Abstract
Upstream open reading frames (uORFs) are initiated by AUG or near-cognate start codons and have been identified in the transcript leader sequences of the majority of eukaryotic transcripts. Functionally, uORFs are implicated in downstream translational regulation of the main protein coding sequence and may serve as a source of non-canonical peptides. Genetic defects in uORF sequences have been linked to the development of various diseases, including cancer. To simplify uORF-related research, the initial release of uORFdb in 2014 provided a comprehensive and manually curated collection of uORF-related literature. Here, we present an updated sequence-based version of uORFdb, accessible at https://www.bioinformatics.uni-muenster.de/tools/uorfdb. The new uORFdb enables users to directly access sequence information, graphical displays, and genetic variation data for over 2.4 million human uORFs. It also includes sequence data of >4.2 million uORFs in 12 additional species. Multiple uORFs can be displayed in transcript- and reading-frame-specific models to visualize the translational context. A variety of filters, sequence-related information, and links to external resources (UCSC Genome Browser, dbSNP, ClinVar) facilitate immediate in-depth analysis of individual uORFs. The database also contains uORF-related somatic variation data obtained from whole-genome sequencing (WGS) analyses of 677 cancer samples collected by the TCGA consortium.
INTRODUCTION
Upstream open reading frames (uORFs) are defined as ORFs initiated in the transcript leader sequence (TLS) preceding the main protein coding sequence (CDS). They are considered to adjust protein expression by translational regulation in response to changing environmental and global translational cellular conditions, as reported for GCN4 in yeasts (1–3), as well as ATF4 (4–6) and ATF5 (7,8) in Metazoa. Furthermore, uORF-associated genetic variability has been attributed to several human diseases, such as Marie Unna hereditary hypotrichosis (9), Cornelia de Lange syndrome (10), and several types of cancer (11,12). The ability of uORFs to regulate translation has been explained by the scanning model (13): after attaching to the 5’-end, ribosomes scan the mRNA towards the 3’-end and start translation at the first suitable initiation codon (13,14). Ribosome profiling and a neural network analysis confirmed that ribosomes may initiate translation at upstream canonical (AUG) and near-cognate (UUG, GUG, CUG, AAG, AGG, ACG, AUA, AUU, AUC) start codons (uStarts) (15), also referred to as alternative translation initiation sites (aTISs). Thus, in case of a transcript containing uORFs, translation would be initiated at the uStart position which alters the expression of the main ORF protein (16). As reviewed by Kozak, the CDS in a uORF-containing transcript can still be translated, if ribosomes are able to scan through the uStart site (leaky scanning) or are able to reinitiate at the main ORF’s start site (17).
The extent of CDS repression by uORFs can be regulated according to cellular conditions by altering the replenishment of lost ribosome co-factors to the pre-initiation complex (18). Furthermore, ribosome stalling may be induced by the nascent uORF peptide or through specific inhibitory sequence contexts at the uORF stop codon. Both result in reduced downstream translation and potentially also in decay of the entire transcript (18). The effect of a uORF on CDS translation is largely dependent on its attributes, such as length (19,20), codon usage (21), peptide sequence (22), position within the transcript (2,4,5,23), the Kozak sequence context surrounding the uORF start codon (24,25), and the sequence context around the uORF stop codon (uStop) (26). Depending on their number, uORFs can also form logic circuits in which individual uORFs influence translation of downstream uORFs, ultimately regulating translation of the CDS (1,2,4–8,20,23). An additional layer of complexity was added only recently by studies showing that active uORF translation may result in the expression of functional peptides (uPeptides) (27–31), which can also be subunits of larger protein complexes (30). As some uPeptides participate in signaling processes associated with cancer (28) and others are contributing to the immunopeptidome (29,31), uPeptides may represent a source of potential future therapeutic targets.
In 2014, we released the initial literature-centric version of uORFdb in order to compile and categorize the current knowledge on uORF biology (32). With the accumulating evidence for uORF and uPeptide functions in (patho-)physiological systems, uORF analyses require either sound bioinformatics skills or a sequence-centric database. Without these, investigators would have to manually scan all three reading frames from all transcripts belonging to their genes of interest by eye; a tedious, but manageable, task for single genes. Using a uORF-centric database, researchers can easily conduct uORF studies of any size. To the best of our knowledge, the plant-focused database uORFlight is the only uORF-centric database that attempts to integrate sequences, variants, and literature (33). However, a comprehensive resource for clinicians and researchers from other fields is still lacking.
Here, we present a substantial update of uORFdb, which is accessible at https://www.bioinformatics.uni-muenster.de/tools/uorfdb. The new uORFdb now provides uORF-related nucleotide and amino acid sequences, information on the sequence context, >1 040 manually curated uORF-related publications, as well as genetic variation data from the general population and somatic variation data from six major types of cancer within one central hub. The current release of uORFdb includes >6.6 million predicted uORFs from 13 eukaryotic species, including >2.4 million human uORFs. Individual and multiple uORFs can be inspected in two types of visualizations, which aim at simplifying functional analyses and predictions: in a streamlined transcript- and reading-frame-specific model of each uORF’s translational context and in a customized link to the UCSC Genome Browser (34). The latter aids with in-depth analyses, such as the comparison between uORFs in uORFdb and established footprints from ribosome profiling. All uORFs are supplemented with downloadable metadata, including uORF sequences, uORF length, position within the transcript, and the Kozak sequence. The graphical displays and the immediately available sequence and context-related information allow for instant preliminary functional predictions by the users in a fraction of the time required for manual data generation.
MATERIALS AND METHODS
Taxonomy data for the database
In order to resolve the taxonomy IDs associated with the genes and publications in the database, we sought to insert the current version of the NCBI taxonomy (35). Thus, we downloaded and decompressed the NCBI taxonomy dump (ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz; accessed May 6, 2022) and extracted the taxonomy IDs and scientific names from the ‘names.dmp’ file. TaxonKit v0.10.1 (36) and a custom script were used to add the full lineage to each entry. If multiple entries had the same lineage and name, only one of the entries was kept (Supplementary Methods).
Literature preparation
We extracted the publications from the database file of the original version of uORFdb. These publications had already been manually curated as described before (32). Briefly, publications had been identified based on a PubMed (35) query and annotated according to a defined set of criteria. For each criterion, publications received either a plus (check mark in the original publication), a minus (cross in the original publication), or no entry (dash in original publication) indicating positive, negative, or no evidence for the feature, respectively (32). This allowed users to search and select for specific features of their interest. Using a custom script, we retrieved additional publication metadata, such as abstract, title, and doi from NCBI’s (35) Literature Citation Exporter application programming interface (API) (https://api.ncbi.nlm.nih.gov/lit/ctxp/v1/pubmed/?format=ris). We queried multiples of the E-utilities (35) for the gene symbol, symbol aliases, gene names, assembly, chromosome, and taxonomy ID of each gene associated with a publication (Supplementary Methods).
Prediction of uORFs
We predicted uORFs using a custom program called uORF_finder. It analyzed genome and transcriptome files from 13 candidate species obtained from the UCSC Genome Browser database (37): Homo sapiens, Drosophila melanogaster, Mus musculus, Danio rerio, Rattusnorvegicus, Bos taurus, Xenopus laevis, Xenopustropicalis, Macaca mulatta, Gallus gallus, Pongo abelii, Sus scrofa, and Pan troglodytes (Supplementary Table S1). The uORF_finder first removed transcripts without an annotated CDS or TLS. Transcripts from non-regular chromosomes (names containing ‘alt’, ‘fix’, ‘random’ and ‘chrUn’) or from chromosomes that were not present in the respective genome FASTA file were filtered out. Additionally, only transcripts with non-redundant IDs were kept. The program then scanned all three reading frames from the TLS of each protein-coding transcript (NM accessions (38)) and predicted uORFs starting with canonical and aTIS codons. The extracted nucleotide sequences (based on the genome, thus they contain thymine instead of uracil) and the translated amino acid sequences were supplemented with metadata supporting estimations on the functionality of each uORF, including uORF length, distance from the 5’-cap structure and the CDS, as well as the Kozak context around the uStart. Moreover, a separate file with transcript and exon metadata was created. We subsequently reformatted the uORF file and the file with the transcripts and exons and annotated them with the gene metadata from NCBI (accessed May 4, 2022). During this process, we discarded any transcript (and all of its exons and uORFs) if the related gene was a pseudo gene or the transcript accession had been withdrawn, suppressed, or renamed by NCBI. A detailed description of the pipeline is available in the Supplementary Methods.
Somatic variant calling in TCGA patients with regard to uORF regions
For patient cohorts of breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), acute myeloid leukemia (LAML), lung adenocarcinoma (LUAD), prostate adenocarcinoma (PRAD), and skin cutaneous melanoma (SKCM) a total of 129 TB of alignments between older assemblies of the human genome and sequencing reads were provided by the TCGA Research Network (https://www.cancer.gov/tcga) and downloaded from the GDC (39) Legacy Archive (https://portal.gdc.cancer.gov/legacy-archive). For every patient, there was at least one BAM file with reads from normal tissue and at least one BAM file with reads from the tumor. First, we selected one of the normal files for each patient, following guidelines adapted from GDC (https://docs.gdc.cancer.gov/Data_Submission_Portal/Users_Guide/Best_Practices/#specifying-tumor-normal-pairs-for-analysis; accessed May 28, 2021). We then used a variant calling workflow that was based on the GDC pipeline (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/DNA_Seq_Variant_Calling_Pipeline/; accessed June 1, 2021) and the Genome Analysis Toolkit's (GATK’s) (40) best practices (https://gatk.broadinstitute.org/hc/en-us/articles/360035894731-Somatic-short-variant-discovery-SNVs-Indels-; accessed June 1, 2021). Briefly, after realignment of the BAM files to GRCh38.p13 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.39; downloaded June 15, 2021), quality control and somatic variant calling in Mutect2 (41) (GATK v4.1.4.1) followed. If a patient had multiple tumor alignments, we performed variant calling on each of them using the same normal file. Subsequently, variants were filtered and annotated with dbSNP (35) and ClinVar (35) (by position and allele or by position only). We only retained variants from uORF regions and kept only the ‘best’ VCF file for every patient, as defined by containing the highest number of variants, being of non-metastatic origin, and being generated based on the largest tumor BAM file of an individual patient (proxy of coverage). We assigned uORF variants with allele counts of the reference populations derived from gnomAD (42) version three (genome study), ExAC (43) version one, and TOPMed (44) version three (version two, if three was not available for the individual variant), which we retrieved from dbSNP. We only did this for uORF variants that matched a variant in dbSNP by position and by allele. Next, we separately predicted the variant effects on the uORF start codon, the Kozak sequence (including the start codon), the uORF sequence (including the start and stop codons), and the uORF stop codon. Moreover, we evaluated the variant effects on the uORF length and the uORF distance from the CDS. To avoid issues with frameshifts, we excluded insertions and deletions and only focused on substitutions of one or multiple base pairs. Additionally, variants were not allowed to overlap with introns. Reference allele frequencies were calculated for gnomAD, ExAC, and TOPMed based on the aforementioned raw allele counts. We calculated the allele frequency of a variant in each cancer type using the tumor genotypes of all patients that were affected by the same variant (defined by chromosome, position, and alleles) and the total allele count for the respective cancer type (Supplementary Table S2). Total allele counts and the allele count derived from the individual patient's genotype were corrected by patient sex for variants on the sex chromosomes (Supplementary Table S2). A detailed description of the pipeline is available in the Supplementary Methods.
Technical implementation
The uORFdb website is hosted on an Apache 2.4.48 server running on FreeBSD 13 and uses in-house JavaScript and CSS. Communication between the website and its users is protected by HTTPS. Internally, the uORF data is retrieved from a PostgreSQL database (version 14beta2) using in-house Perl scripts, making heavy use of the DBI module (https://dbi.perl.org/). To execute our internal Perl scripts based on user selections on the website, we use the Apache module mod_perl (https://perl.apache.org/). The input data (see previous sections) was inserted into the database with custom Perl scripts (Supplementary Methods). The web interface is based on so-called views. Views represent a collection of related data, for example genes, transcripts, uORFs, and variants. Views can be queried from a central search page and users can switch freely between views without following a hard-coded path.
Based on user selection, a custom script extracts the citation metadata for one or multiple publications from uORFdb and creates a RIS citation file. The keyword (KW) section contains the category names from our manual annotations for which positive or negative evidence exists. We tested the RIS citation files with Zotero 6.0.12, Endnote 20.4, and Citavi 6.7.0.0. In order to inform users once new publications are added to the database, we created an RSS feed which contains our manual annotations and is updated once a day (Supplementary Methods). The RSS feed was verified using the W3C Feed Validation Service3 (https://validator.w3.org/feed; accessed August 24, 2022) and the integration of the feed with reference managers was tested using Zotero. We also regularly export the database contents and provide them as separate downloads. For every download, a reduced example file is produced to facilitate exploration of the file format. All downloads are accompanied by MD5 checksum files, so that users are able to verify the downloads (Supplementary Methods).
If the user clicks on a gene, transcript, or uORF, the user is referred to the UCSC Genome Browser, which will display the tracks of all transcripts and uORFs for the gene associated with the selected entry. We create custom tracks in BED detail format on the fly, which contains HTML for custom track details pages with links to the UCSC Table Browser (45) (Supplementary Methods). All features of the web interface were tested on desktop computers with Firefox 91.13.0esr, Edge 105.0.1343.27, Chrome 105.0.5195.102, and Safari 15.6.1.
DATABASE CONTENTS AND FEATURES
Manually curated uORF-related literature
Initially, uORFdb was designed to allow researchers to conveniently find publications addressing specific uORF-related topics by selecting manually curated categories. The first release of uORFdb contained >450 publications (32). Since then, we have continuously updated the literature part of the database, now covering over 1 040 publications. To further improve the usability of uORFdb's indexed collection of publications, users can now download citations in RIS format directly from the uORFdb website. The citations are supplemented with our manual annotations. We also make all publications and manual annotations available as an RSS feed, which sends a notification once a new publication has been added to the database. RSS readers are designed to work well on almost any screen size, allowing users to conveniently explore publications even on the relatively small screens of tablets and mobile phones. Additionally, some reference managers allow users to import publications directly from the RSS feed and to add them to a collection of references.
Genomic position, sequence, and context information for all predicted uORFs
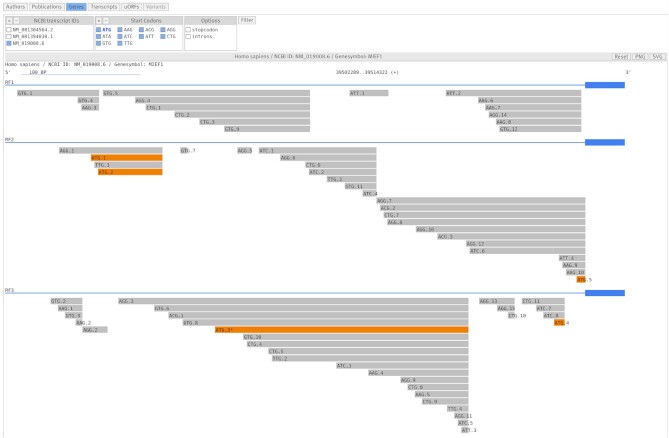
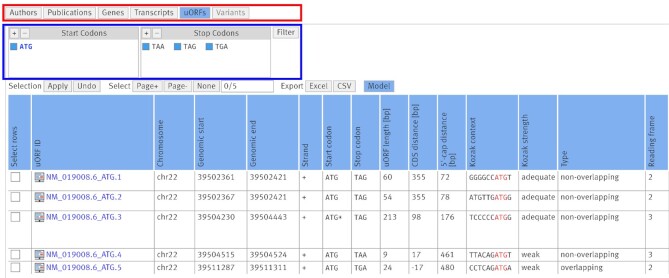
In 2020, we started to extend uORFdb into a central hub for uORF research by adding sequence-based information for all computationally predicted ATG and aTIS uORFs from 13 eukaryotic species, including humans. The uORFs were predicted based on the sequences extracted from the respective genomes, thus the reported sequences contain thymine instead of uracil bases. The extracted nucleotide and the translated amino acid sequences were supplemented with metadata supporting estimations on the functionality of each uORF, including length, distance from the 5’-cap structure and the CDS, as well as the Kozak context around the uStart. After exclusion of uORFs in pseudo genes or withdrawn transcripts, all identified ATG and aTIS uORFs added up to a total of 6 693 228 uORF sequences across all analyzed transcriptomes, including 2 422 112 uORFs identified in the human transcriptome (Supplementary Figure S1). The majority of uORFs were aTIS uORFs (Supplementary Figure S2). The metadata for different uORF-associated features are presented as tables on the website. While this is ideal for an in-depth analysis, it hampers evaluation of uORFs at a glance. Therefore, we implemented two types of graphical displays to aid with quick visual evaluations of the uORF’s context and its potential function. Starting with a selection of genes, transcripts, or uORFs, a click on the ‘Model’ button returns a scaled graphical display of the key features of the uORF’s translational context (Figure 1). At a glance, users can assess and compare the distance between the start of the transcript and the uORF, the distance between the uORF and the CDS, the length of the uORF, and its position related to other surrounding uORFs in a reading-frame-specific manner. Inside the model display, uORFs can be filtered based on the transcript or the start and stop codon (Figure 1). Each transcript variant of a gene is displayed in a separate panel. The panels can be independently enlarged and exported as PNG or SVG files (Figure 1). By clicking on a single uORF, an overlay window opens. It contains a table with nucleotide and peptide sequences, as well as other related metadata, similar to the data contained in the table when opening the ‘uORFs’ view (Figure 2).
Figure 1.
Scaled graphical model for the human MIEF1 TLS and uORFs in the transcript NM_019008.6. The image shows all ATG and aTIS uORFs in uORFdb in the three reading frames (RF1 to RF3) of the transcript. Canonical ATG uORFs with an AUG uStart codon are highlighted in orange by default. The CDS is depicted as a bold blue bar, the TLS as a thin blue bar. The scale of 100 base pairs (BP) is indicated at the top, but only applies to the TLS. The uORF ATG.3 in reading frame three of NM_019008.6 encodes the uPeptide predicted by Vanderperre and coworkers (48). Note that in the MIEF1 transcript NM_001394030.1 the main protein is initiated by the ATG.3 uORF, indicated here by the asterisk symbol. Additionally, the image shows the view buttons and filters at the top that can be used to adjust the display.
Figure 2.
Screenshot of the ‘uORFs’ view showing details for the five ATG uORFs in uORFdb in transcript NM_019008.6 of the human gene MIEF1. Views can be freely changed using the view buttons (red box). The ‘Variants’ view is greyed out, as there were no somatic cancer variants associated with the displayed uORFs. uORFs can be filtered by start and stop codon (blue box). The uORF ATG.3 encodes the uPeptide predicted by Vanderperre and coworkers (48). Note that in the MIEF1 transcript NM_001394030.1, the main protein is initiated by ATG.3, as indicated by the asterisk symbol in the ‘Start codon’ column. The image also shows the export options to Excel and CSV format and the blue ‘Model’ button that can be used to display the uORF model of all uORFs in the current view. The start codon inside each Kozak sequence is highlighted in red. For convenience, we cropped the screenshot after the ‘Reading frame’ column.
The second visualization type in uORFdb uses the UCSC Genome Browser, which allows for additional sequence analyses. Users can inspect all transcripts and uORFs of a single gene by clicking on the respective gene symbol (‘Genes’ and ‘Transcripts’ view) or the uORF ID (‘uORFs’ view). The link in the gene view is only available if the database contains transcripts for the respective gene. If the selected item was a transcript or uORF, it will be automatically highlighted in the Genome Browser. Users can then perform a functional evaluation by comparing the uORF tracks to the multitude of tracks hosted by UCSC, for example ribosome profiling tracks. Clicking on a single uORF or transcript track will open a custom track details page. On this page, we directly link to the UCSC Table Browser where users can either download the sequences (including advanced options, such as repeat masking) or directly import their sequences into the Galaxy platform (46) for easy online analysis. When using our links, all the fields are already filled in, except for the track and table fields, which depend on user selections.
Users who want to perform local analyses can export the entries of the ‘uORFs’ view as CSV or Excel files. Additionally, the nucleotide or amino acid sequences for each uORF, as well as the alternate full sequences for each genetically variant uORF, are available for download as single FASTA files. Database exports are available from the download tab in the website's menu. Exports can be verified using the associated MD5 checksum files and are documented in a README file in the same directory. Since some database exports are several gigabytes in size, we also provide example files, which only contain a limited number of records and can be safely opened in any spreadsheet software.
uORF-related genetic variability
All uORF sequences in uORFdb are connected to genetic variability data obtained from general populations, as well as from disease-related patient cohorts: links in the ‘uORFs’ view will redirect users to variants in the regions of uORF exons listed in dbSNP and ClinVar (‘Exon variants in dbSNP’ and ‘Exon variants in ClinVar’, respectively). Additionally, we analyzed whole-genome sequencing (WGS) data from patient cohorts with six major cancer types provided by the TCGA Research Network and GDC with respect to somatic variants affecting the predicted human uORFs in uORFdb. In total, we detected 129 299 unique putative variants affecting uORF sequences. The ‘Variants’ view reports the variant effect on the respective uORF codon or region. This includes the variant-associated changes with respect to start or stop codon, uORF length, uORF distance from the CDS, and the Kozak sequence. Users can export the resulting alternate uORF sequences, further investigate the somatic variants on dbSNP and ClinVar, and compare the frequencies of the somatic variants in the individual cancer types to variant frequencies in the general population (gnomAD, ExAC, and TOPMed).
Revised more flexible search field and web interface
The new uORFdb can be explored from a central search bar on the uORFdb home page. It now allows a much more flexible search as compared to the initial release, covering numerous categories of search terms including taxon, gene name or symbol, transcript ID, author, PubMed ID, dbSNP or ClinVar IDs, and more. By default, queries address all searchable fields from all views at once. By (un)checking the checkboxes in front of the view and field names, the search can be conveniently adjusted for more specific results. Numbers behind the view names indicate how many hits were found and users can directly access further details for the results in a specific view by clicking on the arrow button next to the view name. In order to ensure good performance for all users, the maximum number of hits per view is 1 000.
PRACTICAL APPLICATION OF uORFdb
In the following section, we give an example of a practical use of uORFdb with a focus on bench scientists and clinicians. In this example, we investigate the human MIEF1 gene (also known as SMCR7L and MID51) and the uPeptide produced by one of its uORFs. The uPeptide SMCR7L (GenBank (47) accession: CCO13821.1) (48) is more abundant than the main protein translated from the CDS (49) and is functional in the context of mitochondrial translation (27). In this exemplary analysis, uORFdb is used to collect more information on this specific uORF and its sequence context.
To start our analysis, we search for the term ‘MIEF1’ in the ‘Genes’ view by using the search bar on the uORFdb start page. By default, all searchable fields from all views listed in the grey panels below the search bar are queried. ‘MIEF1’ is a gene symbol, thus we only choose to search in the ‘Gene symbol’ field of the ‘Genes’ view by deselecting the other checkboxes. In the ‘Genes’ view, we see that genes from nine taxa in uORFdb matched our case-independent search. This was expected, as MIEF1 is an important protein in the context of mitochondrial fission and elongation (50).
We select the human MIEF1 gene by ticking the checkbox in front of the row, followed by a click on ‘Apply.’ From this moment on, all items in the other views are only related to the human MIEF1 gene. Next, we try to identify the target uORF among all 170 uORFs reported in uORFdb for human MIEF1. In uORFdb, uORFs are named following the scheme: ‘[transcript ID]_[type of start codon].[numerical index]’. Starting from one, uORFs on the same transcript variant (TV) and with the same start codon are indexed according to their position on the respective TV, irrespective of reading frame.
According to Vanderperre and coworkers, the ATG uORF encoding the SMCR7L uPeptide was located in the transcript NM_019008 (48). In order to identify the uORF in uORFdb, we first select the respective transcript in the ‘Transcripts" view. Then, we move on to the ‘uORFs’ view, removing all non-ATG uORFs using the filter panel at the top of the page (Figure 2, blue box). By comparing the given uPeptide sequence (48) to the five remaining ATG uORFs in the table (Figure 2), we identify NM_019008.6_ATG.3 as the uORF of interest. Next, we want to compare the transcript NM_019008.6 and the uORF of interest with the other two TVs of MIEF1 to check whether the ATG.3 start site is present in all TVs or if it is exclusively found in NM_019008.6. Therefore, we go back to the ‘Genes’ view and then enter the graphical display via the blue ‘Model’ button at the top of the page. Looking at the graphical model, the ATG.3 uORF appears to be present only in NM_019008.6 and NM_001304564.2 (Figure 1 and Supplementary Figures S3 and S4). The asterisk symbol next to ATG.3 indicates that it appears to be the start site of the CDS in NM_001394030.1 (Figure 1 and Supplementary Figure S3). This is an important feature of the model, as it allows to distinguish between a ‘pure’ uORF and a uORF that is annotated as the CDS in a different TV. By clicking on the tracks of NM_001304564.2_ATG.3 and NM_019008.6_ATG.3, windows with further details similar to the ‘uORFs’ view open and we confirm that both uORFs have identical sequences. Additionally, the uORF model provides a quick overview of the uORF’s context within the transcript: CDS overlap, uORF length, position of the uORF in the TLS, number of additional ATG and aTIS uORFs in the TLS or in the same reading frame are reported (Figure 1 and Supplementary Figures S3 and S4). Comparing NM_019008.6 and NM_001304564.2, the TLS downstream of the ATG.3 uORF appears to be mostly identical, while the upstream sequences are clearly distinct (Figure 1 and Supplementary Figure S3), despite both of them contain two additional (yet distinct) upstream ATG.1 and ATG.2 uORFs. This observation may lead to the assumption that ATG.3 could be differently translated in the two TVs.
By clicking on the uORF ID NM_019008.6_ATG.3 in the ‘uORFs’ view, a separate tab with the UCSC Genome Browser opens. The Genome Browser shows all uORFs from all TVs of the human MIEF1 gene with the selected uORF NM_019008.6_ATG.3 highlighted in red. Next, we set the display of the ribosome profiling track ‘GWIPS-viz Riboseq’ (51) to full. As accessed on August 30, 2022, the track shows ribosome footprint sequence reads over most of the length of all three ATG uORFs mentioned above (ATG.1 to ATG.3). When we now perform the same analysis for NM_001304564.2, we do not find evidence of ribosome binding to ATG.1 and ATG.2.
At this point we may decide that these observations would be worthwhile to be further experimentally investigated, for example by analyzing differential expression of the MIEF1 TVs or by checking the uORFs’ translational impact in a luciferase reporter experiment. To simplify experimental designs, all information required for uORF sequence analysis and expression vector design is immediately available by selecting the respective uORF in the ‘uORFs’ view and exporting the related metadata in Excel or CSV format (Figure 2).
In addition, we now want to look for variants that may affect the translation of the main protein or of the uPeptide itself, both in the general population and in tumor patients. To assess the variant status in the general population, we display all uORFs of NM_019008.6 and NM_001304564.2 in the ‘uORFs’ view. Each uORF has links to dbSNP and ClinVar that query the databases for variants inside the uORF exons (‘Exon variants in dbSNP’ and ‘Exon variants in ClinVar’, respectively). For example, the ATG.3 uORF sequence in both TVs is linked to 94 dbSNP variants, but has no variants annotated in ClinVar (as of August 30, 2022). Two variants are of particular interest, since they affect the start and stop codon, respectively. One variant changes the AUG start codon to an aTIS (rs1367587204: AUG > GUG) and another one deletes the uORF stop codon (rs1235943987: UAG > UGG). The ‘Variants’ view displays a WGS analysis-derived somatic C > G variant in NM_001304564.2 upstream of the ATG.3 uORF detected in 0.43% of alleles in the TCGA BRCA cohort. The table contains further information on this variant, including the effects on the initiation and termination codons, as well as on the Kozak contexts and uORF sequences. There are no dbSNP or ClinVar IDs reported in the ‘Variants’ view, implying that the exact variant (position and allele) has not yet been identified. Furthermore, the links to dbSNP and ClinVar (‘Position-related variants in dbSNP’ and ‘Position-related variants in ClinVar’, respectively) (accessed August 30, 2022) revealed that there was no report of a variant at the same position (irrespective of base change) in neither of the databases. While six uORFs are affected by this variant, only for the NM_001304564.2_ATC.2 uORF the initiation site is altered, leading to the generation of a new ATG uORF, potentially affecting the downstream ATG.3 and MIEF1 main protein translation.
As demonstrated above for the example of the MIEF1 uPeptide reported by Vanderperre and colleagues (48), uORFdb enables users to collect basic and in-depth uORF-related information for further investigations in a fraction of the time compared with manual structure, literature, and variant analysis.
DISCUSSION
Genetic variability of uORFs has been attributed to many diseases (9–12) and current research is just beginning to recognize uORFs as potential treatment options. For example, uORFs contribute to the immunopeptidome (29,31) and the uPeptide uPEP2 from one of the protein kinase C-eta (PKC-η) uORFs has been shown to reduce the growth of breast cancer and the development of metastases in lung and liver, in vivo (28). However, more research needs to be performed to elucidate the role of uORFs in both health and disease. In order to conduct uORF research at scale, dedicated uORF-centric resources for humans and other animal species are needed. Some other databases contain non-canonical proteins, also including uORFs. However, they were not specifically designed for uORF research and lack the ability to make uORF-related publications, graphical visualizations, sequences, sequence contexts, and variations readily accessible from one central hub (52–55). To the best of our knowledge, the only uORF-centric database is uORFlight, but it focuses on plants (33). This motivated us to substantially update uORFdb by adding >6.6 million uORFs from 13 animal species, including >2.4 million human uORFs detected based on individual TVs. uORFs from different TVs may actually represent the same genomic entity, but as explained in the introduction, the uORF function largely depends on its transcript context. In the future, we want to perform comparative genomics on the uORFs in uORFdb and make the results accessible from the web interface. It would be beneficial to know which uORFs are conserved across species, since these are more likely to be functionally relevant (7,15,56,57). In this context, a deeper future integration of sequences, mass spectrometry, and ribosome profiling data in the tables on the website and in the graphical model, may help to select high-confidence uORFs for experimental investigation.
As reviewed by Schuster and Hsieh, ‘untranslated’ regions, such as the TLS, present promising, yet largely unexplored, regions of variation (58). With more variation screenings focusing on uORFs (12,59–61), this is about to change. We aim to support this new development by making uORF-associated somatic mutations and their effects publicly available in uORFdb. Researchers can freely select variants for confirmation in the lab without restrictions imposed by bioinformatic skills or by access to high-performance hardware. Despite the computational power needed to create the data, the cohort size of 677 patients does not compare to resources such as dbSNP and ClinVar. To provide a bigger picture, we added direct links from uORFdb to both databases, allowing users to query variants in the uORF-associated regions.
An accessible and modern web interface integrates the new data with the original set of publications, which we extended to over 1 040 in the last 9 years. In order to facilitate the use of literature outside of the web interface, users can subscribe to an RSS feed or download citations containing our annotations. This can be useful when working with reference managers or on the relatively small screens of mobile phones and tablets. Users can visually collect the relevant information for functional evaluation of uORFs in a fraction of the time needed for manual analyses: Using the uORF model, uORFs can be graphically explored in the context of their specific transcript and reading frames. Using the UCSC Genome Browser, the uORFs can be compared to the multitude of other tracks hosted by UCSC, for instance ribosome profiling tracks. In conclusion, we developed uORFdb to serve as a central hub for uORF research, paving the way for both novices and experts towards a hassle-free experimental analysis of uORF-mediated translational control mechanisms and uORF-encoded peptide functions.
DATA AVAILABILITY
uORFdb is available from https://www.bioinformatics.uni-muenster.de/tools/uorfdb. The scripts described in the Supplementary Methods are available from: https://github.com/IOB-Muenster/uORFdb.
Supplementary Material
ACKNOWLEDGEMENTS
The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We wish to thank the anonymous specimen donors and the investigating research groups for their contributions. We are grateful to the IT department of the University of Münster for access to the WWU Cloud and the PALMA-II High Performance Computing cluster, which were both vital for the variant analyses. Tabea Kischka developed an earlier version of the uORF_finder for human uORF analysis, thus laying the groundwork for the current version.
Contributor Information
Felix Manske, Institute of Bioinformatics, University of Münster, Münster 48149, Germany.
Lynn Ogoniak, Institute of Bioinformatics, University of Münster, Münster 48149, Germany.
Lara Jürgens, Department of Medicine A, Hematology, Oncology, Hemostaseology and Pneumology, University Hospital Münster, Münster 48149, Germany.
Norbert Grundmann, Institute of Bioinformatics, University of Münster, Münster 48149, Germany.
Wojciech Makałowski, Institute of Bioinformatics, University of Münster, Münster 48149, Germany.
Klaus Wethmar, Department of Medicine A, Hematology, Oncology, Hemostaseology and Pneumology, University Hospital Münster, Münster 48149, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Krebshilfe e.V., Bonn, Germany [70113632 to K.W.]. Funding for open access charge: Open Access Publication Fund of the University of Münster.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mueller P.P., Hinnebusch A.G.. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986; 45:201–207. [DOI] [PubMed] [Google Scholar]
- 2. Abastado J.P., Miller P.F., Jackson B.M., Hinnebusch A.G.. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 1991; 11:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sundaram A., Grant C.M.. A single inhibitory upstream open reading frame (uORF) is sufficient to regulate Candida albicans GCN4 translation in response to amino acid starvation conditions. RNA. 2014; 20:559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu P.D., Harding H.P., Ron D.. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004; 167:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vattem K.M., Wek R.C.. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang K., Ryoo H.D., Park J.-E., Yoon J.-H., Kang M.-J.. A Drosophila reporter for the translational activation of ATF4 marks stressed cells during development. PLoS One. 2015; 10:e0126795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Watatani Y., Ichikawa K., Nakanishi N., Fujimoto M., Takeda H., Kimura N., Hirose H., Takahashi S., Takahashi Y.. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region*. J. Biol. Chem. 2008; 283:2543–2553. [DOI] [PubMed] [Google Scholar]
- 8. Zhou D., Palam L.R., Jiang L., Narasimhan J., Staschke K.A., Wek R.C.. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions*. J. Biol. Chem. 2008; 283:7064–7073. [DOI] [PubMed] [Google Scholar]
- 9. Wen Y., Liu Y., Xu Y., Zhao Y., Hua R., Wang K., Sun M., Li Y., Yang S., Zhang X.-J.et al.. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat. Genet. 2009; 41:228–233. [DOI] [PubMed] [Google Scholar]
- 10. Coursimault J., Rovelet-Lecrux A., Cassinari K., Brischoux-Boucher E., Saugier-Veber P., Goldenberg A., Lecoquierre F., Drouot N., Richard A.-C., Vera G.et al.. uORF-introducing variants in the 5′UTR of the NIPBL gene as a cause of Cornelia de Lange syndrome. Hum. Mutat. 2022; 43:1239–1248. [DOI] [PubMed] [Google Scholar]
- 11. Liu L., Dilworth D., Gao L., Monzon J., Summers A., Lassam N., Hogg D.. Mutation of the CDKN2A 5’ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat. Genet. 1999; 21:128–132. [DOI] [PubMed] [Google Scholar]
- 12. Schulz J., Mah N., Neuenschwander M., Kischka T., Ratei R., Schlag P.M., Castaños-Vélez E., Fichtner I., Tunn P.-U., Denkert C.et al.. Loss-of-function uORF mutations in human malignancies. Sci. Rep. 2018; 8:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA. Cell. 1978; 15:1109–1123. [DOI] [PubMed] [Google Scholar]
- 14. Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989; 108:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J.et al.. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012; 22:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calvo S.E., Pagliarini D.J., Mootha V.K.. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002; 299:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip. Rev.: RNA. 2014; 5:765–768. [DOI] [PubMed] [Google Scholar]
- 19. Luukkonen B.G., Tan W., Schwartz S.. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 1995; 69:4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001; 29:5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Col B., Oltean S., Banerjee R.. Translational regulation of human methionine synthase by upstream open reading frames. Biochim. Biophys. Acta, Gene Struct. Expression. 2007; 1769:532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hill J.R., Morris D.R.. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J. Biol. Chem. 1993; 268:726–731. [PubMed] [Google Scholar]
- 23. Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987; 7:3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981; 9:5233–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986; 44:283–292. [DOI] [PubMed] [Google Scholar]
- 26. Miller P.F., Hinnebusch A.G.. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 1989; 3:1217–1225. [DOI] [PubMed] [Google Scholar]
- 27. Rathore A., Chu Q., Tan D., Martinez T.F., Donaldson C.J., Diedrich J.K., Yates III,J.R., Saghatelian A.. MIEF1 microprotein regulates mitochondrial translation. Biochemistry. 2018; 57:5564–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jayaram D.R., Frost S., Argov C., Liju V.B., Anto N.P., Muraleedharan A., Ben-Ari A., Sinay R., Smoly I., Novoplansky O.et al.. Unraveling the hidden role of a uORF-encoded peptide as a kinase inhibitor of PKCs. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2018899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nelde A., Flötotto L., Jürgens L., Szymik L., Hubert E., Bauer J., Schliemann C., Kessler T., Lenz G., Rammensee H.-G.et al.. Upstream open reading frames regulate translation of cancer-associated transcripts and encode HLA-presented immunogenic tumor antigens. Cell. Mol. Life Sci. 2022; 79:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cloutier P., Poitras C., Faubert D., Bouchard A., Blanchette M., Gauthier M.-S., Coulombe B.. Upstream ORF-encoded ASDURF is a novel Prefoldin-like subunit of the PAQosome. J. Proteome Res. 2020; 19:18–27. [DOI] [PubMed] [Google Scholar]
- 31. Erhard F., Dölken L., Schilling B., Schlosser A.. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol. Res. 2020; 8:1018–1026. [DOI] [PubMed] [Google Scholar]
- 32. Wethmar K., Barbosa-Silva A., Andrade-Navarro M.A., Leutz A.. uORFdb—a comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 2014; 42:D60–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niu R., Zhou Y., Zhang Y., Mou R., Tang Z., Wang Z., Zhou G., Guo S., Yuan M., Xu G.. uORFlight: a vehicle toward uORF-mediated translational regulation mechanisms in eukaryotes. Database. 2020; 2020:baaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The Human Genome Browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022; 50:D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen W., Ren H.. TaxonKit: a practical and efficient NCBI taxonomy toolkit. J. Genet. Genomics. 2021; 48:844–850. [DOI] [PubMed] [Google Scholar]
- 37. Lee B.T., Barber G.P., Benet-Pagès A., Casper J., Clawson H., Diekhans M., Fischer C., Gonzalez J.N., Hinrichs A.S., Lee C.M.et al.. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022; 50:D1115–D1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grossman R.L., Heath A.P., Ferretti V., Varmus H.E., Lowy D.R., Kibbe W.A., Staudt L.M.. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 2016; 375:1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M.et al.. The Genome Analysis Toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benjamin D., Sato T., Cibulskis K., Getz G., Stewart C., Lichtenstein L.. Calling somatic SNVs and indels with Mutect2. 2019; bioRxiv doi:2 December 2019, pre-print: not peer-reviewed 10.1101/861054. [DOI]
- 42. Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P.et al.. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020; 581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.et al.. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M.et al.. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature. 2021; 590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. The Galaxy Community The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022; 50:W345–W351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2022; 50:D161–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vanderperre B., Lucier J.-F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.-M., Roucou X.. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013; 8:e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Delcourt V., Brunelle M., Roy A.V., Jacques J.-F., Salzet M., Fournier I., Roucou X.. The protein coded by a short open reading frame, not by the annotated coding sequence, is the main gene product of the dual-coding gene MIEF1*. Mol. Cell. Proteomics. 2018; 17:2402–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Losón O.C., Song Z., Chen H., Chan D.C.. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013; 24:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michel A.M., Fox G., Kiran A.M., De Bo C., O’Connor P.B.F., Heaphy S.M., Mullan J.P.A., Donohue C.A., Higgins D.G., Baranov P.V.. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 2014; 42:D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olexiouk V., Van Criekinge W., Menschaert G.. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018; 46:D497–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y., Zhou H., Chen X., Zheng Y., Kang Q., Hao D., Zhang L., Song T., Luo H., Hao Y.et al.. SmProt: a reliable repository with comprehensive annotation of small proteins identified from ribosome profiling. Genomics, Proteomics Bioinf. 2021; 19:602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brunet M.A., Lucier J.-F., Levesque M., Leblanc S., Jacques J.-F., Al-Saedi H.R.H., Guilloy N., Grenier F., Avino M., Fournier I.et al.. OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res. 2021; 49:D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao W., Zhang S., Zhu Y., Xi X., Bao P., Ma Z., Kapral T.H., Chen S., Zagrovic B., Yang Y.T.et al.. POSTAR3: an updated platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2022; 50:D287–D294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang H., Wang Y., Wu X., Tang X., Wu C., Lu J.. Determinants of genome-wide distribution and evolution of uORFs in eukaryotes. Nat. Commun. 2021; 12:1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kimura M. The neutral theory of molecular evolution. 1983; Cambridge: Cambridge University Press. [Google Scholar]
- 58. Schuster S.L., Hsieh A.C.. The untranslated regions of mRNAs in cancer. Trends Cancer. 2019; 5:245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jürgens L., Manske F., Hubert E., Kischka T., Flötotto L., Klaas O., Shabardina V., Schliemann C., Makalowski W., Wethmar K.. Somatic functional deletions of upstream open reading frame-associated initiation and termination codons in human cancer. Biomedicines. 2021; 9:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Whiffin N., Karczewski K.J., Zhang X., Chothani S., Smith M.J., Evans D.G., Roberts A.M., Quaife N.M., Schafer S., Rackham O.et al.. Characterising the loss-of-function impact of 5’ untranslated region variants in 15,708 individuals. Nat. Commun. 2020; 11:2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee D.S.M., Park J., Kromer A., Baras A., Rader D.J., Ritchie M.D., Ghanem L.R., Barash Y.. Disrupting upstream translation in mRNAs is associated with human disease. Nat. Commun. 2021; 12:1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
uORFdb is available from https://www.bioinformatics.uni-muenster.de/tools/uorfdb. The scripts described in the Supplementary Methods are available from: https://github.com/IOB-Muenster/uORFdb.