase (29).
Interestingly, in the subfamilies S1_4 and S1_7, sequences with fairly different substrate specificities tend to cluster in the same phylogenetic clades. It is more difficult to find a rationale to explain why. However, these particular S1 subfamilies are very large (in August 2022, version 2.3.1: S1_4: 16,944 sequences; S1_7: 8642 sequences) and accumulation of experimental data may support in the future the division of these two huge S1 subfamilies into several smaller, more cohesive subfamilies.
Database growth and creation of new S1 subfamilies
A common challenge to all sequence databases is to keep the content up-to-date. In our initial article (1), SulfAtlas contained the sulfatase sequences identified in UniProt (10) in July 2013 (version 1.0). In the first phase, we had updated the sequence content through a time-consuming, manual approach. New sulfatase sequences were searched in the subsequent releases of UniProt (10) using the SulfAtlas BLAST server. Attribution of a new sequence to an existing (sub)family was based on the conservation of the PROSITE signatures (34) previously defined (1) and on a conservative threshold of identity with version 1.0 sequences (at least 40% over a minimal length compatible with the size of characterized S1 sulfatases (∼400 residues). In February 2019, we thus released a SulfAtlas version 1.1 containing 31 327 sulfatase sequences (∼7 fold the initial size of SulfAtlas).
In version 1.1, the S1 subfamilies were still limited to the first described 73 subfamilies. However, unclassified sequences were accumulating in the ‘storage’ S1_NC subfamily, suggesting that version 1.0 orphan sequences could have found close homologues in subsequent UniProt releases (10). The need to create potential new S1 subfamilies was thus becoming obvious. Due to the huge number of S1 family sequences in the SulfAtlas version 1.1, it was impossible to directly compute a reliable, global phylogenetic tree to determine whether S1_NC sequences could constitute new S1 subfamilies. Instead, we built an internal ‘core’ SulfAtlas dataset limited to 1000 sequences (named Seed_S1_SulfAtlas) and composed of representative sequences of the first 73 S1 subfamilies. To conserve a high sequence diversity, we selected one sequence per eukaryotic or prokaryotic phylum for each S1 subfamily. The Seed_S1_SulfAtlas sequences were aligned with MAFFT (35), with the iterative refinement method L-INS-i and the scoring matrix Blosum62. Each S1_NC sequence and its closest homologues were similarly aligned with MAFFT. Each S1_NC multiple alignment was added to the Seed_S1_SulfAtlas multiple alignment using the Merge option of the MAFFT online version (35). Each resulting multiple alignment was visualized and manually improved using Jalview (36). Phylogenetic trees were derived from these refined alignments with RAxML (37) using Maximum Likelihood method and the substitution model identified by the IQ-TREE web server (38). The reliability of the trees was systematically tested by bootstrap analysis using 100 resamplings of the dataset. The trees were displayed with MEGA 6.06 (39) and an example tree is shown in Supplementary Figure S1. In these trees, the initial 73 subfamilies were well conserved and generally supported by high bootstrap values, validating this approach. In almost all cases, each S1_NC sequence and its closest homologues were indeed forming a new clade. These analyses allowed us to set the limit of these new clades and to keep the remaining orphan sequences in the S1_NC subfamily. Altogether this created 37 new S1 subfamilies (S1_74 – S1_110) and lead to SulfAtlas updates in August 2021 (version 1.2: 35 857 sequences) and in March 2022 (version 1.3: 35 566 sequences). Interestingly, a new sulfatase activity has already been discovered in one of these new S1 subfamilies: [iota-carrageenan] exo-3,6-anhydro-d-galactose-2-sulfate 2-O-sulfatase in the S1_81 subfamily (27).
Considering the ever-increasing number of sequences released in UniProt (10), full manual updating of SulfAtlas is a time-consuming process which is difficult to sustain in the long-term. To cope with this challenge, we have developed another strategy for a semi-automatic update of SulfAtlas. This strategy is detailed in the following sections.
Development of SulfAtlas HHM, an HHM server for SulfAtlas
As an alternative strategy to detect new sulfatase sequences and assign them to their correct (sub)family, we have decided to develop Hidden Markov Models (HMM) specific for each SulfAtlas (sub)family. Beyond our internal need for updating SulfAtlas (see below), it was also an answer to regular external solicitations to analyze complete genomes or even large metagenomes. Indeed, the SulfAtlas BLAST server was well adapted for analyzing a limited number of sequences, but was inadequate for processing large datasets.
In order to detect sulfatase modules, we used as a starting point domain definitions available in the Superfamily database (40). We chose this classification because its HMM library is based on structural protein domains and we were confident to correctly cover the length of the different sulfatase modules. We analyzed the sequences from SulfAtlas (version 1.3) with the Superfamily HHM library using hmmscan (default parameters) from the HMMER package (41) and assigned one superfamily domain for each SulfAtlas family: S1 family = Alkaline phosphatase-like superfamily (NAME = 0042147); S2 family = Clavaminate synthase-like superfamily (NAME = 0041871); S3 family = Metallo-hydrolase/oxidoreductase superfamily (NAME = 0053773); S4 family = Metallo-hydrolase/oxidoreductase (NAME = 0051194). Based on these results, we determined the limits of each sulfatase module within the SulfAtlas full-length sequences. For each (sub)family, sulfatase module sequences were aligned using MAFFT (35) with the globalpair option and the maximum iteration set to 1000. These multiple alignments were manually checked for ensuring their consistency. Finally, we created an HHM for each multiple alignment using the HMMER package (41).
The next step was the definition of rules to reliably assign a sequence to its correct SulfAtlas (sub)family. For this, we have compared all the full-length sequences from SulfAtlas (version 1.3) to the generated HHM library using hmmscan (41) and manually determined in which conditions sequences were correctly assigned. Based on the results, we have defined the following rules: (i) the assignment is reliable when the score is superior to 300 and the coverage is superior to 80% of the HMM length; (ii) when the score is between 200 and 300 (with a coverage > 80%), the sulfatase sequence cannot be reliably automatically assigned to an existing (sub)family and additional manual analyses are needed to determine its status; (iii) a score between 100 and 300 and a coverage inferior to 80% is generally a sulfatase fragment coded by a pseudo-gene or due to an incorrect prediction of an open reading frame (ORF) in a genome.
To benchmark our SulfAtlas HMM library and our defined thresholds, we have created a test library containing our HMMs and all the HMMs from PFAM (version 33.1) (42) and compared it to the sequences from SulfAtlas (version 1.3). Only 10 sequences were not assigned to their correct (sub)family. After manual verification, we found these 10 sequences were in fact sulfatase fragments and their score was just below the threshold. Five other sequences had a better hit with a PFAM HMM (42) than with a SulfAtlas HMM but these sequences were still correctly assigned to their corresponding S1 subfamily.
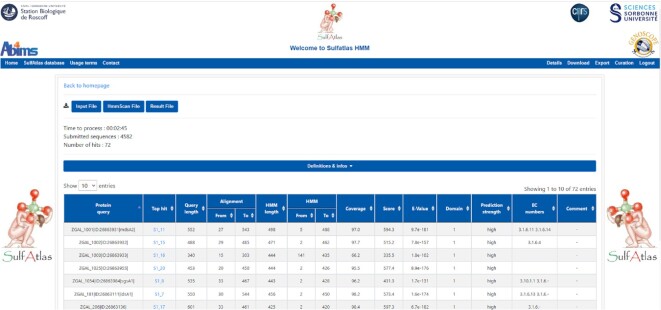
In conclusion, our SulfAtlas HMM library and its associated rules are reliable and have sufficient precision to assign sequences not only at the protein family level but also directly at the subfamily level in the case of the S1 family. On these bases, we have added a new interface to the SulfAtlas database (named ‘SulfAtlas HMM’) to query the SulfAtlas HMM library online using hmmscan (HMMER) on a SLURM-based cluster computing infrastructure (Figure 1). For each protein query, the interface will provide the assigned SulfAtlas (sub)family as well as additional information (e.g. the query and HMM lengths, the coverage, the score, the E-value, etc.). Depending on the load of the underlying computing infrastructure, results can be obtained in a few seconds for individual sequences and in few minutes for complete proteomes (e.g. ∼2 min for a bacterial proteome of ∼4,000 proteins). Queries (protein sequences in FASTA format) can be made by copy-paste or by file upload. The results are obtained directly online or sent by e-mail. We have set a size limit of 50MB for the data submitted online. For larger datasets, users should contact the SulfAtlas team (projet.sulfatlas@sb-roscoff.fr). Importantly, personal data are not saved by SulfAtlas HMM. Scientific data are anonymized and are erased by the server after 7 days. It is noteworthy that the SulfAtlas HMM library has also been incorporated as a routine tool into Microscope (https://www.genoscope.cns.fr/agc/microscope), the microbial genome annotation & analysis platform of Genoscope (43), facilitating the correct annotation of sulfatases in new bacterial genomes.
Figure 1.
Example of a result page of the SulfAtlas HHM server. Data input are protein sequences in FASTA format. They can be copied-pasted or uploaded as a file (with a size limit of 50MB). The results can be obtained directly online or be sent by e-mail. As an example, here results are shown for the complete proteome of Zobellia galactanivorans DsijT (4582 proteins). 72 hits were found (71 sulfatases and 1 pseudo-gene) which is consistent with previous genomic analyses (46). Data processing took 2 minutes and 45 seconds.
Semi-automatic integration of new sulfatases in SulfAtlas
The first releases of SulfAtlas relied only on expert curation to enrich the database with new sulfatases (1). To cope with the increasing flow of sequences, we have decided to develop a custom pipeline to semi-automatically update SulfAtlas, while still maintaining a high quality of information. The heart of this internal pipeline is the SulfAtlas HHM library. The pipeline includes the following steps:
Extraction of new candidate sequences from the Uniprot database
In the first run of the pipeline (T0, done in May 2022), this step builds a local copy of all the sequences of a full Uniprot release (around 214.4 million sequences for Uniprot release 2022_01) (10). Subsequent update runs will query Uniprot to restrict sequence retrieval to entries added and sequences modified since the last Uniprot release.
HMM candidate selection
This step uses HMMER along with the SulfAtlas HMM profiles to compute an HMM score for each candidate. The output of this step is a file describing the HMM matches for each candidate sequence (family and/or subfamily, score, E-value, location and length of the matching fragment or fragments…). Only candidates with an HMM-score of 100 or more are selected for the subsequent stages (189 938 candidates in T0).
HMM full candidate information retrieval
In order to be included in the SulfAtlas database, a sulfatase sequence needs to be supplemented with extra information (taxonomy, NCBI identifier and accession (12), known PDB structures (13), known loci (10)). This step queries Uniprot (10) for each candidate selected in step 2 and completes the initial sequence information with the data retrieved from Uniprot.
BLAST-based candidate consistency checking
To enhance the quality of candidates that will be added to the SulfAtlas database, each of the candidates resulting from step 3 is used as query sequence in a BLAST against the database of sequences from the latest release of SulfAtlas. Information about each candidate is then enriched with the results of its best BLAST hit.
Final candidate selection
The actual set of candidates considered for updating the SulfAtlas database is then built by extracting the candidates from the previous step which comply to two criteria: (i) their HMM-score is 300 or higher (with a coverage >80%) for a given SulfAtlas (sub)family, and (ii) their BLAST significant best hit matches a sequence belonging to the same (sub)family. This double condition is key to ensure the reliability of the (sub)family assignment. Thus, a set of 151 387 candidates complying to these two criteria (referred to as ‘green’ candidates) have been identified in T0. The pipeline also includes a procedure comparing these ‘green’ candidates to the curated sequences already contained in SulfAtlas and thus only the genuine new ‘green’ candidates are automatically added to SulfAtlas (115 831 in T0).
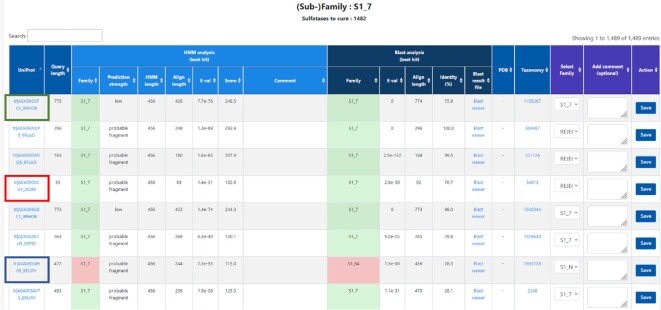
The remaining sequences (38 551 in T0) resulting from stage 2 are considered as ‘gray’ candidates needing manual curation. To analyze these ‘gray’ candidates more efficiently, we have also developed an internal Web based curation tool (Figure 2). This curation tool provides the details of the HHM and BLAST analyses for each sequence and easy commands to either confirm a specific (sub)family or to reject the candidate. These ‘gray’ sulfatase sequences generally fall into three categories: (i) Candidates for which the HMM and BLAST concur to a specific (sub)family, but the HMM score is slightly below 300 (with coverage >80%). The automatic assignment has clearly failed because we have chosen a quite conservative threshold in order to obtain a reliable automatic assignment. In most of these cases, a straightforward verification is sufficient to manually confirm the correct (sub)family assignment. (ii) Candidates with a coverage inferior to 80%. Generally, these sequences are sulfatase fragments corresponding to a pseudo-gene or an incorrectly predicted ORF. We have now decided to definitively reject those sequences from SulfAtlas; (iii) Candidates with a good coverage (>80%) for which the HMM and BLAST assignments are divergent or which have obtained good hits with several HMMs. Generally, these sequences are potential seeds for new subfamilies and we will temporarily assign them to the S1_NC subfamily.
Figure 2.
Home page of the internal web-based curation tool. The updated pipeline automatically feeds the curation tool with candidate sulfatase sequences with uncertain status. Details of HMM and BLAST results are provided for each candidate sequence. The figure shows the results for the S1_7 subfamily. Three sequences are highlighted as example cases: (i) green box (UniProt: A0A086XT05_9RHOB): the HMM and BLAST analyses concur to an S1_7 assignation, but with HMM score slightly below 300 (248.5). Manual verification confirmed that this sequence contained a complete S1_7 sulfatase module but also hemolysin-type calcium-binding regions, explaining its larger size (775 residues). The S1_7 subfamily was thus confirmed in the ‘Select Family’ menu and saved with the ‘Action’ button. (ii) red box (A0A090XCV4_IXORI): the query is a very short sequence (93 residues) and is thus a sulfatase fragment (pseudo-gene or incorrectly predicted ORF) and is definitively rejected. (iii) blue box (A0A0B5GH05_9EURY): this sequence has the correct size to be a functional S1 sulfatase (472 residues) but the HMM and BLAST analyses do not concur on the same subfamily assignment (S1_7 and S1_64). Therefore, this sulfatase is currently orphan and may be a seed for a future new subfamily. For now, it is assigned to the S1_NC subfamily.
The pipeline, which is based on Python and Java programs and shell scripts, has been developed to leverage the power of a SLURM-based cluster computing infrastructure for steps 2 and 4. It can be run stepwise or submitted as a single job. Execution time was around 7 days for the whole 2022_01 UniProt database (T0) (10) and it will be much shorter for the subsequent, incremental updates. The most recent SulfAtlas release (version 2.3.1) is the first including data generated by this pipeline and is significantly larger (162,430 sequences; Table 1).
CONCLUDING REMARKS
The essential added-value of SulfAtlas is to propose a homology-based classification system for sulfatases (into families and subfamilies), allowing a better prediction of substrate specificities. To the best of our knowledge, there is no other database which maintains such a classification system for sulfatases. SulfAtlas mainly provides three types of service: (i) it centralizes the knowledge on these enzymes; (ii) it provides bioinformatic tools for mining sulfatases and (iii) it helps experimentalists in their choice of relevant candidate sulfatases to study. As we hoped in 2016, the SulfAtlas classification has been progressively adopted by the community, as a guide to indeed select candidates for biochemical characterization as shown by numerous recent studies (18,24–26,28,29,32,33), but also as a useful tool for broader metagenomic analyses (44–47). Particularly, the development of SulfAtlas HMM should facilitate and speed up such (meta)genomic studies, allowing a fast assignment of sulfatase sequences at the (sub)family level. Internally, this new tool has already proven to be invaluable to maintain the SulfAtlas database up-to-date. SulfAtlas HMM and our updated pipeline also pave the way for future expansion of SulfAtlas toward other families related to sulfate metabolism, most notably the sulfotransferases.
DATA AVAILABILITY
All data contained in the SulfAtlas database are freely accessible at https://sulfatlas.sb-roscoff.fr/.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Philippe Potin for his support as IDEALG program coordinator. We thank Dr Mirjam Czjzek for her helpful discussions and Dr Elizabeth Ficko-Blean for her help with proof reading of the manuscript.
Contributor Information
Mark Stam, LABGeM, Génomique Métabolique, CEA, Genoscope, Institut François Jacob, CNRS, Université d'Évry, Université Paris-Saclay, 91057, Evry, Ile-de-France, France.
Pernelle Lelièvre, Sorbonne Université, CNRS, Laboratory of Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, Bretagne, France; Sorbonne Université, CNRS, FR2424, ABiMS, Station Biologique de Roscoff, 29680, Roscoff, Bretagne, France.
Mark Hoebeke, Sorbonne Université, CNRS, FR2424, ABiMS, Station Biologique de Roscoff, 29680, Roscoff, Bretagne, France.
Erwan Corre, Sorbonne Université, CNRS, FR2424, ABiMS, Station Biologique de Roscoff, 29680, Roscoff, Bretagne, France.
Tristan Barbeyron, Sorbonne Université, CNRS, Laboratory of Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, Bretagne, France.
Gurvan Michel, Sorbonne Université, CNRS, Laboratory of Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, Bretagne, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
G.M. and M.S. acknowledge support from the Agence National de la Recherche (ANR) with regard to the ‘Blue Enzymes’ project [ANR-14-CE19-0020]; G.M., P.L., M.H. and E.C. are also grateful to ANR for its support with regards to the investment expenditure program IDEALG [ANR-10-BTBR-04]; Roscoff Bioinformatics platform ABiMS (http://abims.sb-roscoff.fr), part of the Institut Français de Bioinformatique [ANR-11-INBS-0013]; BioGenouest network. Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barbeyron T., Brillet-Gueguen L., Carre W., Carriere C., Caron C., Czjzek M., Hoebeke M., Michel G.. Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS One. 2016; 11:e0164846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanson S.R., Best M.D., Wong C.H.. Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed. Engl. 2004; 43:5736–5763. [DOI] [PubMed] [Google Scholar]
- 3. Kahnert A., Kertesz M.A.. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J. Biol. Chem. 2000; 275:31661–31667. [DOI] [PubMed] [Google Scholar]
- 4. Drula E., Garron M.L., Dogan S., Lombard V., Henrissat B., Terrapon N.. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022; 50:D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davison J., Brunel F., Phanopoulos A., Prozzi D., Terpstra P.. Cloning and sequencing of pseudomonas genes determining sodium dodecyl sulfate biodegradation. Gene. 1992; 114:19–24. [DOI] [PubMed] [Google Scholar]
- 6. Barbeyron T., Potin P., Richard C., Collin O., Kloareg B.. Arylsulphatase from Alteromonas carrageenovora. Microbiology. 1995; 141:2897–2904. [DOI] [PubMed] [Google Scholar]
- 7. Knaust A., Schmidt B., Dierks T., von Bulow R., von Figura K.. Residues critical for formylglycine formation and/or catalytic activity of arylsulfatase A. Biochemistry. 1998; 37:13941–13946. [DOI] [PubMed] [Google Scholar]
- 8. Dierks T., Lecca M.R., Schlotterhose P., Schmidt B., von Figura K.. Sequence determinants directing conversion of cysteine to formylglycine in eukaryotic sulfatases. EMBO J. 1999; 18:2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melino S., Capo C., Dragani B., Aceto A., Petruzzelli R.. A zinc-binding motif conserved in glyoxalase II, beta-lactamase and arylsulfatases. Trends Biochem. Sci. 1998; 23:381–382. [DOI] [PubMed] [Google Scholar]
- 10. UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDonald A.G., Boyce S., Tipton K.F.. ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009; 37:D593–D597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2021; 49:D92–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M.et al.. RCSB protein data bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021; 49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lipman D.J., Pearson W.R.. Rapid and sensitive protein similarity searches. Science. 1985; 227:1435–1441. [DOI] [PubMed] [Google Scholar]
- 16. Wilson P.J., Morris C.P., Anson D.S., Occhiodoro T., Bielicki J., Clements P.R., Hopwood J.J.. Hunter syndrome: isolation of an iduronate-2-sulfatase cDNA clone and analysis of patient DNA. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:8531–8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prechoux A., Genicot S., Rogniaux H., Helbert W.. Enzyme-Assisted preparation of furcellaran-like kappa-/beta-Carrageenan. Mar. Biotechnol. 2016; 18:133–143. [DOI] [PubMed] [Google Scholar]
- 18. Ficko-Blean E., Prechoux A., Thomas F., Rochat T., Larocque R., Zhu Y., Stam M., Genicot S., Jam M., Calteau A.et al.. Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria. Nat. Commun. 2017; 8:1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright D.P., Knight C.G., Parkar S.G., Christie D.L., Roberton A.M.. Cloning of a mucin-desulfating sulfatase gene from prevotella strain RS2 and its expression using a bacteroides recombinant system. J. Bacteriol. 2000; 182:3002–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Myette J.R., Soundararajan V., Shriver Z., Raman R., Sasisekharan R.. Heparin/heparan sulfate 6-O-sulfatase from Flavobacterium heparinum: integrated structural and biochemical investigation of enzyme active site and substrate specificity. J. Biol. Chem. 2009; 284:35177–35188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prechoux A., Genicot S., Rogniaux H., Helbert W.. Controlling carrageenan structure using a novel formylglycine-dependent sulfatase, an endo-4S-iota-carrageenan sulfatase. Mar. Biotechnol. 2013; 15:265–274. [DOI] [PubMed] [Google Scholar]
- 22. Grondin J.M., Tamura K., Dejean G., Abbott D.W., Brumer H.. Polysaccharide utilization loci: fueling microbial communities. J. Bacteriol. 2017; 199:e00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cartmell A., Lowe E.C., Basle A., Firbank S.J., Ndeh D.A., Murray H., Terrapon N., Lombard V., Henrissat B., Turnbull J.E.et al.. How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ndeh D., Basle A., Strahl H., Yates E.A., McClurgg U.L., Henrissat B., Terrapon N., Cartmell A.. Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus. Nat. Commun. 2020; 11:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luis A.S., Jin C., Pereira G.V., Glowacki R.W.P., Gugel S.R., Singh S., Byrne D.P., Pudlo N.A., London J.A., Basle A.et al.. A single sulfatase is required to access colonic mucin by a gut bacterium. Nature. 2021; 598:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luis A.S., Basle A., Byrne D.P., Wright G.S.A., London J.A., Jin C., Karlsson N.G., Hansson G.C., Eyers P.A., Czjzek M.et al.. Sulfated glycan recognition by carbohydrate sulfatases of the human gut microbiota. Nat. Chem. Biol. 2022; 18:841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hettle A.G., Hobbs J.K., Pluvinage B., Vickers C., Abe K.T., Salama-Alber O., McGuire B.E., Hehemann J.H., Hui J.P.M., Berrue F.et al.. Insights into the kappa/iota-carrageenan metabolism pathway of some marine pseudoalteromonas species. Commun. Biol. 2019; 2:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robb C.S., Hobbs J.K., Pluvinage B., Reintjes G., Klassen L., Monteith S., Giljan G., Amundsen C., Vickers C., Hettle A.G.et al.. Metabolism of a hybrid algal galactan by members of the human gut microbiome. Nat. Chem. Biol. 2022; 18:501–510. [DOI] [PubMed] [Google Scholar]
- 29. Reisky L., Prechoux A., Zuhlke M.K., Baumgen M., Robb C.S., Gerlach N., Roret T., Stanetty C., Larocque R., Michel G.et al.. A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan. Nat. Chem. Biol. 2019; 15:803–812. [DOI] [PubMed] [Google Scholar]
- 30. Scott H.S., Blanch L., Guo X.H., Freeman C., Orsborn A., Baker E., Sutherland G.R., Morris C.P., Hopwood J.J.. Cloning of the sulphamidase gene and identification of mutations in sanfilippo a syndrome. Nat. Genet. 1995; 11:465–467. [DOI] [PubMed] [Google Scholar]
- 31. Ulmer J.E., Vilen E.M., Namburi R.B., Benjdia A., Beneteau J., Malleron A., Bonnaffe D., Driguez P.A., Descroix K., Lassalle G.et al.. Characterization of glycosaminoglycan (GAG) sulfatases from the human gut symbiont Bacteroides thetaiotaomicron reveals the first GAG-specific bacterial endosulfatase. J. Biol. Chem. 2014; 289:24289–24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silchenko A.S., Rasin A.B., Zueva A.O., Kusaykin M.I., Zvyagintseva T.N., Rubtsov N.K., Ermakova S.P.. Discovery of a fucoidan endo-4O-sulfatase: regioselective 4O-desulfation of fucoidans and its effect on anticancer activity in vitro. Carbohydr. Polym. 2021; 271:118449. [DOI] [PubMed] [Google Scholar]
- 33. Silchenko A.S., Rasin A.B., Zueva A.O., Kusaykin M.I., Zvyagintseva T.N., Kalinovsky A.I., Kurilenko V.V., Ermakova S.P.. Fucoidan sulfatases from marine bacterium Wenyingzhuangia fucanilytica CZ1127(T). Biomolecules. 2018; 8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sigrist C.J., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I.. New and continuing developments at PROSITE. Nucleic Acids Res. 2013; 41:D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katoh K., Rozewicki J., Yamada K.D.. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019; 20:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clamp M., Cuff J., Searle S.M., Barton G.J.. The jalview java alignment editor. Bioinformatics. 2004; 20:426–427. [DOI] [PubMed] [Google Scholar]
- 37. Stamatakis A. Using RAxML to infer phylogenies. Curr. Protoc. Bioinformatics. 2015; 51:6.14.1–6.14.14. [DOI] [PubMed] [Google Scholar]
- 38. Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q.. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016; 44:W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013; 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gough J., Karplus K., Hughey R., Chothia C.. Assignment of homology to genome sequences using a library of hidden markov models that represent all proteins of known structure. J. Mol. Biol. 2001; 313:903–919. [DOI] [PubMed] [Google Scholar]
- 41. Eddy S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009; 23:205–211. [PubMed] [Google Scholar]
- 42. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A.et al.. The pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vallenet D., Calteau A., Dubois M., Amours P., Bazin A., Beuvin M., Burlot L., Bussell X., Fouteau S., Gautreau G.et al.. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020; 48:D579–D589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lapebie P., Lombard V., Drula E., Terrapon N., Henrissat B.. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019; 10:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kappelmann L., Kruger K., Hehemann J.H., Harder J., Markert S., Unfried F., Becher D., Shapiro N., Schweder T., Amann R.I.et al.. Polysaccharide utilization loci of north sea flavobacteriia as basis for using susc/D-protein expression for predicting major phytoplankton glycans. ISME J. 2019; 13:76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barbeyron T., Thomas F., Barbe V., Teeling H., Schenowitz C., Dossat C., Goesmann A., Leblanc C., Oliver Glockner F., Czjzek M.et al.. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans dsijT. Environ. Microbiol. 2016; 18:4610–4627. [DOI] [PubMed] [Google Scholar]
- 47. Priest T., Heins A., Harder J., Amann R., Fuchs B.M.. Niche partitioning of the ubiquitous and ecologically relevant NS5 marine group. ISME J. 2022; 16:1570–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data contained in the SulfAtlas database are freely accessible at https://sulfatlas.sb-roscoff.fr/.