Abstract
The highly polymorphic Major Histocompatibility Complex (MHC) genes are responsible for the binding and cell surface presentation of pathogen or cancer specific T-cell epitopes. This process is fundamental for eliciting T-cell recognition of infected or malignant cells. Epitopes displayed on MHC molecules further provide therapeutic targets for personalized cancer vaccines or adoptive T-cell therapy. To help visualizing, analyzing and comparing the different binding specificities of MHC molecules, we developed the MHC Motif Atlas (http://mhcmotifatlas.org/). This database contains information about thousands of class I and class II MHC molecules, including binding motifs, peptide length distributions, motifs of phosphorylated ligands, multiple specificities or links to X-ray crystallography structures. The database further enables users to download curated datasets of MHC ligands. By combining intuitive visualization of the main binding properties of MHC molecules together with access to more than a million ligands, the MHC Motif Atlas provides a central resource to analyze and interpret the binding specificities of MHC molecules.
INTRODUCTION
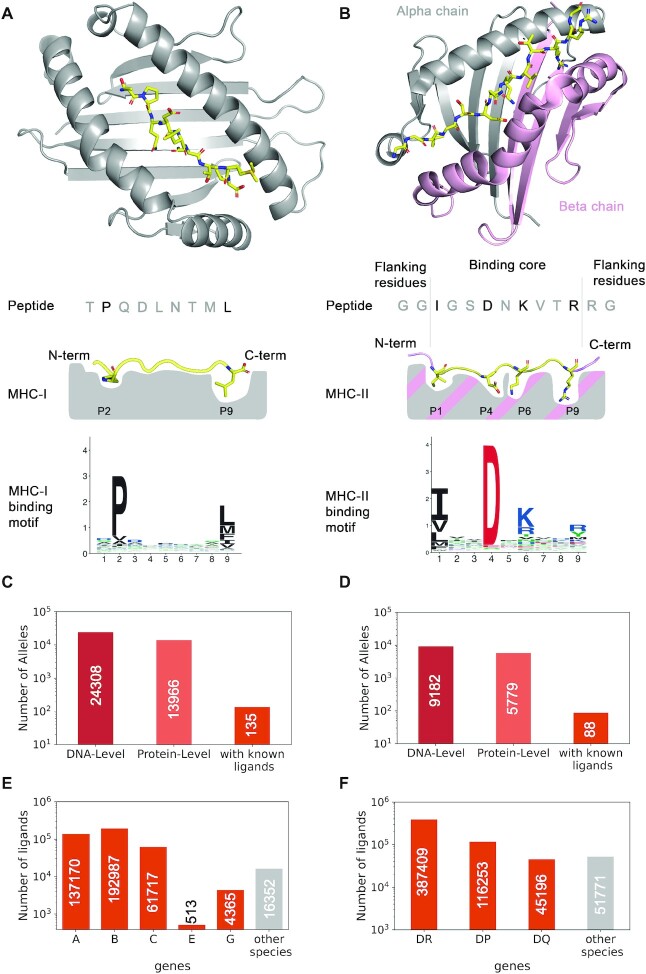
T-cell responses to infected or malignant cells are initiated by the recognition of small peptides displayed on Major Histocompatibility Complex (MHC) molecules. MHC molecules fall into two main classes: MHC class I (MHC-I) recognized by CD8+ T cells and MHC class II (MHC-II) recognized by CD4+ T cells. MHC-I are expressed in most cells (1). They bind short (roughly 8–14 residues, with a preference for 9-mers) peptides derived from intracellular proteins. Primary anchor residues are mainly found at the second and last positions of these peptides (Figure 1A). MHC-I consists of heterodimers with a variable alpha chain and an invariant beta chain (β2-microglobulin). In human, MHC-I alpha chains are encoded by three widely expressed genes (HLA-A, HLA-B and HLA-C) and a few additional ones (e.g. HLA-E and HLA-G) whose expression is restricted to specialized cell types. MHC-I molecules can bind unmodified and post-translationally modified peptides, like phosphorylated peptides (2,3). MHC-II molecules are primarily expressed in antigen presenting cells, like B cells or dendritic cells. They bind longer peptides (roughly 12–25 residues with a preference for 15-mers). Structurally, MHC-II ligands are characterized by a binding core of nine amino acids and flanking residues extending on both sides of the binding core (Figure 1B). MHC-II molecules form heterodimers consisting of an alpha and a beta chain. In human, they are encoded by three sets of genes: (i) HLA-DRA1 dimerizing with HLA-DRB1, HLA-DRB3, HLA-DRB4 or HLA-DRB5, (ii) HLA-DPA1 dimerizing with HLA-DPB1 and (iii) HLA-DQA1 dimerizing with HLA-DQB1.
Figure 1.
Properties of MHC class I and class II molecules. (A) Description of the peptide binding properties of MHC-I molecules. The upper part shows a crystal structure (PDB:4U1H) (48), with the peptide (TPQDLNTML) in yellow and the MHC-I (HLA-B*07:02) in grey. The middle part shows a schematic view of the binding site, with the two main anchor residues at the second and last position (P2 and P9 for 9-mers). The bottom part shows the motif of HLA-B*07:02 for 9-mers. (B) Description of the peptide binding properties of MHC-II molecules. The upper part shows a crystal structure (PDB:7N19) (49), with the peptide (GGIGSDNKVTRRG) in yellow and the MHC-II in grey (alpha chain, HLA-DRA1*01:01) and pink (beta chain, HLA-DRB1*03:01). The middle part shows a schematic view of the binding site, with the main anchor residues (P1, P4, P6 and P9 of the binding core) and flanking residues on both sides of the core. The binding motif is shown in the lower part and was built based on the binding core of the ligands of this allele. (C) Number of documented MHC-I alleles both at the DNA and protein level in IMGT database (50) (data from https://www.ebi.ac.uk/ipd/imgt/hla/about/statistics/, as of July 2022). The third bar shows the number of MHC-I alleles with known naturally presented ligands. (D) Number of documented MHC-II alleles. The third bar shows the number of MHC-II dimers with known naturally presented ligands. (E) Number of known MHC-I ligands for each gene in human and in other species. (F) Number of known MHC-II ligands for each gene in human and in other species.
MHC-I and MHC-II genes show a very high degree of polymorphism, and thousands of different alleles have been documented (Figure 1C, D). In human, MHC alleles are named with two series of digits (e.g. HLA-B*07:02 for class I or HLA-DRB1*03:01 for class II) which unambiguously distinguish each allele at the amino acid level. The first set of digits (i.e. after the ‘*’) indicate broad classes of HLA alleles, while the second set of digits (i.e. after the ‘:’) indicates polymorphisms within each class. Additional polymorphism (either synonymous or intronic) can be found at the DNA level without impacting the MHC protein sequences. Two additional series of digits are used to represent these additional polymorphisms (e.g. HLA-B*07:02:01:01). Non-synonymous polymorphic residues are primarily located in the peptide binding site of MHC molecules. As a result of this polymorphism, different alleles show different binding specificities and bind different repertoires of ligands.
MHC molecules can bind both self and non-self peptides (i.e. peptides coming or absent from the normal proteome respectively). Non-self MHC ligands originating from pathogens or cancer specific non-synonymous genetic alterations (the so-called neo-antigens) can be recognized by T cells via the binding of the T-cell receptor (TCR) to the peptide–MHC complexes. This binding is necessary to initiate and sustain T-cell responses to infections and cancer. For this reason, MHC ligands are promising therapeutic targets that have been widely used in pre-clinical and clinical studies. For instance, in cancer immunotherapy, MHC ligands have been used as personalized vaccines to boost the immune system to recognize neo-antigens (4–6). T cells targeting MHC ligands expressed on the surface of cancer cells (such as tumor associated antigens or neo-antigens) have shown efficacy upon adoptive transfer in multiple tumor types (7,8). Viral peptides presented on MHC molecules have also been used in vaccines against infectious diseases to elicit potent T-cell responses (9).
A widely used approach to identify MHC ligands that could be recognized by T cells is to use in silico predictions (10–15). MHC ligand predictors are machine learning tools trained on large datasets of MHC ligands. Over the last decade, mass spectrometry based MHC peptidomics has become the dominant source of information about MHC binding specificities (16–22). These data enabled researchers to determine binding motifs for hundreds of MHC alleles (11,21–23) (Figure 1C, D). For MHC-I molecules, naturally presented ligands further revealed allele-specific peptide length distributions (24,25). For MHC-II molecules, naturally presented ligands demonstrated specificity in peptide length distributions, position of the binding core with respect to the middle of the peptide (referred to as binding core offset) and N- or C-terminal residues of the ligands (21,26). Unlike for MHC-I alleles, these features are more conserved across MHC-II alleles, though same variability in peptide length distributions was reported (27). In addition, cleavage and processing signals have been reported in the amino acids upstream and downstream of MHC ligands (16,28). Peptides coming from highly expressed genes/proteins also tend to be preferentially displayed on MHC molecules (10,16,29).
To facilitate the understanding of the main binding properties of MHC molecules, we present here the MHC Motif Atlas (http://mhcmotifatlas.org/). This database enables users to visualize binding motifs (including cases of multiple specificity and motifs of phosphorylated ligands) and peptide length distributions for thousands of MHC alleles. In addition, our database can be used to download lists of MHC ligands, MHC sequences and MHC X-ray crystallography structures.
THE MHC MOTIF ATLAS: DATA SOURCES
To derive the binding specificities of MHC molecules, we used naturally presented MHC ligands identified across >500 MHC-I and MHC-II peptidomics samples from human, mouse, cattle and chicken (see Materials and Methods). These include both unmodified and phosphorylated MHC ligands. To remove false-positives, assign allelic restrictions in multi-allelic samples and determine the binding core for MHC-II ligands, motif deconvolution was applied on all samples. Shared motifs across samples sharing the same allele were used to determine the ligands of the different MHC alleles. All motifs were manually verified in all samples. Details about this procedure and the resulting MHC ligand datasets have been previously published for MHC-I ligands (3,11,18,24) and for MHC-II ligands (13,21) (see also Materials and Methods). This enabled us to collect 1 013 733 ligands interacting with 135 MHC-I and 88 MHC-II molecules (Figure 1C–F).
THE MHC MOTIF ATLAS: BUILDING MOTIFS
Motifs for alleles with known ligands were built following the procedure described in (30), which includes renormalization by the background amino acid frequency in the human proteome (see Materials and Methods).
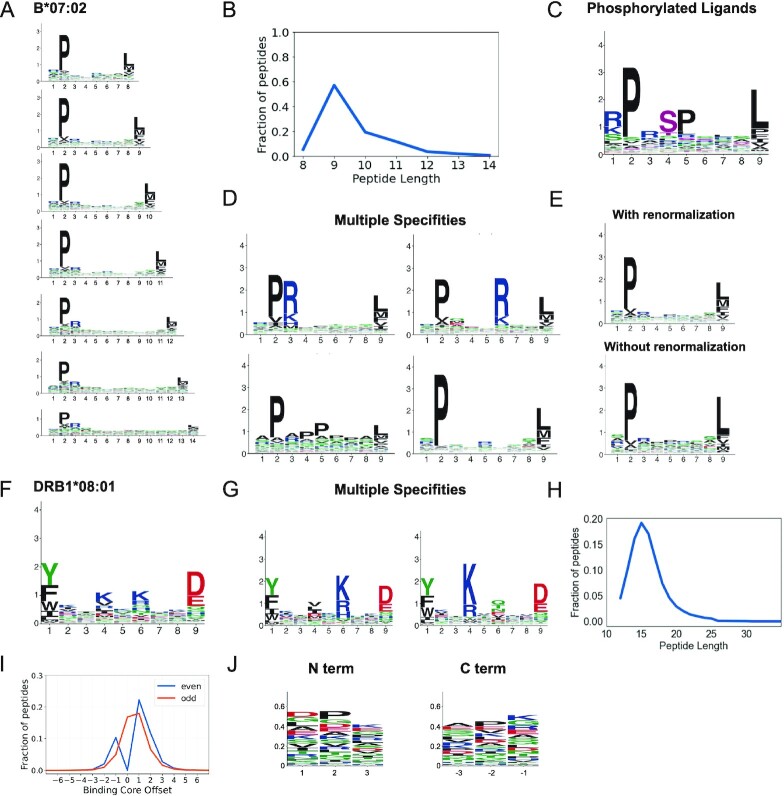
For MHC-I alleles, distinct motifs were built for each length (8- to 14-mers, see example in Figure 2A) since ligands of different lengths display differences in their motifs. The peptide length distributions were also computed for all MHC-I alleles with experimental ligands (see example in Figure 2B). Binding motifs were computed separately for phosphorylated ligands, and the phosphorylated residues are shown in pink (see example in Figure 2C). Multiple specificities, when present, were determined with MixMHCp (31) and all cases were manually evaluated to determine the final number of motifs (Figure 2D). Finally, motifs of raw ligands (i.e. without background amino acid frequency renormalization) were computed (Figure 2E). As it can be seen, background amino acid correction is important to avoid underestimating rare amino acids (e.g. M, 2.1% of the human proteome) or overestimating frequent amino acids (e.g. L, 9.9% of the human proteome).
Figure 2.
Binding specificities of MHC molecules. (A) MHC-I binding motifs for different peptide lengths. (B) Peptide length distribution. (C) Motifs for phosphorylated ligands. (D) MHC-I multiple specificities, including mutual exclusivity of charged amino acids at P3 and P6. (E) Illustration of the difference between motifs with and without background frequency renormalization. (F) MHC-II binding motifs. (G) MHC-II multiple specificities capturing a mutual exclusivity of positively charged amino acids at P4 and P6 (see (13)). (H) Average peptide length distribution for MHC-II ligands. (I) Distribution of peptide binding core offsets for MHC-II ligands of even and odd lengths (0 corresponds to a binding core at the middle of peptides with an odd length, and is not defined for peptides with an even length). (J) Motifs in the first and last three N- and C-terminal residues of MHC-II ligands. Panels A–E are built from HLA-B*07:02 ligands. Panels F–G are built from HLA-DRB1*08:01 ligands. Panels H–J are built from all MHC-II ligands (see (13)).
For MHC-II alleles, motifs were built based on the 9-mer binding core of MHC-II ligands (Figure 2F). Multiple specificities were determined with MoDec (21) and were manually curated (Figure 2G). Peptide length distributions were computed for each allele (see average across alleles in Figure 2H). The distributions of binding core offsets (Figure 2I), as well as the motifs for the three N- and C-terminal residues in the ligands (Figure 2J) were computed based on the entire dataset of MHC-II ligands.
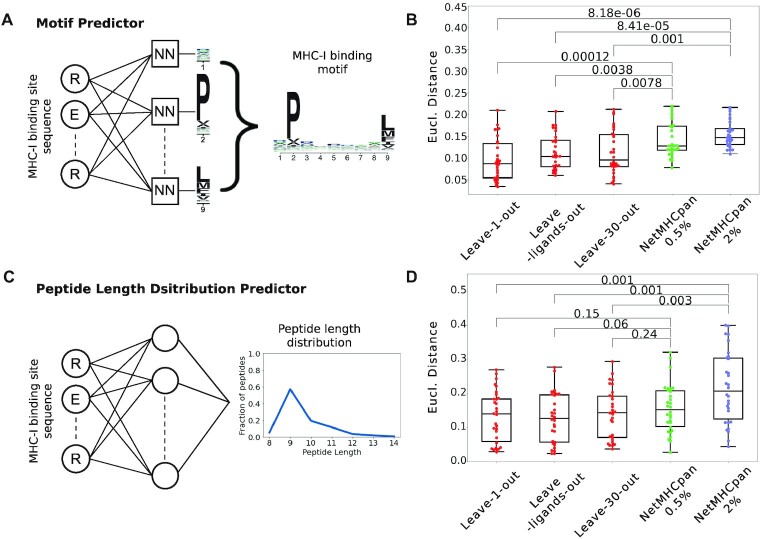
Experimental ligands are only available for a small fraction of MHC molecules (Figure 1C, D). To fill this gap, we developed machine learning predictors of MHC binding motifs based on the neural network framework that we recently introduced for MHC-II alleles (13). For MHC-I alleles, the first set of neural networks uses as input the sequence of the MHC-I binding site and aims at predicting the binding motifs for each peptide length separately (see Materials and Methods and Figure 3A). Another neural network was developed to predict the peptide length distribution based on the sequence of the MHC-I binding site (see Materials and Methods and Figure 3C). To benchmark the accuracy of our predictions with the state-of-the-art NetMHCpan tool (14), we performed multiple cross-validations (see Materials and Methods): (i) a leave-one-allele-out cross-validation, where all data for each allele absent from the training set of NetMHCpan were iteratively removed from the training set (30 alleles in total, see Supplementary Table S1), (ii) a leave-ligands-out cross validation where all peptides with the same sequence as the ligands of the left-out allele where removed, and (iii) a leave-30-alleles-out cross validation where all data from the 30 alleles that are not part of the training of NetMHCpan were removed. The predicted motifs were compared to the experimental ones and those predicted by NetMHCpan (see Materials and Methods). Overall, we observed that binding motifs could be reliably predicted, and the accuracy of our predictions equaled or surpassed the one of NetMHCpan (Figure 3B). Similar results were obtained for our predictions of peptide length distributions of MHC-I alleles (Figure 3D). Motifs for MHC-II molecules without experimental ligands were predicted following the approach described in (13). For these molecules, average peptide length distributions were used, since less variability is observed within MHC-II molecules than within MHC-I molecules.
Figure 3.
Predicting the binding properties of MHC-I molecules. (A) Machine learning framework for the prediction of binding motifs for MHC-I alleles. Distinct neural networks (NN) were built for each peptide length and each position, and the final motif for a given peptide length is built by combining all position. The example illustrates the neural networks for predicting 9-mer binding motifs. (B) Leave-one-allele-out, leave-ligands-out and leave-30-alleles-out cross validation of the predictions of binding motifs for the 30 MHC-I alleles that are not part of the training set of NetMHCpan. Predicted motifs from our method and from NetMHCpan4.1 (at percentile ranks smaller than 0.5% or 2%) were compared to the experimental ones using the Euclidean distance. (C) Architecture of the neural network for the predictions of peptide length distributions for MHC-I alleles without experimental ligands. (D) Leave-one-allele-out, leave-ligands-out and leave-30-alleles-out cross validation of the predictions of peptide length distributions for the 30 MHC-I alleles that are not part of the training set of NetMHCpan. Predicted peptide length distributions from our method and from NetMHCpan4.1 (at percentile ranks smaller than 0.5% or 2%) were compared to the experimental ones using the Euclidean distance. Boxplots indicate the median, upper and lower quartiles. P-values were computed with the paired two-sided Mann–Whitney U-test.
THE MHC MOTIF ATLAS: WEB INTERFACE
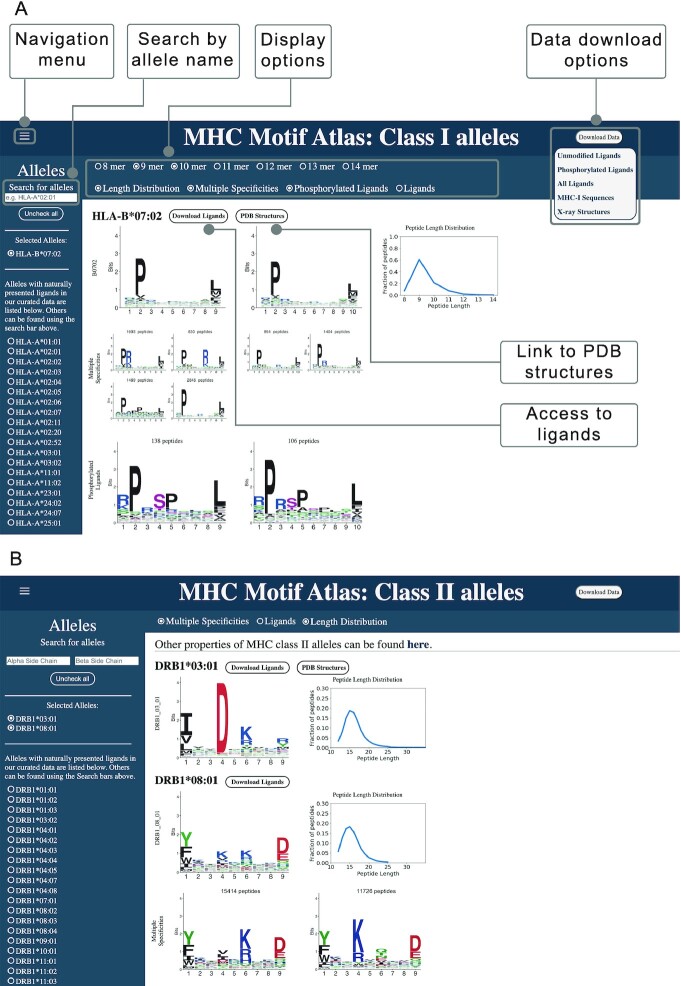
The MHC Motif Atlas provides an intuitive interface to visualize the main peptide binding properties of MHC molecules. For MHC-I, the http://mhcmotifatlas.org/class1 page enables users to display binding motifs and peptide length distributions for ligands of lengths 8–14 (Figure 4A). In addition, we offer the possibility to visualize cases of multiple specificity (e.g. HLA-B*07:02), motifs representing phosphorylated ligands and motifs representing the raw ligands (i.e. without background amino acid frequency corrections). Multiple alleles can be displayed on the same page, which is convenient for comparing the binding motifs and the peptide length distributions. For each allele with known ligands, a link to the list of ligands is provided. Finally, links to known crystal structures are provided when such structures are available. This feature is useful since searches for specific alleles in the Protein Data Bank can be complicated by inconsistencies in the naming of the alleles in publications (e.g. HLA-A*02:01, HLA-A02:01, HLA-A02, HLA-A2, A2).
Figure 4.
MHC Motif Atlas interface of MHC-I and MHC-II. (A) MHC Motif Atlas interface for MHC-I alleles, including visualization of binding motifs, peptide length distributions, multiple specificities and motifs of phosphorylated ligands. The Search field on the top left part enables users to type a part of an allele's name, and all the corresponding alleles will automatically be listed below. By default the alleles listed on the left correspond to those with experimental ligands. The Download Data button allows to download complete lists of MHC-I ligands, as well as MHC-I sequences and X-ray structures PDB identifiers. (B) MHC Motif Atlas interface for MHC-II alleles.
For MHC-II alleles, binding motifs are shown in http://mhcmotifatlas.org/class2 (Figure 4B), including options to show peptide length distributions, multiple specificities and motifs of the raw ligands. When available, links to lists of MHC-II ligands and to known X-ray structures are provided. Other properties are displayed in http://mhcmotifatlas.org/class2_properties.
In both http://mhcmotifatlas.org/class1 and http://mhcmotifatlas.org/class2, the list of alleles shown by default on the left corresponds to those with experimental ligands. To see motifs predicted for other alleles, the user can type the first letters of the allele's name in the search field, and the left menu will automatically list the corresponding alleles.
The MHC Motif Atlas also provides links to different resources to analyze MHC ligands and T-cell epitopes (http://mhcmotifatlas.org/tools). These include links to MHC ligand predictors that can be used through a web interface or as standalone executables, tools for motif deconvolution and allele assignment in MHC peptidomics samples, as well as databases of MHC ligands and T-cell epitopes. An F.A.Q page provides information about MHC molecules and the data presented in the MHC Motif Atlas (http://mhcmotifatlas.org/faq).
DISCUSSION
MHC ligands play a central role in recognition and elimination of infected or malignant cells. To prevent pathogens from optimizing their protein sequences not to bind any MHC molecule, which would make them invisible to T cells, MHC genes have evolved an extremely high degree of polymorphism resulting in a large diversity of binding specificities. These specificities dictate which peptides can bind to a given MHC molecule. The MHC Motif Atlas provides a reliable and interpretable way to understand and visualize the main binding properties of thousands of MHC molecules.
Binding motifs have been widely used to visualize peptides or nucleotides binding to specific proteins, including MHC-I and MHC-II, peptide recognition domains (32) or transcription factors (33). In this framework, each position in the peptide is treated independently. This can mask potential correlations between the amino acids at distinct positions. Such correlations have been reported in multiple instances (34,35), and support the use of machine learning frameworks like neural networks to make predictions of ligands. For MHC molecules, these correlations often reflect different binding modes (e.g. C-terminal extensions in MHC-I ligands (36) or reverse binding of MHC-II ligands (13)) or mutual exclusivity of specific amino acids (e.g. positively charged residues in the ligands pointing to the same residue in the binding site (13)). Because the number of different binding modes is often limited by the structural constraints of the MHC binding site, correlation patterns can often be captured with multiple binding motifs (24), which is why this feature has been included in the MHC Motif Atlas. Another source of correlation specific to MHC-I ligands comes from the different motifs for different peptide lengths. In the MHC Motif Atlas, such correlations have been resolved by displaying motifs for each peptide length separately for MHC-I alleles. For these reasons, (multiple) motifs together with information about peptide length distributions provide a reliable framework to model and visualize the main binding properties of MHC molecules. A frequent question when dealing with multiple motifs for peptide of the same length is how the optimal number of motifs is determined. In the MHCMotifAtlas, cases of multiple specificity are based on our previous studies (13,24), which included a manual curation to focus on cases where the multiple motifs show clear differences and can be linked with structural interpretations.
MHC peptidomics provide a rich source of reliable and biologically relevant information about naturally presented MHC ligands, including properties like peptide length distributions which are not directly available from binding affinity measurements (25). Moreover, these data cover all the most frequent alleles in human. This is why we focused exclusively on such data in the MHC Motif Atlas. MHC-I molecules with only ligands from other sources (e.g. binding affinity measurement) have on average <100 ligands in IEDB (37). For this reason, motifs built for these alleles can be less reliable and we decided not to include these data in our atlas.
The extremely high polymorphism of the MHC locus makes it impossible to have experimental ligands for all alleles. Our ability to predict binding motifs for MHC molecules without ligands is therefore key to cover the repertoire of MHC alleles in the MHC Motifs Atlas. Compared to machine learning pan-allele predictors of MHC-I ligands, like NetMHCpan (14) or MHCflurry (12), our machine learning framework for predicting the binding motifs and peptide length distributions of MHC-I alleles is more interpretable (see (13) for similar results for MHC-II alleles). It is expected that predictions of MHC motifs will be less accurate in species without known MHC ligands (13). This is why we focused on species with known MHC ligands identified by unbiased mass spectrometry based MHC peptidomics.
Compared to existing resources, including the MHCMotifViewer (https://services.healthtech.dtu.dk/service.php?MHCMotifViewer) (38), the SysteMHC Atlas (39), the HLA Ligand Atlas (https://hla-ligand-atlas.org) (40) or the Motif Viewer of NetMHCpan (https://services.healthtech.dtu.dk/service.php?NetMHCpan-4.1) (14), the MHC Motif Atlas provides a more comprehensive characterization and visualization of MHC peptide binding properties. This includes peptide length distributions, cases of multiple specificities, motifs for phosphorylated ligands and the possibility of seeing how MHC-I motifs change with different peptide lengths. Motifs of different alleles can be rapidly compared by displaying multiple alleles on the same page. Moreover, the MHC Motif Atlas provides direct links to the actual data supporting the binding motifs or other properties of MHC molecules. This represents a valuable resource for researchers who want to perform their own analyses or train their own MHC ligand predictors. By providing intuitive visualization of MHC binding properties, the MHC Motif Atlas can also complement machine learning MHC ligand prediction tools, which are often used as black boxes and do not necessarily provide explanations on why a peptide gets a good or bad score.
CONCLUSION
The presentation of peptides on MHC molecules is a necessary condition for T-cell responses against infected or malignant cells. Therefore, a reliable and interpretable visualization of the binding specificities of MHC molecules is useful to better understand why peptides may or may not be presented on MHC molecules in different individuals with different alleles. The MHC Motif Atlas provides a resource to rapidly visualize, analyze and compare the binding properties of both MHC-I and MHC-II molecules. In addition, our atlas provides links to curated datasets of more than a million naturally presented MHC ligands, as well as MHC sequences and MHC X-ray structures. The MHC Motif Atlas represents therefore one of the most comprehensive and integrated resources about MHC molecules and their ligands.
MATERIALS AND METHODS
Sources of MHC ligands
Naturally presented MHC ligands were collected from >500 MHC peptidomics samples from human, mouse, cattle and chicken. These include all samples considered in (11,13). Phosphorylated ligands were retrieved from (3). We further included data from a few recent MHC peptidomics studies (20,40–44). All data were retrieved from the original studies to prevent having filtered data based on MHC ligand predictors. All samples were processed with our motif deconvolution tools (MixMHCp (24) for MHC-I and MoDec (21) for MHC-II) to identify shared motifs across samples sharing the same allele. Details about this procedure and the obtained results have been previously published for MHC-I ligands (18,24) and MHC-II ligands (13,21).
Building MHC motifs and peptide length distributions
For all MHC molecules with naturally presented ligands, Position Probability Matrices (PPMs) were built by computing the frequency of each amino acid at each position in the set of ligands of the given allele, including standard pseudocounts based on BLOSUM62 as described in (21,24). For MHC-I alleles, separate PPMs were built for each ligand length L from 8 to 14. For MHC-II alleles, the PPMs were built based on the 9-mer binding core determined by MoDec for the ligands of each allele. The Position Weight Matrices (PWMs) representing the final motifs were computed by normalizing the PPMs with the amino acid background frequencies of the human proteome, as described in (21,24). For alleles displaying multiple motifs, separate PWMs were also computed for each set of ligands assigned to each motif. Both the final motifs (based on normalized PWMs) and the motifs of the ligands (based on PPMs) were visualized using ggseqlogo (45). PPMs for phosphorylated ligands were computed separately. The phosphorylated residues are shown in purple in the corresponding logos.
Peptide length distributions were determined by computing the fraction of naturally presented MHC ligands of each length (from 8 to 14 for MHC-I and 12 to 25 for MHC-II). For MHC-II alleles, the distribution of the peptide binding core position and the motifs of the three N- and C-terminal residues were computed as in (21).
Predicting MHC motifs
Inspired by our recent work on MHC-II motifs (13), neural networks were used to predict PPMs of MHC-I molecules without known ligands. More precisely, distinct networks were trained for each peptide length (8 to 14) and for each position. The input of each neural network is the list of binding site residues from the MHC-I molecules (34 residues). This binding site was defined as in (46). Each binding site residue was encoded as a 20-dimensional vector based on the BLOSUM62 probability matrix. The output of each network consists of a vector of 20 values, representing the PPM at the corresponding position. Each network is composed of an input layer (34 × 20 nodes), two fully connected hidden layers (128 and 64 nodes, respectively) and an output layer (20 nodes). We used rectified linear unit (ReLU) activation function for the hidden layers and the softmax activation function for the output layer. We used the Kullback Leibler divergence as a loss function, and it was optimized using Adam optimizer with a learning rate of 0.0001. These neural networks were implemented in Python (version 3.7.11), using Keras packages relying on TensorFlow (version 2.2.4-tf). Five hundred epochs were set for the training process. For each allele and each peptide length (8 to 14), we then normalized by background human proteome frequencies and grouped the output of the different networks corresponding to different positions to create the final predicted PWM.
For MHC-II alleles, the 9-mer binding motifs were predicted based on the method described in (13).
Predicting peptide length distributions
A neural network was developed to predict the peptide length distribution of MHC-I molecules. The input layer is the same as for the MHC-I motifs prediction (34 × 20 nodes), followed by one hidden layer (128 nodes) with the rectified linear unit (ReLU) activation function. The output layer is the peptide length distribution (from 8 to 14, i.e. 7 nodes) based on the softmax activation function. We used the Kullback Leibler divergence as a loss function, and it was optimized using Adam optimizer with a learning rate of 0.0001. 125 epochs were set for the training process.
Benchmarking
To benchmark the accuracy of our binding motif and peptide length distribution predictions for MHC-I molecules and compare with the state-of-the-art NetMHCpan, we designed three distinct cross-validation schemes. First, we performed a leave-one-allele-out cross-validation, where each allele absent for the training of NetMHCpan was successively removed from the training set (30 alleles in total, see Supplementary Table S1). Second, we performed leave-ligands-out cross validation, where all peptides found among ligands of the left-out-allele were removed from the training set. Third, we performed a leave-30-alleles-out cross validation by excluding all data from the 30 alleles that are not part of the training set of NetMHCpan. The predicted normalized PWMs (i.e. PPMs divided by the background amino acid frequencies and normalized to one for each position) were then compared to the experimental ones by computing the Euclidean distance for each position on the peptide and averaging these distances. The lower the distance, the closer the predicted motifs are to the experimental ones. Similarly, the predicted peptide length distributions were compared to the experimental ones by computing the Euclidean distance.
To compare these results with NetMHCpan (NetMHCpan4.1 (14)), we created 500 000 random peptides for each length (from 8 to 14) and scored them using NetMHCpan for the 30 MHC-I alleles for which we had experimental data and which are not part of the training set of NetMHCpan. For each allele, peptides with %Rank_EL smaller than 0.5% or smaller than 2% were considered as the ligands to this allele. The ligands were then used to build PWMs (based on a flat background frequency since we used random peptides) for each peptide length and calculate the peptide length distribution for each allele. These PWMs and peptide length distributions were compared to the experimental ones using Euclidean distance.
MHC crystal structures
For MHC-I alleles, X-ray structures were retrieved from the PDB (47) for all class I alleles considered in the MHC Motif Atlas. Only structures with unmodified MHC alleles and a ligand of length 8–14 were considered. Truncated ligands or ligands containing non-natural amino acids were not included. Similarly, X-ray structures were retrieved from the PDB (47) for MHC II alleles.
Website architecture
We created the website using Node.js (also called Node) to run an environment for writing server-side applications in JavaScript alongside the HTML and CSS documents responsible for the website design. The website is implemented as a web application using the Express.js framework to provide a logical routing to different website sections.
DATA AVAILABILITY
All the data used to build the MHC Motif Atlas are available at http://mhcmotifatlas.org/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marthe Solleder for useful feedback about the website.
Author contribution: D.T. performed the new methodological developments, wrote the method section and prepared the figures. S.E. developed the website. J.R. provided data and feedback for the project and the manuscript. D.G. designed the project, supervised the work and wrote the manuscript.
Contributor Information
Daniel M Tadros, Department of Oncology, Ludwig Institute for Cancer Research Lausanne, University of Lausanne, Lausanne, Switzerland; Swiss Institute of Bioinformatics (SIB), Lausanne, Switzerland.
Simon Eggenschwiler, Department of Oncology, Ludwig Institute for Cancer Research Lausanne, University of Lausanne, Lausanne, Switzerland; Swiss Institute of Bioinformatics (SIB), Lausanne, Switzerland.
Julien Racle, Department of Oncology, Ludwig Institute for Cancer Research Lausanne, University of Lausanne, Lausanne, Switzerland; Swiss Institute of Bioinformatics (SIB), Lausanne, Switzerland.
David Gfeller, Department of Oncology, Ludwig Institute for Cancer Research Lausanne, University of Lausanne, Lausanne, Switzerland; Swiss Institute of Bioinformatics (SIB), Lausanne, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss Cancer Research Foundation [KFS-4104-02-2017]. Funding for open access charge: Swiss Cancer Research Foundation [KFS-4104-02-2017].
Conflict of interest statement. None declared.
REFERENCES
- 1. Neefjes J., Jongsma M.L.M., Paul P., Bakke O.. Towards a systems understanding of MHC class i and MHC class II antigen presentation. Nat. Rev. Immunol. 2011; 11:823–836. [DOI] [PubMed] [Google Scholar]
- 2. Cobbold M., De La Peña H., Norris A., Polefrone J.M., Qian J., English A.M., Cummings K.L., Penny S., Turner J.E., Cottine J.et al.. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci. Transl. Med. 2013; 5:203ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Solleder M., Guillaume P., Racle J., Michaux J., Pak H.-S., Müller M., Coukos G., Bassani-Sternberg M., Gfeller D.. Mass spectrometry based immunopeptidomics leads to robust predictions of phosphorylated HLA class I ligands. Mol. Cell. Proteomics. 2020; 19:390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L.et al.. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017; 547:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sahin U., Derhovanessian E., Miller M., Kloke B.-P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B.et al.. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017; 547:222–226. [DOI] [PubMed] [Google Scholar]
- 6. Sahin U., Oehm P., Derhovanessian E., Jabulowsky R.A., Vormehr M., Gold M., Maurus D., Schwarck-Kokarakis D., Kuhn A.N., Omokoko T.et al.. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020; 585:107–112. [DOI] [PubMed] [Google Scholar]
- 7. Leidner R., Sanjuan Silva N., Huang H., Sprott D., Zheng C., Shih Y.-P., Leung A., Payne R., Sutcliffe K., Cramer J.et al.. Neoantigen T-Cell receptor gene therapy in pancreatic cancer. N. Engl. J. Med. 2022; 386:2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tran E., Turcotte S., Gros A., Robbins P.F., Lu Y.-C., Dudley M.E., Wunderlich J.R., Somerville R.P., Hogan K., Hinrichs C.S.et al.. Cancer immunotherapy based on mutation-specific CD4+ t cells in a patient with epithelial cancer. Science (New York, N.Y.). 2014; 344:641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heitmann J.S., Bilich T., Tandler C., Nelde A., Maringer Y., Marconato M., Reusch J., Jäger S., Denk M., Richter M.et al.. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 t cell immunity. Nature. 2022; 601:617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen B., Khodadoust M.S., Olsson N., Wagar L.E., Fast E., Liu C.L., Muftuoglu Y., Sworder B.J., Diehn M., Levy R.et al.. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 2019; 37:1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gfeller D., Schmidt J., Croce G., Guillaume P., Bobisse S., Genolet R., Queiroz L., Cesbron J., Racle J., Harari A.. Predictions of immunogenicity reveal potent SARS-CoV-2 CD8+ T-cell epitopes. 2022; bioRxiv doi:23 May 2022, preprint: not peer reviewed 10.1101/2022.05.23.492800. [DOI] [PMC free article] [PubMed]
- 12. O’Donnell T.J., Rubinsteyn A., Laserson U.. MHCflurry 2.0: improved pan-allele prediction of MHC class I-Presented peptides by incorporating antigen processing. Cell Syst. 2020; 11:42–48. [DOI] [PubMed] [Google Scholar]
- 13. Racle J., Guillaume P., Schmidt J., Michaux J., Larabi A., Lau K., Perez M.A.S., Croce G., Genolet R., Coukos G.et al.. Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes. 2022; bioRxiv doi:29 June 2022, preprint: not peer reviewed 10.1101/2022.06.26.497561. [DOI] [PubMed]
- 14. Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M.. NetMHCpan-4.1 and netmhciipan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020; 48:W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bravi B., Tubiana J., Cocco S., Monasson R., Mora T., Walczak A.M.. RBM-MHC: a semi-supervised machine-learning method for sample-specific prediction of antigen presentation by HLA-I alleles. Cell Syst. 2021; 12:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M.et al.. Mass spectrometry profiling of HLA-Associated peptidomes in Mono-allelic cells enables more accurate epitope prediction. Immunity. 2017; 46:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abelin J.G., Harjanto D., Malloy M., Suri P., Colson T., Goulding S.P., Creech A.L., Serrano L.R., Nasir G., Nasrullah Y.et al.. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity. 2019; 51:766–779. [DOI] [PubMed] [Google Scholar]
- 18. Bassani-Sternberg M., Chong C., Guillaume P., Solleder M., Pak H., Gannon P.O., Kandalaft L.E., Coukos G., Gfeller D.. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 2017; 13:e1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pearson H., Daouda T., Granados D.P., Durette C., Bonneil E., Courcelles M., Rodenbrock A., Laverdure J.-P., Côté C., Mader S.et al.. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest. 2016; 126:4690–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pyke R.M., Mellacheruvu D., Dea S., Abbott C.W., Zhang S.V., Phillips N.A., Harris J., Bartha G., Desai S., McClory R.et al.. Precision neoantigen discovery using Large-scale immunopeptidomes and composite modeling of MHC peptide presentation. Mol. Cell. Proteomics. 2021; 20:100111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Racle J., Michaux J., Rockinger G.A., Arnaud M., Bobisse S., Chong C., Guillaume P., Coukos G., Harari A., Jandus C.et al.. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 2019; 37:1283–1286. [DOI] [PubMed] [Google Scholar]
- 22. Sarkizova S., Klaeger S., Le P.M., Li L.W., Oliveira G., Keshishian H., Hartigan C.R., Zhang W., Braun D.A., Ligon K.L.et al.. A large peptidome dataset improves HLA class i epitope prediction across most of the human population. Nat. Biotechnol. 2019; 38:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alvarez B., Reynisson B., Barra C., Buus S., Ternette N., Connelley T., Andreatta M., Nielsen M.. NNAlign_MA; MHC peptidome deconvolution for accurate MHC binding motif characterization and improved T-cell epitope predictions. Mol. Cell Proteomics. 2019; 18:2459–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gfeller D., Guillaume P., Michaux J., Pak H.-S., Daniel R.T., Racle J., Coukos G., Bassani-Sternberg M.. The length distribution and multiple specificity of naturally presented HLA-I ligands. J. Immunol. 2018; 201:3705–3716. [DOI] [PubMed] [Google Scholar]
- 25. Trolle T., McMurtrey C.P., Sidney J., Bardet W., Osborn S.C., Kaever T., Sette A., Hildebrand W.H., Nielsen M., Peters B.. The length distribution of class I-Restricted t cell epitopes is determined by both peptide supply and MHC allele-specific binding preference. J. Immunol. 2016; 196:1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barra C., Alvarez B., Paul S., Sette A., Peters B., Andreatta M., Buus S., Nielsen M.. Footprints of antigen processing boost MHC class II natural ligand predictions. Genome Med. 2018; 10:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaabinejadian S., Barra C., Alvarez B., Yari H., Hildebrand W.H., Nielsen M.. Accurate MHC motif deconvolution of immunopeptidomics data reveals a significant contribution of DRB3, 4 and 5 to the total DR immunopeptidome. Front. Immunol. 2022; 13:835454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen M., Lundegaard C., Lund O., Keşmir C.. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005; 57:33–41. [DOI] [PubMed] [Google Scholar]
- 29. Bassani-Sternberg M., Pletscher-Frankild S., Jensen L.J., Mann M.. Mass spectrometry of human leukocyte antigen class i peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell Proteomics. 2015; 14:658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gfeller D., Bassani-Sternberg M.. Predicting antigen presentation-what could we learn from a million peptides?. Front. Immunol. 2018; 9:1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bassani-Sternberg M., Gfeller D.. Unsupervised HLA peptidome deconvolution improves ligand prediction accuracy and predicts cooperative effects in Peptide-HLA interactions. J. Immunol. 2016; 197:2492–2499. [DOI] [PubMed] [Google Scholar]
- 32. Kumar M., Gouw M., Michael S., Sámano-Sánchez H., Pancsa R., Glavina J., Diakogianni A., Valverde J.A., Bukirova D., Čalyševa J.et al.. ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2020; 48:D296–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Chèneby J., Kulkarni S.R., Tan G.et al.. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018; 46:D1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gfeller D., Butty F., Wierzbicka M., Verschueren E., Vanhee P., Huang H., Ernst A., Dar N., Stagljar I., Serrano L.et al.. The multiple-specificity landscape of modular peptide recognition domains. Mol. Syst. Biol. 2011; 7:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomovic A., Oakeley E.J.. Position dependencies in transcription factor binding sites. Bioinformatics. 2007; 23:933–941. [DOI] [PubMed] [Google Scholar]
- 36. Guillaume P., Picaud S., Baumgaertner P., Montandon N., Schmidt J., Speiser D.E., Coukos G., Bassani-Sternberg M., Filippakopoulos P., Gfeller D.. The C-terminal extension landscape of naturally presented HLA-I ligands. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B.. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019; 47:D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rapin N., Hoof I., Lund O., Nielsen M.. MHC motif viewer. Immunogenetics. 2008; 60:759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shao W., Pedrioli P.G.A., Wolski W., Scurtescu C., Schmid E., Vizcaíno J.A., Courcelles M., Schuster H., Kowalewski D., Marino F.et al.. The SysteMHC atlas project. Nucleic Acids Res. 2018; 46:D1237–D1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marcu A., Bichmann L., Kuchenbecker L., Kowalewski D.J., Freudenmann L.K., Backert L., Mühlenbruch L., Szolek A., Lübke M., Wagner P.et al.. HLA ligand atlas: a benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer. 2021; 9:e002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lampen M.H., Hassan C., Sluijter M., Geluk A., Dijkman K., Tjon J.M., de Ru A.H., van der Burg S.H., van Veelen P.A., van Hall T.. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol. Immunol. 2013; 53:126–131. [DOI] [PubMed] [Google Scholar]
- 42. DeVette C.I., Andreatta M., Bardet W., Cate S.J., Jurtz V.I., Jackson K.W., Welm A.L., Nielsen M., Hildebrand W.H.. NetH2pan: a computational tool to guide MHC peptide prediction on murine tumors. Cancer Immunol. Res. 2018; 6:636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ebrahimi-Nik H., Michaux J., Corwin W.L., Keller G.L.J., Shcheglova T., Pak H., Coukos G., Baker B.M., Mandoiu I.I., Bassani-Sternberg M.et al.. Mass spectrometry–driven exploration reveals nuances of neoepitope-driven tumor rejection. JCI Insight. 2019; 4:e129152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murphy J.P., Yu Q., Konda P., Paulo J.A., Jedrychowski M.P., Kowalewski D.J., Schuster H., Kim Y., Clements D., Jain A.et al.. Multiplexed relative quantitation with isobaric tagging mass spectrometry reveals class i major histocompatibility complex ligand dynamics in response to doxorubicin. Anal. Chem. 2020; 91:5106–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wagih O. ggseqlogo: a versatile r package for drawing sequence logos. Bioinformatics. 2017; 33:3645–3647. [DOI] [PubMed] [Google Scholar]
- 46. Hoof I., Peters B., Sidney J., Pedersen L.E., Sette A., Lund O., Buus S., Nielsen M.. NetMHCpan, a method for MHC class i binding prediction beyond humans. Immunogenetics. 2009; 61:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M.et al.. RCSB protein data bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021; 49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kløverpris H.N., Cole D.K., Fuller A., Carlson J., Beck K., Schauenburg A.J., Rizkallah P.J., Buus S., Sewell A.K., Goulder P.. A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape. Retrovirology. 2015; 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Greaves S.A., Ravindran A., Santos R.G., Chen L., Falta M.T., Wang Y., Mitchell A.M., Atif S.M., Mack D.G., Tinega A.N.et al.. CD4+ t cells in the lungs of acute sarcoidosis patients recognize an aspergillus nidulans epitope. J. Exp. Med. 2021; 218:e20210785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E.. IPD-IMGT/HLA database. Nucleic Acids Res. 2019; 48:D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used to build the MHC Motif Atlas are available at http://mhcmotifatlas.org/.