Abstract
DrugCentral monitors new drug approvals and standardizes drug information. The current update contains 285 drugs (131 for human use). New additions include: (i) the integration of veterinary drugs (154 for animal use only), (ii) the addition of 66 documented off-label uses and iii) the identification of adverse drug events from pharmacovigilance data for pediatric and geriatric patients. Additional enhancements include chemical substructure searching using SMILES and ‘Target Cards’ based on UniProt accession codes. Statistics of interests include the following: (i) 60% of the covered drugs are on-market drugs with expired patent and exclusivity coverage, 17% are off-market, and 23% are on-market drugs with active patents and exclusivity coverage; (ii) 59% of the drugs are oral, 33% are parenteral and 18% topical, at the level of the active ingredients; (iii) only 3% of all drugs are for animal use only; however, 61% of the veterinary drugs are also approved for human use; (iv) dogs, cats and horses are by far the most represented target species for veterinary drugs; (v) the physicochemical property profile of animal drugs is very similar to that of human drugs. Use cases include azaperone, the only sedative approved for swine, and ruxolitinib, a Janus kinase inhibitor.
INTRODUCTION
DrugCentral has been a public digital database aggregating drug information since 2016 (1). Three major regulatory agencies are continuously monitored: the U.S. Food and Drug Administration—FDA—in the United States (http://www.fda.gov/home), the European Medicines Agency—EMA—in Europe (https://www.ema.europa.eu/en) and the Pharmaceuticals and Medical Devices Agency—PMDA—in Japan (https://www.pmda.go.jp/english/index.html). The resource provides accurate and high-quality data for preclinical research and clinical practice. Chemical structures, molecular physicochemical descriptors, and patent status are linked to bioactivity data and molecular targets. Approved therapeutic drug uses, off-label uses and contraindications are manually curated from drug labels and the scientific literature. Mechanism-of-action targets and bioactivities are annotated where available. Besides pharmacodynamics data, DrugCentral provides several standardized pharmacokinetic descriptors. Statistical signal detection calculations on brute pharmacovigilance data outline post-marketing drug events stored in DrugCentral. Drug products marked in the United States are also available with pharmaceutical formulations, concentrations and administration routes. Altogether, the database acts as a well-rounded drug compendium freely available online and easy to be searched.
Since first introduced and published in the 2017 NAR database issue, DrugCentral has benefited from two major updates, in 2018 (2) and 2021 (3), with expanded functionality toward research areas such as drug repositioning, mining sex-based adverse drug events, and anti-COVID19 chemical identification. Thereby DrugCentral has become an essential resource for the scientific community, firmly linked to well-established resources, e.g. UniProt (4), ChEBI (5), Guide to Pharmacology (6), UniChem (7), Probes & Drugs portal (P&D) (8), PhenCards (9), COVID19db (10), etc. In addition, DrugCentral has become an essential component of the Knowledge Management Center KMC Datasets and Tools (11) in the NIH Common Fund's Illuminating the Druggable Genome (IDG) consortium (https://commonfund.nih.gov/idg).
The current update describes the enrichment in data since the last published version of DrugCentral in 2021 (3) and the addition of new features. First, its primary content was enriched with 285 newly approved drugs up to 31 March 2022, and the approval status was updated as follows: 101 drugs approved by the FDA, 48 by EMA, and 47 by PMDA (Table 1). DrugCentral 2023 adds new features: (i) veterinary drugs, (ii) off-label drug uses not indexed by conventional medical sources and (iii) adverse drug events for pediatric and geriatric medicine.
Table 1.
Differences in data content between DrugCentral 2021 and 2023 (current release)
| Entities (annotated drugs, or active pharmaceutical ingredients) | |||
|---|---|---|---|
| DrugCentral 2021 | DrugCentral 2023 | ||
| Human | Human | Veterinary | |
| Active pharmaceutical ingredients | 4642 | 4773 | 396 |
| FDA drugs | 2220 | 2331 | 396 |
| EMA drugs | 354 | 456 | n/a |
| PMDA drugs | 167 | 435 | n/a |
| Small molecules | 3876 | 3952 | 328 |
| Biologics and peptides | 315 | 364 | 21 |
| Other drugs | 395 | 457 | 47 |
| Parent molecules | 216 (332) | 220 (336) | 25 (35) |
| Off-patent status | 1553 | 1712 | 166 |
| OFP | 996 | 1029 | 119 |
| OFM | 320 | 398 | 46 |
| ONP | 237 | 285 | 1 |
| Drug efficacy targets | 872 (1760) | 927 (1883) | n/a |
| Human protein targets | 659 (1534) | 704 (1640) | n/a |
| Infectious agents targets | 212 (230) | 222 (248) | n/a |
| Protein–drug crystal complex (PDB) | 411 (165) | 608 (271) | n/a |
| All protein–drug crystal complex (PDB) | 5576 (799) | 9979 (1086) | 1270 (111) |
| Bioactivity data points | 16 843 (2052) | 18 830 (2214) | 1628 (193) |
| Human proteins | 12 373 (1837) | 13 9238 (1982) | 1055 (179) |
| Other species | 4470 (1235) | 4892 (1332) | 573 (135) |
| Pharmacological classification | |||
| WHO ATC code | 5067 (3082) | 5025 (3215) | 582 (208) |
| FDA Established Pharmacologic Class | 462 (1256) | 516 (1501) | 86 (141) |
| MeSH pharmacological action | 447 (2661) | 464 (2823) | 234 (301) |
| ChEBI ontology roles | 303 (1607) | 698 (2186) | 268 (242) |
| Drug indications | 2241 (2496) | 2437 (2644) | 1459 (377) |
| Drug contra-indications | 1415 (1399) | 1444 (1427) | n/a |
| Drug off-label uses | 794 (654) | 860 (666) | n/a |
| Pharmaceutical products | 108 035 (1810) | 137 693 (1885) | 1492 (377) |
| Rx pharmaceutical products | 56 515 (1697) | 64 704 (1744) | 849 (305) |
| OTC pharmaceutical products | 51 520 (319) | 72 986 (361) | 660 (130) |
| External identifiers | 63 658 (4639) | 78 928 (4773) | 7651 (396) |
| CAS registry number | 6350 (4642) | 6913 (4773) | 671 (396) |
| PubChem Compound Id | 4399 (4412) | 4590 (4529) | 399 (384) |
| FDA Unique Ingredient Identifier (UNII) | 4505 (4505) | 4844 (4715) | 405 (392) |
| ChEMBL-db id | 6473 (4469) | 6733 (4603) | 643 (375) |
| WHO INN id | 3700 (3700) | 3898 (3939) | 320 (323) |
| SNOMED-CT | 5193 (2910) | 6822 (3160) | 813 (341) |
| KEGG DRUG | 3697 (3698) | 3826 (3827) | 362 (383) |
| NDFRT | 3464 (3314) | 5189 (3595) | 503 (296) |
| RxNorm RxCUI | 3107 (3110) | 3267 (3348) | 346 (352) |
| IUPHAR/BPS ligand id | 1599 (1599) | 1917 (1893) | 157 (156) |
| UMLS CUI | 2835 (2835) | 4801 (4727) | 409 (395) |
| CHEBI | 3855 (3861) | 4045 (4038) | 294 (291) |
| MeSH | 4299 (4056) | 4494 (4279) | 422 (378) |
| DrugBank | 3685 (3699) | 3939 (3933) | 375 (364) |
| Protein Databank ligand id | 695 (659) | 1093 (1154) | 110 (120) |
Veterinary medicine plays a crucial role in human and public health by improving animal health (companion animals, wildlife, exotic animals and food animals), agriculture and food systems, biomedical and comparative medical research, and addressing zoonotic diseases (12). About half of the FDA-approved animal drugs are also approved for humans (13). Nearly all veterinary antibiotics were first approved in humans. However, some were first approved in animals (14). The recent COVID-19 pandemic has emphasized the impact of zoonotic diseases on human health but has also highlighted veterinary drugs as a potential source of viable antivirals (15). Following this path, we indexed a dataset of 395 unique veterinary agents from FDA’s ‘Green Book’, the Center for Veterinary Medicine—Approved Animal Drug Products (https://www.fda.gov/animal-veterinary/products/approved-animal-drug-products-green-book), and curated associated related data to the DrugCentral database.
Prescription of marketed medications not approved by the regulatory agency (e.g. FDA, EMA) is commonly referred to as off-label drug use. Although off-label drug use lacks rigorous regulations, physicians may treat off-label clinical conditions and diseases for patients that respond poorly to conventional treatment, often in specific populations poorly represented in clinical trials, e.g. pediatric, pregnant or psychiatric patients (16). Using pharmaceutical formulations (e.g. oral solution instead of capsules) or dosage (e.g. two tablets instead of one per day) not mentioned in the drug label for an indication also falls into the practice of off-label drug use (16). A study mining US Health National Disease and Therapeutic Index (NDTI) prescription data showed that 21% of overall drug use is off-label, of which 73% had little or no scientific support (17). The authors underlined the need for more efforts to scrutinize under-evaluated off-label uses that compromise patient safety or represent wasteful medication use (17). On the other hand, it is estimated that 57% of drug therapy innovations were discovered by practicing clinicians through field discovery (18). DrugCentral seeks to identify and promote practical off-label uses that improve patient safety and new therapeutic options. Therefore, in this version, we add a set of well-documented off-label uses retrieved from a rather unconventional source, the Reddit medical forum (19).
A third feature of the new DrugCentral version refers to post-marketing drug events in populations up to 17 years of age (pediatric) and over 65 years (geriatric). It is estimated that medical care is responsible for 10 000–444 000 deaths annually (20) and could be the third leading cause of death in the United States (21). Harm rates in pediatrics remain high (19.1 adverse events per 1000 inpatient days) despite nationwide efforts to improve patient safety (22). The newest version of DrugCentral aims to gain more insights into pediatric and geriatric adverse events to optimize medication and improve safety for those patient populations. Thereby, post-marketing surveillance data from FDA’s FAERS has been aggregated based on age, and significant adverse events were identified and incorporated into the database.
CURRENT CONTENT
Active pharmaceutical ingredients—drugs
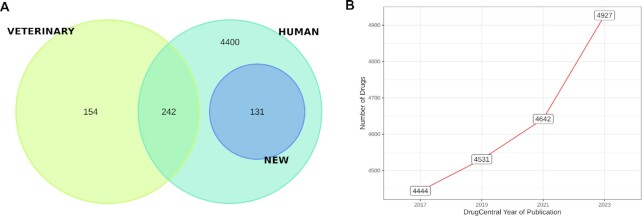
The 2023 version of DrugCentral adds 285 new drugs to the 2021 publication (3). Of these, 131 were approved only for human use and 154 only for veterinary use (Figure 1A). Thereby, 242 drugs already stored in DrugCentral are now associated with human and veterinary approvals. Since its 2016 version, this is the largest increase in new drugs added to a DrugCentral update (Figure 1B).
Figure 1.
Overlap between drugs approved for humans (new entries are plotted as a distinct class) and veterinary use (A), and the number of drugs in DrugCentral NAR publications (B).
Around 73% (255 drugs) of the newly added drugs received their first approval from FDA (including all veterinary drugs), whereas the rest of the drugs were split equally between EMA and PMDA, i.e. 48 and 47 drugs, respectively. As shown in Table 1, there is a ∼2.5 increase in the number of drugs licensed in Japan compared to DrugCentral 2021. The number of new orphan drugs approved to treat rare diseases (23) remains constant at ∼60, as reported in 2021 (Table 1).
In terms of drug types, 197 small organic molecules and 59 biologics, i.e. peptides, monoclonal antibodies (mAb), antibody-drug conjugates (ADCs), proteins, and oligonucleotides, were added to the database. In the latter case, most drugs are represented by mAbs (47%) and peptides (24%). Moreover, almost half of the biologics (46%) received orphan designations and are associated with rare disease therapies.
The 2023 DrugCentral database contains off-patent tags for 1712 drugs, a 10% increase in drug annotations compared to 2021 (24). Around 60% of the drugs are OFP (on-market drugs with expired patent and exclusivity coverage), 17% are OFM (off-market drugs) and 23% are ONP (on-market drugs covered by active patent and exclusivity) status.
DrugCentral entries were identified in several external databases and associated with external mappings, as reported in Table 1. More than 85% (234 drugs) of the newly added drugs have external identifiers in PubChem (25), World Health Organization (WHO) International Nonproprietary Names (INN), DrugBank (26), ChEMBLdb (27), KEGG (28), FDA’s Global Substance Registration System—Unique Ingredient Identifier (UNII; https://bit.ly/3MNMqwQ), Unified Medical Language System, UMLS (29), etc. (Table 1).
Bioactivities and drug targets
The number of bioactivity points increased by 12.6% (to a total of 18 961) compared to 2021 (see Table 1). In total, 487 activity endpoints were captured for newly added drugs. Almost half were manually extracted from drug labels (27%) and the scientific literature (20%). The other half originates from ChEMBLdb (27) and the IUPHAR/BPS Guide to Pharmacology (30). There are 107 targets defined as mechanism-of-action (MoA) targets for 112 new drugs. Only 47 (briefly described in Table 2) of these are new targets compared to the previous DrugCentral version. According to the Target Development Level classification system of human proteins (31) adopted within the IDG consortium, 39 new targets were annotated as Tclin, i.e. MoA targets through which approved drugs exert their therapeutic action (32–34). Currently, 664 Tclin human targets have been identified. According to TDL classification (31), Tchem are proteins that are not Tclin but are known to bind small molecules with high potency; Tbio includes proteins that (i) have Gene Ontology (35) ‘leaf’ (lowest level) term annotations based on experimental evidence, or (ii) meet two of the following three conditions: a fractional publication count (36) >5, three or more Gene RIF, ‘Reference Into Function’ annotations (https://bit.ly/2WDE1oL), or 50 or more commercial antibodies, as counted in the Antibodypedia portal (37). The fourth TDL category, Tdark, includes ∼30% of the human proteins manually curated in UniProt (38) that do not meet Tclin, Tchem or Tbio criteria. The current version of DrugCentral reports 708 Tchem, 415 Tbio and 26 Tdark proteins, mapped onto the Target Central Resource Database (TCRD) and interfaced with the TCRD portal, Pharos, respectively (39,40).
Table 2.
New human drugs with novel mechanisms of action, approved since the DrugCentral 2021 release
| Drug name(s) | Targeta | Target classb | Agency | Indication(s)c |
|---|---|---|---|---|
| inqovi | CDA | Enzyme | FDA | Myelodysplastic syndrome, CMML |
| lonafarnib | FNTA,B | Enzyme | FDA | Hutchinson-Gilford syndrome |
| umbralisib | CSNK1E | Kinase | FDA | Marginal zone & follicular lymphoma |
| fosdenopterin | MOCS1 | Enzyme | FDA | Molybdenum cofactor deficiency A |
| sotorasib | KRAS | Enzyme | FDA, EMA, PMDA | KRAS G12C-mutated locally NSCLC |
| ibrexafungerp | FKS1 | Enzyme | FDA | Vulvovaginal candidiasis |
| belzutifan | EPAS1 | TF | FDA | von Hippel-Lindau disease |
| avacopan | C5AR1 | GPCR | FDA, PMDA, EMA | ANCA-associated vasculitis, microscopic polyangiitis, and granulomatosis with polyangiitis |
| maribavir | UL97 | Kinase | FDA, EMA | Post-transplant CMV infection |
| pafolacianine | FOLR1 | MR | FDA | Imaging ovarian cancer & malignant lesions |
| mitapivat | PKLR | Enzyme | FDA | PK deficiency |
| mavacamten | MYH7 | Other | FDA | Obstructive HCM |
| gefapixant | P2RX3 | Ion channel | PMDA | Refractory chronic cough |
| tapinarof | AHR | TF | FDA | Plaque psoriasis |
| bulevirtide | SLC10A1 | Transporter | EMA | Chronic HBV with delta hepatitis |
| pegcetacoplan | C3 | Macroglobulin | FDA, EMA | Paroxysmal nocturnal hemoglobinuria |
| vosoritide | NPR2 | Enzyme | EMA, FDA | Achondroplasia |
| tirzepatide | GPR119 | GPCR | FDA | Diabetes mellitus type 2 |
| ansuvimab, atoltivimab, maftivimab, odesivimab | EBOV GP | Glycoprotein | FDA | Ebola virus disease |
| evinacumab | ANGPTL3 | Secreted | FDA, EMA | HoFH |
| bamlanivimab, etesevimab, casirivimab, imdevimab, regdanvimab, sotrovimab | S | Glycoprotein | FDA, EMA PMDA, | COVID-19 infection |
| remdesivir | RdRp | RNA polymerase | FDA, EMA PMDA, | COVID-19 infection |
| aducanumab | APP | Unclassified | FDA | Alzheimer's disease |
| bimekizumab | IL17F | Cytokine | EMA, PMDA | Plaque, erythrodermic and pustular psoriasis |
| tralokinumab | IL13 | Cytokine | EMA, FDA | Moderate to severe atopic dermatitis |
| tezepelumab | TSLP | Cytokine | FDA | Severe asthma |
| sutimlimab | C1S | Enzyme | FDA | Cold autoimmune hemolytic anemia |
| relatlimab | LAG3 | TAA | FDA | Unresectable or metastatic melanoma |
| faricimab | ANGPT2 | Secreted | FDA | EMD and diabetic macular edema |
| efgartigimod alfa | FCGRT | MR | FDA, EMA | Immune thrombocytopenia, and MG |
| sacituzumab govitecan | TACSTD2 | TAA | FDA, EMA | Triple negative breast neoplasms |
| belantamab mafodotin | TNFRSF17 | TAA | FDA, EMA | Multiple myeloma |
| tisotumab vedotin | F3 | MR | FDA | Recurrent or metastatic cervical cancer |
| imlifidase | IGHG1-4 | Antibody | EMA | Renal transplant |
| pabinafusp alfa | TFRC | MR | PMDA | Mucopolysaccharidosis type II |
| tebentafusp | HLA-A | Surface antigen | FDA, EMA | Unresectable metastatic uveal melanoma |
| lumasiran | HAO1 | RNA | FDA, EMA | Primary hyperoxaluria,type I |
| flortaucipir F 18 | MAPT | Structural | FDA | Positron emission tomography |
| borofalan (10B) | SLC7A5 | Transporter | PMDA | Neoplasm of head and neck |
| gallium (68Ga) gozetotide, lutetium (177Lu) vipivotide tetraxetan | KLK3 | Surface antigen | FDA | Positron emission tomography, PSMA-positive metastatic castration-resistant prostate cancer |
| piflufolastat F-18 | FOLH1 | Enzyme | FDA | Positron emission tomography |
aAHR, aryl hydrocarbon receptor; ANGPT2, angiopoietin-2; ANGPTL3, angiopoietin-related protein 3; APP, amyloid beta A4 precursor protein; C1S, complement C1s subcomponent; C3, complement C3; C5AR1, C5a anaphylatoxin chemotactic receptor 1; CDA, cytidine deaminase; CSNK1E, casein kinase I isoform epsilon; EPAS1, endothelial PAS domain-containing protein 1; F3, tissue factor; FCGRT, IgG receptor FcRn large subunit p51; FKS1, 1,3-beta-d-glucan-UDP glucosyltransferase; FNTA, protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha; FNTB, protein farnesyltransferase subunit beta; FOLH1, glutamate carboxypeptidase 2; FOLR1, folate receptor alpha; EBOV GP, Ebola virus envelope glycoprotein; GPR119, glucose-dependent insulinotropic receptor; HAO1, hydroxyacid oxidase 1; HLA-A, HLA class I histocompatibility antigen, A-3 alpha chain; IGHG1, Ig gamma-1 chain C region; IGHG2, Ig gamma-2 chain C region; IGHG3, Ig gamma-3 chain C region; IGHG4, Ig gamma-4 chain C region; IL13, interleukin-13; IL17F, interleukin-17F; KLK3, prostate-specific antigen; KRAS, GTPase KRas; LAG3, lymphocyte activation gene 3 protein; MAPT, microtubule-associated protein tau; MOCS1, molybdenum cofactor biosynthesis protein 1; MYH7, Myosin-7; NPR2, atrial natriuretic peptide receptor 2; P2RX3, P2X purinoceptor 3; PKLR, pyruvate kinase PKLR; S, spike glycoprotein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); SLC10A1, sodium/bile acid cotransporter; SLC7A5, large neutral amino acids transporter small subunit 1; RdRp, SARS-CoV-2 RNA-dependent RNA polymerase; TACSTD2, tumor-associated calcium signal transducer 2; TFRC, transferrin receptor protein 1; TNFRSF17, tumor necrosis factor receptor superfamily member 17; TSLP, thymic stromal lymphopoietin; UL97, serine/threonine protein kinase UL97.
bMR, membrane receptor; TAA, tumour-associated antigen; TF, transcription factor.
cANCA, anti-neutrophil cytoplasmic autoantibody; CMML, chronic myelomonocytic leukemia; CMV, cytomegalovirus infection; HBV, viral hepatitis B infection; HCM, hypertrophic cardiomyopathy; HoFH, homozygous familial hypercholesterolemia; NSCLC, non-small cell lung cancer; PK, pyruvate kinase.
The vast majority of the new activity data—76% (359 bioactivities)—is covered by kinases (38%), GPCRs (21%) and enzymes (17%). Kinase activities cover the largest number of protein targets but also the smallest number of drugs among the three protein families. GPCR activities are the highest among these protein families in both human and veterinary drugs, as reflected by the mean values: 8.35 (± 1.56) –log[M] and 7.72 (± 1.54) –log[M], respectively (Table 3).
Table 3.
A brief description of activity data in the main protein target families for the newly added drug set
| Number of activity points | Number of drugs | Number of protein targets | Mean activitya (median) ± SDb | |||||
|---|---|---|---|---|---|---|---|---|
| Protein target family | Human | Veterinary | Human | Veterinary | Human | Veterinary | Human | Veterinary |
| Kinase | 166 | 13 | 21 | 2 | 101 | 8 | 6.79 (6.49) ± 1.41 | 7.65 (7.76) ± 0.82 |
| Enzyme | 42 | 37 | 22 | 14 | 30 | 28 | 7.28 (7.81) ± 1.68 | 5.68 (5.37) ± 1.04 |
| GPCR | 38 | 63 | 19 | 15 | 35 | 43 | 8.35 (8.39) ± 1.56 | 7.72 (7.92) ± 1.54 |
alog[M].
bStandard deviation.
The comparison of the potency values in drugs acting on non-MoA targets versus MoA targets shows a difference of two log units between the median values in favor of the latter. The median for newly added activity values for MoA targets (over the past two years) is higher compared to the median of those indexed in DrugCentral 2021 (Figure 2): at least 80% of the activity values are >7.7 −log[M] (median of 8.4 −log[M]) for the new MoA targets compared to >6.43 −log[M] (median of 8 −log[M]) previously found in DrugCentral. This trend suggests an increase in potency, in particular for novel drugs.
Figure 2.
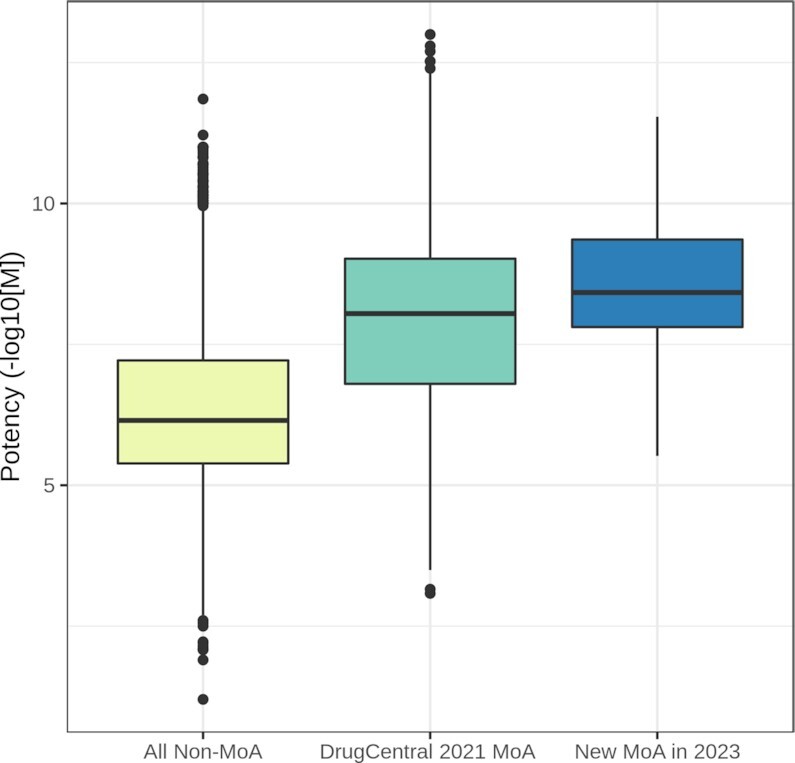
The distribution of bioactivity values in non-MoA targets versus MoA targets in DrugCentral 2021 versus new MoA data in 2023.
The analysis of novel MoA drug targets (targets that an approved drug had not previously perturbed) published annually in the Nature Reviews Drug Discovery series (33,34,41,42) with data pulled from DrugCentral, shows, on average, an enrichment rate of 15 novel drug targets per year (some of the data overlap with Table 2 - in accordance to the time frame reported). These targets are modulated by increasing numbers of mAb and ADC, mainly directed toward cytokines, surface antigens, and membrane receptors. The novel drug targets captured between 2018 and 2021 are modulated by 27 mAb (and ADC) and only 19 small molecules. If this trend persists, in the upcoming years, we expect druggable target discovery to be driven primarily by biologics rather than small-molecule drugs.
Pharmacological classification
DrugCentral entries were mapped into pharmacological classes according to World Health Organization Anatomic, Therapeutic, and Chemical classification system (WHO ATC; WHOCC, https://www.whocc.no/), the Medical Subject Headings (MeSH) (43), the FDA Established Pharmacologic Class (EPC; https://bit.ly/3Tzgwq6), and ChEBI (5). Of 4927 drugs stored in DrugCentral, 65% were successfully mapped into WHO ATC classes and 61% into MeSH data (Table 1). The top three WHO ATC L1 groups (anatomical or pharmacologic) with the largest number of drugs are: the nervous system (522 drugs), the alimentary tract and metabolism (450), and the cardiovascular system (396). In total, 202 new drugs were linked to 404 pharmacologic classification codes.
Pharmaceutical formulations and drug products
FDA drug labels were assessed using DailyMed (44), downloaded on 21 June 2022. A total of 112 359 labels (submitted for human use) were retrieved based on 1885 active ingredients (drugs) mapped into DrugCentral. There are 137 693 drug products, of which 88% are formulated for oral (48%) and topical (40%) administration. However, regarding the number of active ingredients relative to the administration route, we found that 59% of the drugs are oral, followed by 33% parenteral and 18% topical. Further, 81% of the drugs (in 64 704 products) are licensed for human prescription (Rx), and 19% (in 72 986 products) are over-the-counter (OTC) drugs.
NEW DATA AND FUNCTIONALITY
Animal drugs data
Veterinary drugs
We indexed a dataset of 396 unique veterinary drugs from the FDA’s ‘Green Book’ (45). Of these, 242 were already captured in DrugCentral for human use. The other 154 drugs were inserted into the database as new entries (see Figure 1A). Chemical structures were manually assigned to 140 of the latest veterinary drugs, and molecular descriptors and structure depictions were computed per DrugCentral workflow (1).
Most veterinary drugs (83%) are small organic molecules, and only 5.3% are biologics. Regarding mAb-type drugs, there is only one drug licensed for animals, frunevetmab. The absence of ADC and RNA drugs suggests that therapeutic modalities for animal use trail behind human medicines regarding innovation.
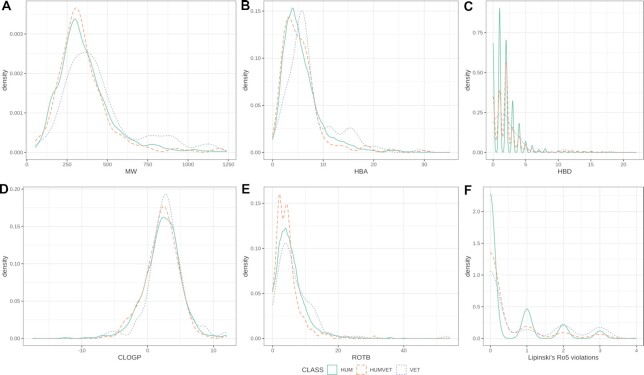
The data set of small molecules with molecular weight (MW) between 50 and 1250 a.m.u. includes 351 animal drugs. In terms of basic physicochemical and structural properties, the middle 80% of the data ranges as follows: 200 ≤ MW ≤ 615, −1 ≤ CLOGP ≤ 5.25 (the calculated 1-octanol/water partition coefficient), 1 ≤ ROTB ≤ 11 (number of rotatable bonds), 2 ≤ HBA ≤ 12 (number of hydrogen bond acceptors), HBD ≤ 4 (number of hydrogen bond donors). The majority of these drugs (75%) are compliant with Lipinski's rules of 5 (Ro5).
A more detailed view of small molecules is provided in Figure 3, which plots density distributions separately for 4078 drugs approved only for human use (HUM), 135 drugs approved only for veterinary use (VET), and 216 drugs approved for both human and veterinary use (HUMVET). The three distributions are very similar, but a slight shift towards larger values can be observed in VET drugs for MW (median of 444.61 a.m.u.) and HBA (median of 7) relative to HUM and HUMVET drugs (MW medians of 358.48 a.m.u. and 345.87 a.m.u., respectively, and HBA medians of 5 and 5, respectively).
Figure 3.
Density plots of molecular properties distributions in drugs approved only for human use (HUM), only for veterinary use (VET), and drugs approved for both human and veterinary use (HUMVET): molecular weight (MW), calculated 1-octanol/water partition coefficient (CLOGP), number of rotatable bonds (RTB), hydrogen bond donors/acceptors (HBD, HBA), violations of Lipinski's rule-of-fives (Ro5).
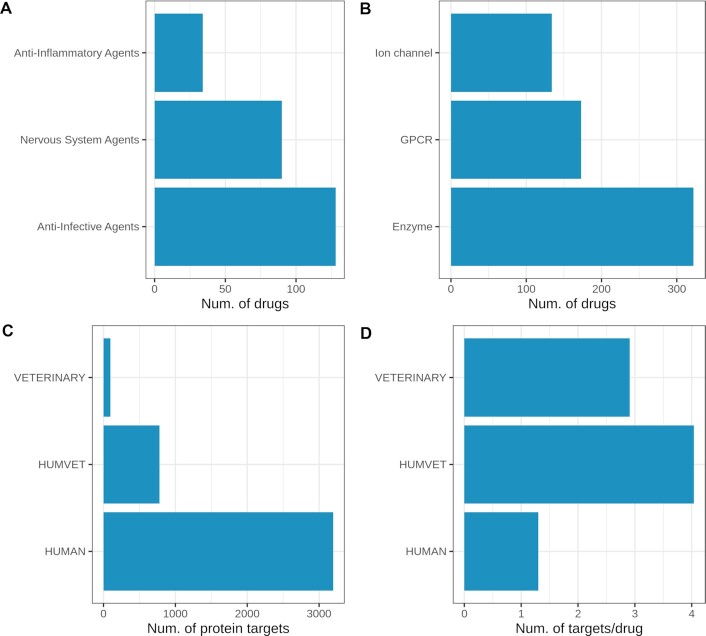
Off-patent classifications were assigned only for veterinary drugs approved for human use since the OrangeBook only covers human drugs. There are 166 drugs mapped into the off-patent schema (24) as follows: 119 OFP, 46 OFM, and only one ONP. The high percentage of OFP drugs, and the similar molecular property profile distribution, suggest that veterinary drugs may be viable candidates in animal drug repositioning programs. The largest pharmacological class in veterinary drugs is defined by anti-infective agents (32% according to the MeSH pharmacological classification), mainly comprising antibiotics and antiparasitics (Figure 4A). Drugs acting on the nervous system (central or peripheral) compose the second largest group with 90 representatives (23%). The third group comprises 34 antiinflammatory drugs: 14 non-steroidal and 20 glucocorticoids. Including veterinary drugs in DrugCentral could inform promiscuity analyses such as BADAPPLE, a chemical pattern detection algorithm (46).
Figure 4.
Statistics on veterinary data: (A) top three pharmaceutical drug classes according to size; (B) the three main target families; (C) the number of protein targets and (D) target per drug in entries shared by veterinary and human data (HUMVET), human and veterinary drugs after excluding HUMVET data.
Bioactivity data in veterinary drugs
The current update adds 1805 bioactivities for 226 veterinary drugs and 804 targets. Nearly half (884) of the activity values are submicromolar. Most bioactivity data (65%) pertain to human targets, followed by 18% determined against rat, mouse, and bovine targets. The main target classes are enzymes (322 targets), GPCRs (173 targets), and ion channels (134 targets), as shown in Figure 4 (B). We compared the bioactivity data from drugs shared by human and veterinary use (HUMVET; 1674 activities) with the remaining data in the two drug categories: HUMAN (18 752 activities) and VETERINARY (131 activities). Unsurprisingly, the HUMAN set covers the largest number of targets (3196 targets and 2462 drugs) compared to HUMVET (780 targets and 193 drugs) and VETERINARY (96 targets and 33 drugs). However, the ratio between the number of targets to the number of drugs is highest in HUMVET (4 targets/drug), followed by VETERINARY (3 targets/drug) and HUMAN (1.3 targets/drug) (Figure 4C and D).
Veterinary drug products
DrugCentral veterinary data contains application numbers and types, trade names, applicant names, and prescription types for 1492 drug products as referenced in FDA’s Green Book. Most of these products (77%) are originals (approved as new animal drug applications; NADA), and the remaining 23% are generics (or ANDA; a copy of an approved new animal drug for which patents or other periods of exclusivity are near expiration). Regarding prescription type, 57% of the veterinary drug products (covering most drugs: 305) are Rx and 43% are OTC.
Veterinary drug uses
Drug label information has been manually extracted and structured as follows: drug names were mapped to DrugCentral synonyms, and indications and contraindications were summarized and separated for each species (if multiple were mentioned). As a result, DrugCentral currently holds 2581 indications (pairs of drug and concept names) for 30 species. Dogs, cattle and cats comprise 62% of the indications covered by 290 veterinary drugs.
Documented off-label uses
The primary sources of information regarding off-label medical use (OLUs) are usually compendia, drug information reference handbooks (47), case studies reported in the scientific literature, and the US National Disease and Therapeutics Index (NDTI), where healthcare providers share information about prescribing patterns and disease treatments. To the 2446 OLUs (651 drugs and 815 diseases/medical conditions) indexed by DrugCentral, we added a set of 66 new off-label uses (40 drugs and 51 unique medical conditions or diseases) extracted from the r/medicine Reddit subforum (https://bit.ly/3MFHIB0).
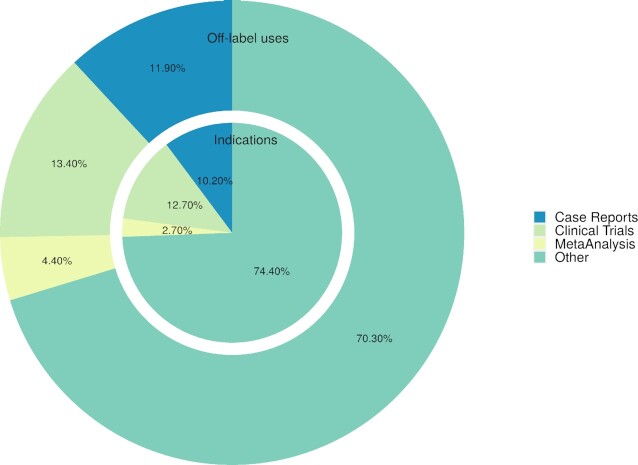
The data set was documented thoroughly using automatic searches in the scientific literature and clinical trials data and compared to 209 approved indications and other off-label uses (OLUs) indexed by DrugCentral (for the same 40 drugs) (19). The results showed that 90% of the new OLUs (pair of drug - medical condition) are mentioned in at least four scientific articles in PubMed, with a median of 36 articles per off-label use. The vast majority (80%) of these clinical trial results have been published. By comparison, the same drug set showed a median of 221 PubMed articles per indication (Table 4). Regarding the publication types associating drug names with off-label uses and approved indications, both sets have similar proportions (Figure 5). However, in the case of meta-analyses, there are two times more publications for approved indications compared to off-label uses.
Table 4.
Summary of the off-label use and approved indications search results in the literature and clinical studies
| Scientific literature | Clinical trials | ||||||
|---|---|---|---|---|---|---|---|
| Dataset | Counts | Total | Range | Median | Total | Range | Median |
| Off-label uses | 66 | 6807 | 0–1061 | 36 | 135 | 454 | 2 |
| On-label uses | 209 | 67 289 | 0–3211 | 221 | 1170 | 6652 | 8 |
Figure 5.
Percentages of scientific publications mentioning off-label and on-label (indication) use according to type.
A second automatic search mapped the OLUs in 8509 public clinical trials provided by the WHO International Clinical Trials Registry Platform (ICTRP; https://www.who.int/clinical-trials-registry-platform). The outcomes showed at least one clinical trial for 73% of the new OLUs and, not surprisingly, for all approved indications (Table 4).
Adverse drug events in pediatric and geriatric patients
Since 2018, DrugCentral has integrated FAERS (Food and Drug Administration Adverse Event Reporting System (https://open.fda.gov/data/faers/), signaling post-marketing adverse drug events based on computed likelihood scores (48). In the 2021 DrugCentral update, we separately calculated FAERS for men and women to enable research on sex differences in drug safety. The 2023 version goes further by computing adverse drug event scores granulated by age: pediatric data (FAERS_PED) covers patients from 1 day to 17 years, and geriatric data (FAERS_GER) focuses on elderly patients over 65 years of age respectively.
As reported in Table 5, the number of significant adverse event signals (the log-likelihood ratio - LLR - exceeds the likelihood threshold, t) found in FAERS_GER is 1753 times larger compared to FAERS_PED. There are 70 drugs and 99 terms describing the adverse event in pediatrics, versus 1724 drugs with 11 832 MedDRA PTs in geriatric data. This comparison used preferred terms PTs from MedDRA, the Medical Dictionary for Regulatory Activities terminology, which is the international medical terminology developed under the auspices of the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (https://www.meddra.org/). This significant difference in reporting might be attributed to increased drug consumption, prolonged treatment periods (chronic diseases), overlapping treatments, and multiple diseases affecting older adults compared to children. For example, the top 3 drugs showing the most AEs are ampicillin, acetylsalicylic acid and ibuprofen (usually used to treat acute diseases) in pediatrics, while in geriatrics, the top drugs are methotrexate, etanercept, and tacrolimus (used in chronic conditions).
Table 5.
Summary of age-specific adverse event data from FAERS, at different LLR levels; t, LLR threshold; API, active pharmaceutical ingredient
| Number of drug-AEa pairs (unique drugs/unique AEs) | ||
|---|---|---|
| Categories | FAERS_PED | FAERS_GER |
| +LLR > t | 136 (70/99) | 238 410(1724/11 832) |
| LLR > 2*t | 13 (12/11) | 126 424 (1437/7964) |
| LLR > 5*t | 2 (2/2) | 50 570 (1092/4284) |
aAdverse event.
DRUGCENTRAL USE CASES
Veterinary drugs
It has been previously noted that half of the FDA-approved animal drugs are approved for human use (13). Current DrugCentral entries show that ∼61.1% of animal drugs are approved for both human and animal use (see also Figure 1). However, only 3.1% of the 4927 drugs indexed in DrugCentral are for animal use only. These statistics suggest that medicines approved for humans are more likely to be repurposed for animal use, not the other way around. This is not always true: the antibiotic moxidectin, initially approved for animal use, was later approved for humans (14). Veterinarians can legally prescribe an approved human drug in animals in certain circumstances, according to the U.S. Federal Food, Drug, and Cosmetic Act (https://bit.ly/3ghtnPf). This suggests that human-to-animal drug repurposing opportunities exist and are supported by ‘extra-label uses’ (approved FDA terminology).
Based on the 395 Green Book medicines captured in DrugCentral, five animal species are predominantly represented concerning drug uses (the number of unique active ingredients targeting that species is in brackets): dogs (211), cats (120), horses (117), cattle (105) and swine (57), respectively. All other species are represented with 43 uses or less. Given this therapeutic arsenal, it appears that humans are more predisposed to provide pharmaceutical care to companion animals (i.e. dogs, cats and horses), than domesticated (food) animals or wild animals. Given this statistic, one can conclude that humans are a dog's best friend.
Use case: Azaperone is a butyrophenone invented by Paul Janssen, as part of a series of neuroleptic drugs (49). Wikipedia states that azaperone is ‘uncommonly used in humans as an antipsychotic drug’ (https://en.wikipedia.org/wiki/Azaperone). However, exhaustive literature searches, including Dutch pharmaceutical references from the 1960s and 1970s, suggest that this drug did not receive regulatory approval for human use (I.M. van Geijlswijk, personal communication). Azaperone (Stresnil®) is used to reduce fear and aggression in recently mixed groups of pigs, based on a 1968 report showing its sedative effect in swine (50). Currently, azaperone is the only sedative approved for pigs. Its DrugCentral record indicates that this is a veterinary drug only (https://drugcentral.org/drugcard/5590).
Human drugs
The lack of approved anti-SARS-CoV-2 drugs during the initial phases of the COVID-19 pandemic led to increased enthusiasm in computer-aided drug repurposing (51). Based on an in-house drug discovery knowledge graph, BenevolentAI scientists proposed baricitinib, at the time approved for rheumatoid arthritis, as a drug repurposing candidate for severe COVID-19 (52,53). Baricitinib was approved (in combination with remdesivir) on November 19 2020 by the US FDA for COVID-19 under the Emergency Use Authorization status (https://bit.ly/3giFeN1). Baricitinib lowers 28-day mortality in hospitalized COVID-19 patients in a significant manner (54).
Use case
According to its in vitro profile, baricitinib is a Janus kinase (JAK) inhibitor that preferentially blocks JAK1 and JAK2, as opposed to other kinases (https://drugcentral.org/drugcard/5202). This specific activity protects severely infected COVID-19 patients from the cytokine storm (55). Ruxolitinib has a similar kinase inhibition profile with a higher JAK1/JAK2 potency (https://drugcentral.org/drugcard/4190). Given their in vitro profile similarity, initial reports suggested that ruxolitinib might be effective against the COVID-19 related cytokine storm (56). However, ruxolitinib failed to show significant protection in the treatment of severe coronavirus disease (57) and the National Institutes of Health recommends against using ruxolitinib for the treatment of COVID-19 (https://bit.ly/3TtgsIe). According to DrugCentral records, baricitinib is approved for atopic dermatitis and rheumatoid arthritis, while ruxolitinib is approved for myeloproliferative disorders, including polycythemia vera. The top 10 FAERS-listed adverse events, regardless of gender or age, are consistent with ruxolitinib affecting the bone marrow (adverse events related to platelets, erythrocytes and white blood cells, and splenomegaly), which probably makes this drug less suitable in severe viral infections.
SUMMARY AND FUTURE DIRECTIONS
DrugCentral 2023 contains human drugs approved up to 31 March 2022. Additional features enhance usability: (i) the addition of veterinary drugs opens new preclinical research opportunities, (ii) new off-label uses were indexed, and iii) adverse drug events have been highlighted in pediatric and geriatric medical fields. The 2023 release also supports ‘Target Card’ and chemical substructure queries (vide infra). We also uncovered several interesting statistics: (i) 60% of the drugs are OFP, 17% are OFM, and 23% are ONP; (ii) based on their administration route, 59% of the drugs are orally formulated, with 33% parenteral and 18% topical; (iii) while 61% of the veterinary drugs are approved for human use, only 3% of all drugs are for animal use – which makes animal-to-human drug repurposing a more difficult prospect; (iv) dogs, cats and horses are the most represented target species in the veterinary pharmaceutical arsenal; (v) regarding their physicochemical property profile (MW, CLOGP, etc.), animal drugs have a similar distribution to human drugs.
In the future, DrugCentral will monitor new drug approvals and continue aggregating its core data: pharmaceutical formulation—drug–drug target–disease association. In addition, we will screen all FDA drug labels to identify repurposed drugs. Future DrugCentral releases will link drugs to clinical trial data and scientific papers using the PubMed Central platform. As noted above regarding off-label uses in humans, it is not trivial to capture ‘extra-label uses’ for veterinary medicine. We invite physicians and veterinarians to contact us concerning validated off-label (human) and extra-label (veterinary) drug uses.
DATA ACCESS
Web interface
The DrugCentral web interface is accessible at https://drugcentral.org/ from multiple platforms (Windows, Linux, Android, Apple OS) via desktops, laptops, tablets, or smartphones. The search bar allows: (i) drug information searches based on drug names, synonyms, brand names and identifiers; (ii) target data search supported by gene names, target names, UniProt accessions, and SwissProt identifiers; (iii) drug use search based on disease names and mappings in SNOMED-CT and OMOP vocabulary terms; (iv) pharmacological action search and (v) drug product search using trade names, FDA National Drug Code (NDC) and active ingredient names. Search results are ranked based on the match between the query term and (a) drug name or synonyms, MoA or drug indication; (b) disease term in drug contra-indications or off-label use, targets listed in the bioactivity profile (but not MoA targets) or pharmacological action description; (c) drug description text; (d) full text in FDA drug labels. The ‘Drugs in the news’ and ‘Featured news’ sections are updated periodically by monitoring drugs widely associated with current events.
Target cards
DrugCentral 2023 now supports ‘Target Cards’ that can be queried using UniProt accession IDs. To perform a ‘Target Card’ search, the user enters a UniProt accession ID at the prompt, e.g. P23975. DrugCentral ‘Target Cards’ depict the ‘Accession,’ ‘SwissProt’, ‘Organism’, ‘Gene’ and ‘Target class’ followed by ‘Drug Relations’ identified by the ‘Bioactivity mechanism-of-action.’ To retrieve all cross-referenced DrugCentral target cards mapped to UniProt Accession Ids, use the following (machine-readable) URL syntax: https://drugcentral.org/static/Drugcentral_uniprot_Mapping.txt.
Chemical structure searching
The current update supports chemical substructure searching. Typing a SMILES (58) string in the DrugCentral Substructure search bar launches a chemical substructure search that finds matching compounds in DrugCentral. The substructure searching is supported by the RDKit (https://www.rdkit.org/) PostgreSQL database cartridge (https://www.rdkit.org/docs/Cartridge.html), which extends PostgreSQL by adding a molecular column type that PostgreSQL can process. To ensure that DrugCentral returns substructure search results quickly, we created a substructure search index for all internal molecular data. The search index allows PostgreSQL to avoid a costly linear scan over each compound in DrugCentrals's PostgreSQL database.
Download
The full database dump is available for download in PostgreSQL format for advanced data query, export, and integration. Structured query language (SQL) examples are provided for user interaction with the local instance, but chemical structures (e.g. SDF, InChI, SMILES), and drug bioactivity profiles are provided in tabular format. The database is available via Docker container (https://dockr.ly/35G46a6), and public instance drugcentral:unmtid-dbs.net:5433. A Python API is also available (https://bit.ly/2RAHRtV).
DATA AVAILABILITY
No new data were generated or analysed in support of this research. The DrugCentral web interface is accessible at https://drugcentral.org/ from multiple platforms (Windows, Linux, Android, Apple OS) via desktops, laptops, tablets or smartphones.
Supplementary Material
ACKNOWLEDGEMENTS
We thank ChemAxon (http://www.chemaxon.com) for the academic license of the software used in this study. We thank Dr I.M. (Inge) van Geijlswijk (Utrecht University, the Netherlands) for pharmaceutical literature queries.
Notes
Present address: Thomas B. Wilson, Lovelace Medical Center, 601 Dr Martin Luther King Jr Ave NE, Albuquerque, NM 87102, USA.
Contributor Information
Sorin Avram, Department of Computational Chemistry, “Coriolan Dragulescu” Institute of Chemistry, 24 Mihai Viteazu Blvd, Timişoara, Timiş 300223, Romania.
Thomas B Wilson, College of Pharmacy, University of New Mexico Health Sciences Center, 2502 Marble Ave, Albuquerque, NM 87106, USA.
Ramona Curpan, Department of Computational Chemistry, “Coriolan Dragulescu” Institute of Chemistry, 24 Mihai Viteazu Blvd, Timişoara, Timiş 300223, Romania.
Liliana Halip, Department of Computational Chemistry, “Coriolan Dragulescu” Institute of Chemistry, 24 Mihai Viteazu Blvd, Timişoara, Timiş 300223, Romania.
Ana Borota, Department of Computational Chemistry, “Coriolan Dragulescu” Institute of Chemistry, 24 Mihai Viteazu Blvd, Timişoara, Timiş 300223, Romania.
Alina Bora, Department of Computational Chemistry, “Coriolan Dragulescu” Institute of Chemistry, 24 Mihai Viteazu Blvd, Timişoara, Timiş 300223, Romania.
Cristian G Bologa, Translational Informatics Division, Department of Internal Medicine, University of New Mexico Health Sciences Center, 700 Camino de Salud NE, Albuquerque, NM 87106, USA.
Jayme Holmes, Translational Informatics Division, Department of Internal Medicine, University of New Mexico Health Sciences Center, 700 Camino de Salud NE, Albuquerque, NM 87106, USA.
Jeffrey Knockel, Department of Computer Science, University of New Mexico, 1901 Redondo S Dr, Albuquerque, NM 87106, USA.
Jeremy J Yang, Translational Informatics Division, Department of Internal Medicine, University of New Mexico Health Sciences Center, 700 Camino de Salud NE, Albuquerque, NM 87106, USA.
Tudor I Oprea, Translational Informatics Division, Department of Internal Medicine, University of New Mexico Health Sciences Center, 700 Camino de Salud NE, Albuquerque, NM 87106, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) Common Fund [U24 CA224370 to S.A., C.G.B., J.H., T.B.W., R.C., L.H., ABora, ABorota, J.J.Y., J.K., T.I.O.]. Funding for open access charge: NIH [U24 CA224370].
Conflict of interest statement. T.I.O. and C.B. were full-time employees of Roivant Sciences Inc. T.I.O. received honoraria or consulted for Abbott, AstraZeneca, Chiron, Genentech, Infinity Pharmaceuticals, Merz Pharmaceuticals, Merck KGaA, Mitsubishi Tanabe, Novartis, Ono Pharmaceuticals, Pfizer, Roche, Roivant Discovery, Sanofi and Wyeth. He served on the Scientific Advisory Board of ChemDiv Inc. and InSilico Medicine. The other authors declare no competing interests.
REFERENCES
- 1. Ursu O., Holmes J., Knockel J., Bologa C.G., Yang J.J., Mathias S.L., Nelson S.J., Oprea T.I.. DrugCentral: online drug compendium. Nucleic Acids Res. 2016; 45:D932–D939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ursu O., Holmes J., Bologa C.G., Yang J.J., Mathias S.L., Stathias V., Nguyen D.-T., Schürer S., Oprea T.. DrugCentral 2018: an update. Nucleic Acids Res. 2019; 47:D963–D970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avram S., Bologa C.G., Holmes J., Bocci G., Wilson T.B., Nguyen D.-T., Curpan R., Halip L., Bora A., Yang J.J.et al.. DrugCentral 2021 supports drug discovery and repositioning. Nucleic Acids Res. 2021; 49:D1160–D1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alexander S.P.H., Kelly E., Mathie A., Peters J.A., Veale E.L., Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., Sharman J.L.et al.. THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: introduction and other protein targets. Br. J. Pharmacol. 2019; 176:S1–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambers J., Davies M., Gaulton A., Hersey A., Velankar S., Petryszak R., Hastings J., Bellis L., McGlinchey S., Overington J.P.. UniChem: a unified chemical structure cross-referencing and identifier tracking system. J. Cheminform. 2013; 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Škuta C., Southan C., Bartůněk P.. Will the chemical probes please stand up. RSC Med Chem. 2021; 12:1428–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Havrilla J.M., Liu C., Dong X., Weng C., Wang K.. PhenCards: a data resource linking human phenotype information to biomedical knowledge. Genome Med. 2021; 13:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang W., Zhang Y., Min Z., Mo J., Ju Z., Guan W., Zeng B., Liu Y., Chen J., Zhang Q.et al.. COVID19db: a comprehensive database platform to discover potential drugs and targets of COVID-19 at whole transcriptomic scale. Nucleic Acids Res. 2022; 50:D747–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kropiwnicki E., Binder J.L., Yang J.J., Holmes J., Lachmann A., Clarke D.J.B., Sheils T., Kelleher K.J., Metzger V.T., Bologa C.G.et al.. Getting started with the IDG KMC datasets and tools. Curr. Protoc. 2022; 2:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King L.J., Centers For Disease Control And Prevention Veterinary medicine and public health at CDC. MMWR Suppl. 2006; 55:7–9. [PubMed] [Google Scholar]
- 13. Scott K.A., Qureshi M.H., Cox P.B., Marshall C.M., Bellaire B.C., Wilcox M., Stuart B.A.R., Njardarson J.T.. A structural analysis of the FDA green book-approved veterinary drugs and roles in human medicine. J. Med. Chem. 2020; 63:15449–15482. [DOI] [PubMed] [Google Scholar]
- 14. Milton P., Hamley J.I.D., Walker M., Basáñez M.-G.. Moxidectin: an oral treatment for human onchocerciasis. Expert Rev. Anti. Infect. Ther. 2020; 18:1067–1081. [DOI] [PubMed] [Google Scholar]
- 15. Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y.et al.. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020; 30:678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wittich C.M., Burkle C.M., Lanier W.L.. Ten common questions (and their answers) about off-label drug use. Mayo Clin. Proc. 2012; 87:982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Radley D.C., Finkelstein S.N., Stafford R.S.. Off-label prescribing among office-based physicians. Arch. Intern. Med. 2006; 166:1021–1026. [DOI] [PubMed] [Google Scholar]
- 18. Demonaco H.J., Ali A., Hippel E.von. The major role of clinicians in the discovery of off-label drug therapies. Pharmacotherapy. 2006; 26:323–332. [DOI] [PubMed] [Google Scholar]
- 19. Avram S., Halip L., Curpan R., Borota A., Bora A., Oprea T.I.. Annotating off-label drug usage from unconventional sources. 2022; medRxiv doi:09 September 2022, preprint: not peer reviewed 10.1101/2022.09.08.22279709. [DOI]
- 20. James J.T. A new, evidence-based estimate of patient harms associated with hospital care. J. Patient Saf. 2013; 9:122–128. [DOI] [PubMed] [Google Scholar]
- 21. Makary M.A., Daniel M.. Medical error-the third leading cause of death in the uS. BMJ. 2016; 353:i2139. [DOI] [PubMed] [Google Scholar]
- 22. Stockwell D.C., Landrigan C.P., Toomey S.L., Loren S.S., Jang J., Quinn J.A., Ashrafzadeh S., Wang M.J., Wu M., Sharek P.J.et al.. Adverse events in hospitalized pediatric patients. Pediatrics. 2018; 142:2017–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tambuyzer E., Vandendriessche B., Austin C.P., Brooks P.J., Larsson K., Miller Needleman K.I., Valentine J., Davies K., Groft S.C., Preti R.et al.. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat. Rev. Drug Discov. 2020; 19:93–111. [DOI] [PubMed] [Google Scholar]
- 24. Avram S., Curpan R., Halip L., Bora A., Oprea T.I.. Off-Patent drug repositioning. J. Chem. Inf. Model. 2020; 60:5746–5753. [DOI] [PubMed] [Google Scholar]
- 25. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J.. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006; 34:D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M.et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004; 32:D267–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harding S.D., Armstrong J.F., Faccenda E., Southan C., Alexander S.P.H., Davenport A.P., Pawson A.J., Spedding M., Davies J.A.NC-IUPHAR . The IUPHAR/BPS guide to PHARMACOLOGY in 2022: curating pharmacology for COVID-19, malaria and antibacterials. Nucleic Acids Res. 2022; 50:D1282–D1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oprea T.I., Bologa C.G., Brunak S., Campbell A., Gan G.N., Gaulton A., Gomez S.M., Guha R., Hersey A., Holmes J.et al.. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 2018; 17:317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T.I.et al.. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017; 16:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ursu O., Glick M., Oprea T.. Novel drug targets in 2018. Nat. Rev. Drug Discov. 2019; 18:328. [DOI] [PubMed] [Google Scholar]
- 34. Avram S., Halip L., Curpan R., Oprea T.I.. Novel drug targets in 2019. Nat. Rev. Drug Discov. 2020; 19:300. [DOI] [PubMed] [Google Scholar]
- 35. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pafilis E., Frankild S.P., Fanini L., Faulwetter S., Pavloudi C., Vasileiadou A., Arvanitidis C., Jensen L.J.. The SPECIES and ORGANISMS resources for fast and accurate identification of taxonomic names in text. PLoS One. 2013; 8:e65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Björling E., Uhlén M.. Antibodypedia, a portal for sharing antibody and antigen validation data. Mol. Cell. Proteomics. 2008; 7:2028–2037. [DOI] [PubMed] [Google Scholar]
- 38. Consortium TheUniProt, Bateman A., Martin M.-J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R.et al.. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen D.-T., Mathias S., Bologa C., Brunak S., Fernandez N., Gaulton A., Hersey A., Holmes J., Jensen L.J., Karlsson A.et al.. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017; 45:D995–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheils T.K., Mathias S.L., Kelleher K.J., Siramshetty V.B., Nguyen D.-T., Bologa C.G., Jensen L.J., Vidović D., Koleti A., Schürer S.C.et al.. TCRD and pharos 2021: mining the human proteome for disease biology. Nucleic Acids Res. 2021; 49:D1334–D1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Avram S., Halip L., Curpan R., Oprea T.I.. Novel drug targets in 2020. Nat. Rev. Drug Discov. 2021; 20:333. [DOI] [PubMed] [Google Scholar]
- 42. Avram S., Halip L., Curpan R., Oprea T.I.. Novel drug targets in 2021. Nat. Rev. Drug Discov. 2022; 21:328. [DOI] [PubMed] [Google Scholar]
- 43. Nelson S.J. Medical terminologies that work: the example of MeSH. 2009 10th International Symposium on Pervasive Systems, Algorithms, and Networks. 2009; 380–384. [Google Scholar]
- 44. Shingjergji K., Celebi R., Scholtes J., Dumontier M.. Relation extraction from DailyMed structured product labels by optimally combining crowd, experts and machines. J. Biomedical Semantics. 2021; 122:103902. [DOI] [PubMed] [Google Scholar]
- 45. Center for Veterinary Medicine Approved Animal Drug Products (Green Book) U.S. Food and Drug Administration. 2022; https://www.fda.gov/animal-veterinary/products/approved-animal-drug-products-green-book. [Google Scholar]
- 46. Yang J.J., Ursu O., Lipinski C.A., Sklar L.A., Oprea T.I., Bologa C.G.. Badapple: promiscuity patterns from noisy evidence. J. Cheminform. 2016; 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ventola C.L. Off-label drug information: regulation, distribution, evaluation, and related controversies. P T. 2009; 34:428–440. [PMC free article] [PubMed] [Google Scholar]
- 48. Huang L., Zalkikar J., Tiwari R.C.. A likelihood ratio test based method for signal detection with application to FDA’s drug safety data. J. Am. Stat. Assoc. 2011; 106:1230–1241. [Google Scholar]
- 49. Janssen P.A.A. Heterocyclic derivatives of 1-phenyl-omega-(piperazine)alkanols. 1961; US Patent 2979508. [Google Scholar]
- 50. Marsboom R., Symoens J.. Ervaringen met azaperone (R1929*) als sedativum bij het varken. Tijdschr. Diergeneeskd. 1968; 93:3–15. [Google Scholar]
- 51. Levin J.M., Oprea T.I., Davidovich S., Clozel T., Overington J.P., Vanhaelen Q., Cantor C.R., Bischof E., Zhavoronkov A.. Artificial intelligence, drug repurposing and peer review. Nat. Biotechnol. 2020; 38:1127–1131. [DOI] [PubMed] [Google Scholar]
- 52. Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J.. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020; 395:e30–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith D.P., Oechsle O., Rawling M.J., Savory E., Lacoste A.M.B., Richardson P.J.. Expert-Augmented computational drug repurposing identified baricitinib as a treatment for COVID-19. Front. Pharmacol. 2021; 12:709856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Selvaraj V., Finn A., Lal A., Khan M.S., Dapaah-Afriyie K., Carino G.P.. Baricitinib in hospitalised patients with COVID-19: a meta-analysis of randomised controlled trials. EClinicalMedicine. 2022; 49:101489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fajgenbaum D.C., June C.H.. Cytokine storm. N. Engl. J. Med. 2020; 383:2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huarte E., Peel M.T., Verbist K., Fay B.L., Bassett R., Albeituni S., Nichols K.E., Smith P.A.. Ruxolitinib, a JAK1/2 inhibitor, ameliorates cytokine storm in experimental models of hyperinflammation syndrome. Front. Pharmacol. 2021; 12:650295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cao Y., Wei J., Zou L., Jiang T., Wang G., Chen L., Huang L., Meng F., Huang L., Wang N.et al.. Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J. Allergy Clin. Immunol. 2020; 146:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988; 28:31–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analysed in support of this research. The DrugCentral web interface is accessible at https://drugcentral.org/ from multiple platforms (Windows, Linux, Android, Apple OS) via desktops, laptops, tablets or smartphones.