Abstract
PubChem (https://pubchem.ncbi.nlm.nih.gov) is a popular chemical information resource that serves a wide range of use cases. In the past two years, a number of changes were made to PubChem. Data from more than 120 data sources was added to PubChem. Some major highlights include: the integration of Google Patents data into PubChem, which greatly expanded the coverage of the PubChem Patent data collection; the creation of the Cell Line and Taxonomy data collections, which provide quick and easy access to chemical information for a given cell line and taxon, respectively; and the update of the bioassay data model. In addition, new functionalities were added to the PubChem programmatic access protocols, PUG-REST and PUG-View, including support for target-centric data download for a given protein, gene, pathway, cell line, and taxon and the addition of the ‘standardize’ option to PUG-REST, which returns the standardized form of an input chemical structure. A significant update was also made to PubChemRDF. The present paper provides an overview of these changes.
INTRODUCTION
PubChem (https://pubchem.ncbi.nlm.nih.gov) (1,2) is a public chemical database at the National Center for Biotechnology Information (NCBI) of the National Library of Medicine (NLM), an institute within the U.S. National Institutes of Health (NIH). With millions of users every month (Figure 1), PubChem is a popular resource that serves a wide range of audiences, including researchers, chemical health and safety officers, patent agents, educators, and students. Importantly, PubChem data is commonly used in many artificial intelligence and machine learning studies (3–17).
Figure 1.
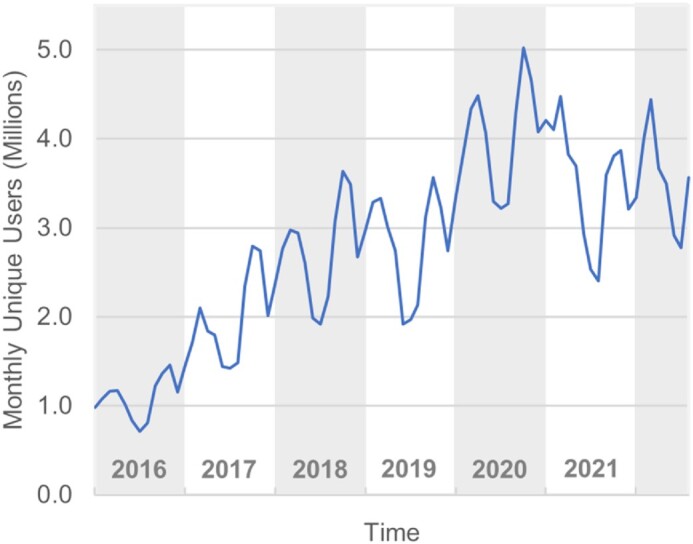
Monthly unique users who visited PubChem. The statistics in this Figure include interactive users only and exclude programmatic users.
PubChem is a data aggregator, which collects chemical information from hundreds of data sources (872 sources as of 6 September 2022). The majority of chemicals contained in PubChem are small molecules, but PubChem also contains other chemical entities, such as siRNA, miRNA, lipids, carbohydrates, and chemically-modified biopolymers. This data is organized into multiple data collections (18,19), including Substance, BioAssay, Compound, Protein, Gene, Pathway, Cell Line, Taxonomy, and Patent. While Substance archives depositor-provided chemical descriptions, Compound contains unique chemical structures extracted from Substance (19). BioAssay archives the depositor-provided description and test results of biological assay experiments (20). Protein, Gene, Pathway, Cell Line and Taxonomy provide a target-centric view of chemical information for a given protein, gene, pathway, cell line, and taxon, respectively (18). The Patent collection provides information on chemicals, proteins, genes, and taxons mentioned in each patent document. Table 1 shows the number of records in PubChem's data collections (as of 6 September 2022). The up-to-date record counts can be found at the PubChem Statistics page (https://pubchemdocs.ncbi.nlm.nih.gov/statistics).
Table 1.
PubChem data counts (as of 6 September 2022). The up-to-date record counts are available at the PubChem Statistics page (https://pubchemdocs.ncbi.nlm.nih.gov/statistics)
| Type | Count | Description |
|---|---|---|
| Compound | 111 892 547 | Unique chemical structures extracted from contributed PubChem Substance records |
| Substance | 296 900 675 | Information about chemical entities provided by PubChem contributors |
| BioAssay | 1 506 765 | Biological experiments provided by PubChem contributors |
| Bioactivity | 296 804 899 | Biological activity data points reported in PubChem BioAssays |
| Protein | 185 153 | Proteins tested in PubChem BioAssays and those involved in PubChem Pathways and identified in PubChem Patents |
| Gene | 103 988 | Genes tested in PubChem BioAssays and those involved in PubChem Pathways and identified in PubChem Patents |
| Pathway | 239 183 | Interactions between chemicals, genes, and proteins |
| Cell Line | 1964 | Cell lines tested in PubChem BioAssays |
| Taxonomy | 112 531 | Organisms of proteins/genes tested in PubChem BioAssays and those involved in PubChem Pathways and identified in PubChem Patents |
| Patent | 42 395 312 | Patents with links in PubChem |
| Data Sources | 872 | Organizations contributing data to PubChem |
Various aspects of PubChem have been described in our previous papers, including those published in the Nucleic Acid Research Database issues (1,19,21) and Webserver issues (22,23). The present paper describes general updates to PubChem for the past two years, including new data content, the creation of Cell Line and Taxonomy data collections, bioassay data model change, improved programmatic access, and the expansion of PubChemRDF data.
NEW DATA CONTENT
A list of PubChem data sources can be accessed through the PubChem Sources page (https://pubchem.ncbi.nlm.nih.gov/sources). For the past two years, data from about 120 new sources have been added to PubChem, which now provides information from more than 870 data sources. While some of the new data are submitted by individual sources and archived in the Substance and BioAssay collections, other data are integrated by the PubChem team to annotate the existing PubChem records. Below are some noteworthy sources.
Drug information
Many of the annotations newly added to PubChem are about drugs. Notably, PubChem integrated information about human and animal drug products from the U.S. Food & Drug Administration (FDA) National Drug Code (NDC) Directory (https://www.accessdata.fda.gov/scripts/cder/ndc/index.cfm) and the FDA Green Book (https://www.fda.gov/animal-veterinary/products/approved-animal-drug-products-green-book), respectively. The FDA’s Drug-Induced Liver Injury Rank (DILIrank) data set (24) was also integrated into PubChem. In addition, PubChem now provides information on drugs used for the treatment of HIV/AIDS and opportunistic infection, from the drug database at the clinicalinfo.hiv.gov website (https://clinicalinfo.hiv.gov/en/drugs).
Chemical health and safety information
Annotations related to chemical health and safety were also added to PubChem. Examples are occupational health information (e.g. adverse effects of workplace exposures to chemical agents) from Haz-Map (https://haz-map.com/) and the carcinogen classification and related monograph links from the International Agency for Research on Cancer (IARC) (https://www.iarc.who.int/). Other noteworthy annotations are the acute exposure guideline levels (AEGLs) from the U.S. Environmental Protection Agency (EPA) (https://www.epa.gov/aegl). The AEGLs describe the human health effects of once-in-a-lifetime, or rare, exposure to airborne chemicals and are used by emergency responders when dealing with chemical spills or other catastrophic exposures.
CAS registry numbers
The Chemical Abstracts Service (CAS) of the American Chemical Society (ACS) provided PubChem with the CAS registration numbers (RNs) for more than 400,000 chemicals available at the CAS Common Chemistry website (https://commonchemistry.cas.org/) (25). This helps PubChem to identify and validate authoritative CAS RNs scattered across many chemical information resources on the internet, enabling more accurate data exchange and integration between the resources.
Patent information
PubChem imported patent information from Google Patents (https://patents.google.com/) (26). This data integration involves two public data sets available in Google BigQuery (https://cloud.google.com/bigquery):
-
Google Patents Research Data
-
Google Patents Public Data (provided by IFI CLAIMS Patent Services)
The addition of Google Patents data substantially expanded PubChem's patent data collection, which now contains 767 million links between 37 million chemical structures and the 25 million patent documents that mention them. The imported data also includes links between patents and other PubChem entities like genes, proteins, and taxons. These data cover patent applications and grants from international patent offices like the World Intellectual Property Organization (WIPO) and European Patent Office (EPO), as well as those from many national patent offices, including the US Patent and Trademark Office (USPTO), Japan Patent Office (JPO), and Korean Intellectual Property Office (KIPO).
Data from Google Patents are used to generate Patent Summary pages, which present information for individual patent documents. As an example, the following URL directs you to the Patent Summary for the US Patent document, US-2005209186-A1:
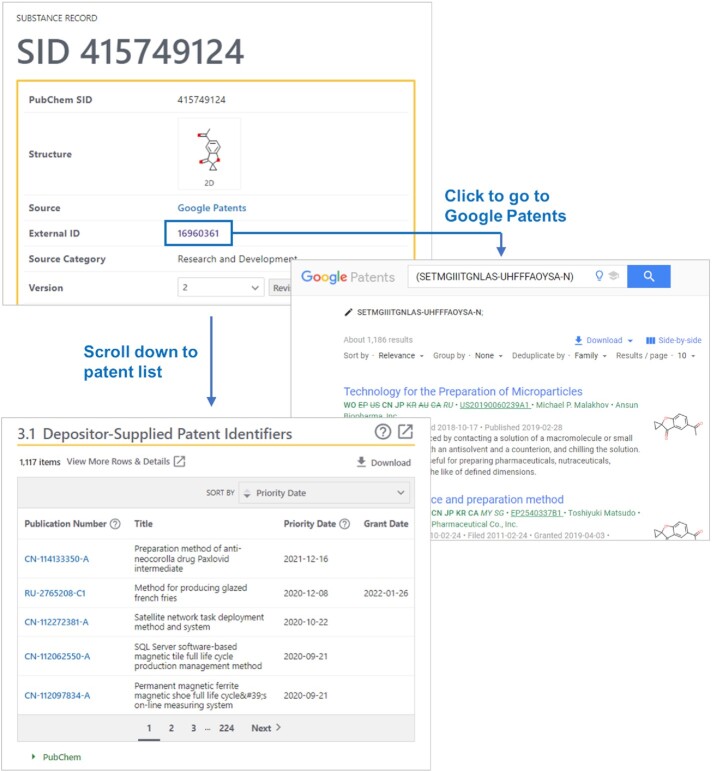
The Google Patents data includes a list of patent documents mentioning a particular chemical. This list is accessible through a Substance Record page for that chemical. For example, the following URL corresponds to the Record page for SID 415749124 (spizofurone) from Google Patents (Figure 2):
Figure 2.
The Substance Record page for SID 415749124 (spizofurone) (https://pubchem.ncbi.nlm.nih.gov/substance/415749124). The patent identifiers associated with this chemical are listed in the ‘Depositor-Supplied Patent Identifiers’ section. The data presented in this section can be downloaded using the ‘Download’ button above the top-right corner of the patent table. Clicking the External ID link (16960361) directs to the Google Patents site, where the user can find additional information about these patents.
As depicted in Figure 2, this page has the ‘Depositor-Supplied Patent Identifiers’ section, showing a list of the patent documents for spizofurone. In addition, clicking the External ID link on this page directs to the Google Patents site, where the user can find additional information.
The chemical-patent links from Google Patents are combined with those from other sources and presented in the Depositor-Supplied Patent Identifiers section of the Compound Summary page, as shown on the following page:
The integration of Google Patents information with PubChem represents a significant update to the existing patent data content in PubChem. It is worth mentioning that chemicals listed on the Summary page of a patent are not necessarily subject matters claimed in the patent. They could be mentioned as prior arts or as reactants, catalysts, and other reagents used to generate patent-protected chemicals. As explained in other studies (26–28), it is challenging to distinguish claimed subject-matter chemicals from others in patent documents. This is one of the areas that need improvement in PubChem.
CELL LINE AND TAXONOMY DATA COLLECTIONS
As explained in the Introduction, PubChem has multiple data collections. Among them, Cell Line and Taxonomy (18) are the newest ones. Each record in these collections contains chemical information related to a given cell line or taxon (organism) and additional annotations about that cell line or taxon. These annotations are collected from authoritative and curated data sources.
The Cell Line collection contains 1,964 cell lines that are tested against in any bioassays archived in PubChem. The summary page for each cell line is accessible via the URL containing the cell line abbreviation, Cellosaurus ID (29), or ChEMBL Cell Line ID (30). For example, these are the Summary page for the Michigan Cancer Foundation-7 (MCF-7) cell line (Cellosaurus ID: CVCL_0031; ChEMBL ID: CHEMBL3308403), which is a human breast cancer cell line commonly tested in PubChem bioassays:
The Cell Line Summary page displays the bioactivity data of the chemicals tested against the cell line, along with synonyms, diseases, references, classifications, and so on. These annotations are collected from authoritative and curated data sources, including the Medical Subject Headings (MeSH) (https://www.nlm.nih.gov/mesh/), Cellosaurus (29), ChEMBL (30), Cell Line Ontology (CLO) (31), NCI Thesaurus (NCIt) (32), the Library of Integrated Network-Based Cellular Signatures (LINCS) Data Portal (33), and Harvard Medical School (HMS) LINCS Database (http://lincs.hms.harvard.edu/db/).
The Taxonomy collection covers over 110 000 taxa associated with bioassay or pathway records in PubChem. The Taxonomy Summary page for a taxon is accessible through an URL containing its scientific name, common name, or NCBI Taxonomy ID, as shown in the following examples (for Danio rerio (zebrafish), whose NCBI Taxonomy ID is 7655):
This Summary page shows the chemicals tested against zebrafish in PubChem bioassays and the proteins targeted in those assays. Importantly, this page presents a list of the whole-organism bioassays performed on this taxon, which is accessible through the following URL:
This list provides users with a quick and convenient way to identify the whole-organism bioassays that do not have a specific gene or protein target. The Taxonomy Summary page also presents the chemicals and proteins involved in the biological pathways for zebrafish. Additionally, this page contains information on the taxon, collected from various primary data sources such as NCBI Taxonomy (34), Integrated Taxonomic Information System (ITIS) (https://www.itis.gov/), Catalogue of Life (COL) (https://www.catalogueoflife.org/), NCIt (32), GlyCosmos Glycoscience Portal (35), UniProt (36), and LOTUS (https://lotus.naturalproducts.net/) (37).
BIOASSAY DATA MODEL CHANGE
As of 6 September 2022, PubChem contains 297 million bioactivity data points from 1.5 million bioassays. The data model used to store these bioactivity data was updated, changing the format of the bioassay data uploaded to or downloaded from PubChem. Therefore, new assay data submitted to PubChem must be formatted based on the new data model and the existing software programs that download and read PubChem's bioassay data should also be updated accordingly.
The specification of the new bioassay data model is available at the PubChem FTP site (https://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/pcassay2.asn). One of the major changes in the data model involves panel assays, which contain bioactivity data for multiple targets (sometimes up to thousands). In the old data model, the data for each target of a panel assay spanned over multiple columns in the data table. This resulted in data tables with varying column widths (up to tens of thousands of columns), making it difficult to manage panel assay data. In the new data model, the format for panel assays is no longer column-based, and each data point is stored in a row. All existing panel assays were converted into this new, row-based format.
Another important change was the inclusion of endpoint qualifiers (e.g., >, >=, <, <= and =) in the data specification. Without these qualifiers, bioactivity data could be misinterpreted. For instance, although a compound with EC50 = 1 μM has different bioactivity from those with EC50 < 1 μM or EC50 > 1 μM, they all appear identical without endpoint qualifiers. While about 6 million bioactivity data points (corresponding to 2% of all bioactivity data points) have this qualifier information, users often overlooked it when examining the assay data. The new bioassay data model explicitly includes endpoint qualifiers (e.g. the ‘Standard Relation’ fields shown in the following example):
Textual data representation was also improved. PubChem assay data often contains UTF-8 characters that was not properly displayed as a text file. Examples are Greek letters commonly used in protein/gene names (e.g., β-lactamase) or units often found in experimental protocols (e.g., °C or °F). The updated bioassay data model now supports UTF-8 characters.
In the past, numeric identifiers called GI numbers were used to specify proteins or nucleotides associated with bioassays in PubChem (e.g., as assay targets or cross-references). However, as described elsewhere (38–40), the NCBI phased out the use of GI numbers in its databases (including PubChem) and replaced them with accession identifiers. Accordingly, the new data model uses accession identifiers only. As a result, only accessions can be used for new assay submissions and all GI numbers in the existing assays were converted to accessions.
PROGRAMMATIC ACCESS
PubChem provides multiple programmatic access routes, including E-Utilities (22), Power User Gateway (PUG) (22), PUG-SOAP (22), PUG-REST (22,23,41), PUG-View (42), and PubChemRDF REST interface (43). For the past two years, new functionalities were added mostly to PUG-REST and PUG-View. An example is the ‘standardize’ operation added to PUG-REST. This operation returns the standardized form of an input chemical structure, which can be specified with the Simplified Molecular-Input Line-Entry System (SMILES) string (44–46), IUPAC International Chemical Identifier (InChI) (47), or structure-data file (SDF). The valid output formats for this operation are SDF, XML, JSON(P) and ASNT/B, as shown in these examples (Figure 3):
Figure 3.
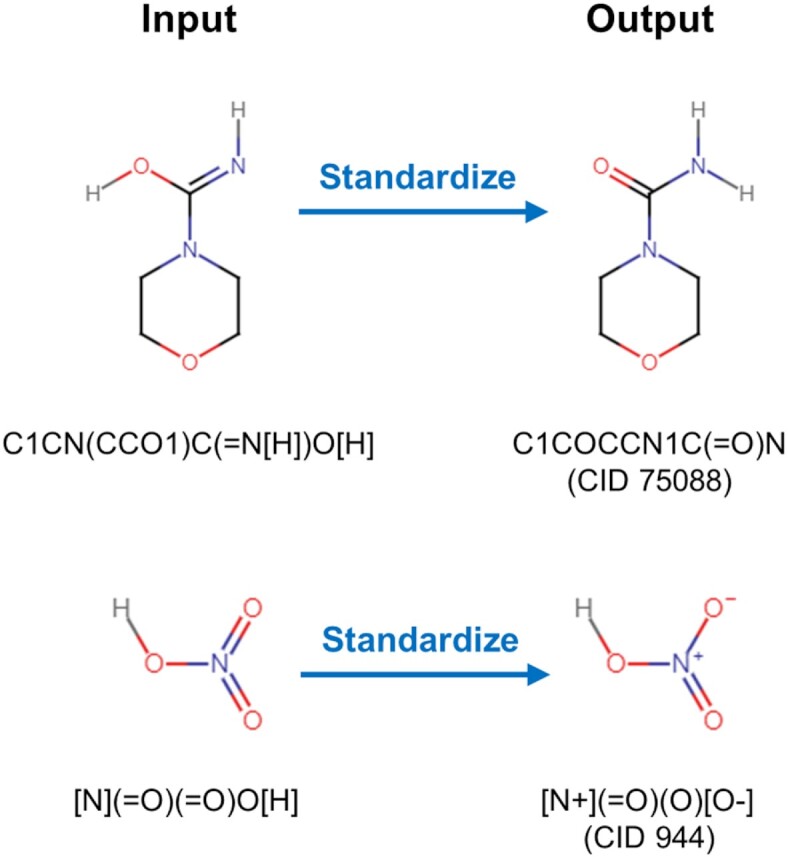
Effects of the ‘standardize’ operation of PUG-REST. This operation takes a chemical structure (given in a SMILES, InChI, or SDF) as an input, and returns its standardized form.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/standardize/smiles/C1CN(CCO1)C(=N[H])O[H]/SDF
https://pubchem.ncbi.nlm.nih.gov/rest/pug/standardize/smiles/[N](=O)(=O)O[H]/JSON
Note here that the input structures go through PubChem's chemical structure standardization process (48) and are modified to the structures of CIDs 75088 and 944, respectively (see Figure 3). The ‘standardize’ option allows users to check how their chemical structures would be processed and modified through PubChem's chemical structure standardization process. By default, the output from the standardize operation includes components and neutralized forms, unless ‘include_component=false’ is specified. As an example, consider the following PUG-REST requests:
Both requests take the acetate anion ‘CC(=O)[O-]’ as an input, but the first one returns the acetate anion as well as its neutralized form (acetic acid), while the second one returns only the acetate. Now consider the following requests takes sodium acetate as an input:
The first one returns sodium acetate as well as the neutral forms of its components (i.e., acetic acid and the sodium atom), while the second request returns only sodium acetate.
It is noteworthy that, because some isomeric SMILES and InChI strings contain special characters incompatible with the URL syntax (e.g., the forward slash (‘/’)), those identifiers should be provided as a URL parameter, as shown in this example:
Another important functionality added to PUG-REST is programmatic access to target-centric data (i.e., data for a given protein, gene, cell line, or taxon). For example, the following PUG-REST request returns a summary of the A-549, MCF-7, and HCT-116 cell lines (cell line accessions: CVCL_0023, CVCL_0031, and CVCL_0291, respectively):
The resulting output contains the cell accession, name, sex, category, source tissue, source organism, and list of synonyms for the input cell lines.
It is also possible to get a list of bioassays tested for a given cell line, as shown in this example:
Note that the input cell line in the above request is specified with its name (MCF-7). The bioassay list from this request can be used to programmatically download the bioactivity data for the MCF-7 cell line through PUG-REST:
In addition, PUG-View (42) can be used to download the annotations displayed on the Summary pages of a cell line or taxon. The following examples are the PUG-View requests that download annotations for the MCF-7 cell line and Sus scrofa (pig, NCBI Taxonomy ID: 9823), respectively:
https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/data/cell/MCF-7/JSON/
https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/data/taxonomy/9823/JSON/
PubChemRDF
PubChemRDF (43) refers to machine-readable PubChem data formatted using the resource description framework (RDF) (https://www.w3.org/RDF/). In RDF, knowledge is expressed as a ‘triple’, which consists of a subject, predicate, and object, where the predicate defines the relationship between the subject and object. A significant update was made to PubChemRDF and the current release of PubChemRDF (version 1.7.2-beta) now contains 120 billion triples, grouped into 16 subdomains based on the type of subject (compound, substance, bioassay, descriptor, protein, gene, reference, endpoint, measure group, etc.) (https://pubchemdocs.ncbi.nlm.nih.gov/rdf-statistics).
One of the major changes to PubChemRDF is the addition of a new subdomain, called Pathway, which encodes information on biological pathways and their relationship with genes, proteins, and chemicals. This Pathway subdomain supersedes the BioSystem subdomain used in the previous versions of PubChemRDF.
Another important change is the update of predicates that define the semantic relationships between entities (i.e., subjects and objects). Whenever possible, PubChemRDF describes relationships between entities by using pre-existing, domain-specific ontological frameworks (rather than creating new ones), including Chemical Entities of Biological Interest (ChEBI) (49), CHEMical INFormation ontology (CHEMINF) (50), Protein Ontology (PRO) (51), Gene Ontology (GO) (52,53), BioAssay Ontology (BAO) (54), Semanticscience Integrated Ontology (SIO) (55) and many others. Since the initial release of PubChemRDF, some terms in these ontologies were deprecated or replaced with new ones. These changes are reflected in the new release of PubChemRDF. In addition, some predicates from PubChem's internal vocabulary were replaced in favor of external ones (e.g. ‘vocab:geneSymbol’ replaced with ‘bao:BAO_0002870’).
The initial version of PubChemRDF used GI numbers to denote proteins, but the NCBI phased out the use of GI numbers, as mentioned previously. Accordingly, changes were made to allow users to access PubChemRDF data using the protein accession identifiers.
SUMMARY
In the past 2 years, significant changes were made to PubChem. With the integration of data from over 120 new data sources, PubChem now provides chemical information from >870 data sources (as of September 6, 2022). The newly added data include drug information (from the clinicalinfo.hiv.gov drug database as well as FDA’s NDC Directory, Green Book, and DILIrank data set) and chemical health and safety information from Haz-Map, IARC and EPA. Patent information from Google Patents greatly expanded the coverage of the Patent data collection. The CAS Common Chemistry provided PubChem with the CAS RNs for 400 000 chemicals, making it possible to identify and validate correct CAS RNs from other data sources.
New data collections, called Cell Line and Taxonomy, were created to help users quickly access PubChem data specific to a given cell line and taxon, respectively. The Summary page of each record in these data collections contains relevant bioactivity data archived in PubChem BioAssay as well as annotations about the cell line or taxon, collected from authoritative data sources. Programmatic access to the two new data collections is available.
PubChem updated its data model used to store bioactivity data in BioAssay. In the updated bioassay data model, each data point for panel assays is stored in a row, making it easier to manage the panel assay data. The new data model explicitly includes the endpoint qualifiers (e.g., >, <, >=, <= and =) and supports UTF-8 characters. The GI numbers for target proteins are replaced with NCBI accessions. Assay data depositors are required to format their data based on the new model, and software developers should update their code accordingly to correctly load bioassay data downloaded from PubChem.
New functionalities have been added to PUG-REST and PUG-View. Now PUG-REST has the ‘standardize’ operation, which returns the standardized structures of an input structure, helping users to programmatically check how their structures are processed and modified when submitted to PubChem. In addition, PUG-REST and PUG-View support programmatic access to target-centric data (i.e., for a given protein, gene, pathway, cell line, or taxon).
Major changes were also made to PubChemRDF. The deprecated or outdated terms from external ontological frameworks were updated in the new release. The Pathway subdomain was added to encode semantic relationships of biological pathways with chemicals, genes, and proteins. NCBI accessions are now used to denote proteins instead of GI numbers.
The overall goal of PubChem is to provide rapid access to chemical information for its broad audience within the biomedical research community and beyond. To achieve this goal, PubChem continues to improve the breadth and depth of data, by identifying and integrating high-quality data from authoritative and curated sources. In addition, PubChem will keep up with ever-evolving technologies to improve existing tools and services and develop new ones that enable rapid access to information. In this spirit, upcoming improvements are, among others, putting a special emphasis on further improving data quality, modernizing interfaces, and enabling enhanced handling of chemicals without discrete structures (e.g., polymers and mixtures). This also includes efforts to broaden the coverage of the knowledge panels (21,56), which summarize the relationship between chemicals, genes, proteins, and diseases by analyzing their co-occurrences in scientific articles and patent documents.
DATA AVAILABILITY
All PubChem data, tools, and services are provided to the public free of charge. They are accessible from the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov).
ACKNOWLEDGEMENTS
We appreciate the hundreds of data contributors for making their data openly accessible within PubChem. Special thanks go to the entire NCBI staff (especially to the help desk and systems support teams).
Contributor Information
Sunghwan Kim, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Jie Chen, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Tiejun Cheng, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Asta Gindulyte, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Jia He, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Siqian He, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Qingliang Li, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Benjamin A Shoemaker, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Paul A Thiessen, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Bo Yu, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Leonid Zaslavsky, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Jian Zhang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Evan E Bolton, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
FUNDING
National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health; Funding for open access charge: The National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim S., Chen J., Cheng T.J., Gindulyte A., He J., He S.Q., Li Q.L., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim S. Getting the most out of PubChem for virtual screening. Expert Opin Drug Discov. 2016; 11:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Himmetoglu B. Tree based machine learning framework for predicting ground state energies of molecules. J. Chem. Phys. 2016; 145:134101. [DOI] [PubMed] [Google Scholar]
- 4. Stork C., Wagner J., Friedrich N.O., Kops C.D., Sicho M., Kirchmair J.. Hit Dexter: a machine-learning model for the prediction of frequent hitters. ChemMedChem. 2018; 13:564–571. [DOI] [PubMed] [Google Scholar]
- 5. Ludwig M., Duhrkop K., Bocker S.. Bayesian networks for mass spectrometric metabolite identification via molecular fingerprints. Bioinformatics. 2018; 34:i333–i340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dias T., Gaudencio S.P., Pereira F.. A computer-driven approach to discover natural product leads for methicillin-resistant staphylococcus aureus infection therapy. Mar Drugs. 2019; 17:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ogura K., Sato T., Tuki H., Honma T.. Support vector machine model for hERG inhibitory activities based on the integrated hERG database using descriptor selection by NSGA-II. Sci. Rep. 2019; 9:12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh N., Chaput L., Villoutreix B.O.. Fast rescoring protocols to improve the performance of structure-based virtual screening performed on protein-protein interfaces. J. Chem. Inf. Model. 2020; 60:3910–3934. [DOI] [PubMed] [Google Scholar]
- 9. Tran-Nguyen V.K., Jacquemard C., Rognan D.. LIT-PCBA: an unbiased data set for machine learning and virtual screening. J. Chem. Inf. Model. 2020; 60:4263–4273. [DOI] [PubMed] [Google Scholar]
- 10. Korkmaz S. Deep learning-based imbalanced data classification for drug discovery. J. Chem. Inf. Model. 2020; 60:4180–4190. [DOI] [PubMed] [Google Scholar]
- 11. Wen M.J., Blau S.M., Spotte-Smith E.W.C., Dwaraknath S., Persson K.A.. BonDNet: a graph neural network for the prediction of bond dissociation energies for charged molecules. Chem. Sci. 2021; 12:1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jia X.L., Ciallella H.L., Russo D.P., Zhao L.L., James M.H., Zhu H.. Construction of a virtual opioid bioprofile: a data-driven QSAR modeling study to identify new analgesic opioids. ACS Sustain. Chem. Eng. 2021; 9:3909–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuo Z.R., Wang P.L., Chen X.W., Li T., Ge H., Qian D.H.. SWnet: a deep learning model for drug response prediction from cancer genomic signatures and compound chemical structures. BMC Bioinf. 2021; 22:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Handsel J., Matthews B., Knight N.J., Coles S.J.. Translating the InChI: adapting neural machine translation to predict IUPAC names from a chemical identifier. J Cheminform. 2021; 13:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dey V., Machiraju R., Ning X.. Improving compound activity classification via deep transfer and representation learning. ACS Omega. 2022; 7:9465–9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Isigkeit L., Chaikuad A., Merk D.. A consensus compound/bioactivity dataset for data-driven drug design and chemogenomics. Molecules. 2022; 27:2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maki J., Oshimura A., Tsukano C., Yanagita R.C., Saito Y., Sakakibara Y., Irie K.. AI and computational chemistry-accelerated development of an alotaketal analogue with conventional PKC selectivity. Chem. 2022; 58:6693–6696. [DOI] [PubMed] [Google Scholar]
- 18. Kim S., Cheng T.J., He S.Q., Thiessen P.A., Li Q.L., Gindulyte A., Bolton E.E.. PubChem Protein, Gene, Pathway, and Taxonomy data collections: bridging biology and chemistry through Target-Centric Views of PubChem data. J. Mol. Biol. 2022; 434:167514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L.Y., He J.E., He S.Q., Shoemaker B.A.et al.. PubChem Substance and Compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y.L., Bryant S.H., Cheng T.J., Wang J.Y., Gindulyte A., Shoemaker B.A., Thiessen P.A., He S.Q., Zhang J.. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017; 45:D955–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S., Chen J., Cheng T.J., Gindulyte A., He J., He S.Q., Li Q.L., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S., Thiessen P.A., Bolton E.E., Bryant S.H.. PUG-SOAP and PUG-REST: web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015; 43:W605–W611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim S., Thiessen P.A., Cheng T.J., Yu B., Bolton E.E.. An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Res. 2018; 46:W563–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen M.J., Suzuki A., Thakkar S., Yu K., Hu C.C., Tong W.D.. DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov Today. 2016; 21:648–653. [DOI] [PubMed] [Google Scholar]
- 25. Jacobs A., Williams D., Hickey K., Patrick N., Williams A.J., Chalk S., McEwen L., Willighagen E., Walker M., Bolton E.et al.. CAS Common Chemistry in 2021: expanding access to trusted chemical information for the scientific community. J. Chem. Inf. Model. 2022; 62:2737–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnabas S.J., Böhme T., Boyer S.K., Irmer M., Ruttkies C., Wetherbee I., Kondić T., Schymanski E.L., Weber L.. Extraction of chemical structures from literature and patent documents using open access chemistry toolkits: a case study with PFAS. Digital Discov. 2022; 1:490–501. [Google Scholar]
- 27. Akhondi S.A., Rey H., Schworer M., Maier M., Toomey J., Nau H., Ilchmann G., Sheehan M., Irmer M., Bobach C.et al.. Automatic identification of relevant chemical compounds from patents. Database. 2019; 2019:baz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Falaguera M.J., Mestres J.. Identification of the core chemical structure in SureChEMBL patents. J. Chem. Inf. Model. 2021; 61:2241–2247. [DOI] [PubMed] [Google Scholar]
- 29. Bairoch A. The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrian-Uhalte E.et al.. The ChEMBL database in 2017. Nucleic Acids Res. 2017; 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarntivijai S., Lin Y., Xiang Z.S., Meehan T.F., Diehl A.D., Vempati U.D., Schurer S.C., Pang C., Malone J., Parkinson H.et al.. CLO: the cell line ontology. J. Biomed. Semant. 2014; 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sioutos N., de Coronado S., Haber M.W., Hartel F.W., Shaiu W.L., Wright L.W.. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J. Biomed. Inform. 2007; 40:30–43. [DOI] [PubMed] [Google Scholar]
- 33. Stathias V., Turner J., Koleti A., Vidovic D., Cooper D., Fazel-Najafabadi M., Pilarczyk M., Terryn R., Chung C., Umeano A.et al.. LINCS data portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020; 48:D431–D439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamada I., Shiota M., Shinmachi D., Ono T., Tsuchiya S., Hosoda M., Fujita A., Aoki N.P., Watanabe Y., Fujita N.et al.. The GlyCosmos Portal: a unified and comprehensive web resource for the glycosciences. Nat. Methods. 2020; 17:649–650. [DOI] [PubMed] [Google Scholar]
- 36. Bateman A., Martin M.J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bursteinas B.et al.. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rutz A., Sorokina M., Galgonek J., Mietchen D., Willighagen E., Gaudry A., Graham J.G., Stephan R., Page R., Vondrášek J.et al.. The LOTUS initiative for open knowledge management in natural products research. Elife. 2022; 11:e70780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Agarwala R., Barrett T., Beck J., Benson D.A., Bollin C., Bolton E., Bourexis D., Brister J.R., Bryant S.H., Canese K.et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018; 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2017; 45:D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2016; 44:D67–D72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim S., Thiessen P.A., Bolton E.E.. Programmatic retrieval of small molecule information from PubChem using PUG-REST. Methods in Pharmacology and Toxicology. 2018; Totowa, NJ: Humana Press; 1–24. [Google Scholar]
- 42. Kim S., Thiessen P.A., Cheng T., Zhang J., Gindulyte A., Bolton E.E.. PUG-View: programmatic access to chemical annotations integrated in PubChem. J Cheminform. 2019; 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fu G., Batchelor C., Dumontier M., Hastings J., Willighagen E., Bolton E.. PubChemRDF: towards the semantic annotation of PubChem Compound and Substance Databases. J Cheminform. 2015; 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weininger D. SMILES. 3. DEPICT - graphical depiction of chemical structures. J. Chem. Inf. Comput. Sci. 1990; 30:237–243. [Google Scholar]
- 45. Weininger D., Weininger A., Weininger J.L. SMILES. 2. algorithm for generation of unique smiles notation. J. Chem. Inf. Comput. Sci. 1989; 29:97–101. [Google Scholar]
- 46. Weininger D. SMILES, a chemical language and information-system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988; 28:31–36. [Google Scholar]
- 47. Heller S.R., McNaught A., Pletnev I., Stein S., Tchekhovskoi D.. InChI, the IUPAC International Chemical Identifier. J Cheminform. 2015; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hähnke V.D., Kim S., Bolton E.E.. PubChem chemical structure standardization. J. Cheminform. 2018; 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hastings J., Chepelev L., Willighagen E., Adams N., Steinbeck C., Dumontier M.. The Chemical Information Ontology: provenance and disambiguation for chemical data on the biological semantic web. PLoS One. 2011; 6:e25513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Natale D.A., Arighi C.N., Blake J.A., Bona J., Chen C.M., Chen S.C., Christie K.R., Cowart J., D’Eustachio P., Diehl A.D.et al.. Protein Ontology (PRO): enhancing and scaling up the representation of protein entities. Nucleic Acids Res. 2017; 45:D339–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carbon S., Douglass E., Good B.M., Unni D.R., Harris N.L., Mungall C.J., Basu S., Chisholm R.L., Dodson R.J., Hartline E.et al.. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Visser U., Abeyruwan S., Vempati U., Smith R.P., Lemmon V., Schurer S.C.. BioAssay ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinf. 2011; 12:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dumontier M., Baker C.J.O., Baran J., Callahan A., Chepelev L., Cruz-Toledo J., Del Rio N.R., Duck G., Furlong L.I., Keath N.et al.. The semanticscience integrated ontology (SIO) for biomedical research and knowledge discovery. J. Biomed. Semant. 2014; 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zaslavsky L., Cheng T., Gindulyte A., He S., Kim S., Li Q., Thiessen P., Yu B., Bolton E.E.. Discovering and summarizing relationships between chemicals, genes, proteins, and diseases in PubChem. Front. Res. Metr. Anal. 2021; 6:689059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All PubChem data, tools, and services are provided to the public free of charge. They are accessible from the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov).