Abstract
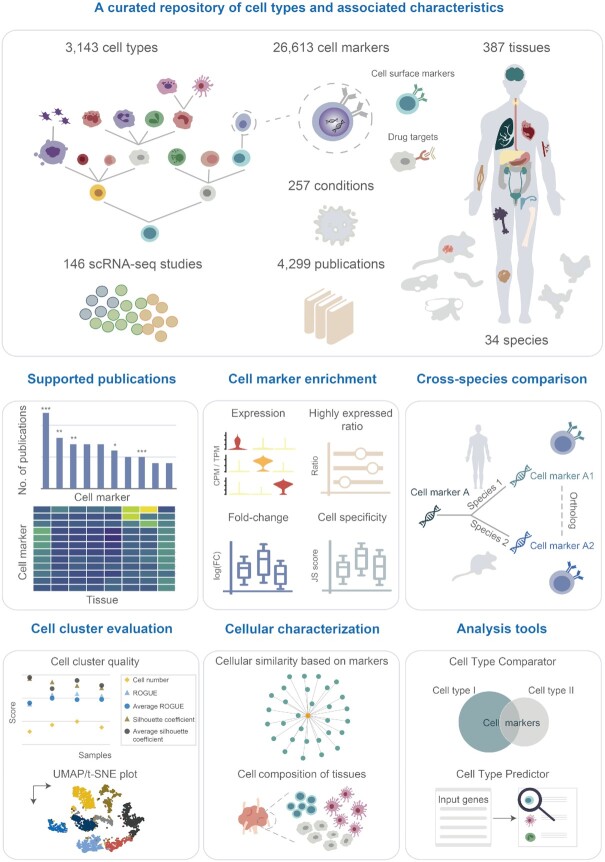
Single-cell studies have delineated cellular diversity and uncovered increasing numbers of previously uncharacterized cell types in complex tissues. Thus, synthesizing growing knowledge of cellular characteristics is critical for dissecting cellular heterogeneity, developmental processes and tumorigenesis at single-cell resolution. Here, we present Cell Taxonomy (https://ngdc.cncb.ac.cn/celltaxonomy), a comprehensive and curated repository of cell types and associated cell markers encompassing a wide range of species, tissues and conditions. Combined with literature curation and data integration, the current version of Cell Taxonomy establishes a well-structured taxonomy for 3,143 cell types and houses a comprehensive collection of 26,613 associated cell markers in 257 conditions and 387 tissues across 34 species. Based on 4,299 publications and single-cell transcriptomic profiles of ∼3.5 million cells, Cell Taxonomy features multifaceted characterization for cell types and cell markers, involving quality assessment of cell markers and cell clusters, cross-species comparison, cell composition of tissues and cellular similarity based on markers. Taken together, Cell Taxonomy represents a fundamentally useful reference to systematically and accurately characterize cell types and thus lays an important foundation for deeply understanding and exploring cellular biology in diverse species.
INTRODUCTION
Single-cell sequencing technologies have emerged as a powerful approach to delineate cellular composition diversity (1), trace cell lineages (2), characterize tumor microenvironment (3) and elucidate complex mechanisms of organ development and diseases at single-cell scale (4). Particularly, single-cell multi-omics studies have uncovered a large variety of previously uncharacterized cell populations, thereby providing unprecedented opportunities to capture a whole picture of cellular composition diversity (5–7). Therefore, synthesizing our growing knowledge of cellular characteristics by accounting for diverse species, tissues and biological conditions is critical for revealing cellular heterogeneity, developmental processes and tumorigenesis at single-cell resolution.
In the past several years, valuable efforts have been made to establish databases for cell types and/or cell markers (8–14). Among them, representative databases are Cell Ontology (8), CellFinder (14), CellMarker (9), SHOGoiN (10) and PanglaoDB (11). Specifically, Cell Ontology, a widely used ontology for the representation of cell types, provides hierarchical relationships for approximately 2,600 cell types (8). CellFinder characterizes 3,394 mammalian cell types and contains ∼200 microarray and bulk RNA-seq profiles (14). Considering that one cell type may have multiple cell markers, CellMarker constructs a curated compendium of 15,778 cell markers for 1,702 cell types in human and 11,388 cell makers for 1,421 cell types in mouse (9). Similarly, SHOGoiN integrates diverse human cell information including 23,000 entries of 355 cell types and 5,630 cell markers along with 7,161 single-cell RNA-sequencing (scRNA-seq) profiles (10). In addition, PanglaoDB collects 178 cell types and 4,681 markers as well as scRNA-seq data for human and mouse (11). However, these existing databases, albeit widely used, have several limitations. First, most of cell types in Cell Ontology are not associated with available cell markers, whereas CellMarker, SHOGoiN and PanglaoDB contain limited cell types. Second, Cell Ontology, CellFinder and CellMarker do not provide single-cell transcriptome evaluation for cell markers. Third, existing databases cover limited species and none of them enable comparison of cell types across species. Last but not least, multiple evidence-based assessment for cell markers and cell clusters is not adequate in these databases, thus presenting substantial challenges in cell type delineation.
To address these issues, here we present Cell Taxonomy (https://ngdc.cncb.ac.cn/celltaxonomy), a comprehensive and curated repository of cell types and cell markers covering a wide range of species, tissues and conditions. Based on 4,299 literature as well as integration from relevant resources, Cell Taxonomy houses a comprehensive collection of 3,143 cell types and 26,613 associated cell markers in 257 conditions and 387 tissues across 34 species. Additionally, it incorporates single-cell RNA-seq profiles comprising approximately 3.5 million cells to enable single-cell validation for cell markers. Furthermore, Cell Taxonomy performs cross-species comparison of cell types and cell markers based on orthologous information. Importantly, to help users to select robust cell markers, Cell Taxonomy provides quality evaluations based on multiple evidence of expression enrichment, supported literature and conservation across species. And extensive assessments of cell clusters in scRNA-seq studies are provided to facilitate users to choose high-quality expression profiles for cell annotation. Moreover, cell composition of tissues, cellular similarity based on markers and cell surface marker information are characterized in Cell Taxonomy. And a set of easy-to-use tools is deployed to predict cell types by user-provided gene list and to compare cell markers between cell types.
MATERIALS AND METHODS
Data collection and curation
A comprehensive collection of cell types, cell markers, tissues and conditions were manually curated from 1,555 single-cell relevant publications by Cell Taxonomy curation team as well as integrated from multiple resources including Cell Ontology (8), CellMarker (9), SHOGoiN (10), PanglaoDB (11), CellMatch (12), scTyper.db (13), Human Cell Landscape (HCL) (15), OnClass (16), Human Cell Atlas (17), tinyatlas (https://github.com/hbc/tinyatlas) and Invitrogen Immune Cell Guide (https://www.thermofisher.com/hk/en/home/global/forms/download-immune-cell-guide.html). The cell types labeled as ‘obsolete’ in Cell Ontology were deprecated. Specifically, full texts of these publications were manually surveyed to extract essential information of cell types, species, tissues, conditions, cellular annotation, cell markers and cellular hierarchical relationships based on controlled vocabularies (https://ngdc.cncb.ac.cn/celltaxonomy/help#curation_model). Since most single-cell studies named cell types without controlled vocabularies, all cell types collected in Cell Taxonomy were manually curated based on standardized terms and assigned with a unique identifier prefixed with CT. To remove the redundant entries, cell markers from the same species, tissues, cell types, conditions and publications were merged. Cell surface markers in human were collected and filtered from the Human Protein Atlas (https://v13.proteinatlas.org/humanproteome/secretome) by selecting genes that code for membrane proteins or membrane and secreted proteins with supported evidence at protein or transcript level. Gene entities were standardized using unique identifiers from Entrez as well as Ensembl (18), UniPort (19) and Pfam (20). The descriptive terms for tissues and diseases were adopted from Uber-anatomy ontology (UBERON) (21) and Disease Ontology (DO) (22), and cellular images were downloaded from Cell Image Library (23).
scRNA-seq data collection and analysis
To explore expression profiles for cell types and cell markers, a total of 146 scRNA-seq studies for human and mouse were integrated from Human Cell Atlas (17,24–27), CellBlast (28), GEO (29), Mouse Cell Atlas (30), Tabula Muris (31), PanglaoDB (11) and Single Cell Portal (https://singlecell.broadinstitute.org/single_cell). The single-cell samples were selected by the following two criteria: (i) available annotation of cell types and (ii) the number of cell types ≥ 2. These studies comprised libraries of 10X Chromium (32), SMART-seq2 (33), Drop-seq (34), Microwell-seq (30) and Seq-Well (35). Cells were labeled based on cell types classified in the original publications, and the cell types, tissues and conditions in single-cell studies were manually annotated with standardized names in Cell Taxonomy. To visualize high-dimensional expression profiles, t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) were implemented by RunTSNE and RunUMAP in R Seurat v4.0 package (parameters’ setting: -n.components = 2 or 3, -seed.use = 100, and perplexity = 30) (36–39).
Quality evaluation of cell markers
Multiple metrics for evaluating cell markers were calculated, including expression enrichment (fold-change [FDR < 0.05], highly expressed ratio, cell specificity score), supported-publication count and conservation score. Differential pattern of cell markers between one cell type and others was calculated by FindMarkers in Seurat v4.0 (log2FC > 0.25, FDR < 0.05) (36). Highly expressed ratio was the number of samples in which genes are more highly expressed in specific cell type compared to other cell types (log2FC > 0.25, FDR < 0.05) divided by the number of total samples for this cell type and gene. In addition, Jensen-Shannon (JS) divergence score, ranging from 0 to 1, was calculated to quantify the cell type specificity of cell markers (40,41). The number of supported publications for cell markers in various species and tissues was counted, and the Fisher’s exact test was performed to examine whether cell markers are specifically associated with one certain cell type by publication count. In addition, conservation score was the number of orthologous genes that were also reported as cell markers for this cell type in other species.
Assessment of cell clusters in scRNA-seq studies
To measure the accuracy or goodness of cell clusters, five indexes were calculated including cell number, silhouette coefficient (42), ROGUE purity score (43), average silhouette coefficient and average ROGUE of all cell clusters. Silhouette coefficient ranges from -1 to 1, indicating the difference between intra-cluster distance and inter-cluster distance, where 1 means clusters are well apart from each other, 0 means clusters are indifferent, and -1 means clusters are defined in the wrong way (42). ROGUE purity score ranges from 0 to 1 and a greater value indicates a cell cluster with higher purity (43). All statistical analyses were performed by R 4.0.3.
Similarity analysis of cell types based on cell markers
The similarity between two cell types was measured according to their cell markers, which can be estimated as
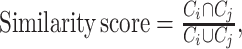 where
where 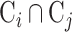 is the intersection of cell markers between cell type i and cell type j and
is the intersection of cell markers between cell type i and cell type j and 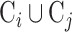 is the union of cell markers of the two cell types. The similarity score ranges from 0 to 1, where 1 indicates that the two cell types have the completely same cell markers and 0 means the two cell types share no common cell marker.
is the union of cell markers of the two cell types. The similarity score ranges from 0 to 1, where 1 indicates that the two cell types have the completely same cell markers and 0 means the two cell types share no common cell marker.
Cell marker orthology and cross-species comparison
Considering that some cell types may lack reported cell markers in certain species, orthologous genes of cell markers in other species may provide potential clues. We downloaded and extracted the orthologous relationships from NCBI HomoloGene (https://ftp.ncbi.nih.gov/pub/HomoloGene). The orthologous information for cell markers in multiple species is presented in the browse page of cell markers as well as individual cell marker page. To compare the same cell type among different species, we identified orthologous genes that were also reported as cell markers for the same cell type. The cell-type similarity across species was evaluated by the number of orthologous cell markers divided by the union of cell markers in the two species.
Database implementation
Cell Taxonomy was built by SpringBoot (https://spring.io/projects/spring-boot) and Mybatis (https://mybatis.org/mybatis-3) as backend web framework and MySQL (https://www.mysql.com) as database engine. Web interfaces were developed by HTML (HyperText Markup Language), CSS (Cascading Style Sheets), Thymeleaf (https://www.thymeleaf.org) and AJAX (Asynchronous JavaScript and XML). Bootstrap (https://getbootstrap.com) was adopted as a frontend framework, offering consistent templates, components and interfaces for facilitating the development of web pages. Also, data visualization was rendered by HighCharts (https://www.highcharts.com.cn), ECharts (http://echarts.apache.org), Plotly.js (https://plotly.com/javascript) and DataTables (https://datatables.net).
DATABASE CONTENTS AND FEATURES
Cell Taxonomy provides a comprehensive and curated repository for cell types and associated cell markers in combination with multifaceted cellular characterization (Figure 1). A standardized taxonomy is constructed for 3,143 cell types by literature curation and comprehensive integration, and 26,613 cell markers are collected based on 4,299 publications, covering 257 conditions and 387 tissues across 34 species. In addition, single-cell transcriptomic profiles of ∼3.5 million cells from 146 scRNA-seq studies are incorporated to help users explore expression profiles of cell types and cell markers. Importantly, Cell Taxonomy provides multi-evidence assessment of cell markers and cell clusters, facilitating users to select high-quality cell markers and expression profiles for cell types under investigation. Also, extensive cellular characterization is provided, including cross-species comparison of cell types and cell markers, cell composition of tissues and cellular similarity based on cell markers. Moreover, two interactive tools are developed to enable users to predict cell types given a user-defined gene list and compare cell markers of different cell types.
Figure 1.
Schematic overview of the contents and functionalities in Cell Taxonomy. Data are organized and browsed in terms of cell types, cell markers, tissues, species, conditions, studies and publications. Multifaceted cellular characterization is provided including literature supported evidence, cell marker enrichment, cross-species comparison, cell cluster evaluation and cellular characterization. Two analysis tools are deployed online for comparing and predicting cell types.
A standardized taxonomy for cell types
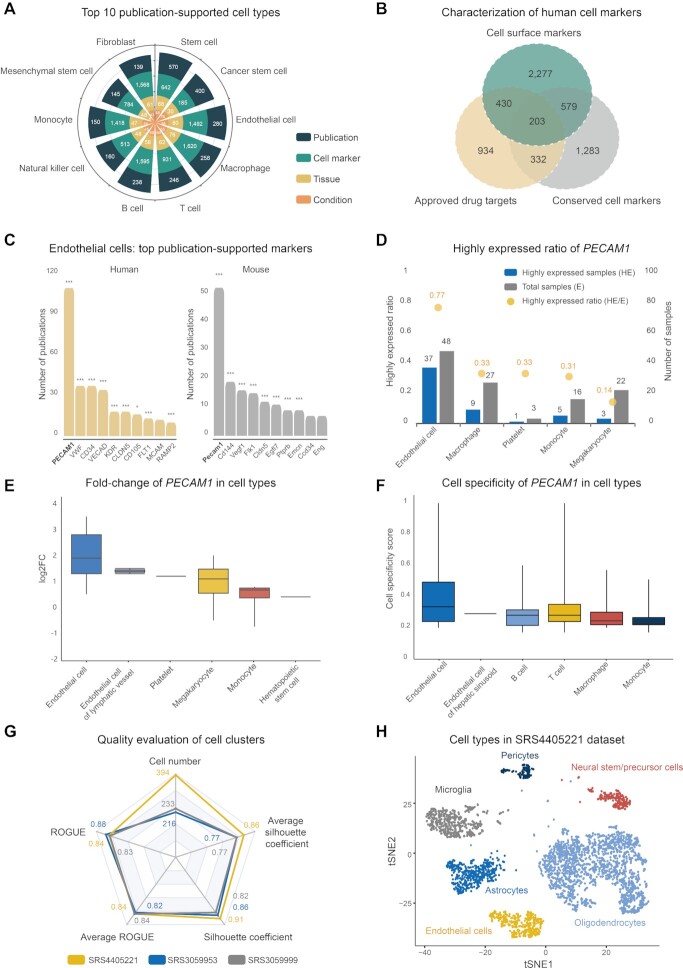
To structurally describe diverse cell types, Cell Taxonomy constructs a standardized taxonomy for cell types based on manual curation and comprehensive integration (see details in Materials and Methods). Thus, each cell type in Cell Taxonomy is assigned with a unique taxonomy ID and associated with an abundance of relevant information based on a curation model (https://ngdc.cncb.ac.cn/celltaxonomy/help#curation_model/). As a result, the current version of Cell Taxonomy presents a comprehensive and well-structured taxonomy for 3,143 cell types, and 82.8% (2,601) are annotated with available cell markers, giving rise to a total of 226,222 entries collected from 4,299 publications. We find that stem cells, cancer stem cells and endothelial cells top the ranking of publication-supported cell types; stem cells are associated with 642 cell markers in 68 tissues and 48 conditions (Figure 2A). Importantly, 438 cell types previously uncharacterized in other databases are curated from 1,555 publications with available cell markers. And 397 cell types currently undescribed in Cell Ontology are incorporated into an expanded cell topology tree with standardized Cell Taxonomy IDs and descriptive terms, expanding the compendium of Cell Ontology by 15.3%. Collectively, a standardized taxonomy for curated cell types is of fundamental significance for characterizing cell types and disentangling cellular heterogeneity.
Figure 2.
Statistics of Cell Taxonomy and assessment of cell markers and cell clusters. (A) Top 10 publication-supported cell types as well as their statistics in terms of cell markers, tissues, conditions and publications. (B) Characterization of cell markers in aspect of cell surface markers, approved drug targets and conservation across species. (C) Top 10 publication-supported cell markers of endothelial cells in human and mouse. Statistical significance levels by p-values are calculated by the Fisher’s exact test with FDR correction (BH) and coded by: *** for < 0.001, ** for [0.001, 0.01) and * for [0.01, 0.05) (https://ngdc.cncb.ac.cn/celltaxonomy/celltype/CT:00000153#cell_marker/). (D) Highly expressed ratio of PECAM1 in human cell types (https://ngdc.cncb.ac.cn/celltaxonomy/celltype/CT:00000153#cell_marker_expression_enrichment/). (E) The distribution of expression fold-change of PECAM1 in various human cell types across samples. (F) Cell specificity score of PECAM1 in various human cell types across samples. (G) Quality evaluation of cell clusters for endothelial cells based on silhouette coefficient, ROGUE purity score and their averaged estimates as well as cell number. (H) t-SNE graph of cell types in SRS4405221 (https://ngdc.cncb.ac.cn/celltaxonomy/study/31270459#dimensional_reduction/).
A comprehensive compendium of cell markers
To date, Cell Taxonomy houses 26,613 available cell markers for 2,601 cell types in 34 species supported by 4,299 publications. Cell surface markers are commonly used in pre-classifying certain cell types via fluorescence-activated cell sorting (FACS) and clinical use in drug discovery for extracellular accessibility for pharmacological intervention (44). Among 15,316 human cell markers in Cell Taxonomy, 3,489 genes are predicted to be cell surface markers for 758 cell types, which code for membrane-bound protein products, and 93.8% (3,271) are supported by evidence at protein level and 88.0% (3,071) are annotated with well-characterized antibodies (45) (Figure 2B). In addition, 1,899 human cell markers are targeted by approved drugs listed in DrugBank (46), which are mostly inhibitor targets (876) and antagonist targets (288) (Figure 2B). Specifically, 633 human cell surface markers are approved drug targets mostly by inhibitor and antagonist (Supplementary Figure S1A). Moreover, according to cross-species comparison of cell markers, 2,397 human cell markers are conserved across various species in 937 cell types (Figure 2B). For example, PECAM1 is the classic cell marker for human endothelial cells, and consistently, Pecam1, its ortholog in mouse, is also extensively reported as cell marker for mouse endothelial cells (Figure 2B and Supplementary Figure S1B–E). As a result, there are 203 human cell surface markers that are targeted by approved drugs and conserved across various species (Figure 2B).
Extensive assessment of cell markers
High-quality cell markers are essential for cell type identification. To help users select high-quality cell markers, Cell Taxonomy provides quality assessment metrics based on multiple evidences from supported literature, expression enrichment and cross-species conservation. Based on the above metrics, thus, high-confidence cell markers can be prioritized and obtained. Taking human endothelial cells as an example, among 1,492 cell markers reported by 280 publications and databases, PECAM1 is prioritized as high-confidence cell marker (Supplementary Figure S1F). In specific, PECAM1 is extensively reported as cell marker for human endothelial cells in Cell Taxonomy, which is supported by 109 publications and specifically associated with endothelial cells relative to other cell types as shown in the panel of ‘Cell marker’ (FDR = 2.1e-75, Figure 2C). Moreover, based on the comparison across species, the orthologous genes of PECAM1 in Mus musculus, Rattus norvegicus and Gallus gallus are also reported as cell markers for endothelial cells (Supplementary Figure S1E). Additionally, Cell Taxonomy also evaluates the enrichment pattern of cell markers in 146 scRNA-seq studies containing ∼3.5 million single cells in the panel of ‘Cell marker expression enrichment’. For example, among single-cell samples for endothelial cells, PECAM1 is more highly expressed in endothelial cells compared to other cell types (log2FC > 0.25, FDR < 0.05) in 77% of these samples (highly expressed ratio at 0.77, Figure 2D), along with high median value of log2FC at 2.0 (Figure 2E) and cell specificity score at 0.34 (Figure 2F). Moreover, Cell Taxonomy enables users to investigate cell marker enrichment pattern in specific scRNA-seq study (Supplementary Figure S2A–C). Together, Cell Taxonomy serves as a useful reference to ease users to choose high-quality cell markers for cell types and determine highly relevant cell types for specific genes.
Evaluation of cell clusters based on scRNA-seq profiles
Cell type annotation highly depends on reference expression profiles for accurate assignment of cell types (47). Therefore, based on scRNA-seq profiles of ∼3.5 million cells from 679 samples in 146 studies, Cell Taxonomy offers the quality evaluation of cell clusters to ease users to select high-quality expression profiles for cell type assignment. To measure the goodness of cell clusters, five indexes are provided, including silhouette coefficient, ROGUE purity score, their averaged estimates over all cell clusters, as well as cell number (Supplementary Figure S3A). For example, among scRNA-seq samples for mouse endothelial cells, there are three samples (SRS4405221, SRS3059953 and SRS3059999; by setting the filters of cell number, ROGUE, its averaged estimate at top 25% and silhouette coefficient, its averaged estimate at top 50%) that have expression profiles of higher quality for endothelial cells (Figure 2G). As for SRS4405221, the endothelial cell clusters are of high silhouette coefficient at 0.91 and ROGUE purity score at 0.84 (Figure 2G). It means that endothelial cells are considerably apart from other cell clusters and relatively of high purity, which is also consistent well with the t-SNE graph in the panel of ‘Dimensional reduction’ in the scRNA-seq study page (Figure 2H). In addition, the overall quality of cell clusters in SRS4405221 is relatively high with average silhouette coefficient at 0.86 and average ROGUE value at 0.84. Therefore, these results indicate that this sample is a potential high-quality expression profile for endothelial cells. Similar pattern is observed for another two samples of SRS3059953 and SRS3059999. Collectively, Cell Taxonomy performs extensive assessment of cell clusters in large-scale scRNA-seq studies and bears wide utility for selecting high-quality expression profiles for cell type annotation.
Cellular composition of tissues and similarity analysis
To systematically explore cell heterogeneity across tissues, Cell Taxonomy infers the cell composition in various tissues based on representative large-scale scRNA-seq studies in the panel of ‘cellular composition of tissues’. For example, endothelial cells are widely distributed in 13 human tissues in Tabula sapiens and 10 mouse tissues in Tabula Muris, while type II pneumocytes are specifically abundant in lung in both Tabula sapiens and Tabula Muris (Supplementary Figure S3B–E). Additionally, Cell Taxonomy is capable to estimate the cellular similarity based on cell markers, which can be used to characterize cell types and reveal potential cellular functions. In human liver, for instance, central memory CD4+ T cells are very similar with central memory CD8+ T cells in terms of their marker component (similarity score = 0.90) as shown in the ‘cellular similarity based on cell marker’ panel (Supplementary Figure S3F). Moreover, for the same cell type, Cell Taxonomy provides the similarity comparison among different species in light of orthologous cell markers, as demonstrated in the panel of ‘cellular comparison across species’ (Supplementary Figure S3G).
Content organization and access
Cell Taxonomy organizes and presents all contents (including curated information and data) in terms of cell types, cell markers, species, tissues, conditions, publications and studies, corresponding to seven different modules, respectively (Figure 1). The cell type module contains cell marker assessment (publication, expression enrichment, cross-species conservation and cell surface marker), cell cluster evaluation and cellular characterization (cell composition of tissues and cellular similarity based on markers). The cell marker module prioritizes highly relevant cell types for specific genes based on supported publications, expression enrichment and cross-species conservation. The tissue module visualizes all relevant cell types and cell markers supported by literature. The species and condition modules list all associated cell types, cell markers, tissues and publications. The scRNA-seq study module enables users to search and filter specific scRNA-seq studies that match certain criteria (such as species, cell type and tissue) and interactively visualize single-cell expression profiles, facilitating in-depth exploration of cell marker enrichment and cell cluster distance. The publication module displays a full list of relevant publications in association with specific curated cell types and cell markers. In addition, Cell Taxonomy deploys two useful tools for comparing cell types based on cell markers and predicting cell types according to user-provided gene list, respectively (Figure 1).
DISCUSSION AND FUTURE DEVELOPMENTS
Single-cell multi-omics sequencing technologies have led to a large number of single-cell studies conducted throughout the world, yielding an unprecedented compendium of cell types (48–52). Here, we construct Cell Taxonomy, a comprehensive and curated repository that features comprehensive literature curation, single-cell data integration, multifaceted cellular characterization, cross-species comparison and extensive evaluation of cell markers and cell clusters. Currently, it houses a total of 3,143 cell types organized in a standardized taxonomy with 26,613 associated cell markers covering 257 conditions and 387 tissues across 34 species. Additionally, Cell Taxonomy delivers interactive online services to facilitate users to predict cell types based on customized gene list and perform cell type comparison. Continuous efforts for upgrading Cell Taxonomy include: (i) manual curation of cell types and cell markers from newly published studies, (ii) integration of newly released datasets covering more single-cell omics, species, tissues and conditions, (iii) frequent updates of web interfaces to improve data presentation and visualization, (iv) enhancing external links to related database resources and (v) adding a submission functionality to accept user-curated information. Taken together, Cell Taxonomy provides a thoroughly characterized landscape of cell types and cell markers in diverse tissues across multiple species and thus bears great potential to serve as a broadly useful cellular reference for the global scientific communities.
DATA AVAILABILITY
Cell Taxonomy can be accessed at https://ngdc.cncb.ac.cn/celltaxonomy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Zhao Li, Qiang Du and Zhuojing Fan for their help on the web design. We also thank a number of users for reporting bugs and providing suggestions as well as anonymous reviewers for their valuable comments on this work.
Contributor Information
Shuai Jiang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Qiheng Qian, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Tongtong Zhu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wenting Zong, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yunfei Shang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Tong Jin, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yuansheng Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Ming Chen, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Zishan Wu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yuan Chu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Rongqin Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Sicheng Luo, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wei Jing, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dong Zou, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Yiming Bao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jingfa Xiao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Zhang Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese Academy of Sciences [XDA19050302; XDB38030400]; Youth Innovation Promotion Association of Chinese Academy of Science [2018134]; National Key Research and Development Program of China [2020YFA0907001; 2017YFC0907502]; Chinese Academy of Sciences [153F11KYSB20160008]; Chinese Academy of Sciences [WX145XQ07-04]; National Natural Science Foundation of China [32100520; 32030021; 31871328; 31100915; 31970634; 31681595]; National Key Research and Development Program of China [2016YFC0901903]; The Open Biodiversity and Health Big Data Programme of IUBS. Funding for open access charge: National Key Research and Development Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang Y., Navin N.E.. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015; 58:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kester L., van Oudenaarden A.. Single-Cell transcriptomics meets lineage tracing. Cell Stem Cell. 2018; 23:166–179. [DOI] [PubMed] [Google Scholar]
- 3. Cheng S., Li Z., Gao R., Xing B., Gao Y., Yang Y., Qin S., Zhang L., Ouyang H., Du P.et al.. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021; 184:792–809. [DOI] [PubMed] [Google Scholar]
- 4. Potter S.S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018; 14:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tosti L., Hang Y., Debnath O., Tiesmeyer S., Trefzer T., Steiger K., Ten F.W., Lukassen S., Ballke S., Kuhl A.A.et al.. Single-Nucleus and in situ RNA-Sequencing reveal cell topographies in the human pancreas. Gastroenterology. 2021; 160:1330–1344. [DOI] [PubMed] [Google Scholar]
- 6. Sanmarco L.M., Wheeler M.A., Gutierrez-Vazquez C., Polonio C.M., Linnerbauer M., Pinho-Ribeiro F.A., Li Z., Giovannoni F., Batterman K.V., Scalisi G.et al.. Gut-licensed IFNgamma(+) NK cells drive LAMP1(+)TRAIL(+) anti-inflammatory astrocytes. Nature. 2021; 590:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D.et al.. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019; 179:829–845. [DOI] [PubMed] [Google Scholar]
- 8. Diehl A.D., Meehan T.F., Bradford Y.M., Brush M.H., Dahdul W.M., Dougall D.S., He Y., Osumi-Sutherland D., Ruttenberg A., Sarntivijai S.et al.. The cell ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics. 2016; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M.et al.. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019; 47:D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hatano A., Chiba H., Moesa H.A., Taniguchi T., Nagaie S., Yamanegi K., Takai-Igarashi T., Tanaka H., Fujibuchi W.. CELLPEDIA: a repository for human cell information for cell studies and differentiation analyses. Database (Oxford). 2011; 2011:bar046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franzen O., Gan L.M., Bjorkegren J.L.M.. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford). 2019; 2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shao X., Liao J., Lu X., Xue R., Ai N., Fan X.. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. Iscience. 2020; 23:100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi J.H., In Kim H., Woo H.G. scTyper: a comprehensive pipeline for the cell typing analysis of single-cell RNA-seq data. BMC Bioinf. 2020; 21:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stachelscheid H., Seltmann S., Lekschas F., Fontaine J.F., Mah N., Neves M., Andrade-Navarro M.A., Leser U., Kurtz A.. CellFinder: a cell data repository. Nucleic Acids Res. 2014; 42:D950–D958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W.et al.. Construction of a human cell landscape at single-cell level. Nature. 2020; 581:303–309. [DOI] [PubMed] [Google Scholar]
- 16. Wang S., Pisco A.O., McGeever A., Brbic M., Zitnik M., Darmanis S., Leskovec J., Karkanias J., Altman R.B.. Leveraging the cell ontology to classify unseen cell types. Nat. Commun. 2021; 12:5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M.et al.. The human cell atlas. Elife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R.et al.. Ensembl 2022. Nucleic Acids Res. 2022; 50:D988–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mungall C.J., Torniai C., Gkoutos G.V., Lewis S.E., Haendel M.A.. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012; 13:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schriml L.M., Munro J.B., Schor M., Olley D., McCracken C., Felix V., Baron J.A., Jackson R., Bello S.M., Bearer C.et al.. The human disease ontology 2022 update. Nucleic Acids Res. 2022; 50:D1255–D1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orloff D.N., Iwasa J.H., Martone M.E., Ellisman M.H., Kane C.M.. The cell: an image library-CCDB: a curated repository of microscopy data. Nucleic Acids Res. 2013; 41:D1241–D1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tabula Sapiens C., Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P.et al.. The tabula sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022; 376:eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eraslan G., Drokhlyansky E., Anand S., Fiskin E., Subramanian A., Slyper M., Wang J., Van Wittenberghe N., Rouhana J.M., Waldman J.et al.. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science. 2022; 376:eabl4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dominguez Conde C., Xu C., Jarvis L.B., Rainbow D.B., Wells S.B., Gomes T., Howlett S.K., Suchanek O., Polanski K., King H.W.et al.. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science. 2022; 376:eabl5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suo C., Dann E., Goh I., Jardine L., Kleshchevnikov V., Park J.E., Botting R.A., Stephenson E., Engelbert J., Tuong Z.K.et al.. Mapping the developing human immune system across organs. Science. 2022; 376:eabo0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao Z.J., Wei L., Lu S., Yang D.C., Gao G.. Searching large-scale scRNA-seq databases via unbiased cell embedding with cell BLAST. Nat. Commun. 2020; 11:3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F.et al.. Mapping the mouse cell atlas by microwell-seq. Cell. 2018; 172:1091–1107. [DOI] [PubMed] [Google Scholar]
- 31. Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group and Principal investigators Single-cell transcriptomics of 20 mouse organs creates a tabula muris. Nature. 2018; 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J.et al.. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R.. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014; 9:171–181. [DOI] [PubMed] [Google Scholar]
- 34. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M.et al.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gierahn T.M., Wadsworth M.H. 2nd, Hughes T.K., Bryson B.D., Butler A., Satija R., Fortune S., Love J.C., Shalek A.K.. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017; 14:395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hao Y., Hao S., Andersen-Nissen E., Mauck W.M. 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M.et al.. Integrated analysis of multimodal single-cell data. Cell. 2021; 184:3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of single-cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A.. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015; 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang S., Cheng S.J., Ren L.C., Wang Q., Kang Y.J., Ding Y., Hou M., Yang X.X., Lin Y., Liang N.et al.. An expanded landscape of human long noncoding RNA. Nucleic Acids Res. 2019; 47:7842–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. J.Rousseeuw P Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987; 20:53–65. [Google Scholar]
- 43. Liu B., Li C., Li Z., Wang D., Ren X., Zhang Z.. An entropy-based metric for assessing the purity of single cell populations. Nat. Commun. 2020; 11:3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bausch-Fluck D., Goldmann U., Muller S., van Oostrum M., Muller M., Schubert O.T., Wollscheid B.. The in silico human surfaceome. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E10988–E10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A.et al.. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 46. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pasquini G., Rojo Arias J.E., Schafer P., Busskamp V.. Automated methods for cell type annotation on scRNA-seq data. Comput. Struct. Biotechnol. J. 2021; 19:961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A.. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015; 58:610–620. [DOI] [PubMed] [Google Scholar]
- 49. Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018; 50:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., He Y., Wang L., Zhang Q., Kim A.et al.. Single-Cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020; 181:442–459. [DOI] [PubMed] [Google Scholar]
- 51. Bian S., Hou Y., Zhou X., Li X., Yong J., Wang Y., Wang W., Yan J., Hu B., Guo H.et al.. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018; 362:1060–1063. [DOI] [PubMed] [Google Scholar]
- 52. Chen L., Fan R., Tang F.. Advanced Single-cell Omics Technologies and Informatics Tools for Genomics, Proteomics, and Bioinformatics Analysis. Genom. Proteom. Bioinf. 2021; 19:343–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cell Taxonomy can be accessed at https://ngdc.cncb.ac.cn/celltaxonomy.