Abstract
Gut microbiota plays a significant role in maintaining host health, and conversely, disorders potentially lead to dysbiosis, an imbalance in the composition of the gut microbial community. Intervention approaches, such as medications, diets, and several others, also alter the gut microbiota in either a beneficial or harmful direction. In 2020, the gutMDisorder was developed to facilitate researchers in the investigation of dysbiosis of gut microbes as occurs in various disorders as well as with therapeutic interventions. The database has been updated this year, following revision of previous publications and newly published reports to manually integrate confirmed associations under multitudinous conditions. Additionally, the microbial contents of downloaded gut microbial raw sequencing data were annotated, the metadata of the corresponding hosts were manually curated, and the interactive charts were developed to enhance visualization. The improvements have assembled into gutMDisorder v2.0, a more advanced search engine and an upgraded web interface, which can be freely accessed via http://bio-annotation.cn/gutMDisorder/.
INTRODUCTION
Gut microbes, including commensal and pathogenic microbes, colonize in intestinal tract of hosts, and have a crucial impact on gut homeostasis and host health by producing various metabolites with the ability to affect the gut barrier and immunity (1–3). Disease onset and progression routinely correlate with an increase or decrease of pathogenic or commensal microbes (4,5). Additionally, many intervention measures predispose to an altered composition of the gut microbial community (6,7). Recently, advanced sequencing technologies allow for convenient investigation of the confirmed associations involved in gut microbial dysbiosis as occurs with disorders and interventions. Benefiting from the improved sampling methods, the composition and properties of gut microbes related with different disease signatures and treatment programs are increasingly evaluated (8,9).
In 2020, gutMDisorder v1.0 was initially released following a thorough review of publications reporting experimentally validated associations between gut microbes and disorders or interventions (10). As the gut microbial knowledge base is increased, the focus has shifted to gut microbial dysbiosis under a variety of conditions, which is equally critical to the understanding of microbes, hosts, and their interactions, and does not limit to the healthy controls. Ma et al. reported microbial dysbiosis in patients with Crohn's disease (CD), and they also observed that the abundance of Bacteroidetes was significantly decreased in patients with active CD, as compared with the inactive CD group, illustrating the composition variation of gut microbes at different stages of the same disorder (11). Loomba et al. observed patients with non-alcoholic steatohepatitis to whom varied doses of aldafermin were administrated, manifested variations in the Veillonella genus (12). Vemuri et al. indicated that young and aging mice differed dramatically in the relative abundance of Bacteroidetes and Firmicutes, similarly, the addition of probiotics resulted in different microbial alterations for both groups (13).
The sequencing data of gut microbiome continuously emerges, and the analysis of raw sequencing data using unified pipeline can contribute to comprehensively and intuitively verifying or confirming gut microbial dysbiosis under a range of conditions. Therefore, gutMDisorder was updated by repeating the reviews of all the publications consulted in v1.0 and reviewing new publications, thereby manually integrating associations under multitudinous conditions. The microbial content of raw microbial sequencing data was then downloaded and annotated, manually curating the metadata of the corresponding hosts and providing interactive charts for viewing. Advanced filtering options were then added to optimize the searching system. Finally, the system interface was upgraded for aesthetic and minimalist design to enhance the user experience.
DATA COLLECTION AND DATABASE CONTENT
To update gutMDisorder, the publications referred to in v1.0 were re-evaluated and new reports from 31 October 2019 to 31 October 2021 were included by searching the PubMed database with the following keywords: ‘gut’, ‘intestinal’, ‘microbiota’, ‘microbiome’. All the publications were downloaded and gut microbial alteration information manually extracted across multiple control conditions, not just limited to the healthy control group alone. Microbial alterations are not limited to statistically significant increase and decrease, but also include the presence and absence of microbes without statistical significance, in the studies published. The total number of literature-based associations increased considerably when compared with the previous version. For example, gutMDisorder v1.0 contained 2,263 associations between 579 gut microbes, and 123 disorders and 77 interventions in human reports, as well as 930 associations between 273 gut microbes, and 33 disorders and 151 interventions in mouse experimental studies. In the currently updated version of gutMDisorder, 4,164 experimentally validated associations between 774 gut microbes, and 346 phenotype and 316 intervention comparisons in human; and 2,251 associations between 425 gut microbes, and 55 phenotype and 285 intervention comparisons in mouse experiments were collected from 548 publications, as presented in Table 1.
Table 1.
Number of gut microbes, phenotype comparisons, intervention comparisons and their associations in humans and mice
| Version | Associations source | Host species | No. of gut microbes | No. of phenotype comparisons | No. of intervention comparisons (chemicals and drugs/diet, food, and nutrition/others) | No. of associations |
|---|---|---|---|---|---|---|
| 1.0 | Literature-based associations | Human | 579 | 123 | 77 (46/ 15/ 16) | 2,263 |
| 1.0 | Literature-based associations | Mouse | 273 | 33 | 151 (66/ 52/ 33) | 930 |
| 2.0 | Literature-based associations | Human | 774 | 346 | 316 (132/ 132/ 52) | 4164 |
| 2.0 | Literature-based associations | Mouse | 425 | 55 | 285 (148/ 94/ 43) | 2251 |
| 2.0 | Raw data-based associations | Human | 1009 | 113 | 19 (9/ 3/ 7) | 8410 |
| 2.0 | Raw data-based associations | Mouse | 358 | 9 | 35 (13/ 16/ 6) | 1720 |
Additionally, gut microbial raw sequencing data were extracted from electronic databases, Sequence Read Archive (SRA) (14) and European Bioinformatics Institute (EBI) (15), based on the keywords: ‘gut microbiome’, ‘fecal’, ‘multispecies’. A total of 105 gut microbial datasets involving 16s rRNA gene sequencing and whole metagenomic sequencing were obtained, and then the metadata of the corresponding hosts was manually curated. The datasets were then processed using unified pipelines, QIIME 2 (16) and MetaPhIAn3 (17), for 16s rRNA gene and whole metagenomic sequencing data, respectively. Once taxonomic tables were obtained, alpha-diversity analysis including Shannon and Simpson index, and beta-diversity analysis including non-metric multidimensional scaling and principal co-ordinates analysis were performed. Linear discriminant analysis effect size method (18) was subsequently used to identify marker microbes with linear discriminant analysis scores of 2 were used as the cutoff. As a result, 8410 calculated associations between 1009 gut microbes, and 113 phenotype and 19 intervention comparisons in human reports, and 1720 associations between 358 gut microbes, and 9 phenotype and 35 intervention comparisons in mouse experiments were collected from 105 studies involving analysis of 14 581 samples (Table 1). Figure 1A and B are pie charts depicting the distribution of gut microbes among the classifications of the National Center for Biotechnology Information (NCBI) Taxonomy database (19); numerous microbes are displayed at genus and species level. Figure 1C and D delineates the distribution of literature-based associations among host species in different years and raw data-based associations among sequencing technologies in different years; 16s rRNA gene sequencing technology is the widely applicated in the field of gut microbiota.
Figure 1.
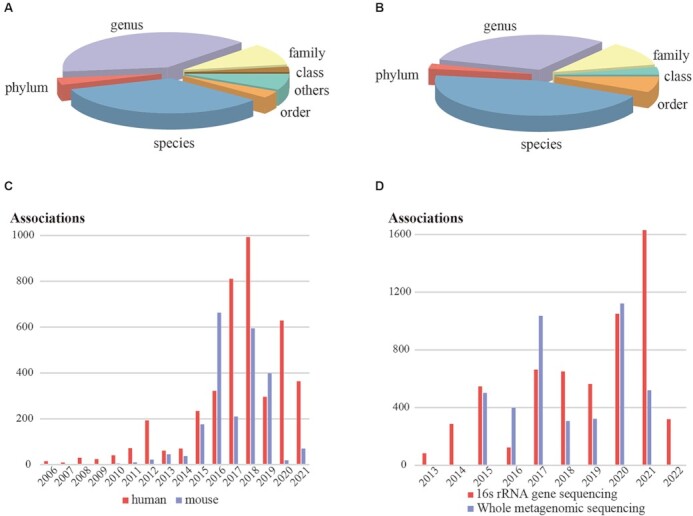
Distribution of gut microbes and associations in gutMDisorder v2.0. (A) The distribution of gut microbes among different taxa from literature-based associations. (B) The distribution of gut microbes among different taxa from raw data-based associations. (C) The distribution of literature-based associations among host species in different years. (D) The distribution of raw data-based associations among sequencing technologies in different years.
USER INTERFACE
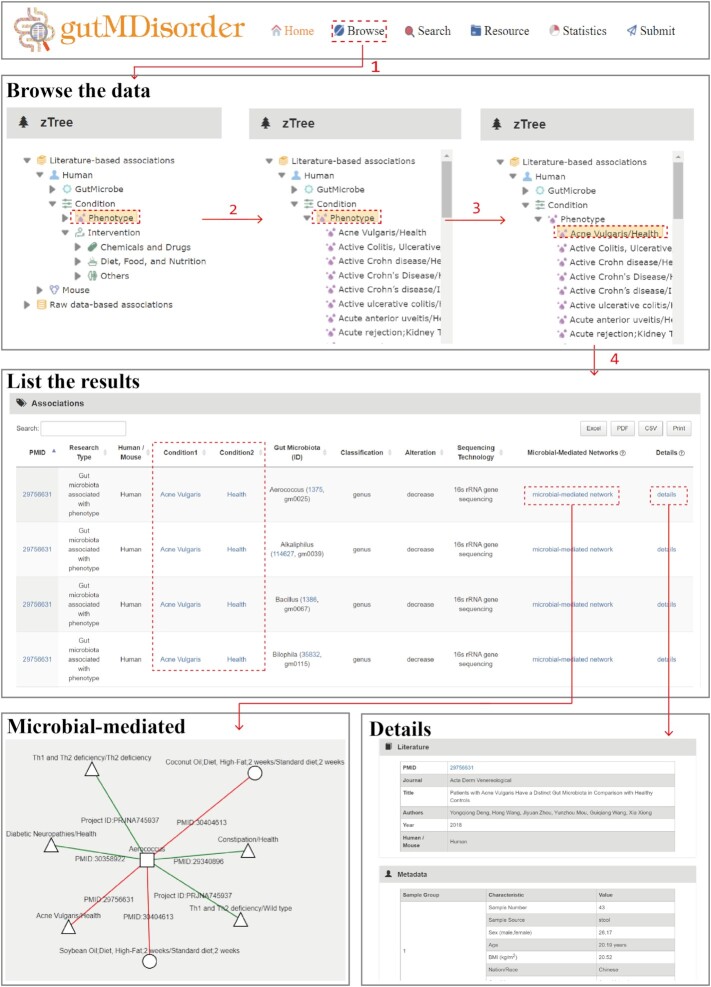
The updated version of gutMDisorder has additional features, such as a tree browser in the ‘Browse’ interface and incorporates search engines in the ‘Search’ interface to query the details regarding the relationships between gut microbes, phenotypes and interventions. The schematic workflows based on published reports and raw data were depicted in Figures 2 and 3, respectively.
Figure 2.
Schematic workflow for literature-based associations in gutMDisorder v2.0.
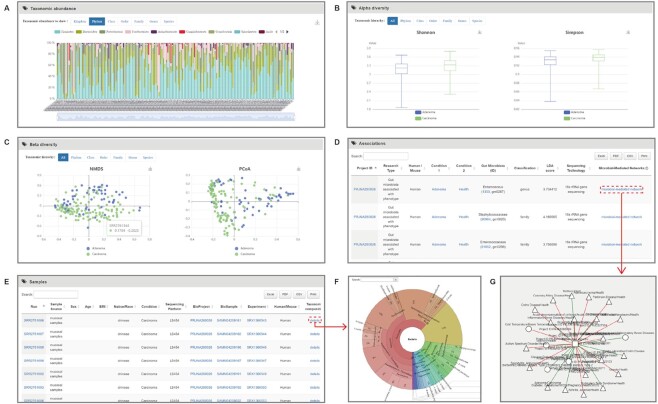
Figure 3.
Results of raw data-based associations in gutMDisorder v2.0. (A) Taxonomic abundances plot displaying the abundance per sample in the related project. Each color represents a different gut microbe. (B) Boxplots displaying alpha diversity. (C) Scatterplots depicting beta diversity. (D) Table demonstrating associations in this comparison. (E) Table detailing sample descriptions. (F) Pie chart depicting the taxonomic abundance. (G) The network illustrating the microbe relationships and comparisons.
The tree browser is initially divided into the literature-based and raw data-based associations according to the association source, these are taken as the root categories. Next, according to host species, gutMDisorder v2.0 bisects into ‘human’ and ‘mouse’ as sub-categories, within each contains two groups: ‘GutMicrobe’ and ‘Condition’. ‘Phenotype’ and ‘Intervention’ are collectively referred to as ‘Condition’. Users can click the ‘GutMicrobe’ or ‘Phenotype’ category to list the names of all gut microbes or phenotype comparisons belonging to the corresponding host species as leaf nodes. The ‘Intervention’ is likewise divided into three groups: ‘Chemical and Drug’, ‘Diet, Food, and Nutrition’, and ‘Others’. Specific intervention comparisons are displayed by clicking each category.
Figure 2 indicates a partial list of ‘human’ species in literature-based associations. For instance, after selecting the phenotype comparison ‘Acne Vulgaris/Health’, the information and alterations of gut microbes are retrieved and displayed in the table where an association with a brief introduction is represented in one row. The microbe identifier and conditions may be linked with the NCBI taxonomy database, Medical Subject Headings, and PubMed for detailed descriptions of the entities. The network diagram of the gut microbe in this line is obtained by clicking ‘microbial-mediated’. With this microbe as the central node, the comparisons related to this gut microbe are connected around, and distinguished by varied graphic shapes. Alterations of the gut microbe are distinguished by different colors. Finally, clicking ‘details’ leads users to the peer-reviewed information obtained from the publications, relevant information regarding the samples, and the detailed information describing the association.
Figure 3 includes a partial list of ‘human’ species in raw data-based associations. For example, after selecting a phenotype comparisons ‘Adenoma/Carcinoma’, the five results of each project: ‘Taxonomic abundance’, ‘Alpha diversity’, ‘Beta diversity’, ‘Associations’ and ‘Sample’ are retrieved.
Taxonomic abundances: The taxonomic abundances of each project are presented in the form of histograms, which can be downloaded at the seven levels of kingdom, phylum, class, order, family, genus and species.
Alpha diversity: Alpha diversity reflects the gut microbial richness of each phenotype. It is measured by the Shannon index and Simpson index, and graphically presented by boxplots.
Beta diversity: Beta diversity illustrates the differences in gut microbial composition of different groups and is graphically presented by scatterplots.
Associations: In the association section, the identifier of ‘microbe’, ‘condition’, and ‘project ID’ are linked to the NCBI database, and by clicking ‘microbial-mediated network’, the relationships of microbe and comparisons are displayed in the network.
Samples: The identifiers ‘Run’, ‘BioProject’, ‘BioSample’ and ‘Experiment’ are additionally linked to the NCBI database. Users can click the ‘details’ icon to obtain a pie chart generated by Krona (20) demonstrating relative abundances of gut microbes for each sample.
In the current version of the search engine, many new filters have been added to meet the requirements of researchers from many disciplines, and users are able to restrict filters through multiple dimensions, thereby achieving more direct access to the associated data. The gutMDisorder search engine provides a ‘Resource’ interface to download an abundance of profiles of raw data-based studies. Finally, researchers are encouraged to submit novel information regarding associations or raw data via the ‘Submit’ page.
CONCLUSION
Given the high demand and significance of gut microbiota associated with disorders and interventions, gutMDisorder was developed and improved. To update this database, we concentrate on the gut microbial dysbiosis under varied conditions, not only those of healthy controls. Moreover, the database is no longer confined to manual extraction from publications, but includes annotated microbial contents of downloaded gut microbial raw sequencing data. The search engine and system interface were also updated. The current gutMDisorder v2.0 contains 4164 literature-based associations between 774 gut microbes, and 346 phenotype and 316 intervention comparisons from human reports, and 2251 associations between 425 gut microbes, and 55 phenotype and 285 intervention comparisons from mouse studies collated from 548 papers. Additionally, the database documents 8410 raw data-based associations between 1009 gut microbes, and 113 phenotype and 19 intervention comparisons in humans, and 1720 associations between 358 gut microbes, and 9 phenotype and 35 intervention comparisons in mice, obtained from 105 gut microbial datasets.
gutMDisorder v1.0 has provided an unprecedented benchmark set of associations between gut microbiota and disorders for the study of human health and diseases, while v2.0 further expands and refines the benchmark set. In the future, our team will devote maximum efforts to establish a comprehensive framework in order to predict the gut microbe-disease associations, thus provide guidance for subsequent experimental verification. Additionally, raw data processing make it possible to perform meta-analysis in the application of multiple datasets of different diseases, leading to more instructive achievements that can be integrated into our database. And beyond that, abundant tools for metabolic pathway enrichment analysis and biomarker identification through machine learning will be also included. Indeed, with the emergence of a large number of publications and various sequencing data, gutMDisorder will continuously update its data, search engine and connotation.
DATA AVAILABILITY
gutMDisorder v2.0 is freely accessible at http://bio-annotation.cn/gutMDisorder/.
Contributor Information
Changlu Qi, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Yiting Cai, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Kai Qian, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Xuefeng Li, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Jialiang Ren, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Ping Wang, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Tongze Fu, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Tianyi Zhao, School of Medicine and Health, Harbin Institute of Technology, Harbin 150001, Heilongjiang, China.
Liang Cheng, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China; NHC Key Laboratory of Molecular Probes and Targeted Diagnosis and Therapy, Harbin Medical University, Harbin 150028, Heilongjiang, China.
Lei Shi, NHC Key Laboratory of Molecular Probes and Targeted Diagnosis and Therapy, Harbin Medical University, Harbin 150028, Heilongjiang, China.
Xue Zhang, NHC Key Laboratory of Molecular Probes and Targeted Diagnosis and Therapy, Harbin Medical University, Harbin 150028, Heilongjiang, China; McKusick-Zhang Center for Genetic Medicine, State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China.
FUNDING
Tou-Yan Innovation Team Program of the Heilongjiang Province [2019-15]; National Natural Science Foundation of China [62222104, 61871160, 62172130]; Heilongjiang Postdoctoral Fund [LBH-Q20030]. Funding for open access charge: Tou-Yan Innovation Team Program of the Heilongjiang Province [2019-15]; National Natural Science Foundation of China [62222104, 61871160, 62172130]; Heilongjiang Postdoctoral Fund [LBH-Q20030].
Conflict of interest statement. None declared.
REFERENCES
- 1. Thursby E., Juge N.. Introduction to the human gut microbiota. Biochem. J. 2017; 474:1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Postler T.S., Ghosh S.. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017; 26:110–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng L., Qi C., Yang H., Lu M., Cai Y., Fu T., Ren J., Jin Q., Zhang X.. gutMGene: a comprehensive database for target genes of gut microbes and microbial metabolites. Nucleic Acids Res. 2022; 50:D795–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Young V.B. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017; 356:j831. [DOI] [PubMed] [Google Scholar]
- 5. Manor O., Dai C.L., Kornilov S.A., Smith B., Price N.D., Lovejoy J.C., Gibbons S.M., Magis A.T.. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020; 11:5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hughes R.L., Holscher H.D.. Fueling gut microbes: a review of the interaction between diet, exercise, and the gut microbiota in athletes. Adv. Nutr. 2021; 12:2190–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi C., Wang P., Fu T., Lu M., Cai Y., Chen X., Cheng L.. A comprehensive review for gut microbes: technologies, interventions, metabolites and diseases. Brief. Funct. Genomics. 2021; 20:42–60. [DOI] [PubMed] [Google Scholar]
- 8. Song E.J., Lee E.S., Nam Y.D.. Progress of analytical tools and techniques for human gut microbiome research. J. Microbiol. 2018; 56:693–705. [DOI] [PubMed] [Google Scholar]
- 9. Zhao Y., Wang C.C., Chen X.. Microbes and complex diseases: from experimental results to computational models. Brief Bioinform. 2021; 22:bbaa158. [DOI] [PubMed] [Google Scholar]
- 10. Cheng L., Qi C., Zhuang H., Fu T., Zhang X.. gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2020; 48:D554–D560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma H.Q., Yu T.T., Zhao X.J., Zhang Y., Zhang H.J.. Fecal microbial dysbiosis in Chinese patients with inflammatory bowel disease. World J. Gastroenterol. 2018; 24:1464–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loomba R., Ling L., Dinh D.M., DePaoli A.M., Lieu H.D., Harrison S.A., Sanyal A.J.. The commensal microbe veillonella as a marker for response to an FGF19 Analog in NASH. Hepatology. 2021; 73:126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vemuri R., Shinde T., Gundamaraju R., Gondalia S.V., Karpe A.V., Beale D.J., Martoni C.J., Eri R.. Lactobacillus acidophilus DDS-1 modulates the gut microbiota and improves metabolic profiles in aging mice. Nutrients. 2018; 10:1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database C.. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cantelli G., Bateman A., Brooksbank C., Petrov A.I., Malik-Sheriff R.S., Ide-Smith M., Hermjakob H., Flicek P., Apweiler R., Birney E.et al.. The European Bioinformatics Institute (EMBL-EBI) in 2021. Nucleic Acids Res. 2022; 50:D11–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F.et al.. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019; 37:852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beghini F., McIver L.J., Blanco-Miguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M.et al.. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021; 10:e65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C.. Metagenomic biomarker discovery and explanation. Genome Biol. 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B.et al.. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ondov B.D., Bergman N.H., Phillippy A.M.. Interactive metagenomic visualization in a Web browser. BMC Bioinf. 2011; 12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
gutMDisorder v2.0 is freely accessible at http://bio-annotation.cn/gutMDisorder/.