Abstract
NLM’s conserved domain database (CDD) is a collection of protein domain and protein family models constructed as multiple sequence alignments. Its main purpose is to provide annotation for protein and translated nucleotide sequences with the location of domain footprints and associated functional sites, and to define protein domain architecture as a basis for assigning gene product names and putative/predicted function. CDD has been available publicly for over 20 years and has grown substantially during that time. Maintaining an archive of pre-computed annotation continues to be a challenge and has slowed down the cadence of CDD releases. CDD curation staff builds hierarchical classifications of large protein domain families, adds models for novel domain families via surveillance of the protein ‘dark matter’ that currently lacks annotation, and now spends considerable effort on providing names and attribution for conserved domain architectures. CDD can be accessed at https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
INTRODUCTION AND OVERVIEW
CDD aims to collect a comprehensive set of protein and domain family models, and it does allow for considerable redundancy in the model set, to ensure good coverage of the protein space. Models that provide significantly overlapping annotation are clustered into protein domain superfamilies, and when domain annotation fails to exceed critical model-specific score thresholds, CDD by default reports superfamily annotation rather than individual model hits. For each model, we compute a consensus sequence, which is used for display purposes only, and reflects the length of the position-specific score matrix (PSSM). While consensus sequences are visible and made available, CDD is not a sequence collection, but is rather meant to enrich the annotation of existing sequence collections. The current CDD version, v3.20, contains 59 693 protein- and protein domain-models obtained from Pfam (1) version 34, SMART (2), the COGs collection (3), TIGRFAMS (4), the NCBI Protein Clusters collection (5), NCBIfam (6) and CDD’s in-house data curation effort (7) (about 1600 new or updated models). For CDD v3.20, the fixed assumed size of the domain model database has again been increased to match the current size of the model collection, resulting in marginally higher E-values reported by RPS-BLAST (8). The upcoming CDD release v3.21 will contain a revised COGs collection, Pfam version 35 and around 500 new or updated models from CDD’s in-house curation. Table 1 details the composition of CDD release v3.20 and the contributions from each source database.
Table 1.
Composition of the CDD model collection
| Data source | Version | Number of models |
|---|---|---|
| Pfam | 34 | 19 178 |
| CDD in-house curation | 3.20 | 18 882 |
| Protein Clusters | (25 October 2021)a | 10 140 |
| COGs | 1 | 4871 |
| TIGRFAM | 15 | 4488 |
| NCBIfams | (25 October 2021) | 1125 |
| SMART | 6* | 1009 |
| CDD superfamily clusters | 3.20 | 4541 |
aRecent changes to the protein clusters and SMART collections were the removal of models considered redundant.
The sizes of these source databases vary considerably, as they were assembled for distinct purposes, and may only consider subsets of the protein space, such as limited by taxonomy. Neither of them is a complete accounting of protein and protein domain families, and neither is the aggregate of many resources, as (1) available protein sequence collections are expanding rapidly and becoming more diverse and (2) limited curation resources impact further growth.
Table 2 shows the 20 largest classifications for common and functionally diverse domain families that have recently been updated or added to CDD. Hierarchical classifications are revisited as curation resources permit. Quite commonly, the analysis of novel ‘dark matter’ families suggests membership in a previously established superfamily classification, for example, or the availability of newly determined 3D structures suggests changes to the multiple sequence alignments that may have an impact on classification details. Over 4700 models curated by the CDD group have been newly published or updated since CDD release v3.16. As of now, a total of 42 937 site annotations are available on 15 943 out of 18 882 CDD staff-curated domain models. Sequence patterns have been recorded for 4971 of these site annotations, so that pattern matching can be used to decide whether annotation can be mapped on to individual sequences.
Table 2.
The largest domain family hierarchies created or updated since CDD release v3.17
| Root | Models | Name |
|---|---|---|
| cd14964 | 592 | seven-transmembrane G protein-coupled receptor superfamily |
| cd00590 | 586 | RNA recognition motif (RRM) superfamily |
| cd13968 | 585 | Catalytic domain of the Protein Kinase superfamily |
| cd00162 | 417 | RING finger (Really Interesting New Gene) domain and U-box domain superfamily |
| cd00174 | 328 | Src Homology 3 domain superfamily |
| cd00900 | 327 | Pleckstrin homology-like domain |
| cd00196 | 311 | Beta-grasp ubiquitin-like fold |
| cd00096 | 291 | Immunoglobulin domain |
| cd01165 | 242 | BTB/POZ domain superfamily |
| cd00648 | 239 | Type 2 periplasmic binding fold superfamily |
| cd15489 | 221 | PHD finger superfamily |
| cd00083 | 202 | basic Helix Loop Helix (bHLH) domain superfamily |
| cd01391 | 191 | Type 1 periplasmic binding fold superfamily |
| cd06174 | 185 | Major Facilitator Superfamily |
| cd14494 | 171 | Cys-based protein tyrosine phosphatase and dual-specificity phosphatase superfamily |
| cd17912 | 170 | N-terminal helicase domain of the DEAD-box helicase superfamily |
| cd08368 | 159 | LIM domain superfamily |
| cd00014 | 158 | calponin homology (CH) domain superfamily |
| cd00194 | 157 | UBA domain-like superfamily |
| cd00105 | 153 | K homology (KH) RNA-binding domain, type I |
The table lists the root node of each hierarchy, the number of models in the hierarchy (including the root node and intermediate nodes if present), and the name of the protein domain (super) family.
We identify as ‘dark matter’ those protein sequences in a collection of non-redundant representatives from a taxonomically diverse set, which do not yet have conserved domain annotation via CDD. These are clustered into sequence-similar groups, and multiple sequence alignment models are created for clusters that contain either (1) sequences obtained from experimentally determined 3D structure, or (2) sequences associated with publications, unless these publications describe very large sequence sets such as complete genomes. These alignment models together with the sources that may provide functional information are triaged by curation staff and selected for inclusion in CDD if generic functional information can be provided, as a minimum.
SARS-CoV-2
In early 2020, CDD version v3.18 was released and re-released a short time later, as new curated models were added to provide complete annotation of the SARS-CoV-2 proteins. A list of those proteins mapped to domain models and experimentally determined 3D structures is available via https://www.ncbi.nlm.nih.gov/Structure/SARS-CoV-2.html. As a result of this work, a detailed classification of the conserved catalytic core domain of the RNA-dependent RNA polymerase (RdRp) from the positive-sense single-stranded RNA [(+)ssRNA] viruses and closely related viruses has been provided under the root accession cd23167.
Model-specific word score thresholds
Beginning with the release of CDD v3.19, we have been optimizing RPS-BLAST (8) search databases for performance. The BLAST heuristics implemented in RPS-BLAST uses initial matches of query three-letter words via a large lookup table that records matches on individual database PSSMs (position-specific score matrices) crossing a word-score threshold. Initially, a uniform word-score threshold was used across the entire search database. The selection of the word-score threshold implies a trade-off between search speed and sensitivity. The threshold affects the size of the lookup table, which is directly linked to search speed. A large fraction of the time spent by the BLAST heuristic goes into exploring initial word hits and resulting local alignments that will eventually be abandoned and not reported. By raising the word-score threshold, many of these futile cycles can be avoided. The majority of the models included in the CDD collection do not require a very low word-score threshold in order to be identified as matches by members of the respective protein (domain) family. We have implemented a simple procedure to determine suitable model-specific thresholds, which uses a list of bona-fide family members, either as provided by the model data source (such as Pfam or COGs), or as detected by RPS-BLAST with significant score when using a low default word-score threshold. We then titrate the word-score threshold up to a point where RPS-BLAST will still detect matches for >99% of those family members. Overall, this change had very little impact on concise domain annotation (which favors the best-scoring hits) and derived domain architectures. We were able to reduce RPS-BLAST runtime for typical searches by a factor of 3, approximately. This has helped to keep up with the rapid growth of the sequence databases while still providing pre-computed results for a large set of representative sequences.
SPARCLE superfamily architectures
We define protein domain architectures as the sequential (N- to C-terminal) list of one or more domain footprints annotated on a protein sequence. SPARCLE, which stands for ‘Subfamily Protein Architecture Labeling Engine’, groups proteins by domain architecture, and now considers both specific and superfamily domain architectures. Superfamily architectures collect proteins that have significant hits to one or more domains that do not exceed a model-specific bitscore thresholds that labels them as high-confidence or ‘specific’. Typically, they will be assigned more generic names, while specific domain architectures may group more closely releated proteins and be assigned more precise and informative names and functional descriptions. To date, CDD curators have reviewed and assigned names and functional labels to about 40,000 well-represented architectures, with a focus on those common in bacterial genomes and those providing coverage for the genomes of eukaryotes such as fungi, protists, and nematodes, in work aimed at supporting the NIH Comparative Genome Resource (CGR, https://www.ncbi.nlm.nih.gov/comparative-genomics-resource/). A publicly accessible Entrez database supports text queries into protein family models that include not only architectures, but also HMMs, and NCBI BLAST rules, and points to summary information as well as links to other databases and sources of attribution.
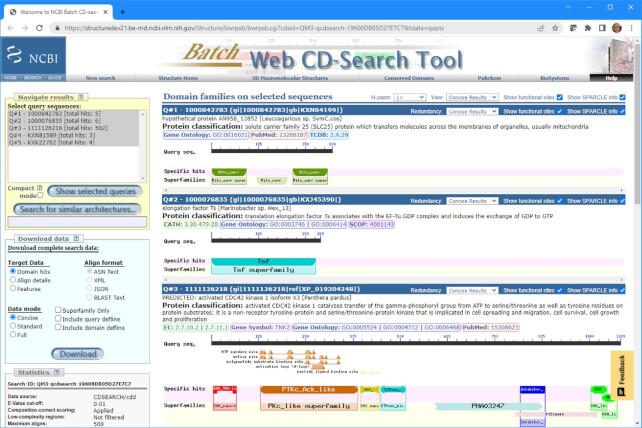
For the past year, we added attribution to conserved domain architecture, which can be mapped to individual architecture member sequences. Attribution sources are (i) citations, recorded as PubMed IDs, (ii) E.C. numbers (9), (iii) GO terms (10), (iv) gene symbols, (v) TCDB identifiers (11), (vi) CAZy identifiers (12), (vii) MEROPS identifiers (13), (viii) SCOP (14) and/or CATH (15) identifiers. These are assigned and validated by curation staff and are now displayed on CD-Search and BATCH CD-Search pages if the user query matches the corresponding curated domain architecture (see Figure 1).
Figure 1.
BATCH CD-Search results formatted for a few query sequences demonstrate the availability of domain architecture information (under the heading ‘Protein classification’), as well as transferable attributes assigned to each architecture, on top of domain footprint annotation and functional sites associated with some of the domain models. The protein classification information and site annotations can be toggled off for a sparse display focusing on domain footprints only.
SPARCLE architecture curation supports the automated, evidence-based assignment of names to proteins in RefSeq and the Prokaryotic Genome Annotation Pipeline (PGAP) (6). At this time, about 60 million bacterial RefSeq proteins are named via SPARCLE (out of 195 million total bacterial proteins and 160 million proteins with naming evidence provided).
DATA AVAILABILITY
Table 3 lists URLs for major services, tools, and data collections provided by CDD. RPS-BLAST is a part of NCBI’s BLAST software distribution, while CDD provides pre-formatted RPS-BLAST search databases, so that conserved domain searches can be run locally. The rpsbproc stand-alone utility uses additional data to format RPS-BLAST results so that they agree with reports generated by CD-Search (16) and BATCH CD-Search, including domain superfamily assignments and site annotations. The sparclbl (SparcleLabel) utility processes results from local RPS-BLAST searches and provides suggestions for protein names based on domain architecture and is used in NCBI’s prokaryotic genome annotation pipeline (PGAP) (6).
Table 3.
URLs and other resources associated with the CDD project
| URL | Description |
|---|---|
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation |
| https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | BATCH CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation. Up to 1000 protein queries may be submitted per request, and the size of queries is restricted to no more than 40 000 residues. |
| https://www.ncbi.nlm.nih.gov/cdd | Entrez search interface to CDD |
| https://ftp.ncbi.nih.gov/pub/mmdb/cdd | CDD FTP site, see README file for content |
|
https://ftp.ncbi.nlm.nih.gov/toolbox
executables can be obtained from: https://www.ncbi.nlm.nih.gov/BLAST/download.shtml |
RPS-BLAST stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution |
| https://www.ncbi.nlm.nih.gov/protfam | Entrez interface to the Protein Families collection, which includes SPARCLE domain architectures |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/rpsbproc | Standalone utility for enriching and formatting RPS-BLAST results |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/SparcleLabel/ | Standalone utility for naming/labeling proteins using SPARCLE |
CONCLUSION
The major focus of ongoing work at CDD is to ensure sustainability of the data flow and public services. We strive to keep up with the growth of the sequence collections and to provide a source of annotation that maintains relatively high coverage while improving accuracy. We continue to add novel domain family models to the collection and to establish hierarchical classifications of selected protein domain families where they will have a significant impact on protein naming by domain architecture. We are investigating whether we’ll be able to provide meaningful filtering of search results by taxonomy, for cases where the taxonomic source of the user query sequence is known, and how this knowledge could be used to speed up searching.
ACKNOWLEDGEMENTS
We are grateful for project management assistance by Emily Davis and Megdelawit Mersha; we thank the NCBI Information Engineering Branch, specifically the BLAST and the NCBI RefSeq teams, for continuing support with software and database development, the authors of Pfam, SMART, COGs, TIGRFAMs, NCBIfam, and NCBI’s Protein Clusters database for providing access to resources and data, and the InterPro resource as well as CDD users for valuable feedback. Comments, suggestions, and questions are welcome and should be directed to: info@ncbi.nlm.nih.gov.
Contributor Information
Jiyao Wang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Farideh Chitsaz, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Myra K Derbyshire, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Noreen R Gonzales, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Marc Gwadz, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Shennan Lu, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Gabriele H Marchler, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
James S Song, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Narmada Thanki, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Roxanne A Yamashita, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Mingzhang Yang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Dachuan Zhang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Chanjuan Zheng, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Christopher J Lanczycki, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Aron Marchler-Bauer, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bldg. 38 A, Room 8N805, 8600 Rockville Pike, Bethesda, MD 20894, USA.
FUNDING
National Center for Biotechnology Information of the National Library of Medicine, National Institutes of Health. Funding for open access charge: National Center for Biotechnology Information, National Library of Medicine, NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Letunic I., Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018; 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V.. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001; 29:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haft D.H., Selengut J.D., Richter A.R., Harkins D., Basu M.K., Beck E.. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013; 41:D387–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klimke W., Agarwala R., Badretdin A., Chetvernin S., Ciufo S., Fedorov B., Kiryutin B., O’Neill K., Resch W., Resenchuk S.et al.. The national center for biotechnology information's protein clusters database. Nucleic Acids Res. 2009; 37:D216–D223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W., O’Neill K., Haft D.H, DiCuccio M., Chetvernin V., Badretdin A., Coulouris G., Chitsaz F., Derbyshire M.K., Durkin A.S.et al.. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021; 49:D1020–D1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S.et al.. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020; 48:D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchler-Bauer A., Panchenko A.R., Shoemaker B.A., Thiessen P.A., Geer L.Y., Bryant S.H.. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002; 30:281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Webb E.C. Enzyme nomenclature 1992: recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classifications of Enzymes. 1992; Academic Press. [Google Scholar]
- 10. Gene Ontology Consortium The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saier M.H., Reddy V.S., Moreno-Hagelsieb G., Hendargo K.J., Zhang Y., Iddamsetty V., Lam K.J.K., Tian N., Russum S., Wang J.et al.. The transporter classification database (TCDB): 2021 update. Nucleic Acids Res. 2021; 49:D461–D467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drula E., Garron M.-L., Dogan S., Lombard V., Henrissat B., Terrapon N.. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022; 50:D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D.. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018; 46:D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andreeva A., Kulesha E., Gough J., Murzin A.G.. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020; 48:D376–D382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sillitoe I., Bordin N., Dawson N., Waman V.P., Ashford P., Scholes H.M., Pang C.S.M., Woodridge L., Rauer C., Sen N.et al.. CATH: increased structural coverage of functional space. Nucleic Acids Res. 2021; 49:D266–D273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchler-Bauer A., Bryant S.H.. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2005; 32:W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table 3 lists URLs for major services, tools, and data collections provided by CDD. RPS-BLAST is a part of NCBI’s BLAST software distribution, while CDD provides pre-formatted RPS-BLAST search databases, so that conserved domain searches can be run locally. The rpsbproc stand-alone utility uses additional data to format RPS-BLAST results so that they agree with reports generated by CD-Search (16) and BATCH CD-Search, including domain superfamily assignments and site annotations. The sparclbl (SparcleLabel) utility processes results from local RPS-BLAST searches and provides suggestions for protein names based on domain architecture and is used in NCBI’s prokaryotic genome annotation pipeline (PGAP) (6).
Table 3.
URLs and other resources associated with the CDD project
| URL | Description |
|---|---|
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation |
| https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | BATCH CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation. Up to 1000 protein queries may be submitted per request, and the size of queries is restricted to no more than 40 000 residues. |
| https://www.ncbi.nlm.nih.gov/cdd | Entrez search interface to CDD |
| https://ftp.ncbi.nih.gov/pub/mmdb/cdd | CDD FTP site, see README file for content |
|
https://ftp.ncbi.nlm.nih.gov/toolbox
executables can be obtained from: https://www.ncbi.nlm.nih.gov/BLAST/download.shtml |
RPS-BLAST stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution |
| https://www.ncbi.nlm.nih.gov/protfam | Entrez interface to the Protein Families collection, which includes SPARCLE domain architectures |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/rpsbproc | Standalone utility for enriching and formatting RPS-BLAST results |
| https://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/SparcleLabel/ | Standalone utility for naming/labeling proteins using SPARCLE |