Abstract
The Tumor Immune Single Cell Hub 2 (TISCH2) is a resource of single-cell RNA-seq (scRNA-seq) data from human and mouse tumors, which enables comprehensive characterization of gene expression in the tumor microenvironment (TME) across multiple cancer types. As an increasing number of datasets are generated in the public domain, in this update, TISCH2 has included 190 tumor scRNA-seq datasets covering 6 million cells in 50 cancer types, with 110 newly collected datasets and almost tripling the number of cells compared with the previous release. Furthermore, TISCH2 includes several new functions that allow users to better utilize the large-scale scRNA-seq datasets. First, in the Dataset module, TISCH2 provides the cell–cell communication results in each dataset, facilitating the analyses of interacted cell types and the discovery of significant ligand–receptor pairs between cell types. TISCH2 also includes the transcription factor analyses for each dataset and visualization of the top enriched transcription factors of each cell type. Second, in the Gene module, TISCH2 adds functions for identifying correlated genes and providing survival information for the input genes. In summary, TISCH2 is a user-friendly, up-to-date and well-maintained data resource for gene expression analyses in the TME. TISCH2 is freely available at http://tisch.comp-genomics.org/.
INTRODUCTION
Tumor forms through a series of critical transitions, including from normal to pre-cancerous lesions and from pre-malignant to malignant states. The multistates were associated with dynamic cellular components and cell communication in the tumor microenvironment (TME) (1–3). Cancer immunotherapy has brought a paradigm shift to cancer treatment in recent years (4). Due to the striking genetic and cellular heterogeneity within the TME, only a fraction of patients could benefit from immunotherapy (5,6). Additionally, some patients showing good initial responses to treatment ultimately develop drug resistance during the immunotherapy (7). Therefore, investigating the change of cell type compositions under different conditions and cell–cell interaction in the TME could potentially overcome drug resistance and discover novel clinical implications.
Single-cell RNA-seq (scRNA-seq) has been a powerful technology to investigate the heterogeneity of the cell type compositions in the TME (8). It has been widely used to discover the novel cell types that are involved in tumor progression and identify cells that contributed to drug resistance (9,10). However, the rapidly accumulated tumor scRNA-seq data have also posed a critical barrier to widespread data re-use and exploration. Although there are several existing scRNA-seq databases that try to collect and display the tumor scRNA-seq datasets, including CancerSCEM (11), DISCO (12) and CancerSEA (13), most of them are limited in the number of datasets included and the coverage of covered cancer types, and have few downstream analysis functions. Previously, we developed TISCH (14), an scRNA-seq database focused on the scRNA-seq datasets from the TME. TISCH uniformly processed scRNA-seq data with a standardized workflow that removes batches between samples, uniformly annotates the cell types and identifies the malignant cells. The online visualization and analysis function of TISCH allows the biomedical research community to explore gene expression in the TME at single-cell resolution.
Here, we present an updated version of TISCH, which includes >6 million cells from 190 tumor scRNA-seq datasets across 50 cancer types, almost triple the number of cells and datasets of the previous version. To increase the utility of this resource, we have also provided several new functions that allow users to explore the driver transcriptional regulators of each cell type, find the communications between different cell types, query a gene's survival and identify its co-expressed genes across different datasets.
MATERIALS AND METHODS
Data collection and pre-processing in TISCH2
We applied a text mining-based data parsing workflow to collect datasets in the TISCH2 database. The workflow searches the scRNA-seq data based on single-cell and tumor-related keywords in the Gene Expression Omnibus (GEO) and ArrayExpress. After the web parser, we manually curated the datasets and then collected sample information from databases or the original studies, including patient ID, platform, source, tissue, cell number, species, cancer type, publication, etc. We applied the MAESTRO workflow to process all the collected datasets, including quality control, batch effect removal, cell clustering, differential expression analysis, cell type annotation, malignant cell classification and gene set enrichment analysis (GESA) (15–23). After quality control, the TISCH2 database contains 190 datasets from 50 cancer types and 20 tissue types in total (Supplementary Table S1).
Cell–cell interaction analysis
To evaluate the cell–cell interactions between different cell type clusters, we performed CellChat analyses based on the expression of known ligand–receptor (L–R) pairs in different clusters (24). For each dataset, we followed the official workflow and ran CellChat with standard parameters set to infer the cellular communication network. The number of significant L–R interaction pairs and the communication probability between two clusters were calculated and displayed by pheatmap R packages and the ‘netVisual_circle’ function in CellChat R packages. In addition, we also provide the significant L–R interaction pairs for each cluster as the source or target cells, with the P-value threshold set as 0.05. All these analyses were shown on the CCI (cell–cell interaction) page under the Dataset module.
TF enrichment analysis
Identifying the transcriptiion factors (TFs) is crucial to understanding the underlying gene regulatory network in different cell types. LISA builds an epigenetic model based on histone mark ChIP-seq and chromatin accessibility profiles to find factors that are most likely to regulate the input genes (25). Here, we performed LISA analyses for each cell type cluster, with the number of the background gene set as 3000, and the number of differentially expressed genes (DEGs) for each cluster set as 500; all of the DE genes were used in the LISA analyses if the total number of DEGs is less than 500.
For each dataset, the TF enrichment result of each cluster was ranked based on the combined P-value after the normalization and log10 transformations, and the top enriched TFs from different cell type clusters were visualized by heatmap (26). Due to the heterogeneity within malignant cells and the difference between the malignant cell and other cells, we process and visualize malignant cells with other cells separately. Additionally, for each cluster, we provided dot plot visualization to display the top 10 TFs with their expression levels, and the top 50 ranked TFs were shown in the table of each dataset.
Survival analysis
To facilitate users evaluating the clinical effect of the specific gene, we added the survival analysis in the Gene module. The expression and clinical data of TCGA were downloaded from the GDC TCGA data portal (27,28). For each gene, we calculated the hazard ratio (HR) (3) and generated significant P-values in 33 cancer types separately.
Gene-gene correlation
Co-expressed genes are potentially associated with the same functions and biological processes. Due to the large number of cells in TISCH2, the correlation computation will be time and memory consuming, and could be affected by the high drop-out rate in scRNA-seq data. Thus, we partitioned single cells into small groups (called mini-clusters hereafter) based on their similarity in the KNN graph; each of the mini-clusters contained 30 similar cells and the expression levels were averaged. This strategy is similar to MetaCell, and Zheng et al. characterized mini-cluster methods (29,30). The correlation was calculated with the R package Hmisc using the rcorr function at the mini-cluster-level expression matrix (https://CRAN.R-project.org/package=Hmisc). Considering the diversity of gene expression patterns in different cell types, besides the global correlation, we also calculated the gene–gene correlation within specific cell lineages for each dataset. To decrease the noise of genes that are always expressed at a low level but keep genes which are highly expressed in rare cell types, for each dataset, we only calculated the correlation between genes that have an averaged logTPM (transcripts per million) ≥0.5 or max logTPM ≥2. Gene pairs with correlation test P-value >0.05 or absolute correlation coefficient <0.2 were regard as not significantly correlated genes and were also removed from the correlation results. If users search for one gene that was filtered in the selected datasets in the gene correlation tab, TISCH2 will return ‘No significant correlated genes’. Finally, a correlation table as well as a heatmap for the top correlated genes will be displayed for the input gene in the Gene module. To reduce the size of the correlation heatmap, only the top 50 correlated genes that appear in more than half of the selected datasets will be displayed, and only the top 500 correlated genes for each cell type lineage will be displayed in the table.
RESULTS
Data summary
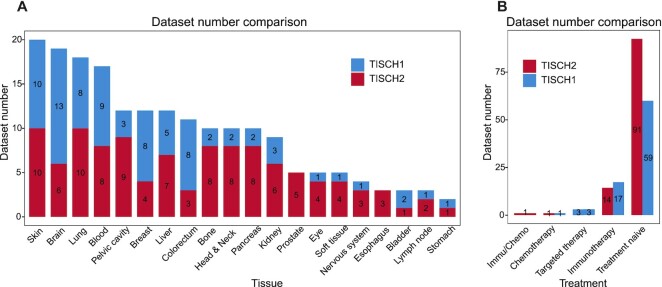
The TISCH2 database contains ∼6 297 320 cells from 190 datasets from 50 cancer types in 20 different tissues, compared with the previous version of TISCH which includes ∼2 045 746 cells from 79 datasets (Figure 1A; Supplementary Figure S1); the total number of cells and datasets is almost three times as many as the size of the previous version. As for the tumor scRNA-seq datasets with treatment, 40 datasets under different treatment conditions were provided in the current TISCH2 (35 human datasets and 5 mouse datasets), which contains immunotherapy, chemotherapy, targeted therapy and combined therapies (Figure 1B). On average, each dataset has 32 210 cells, with the largest dataset from NSCLC having 816 654 cells (31).
Figure 1.
Comparison of the data in TISCH1 and TISCH2. (A and B) The dataset number comparison between the TISCH1 (blue) and TISCH2 (red) at the tissue level (A) and the treatment strategy level (B).
New functions of TISCH2
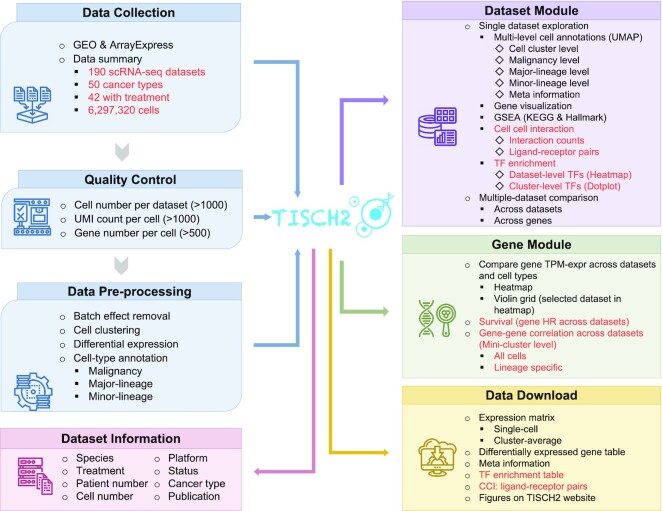
TISCH2 is not only a comprehensive resource of single-cell gene expression in the TME but also provides many useful analysis functions. The previous version of TISCH mainly includes two modules, the Dataset module and the Gene module. The updated TISCH2 includes two new functions, CCIs and TF enrichment analyses, in the Dataset module, and two new functions, survival and correlated gene analyses, in the Gene module (Figure 2).
Figure 2.
Overview of the current TISCH2 workflow and features. The words marked in red are updated datasets, newly added functions and features. TISCH2 automatically parsed and collected tumor scRNA-seq datasets from GEO or ArrayExpress databases. All datasets were then uniormly processed with a standardized workflow, including quality control, batch effect removal, cell clustering, differential expression analysis and cell type annotation at multiple levels. TISCH2 displays datasets with relevant study information, including species, treatment, the number of patients and cells, technology platform and the original study. In the Dataset module, TISCH2 provides cell type annotation visualization, multiple gene expression visualization, DEGs, functional enrichment analysis for single or multiple datasets, CCI exploration and TF enrichment analysis for cluster levels. In the Gene module, TISCH2 provides single gene expression visualization across multiple datasets and cell types, the clinical effect of the gene by survival analysis and gene–gene correlation visualization. TISCH2 provides a download page for users to access all the above data and analysis results.
Dataset module functions
In the Dataset module, TISCH2 supports the detailed exploration of a single dataset or between multiple datasets. For each dataset, TISCH2 will display its cancer type, species, platform information, treatment, stage and related publications. In particular, for the immunotherapy datasets, TISCH2 provides additional therapy-related analyses on the dataset page, including cell type proportion comparison and DEG analyses between patients in different treatment or response groups, and immunotherapy-associated signature visualization. The detailed analysis results will be shown in six different tabs, including an overview page for clustering, cell type annotation and DEGs, a gene page for expression visualization, a GSEA page for functional analyses of each cell type, a TF page for transcription regulator analyses, a CCI page for cell–cell interaction analyses and a download page for all the above data and analyses. We will mainly introduce the new TF and CCI functions.
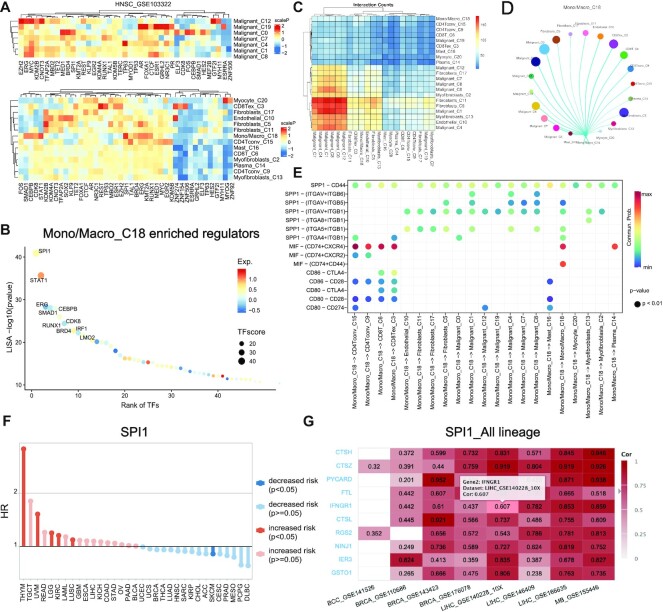
In the TF tab, the pre-calculated TF enrichment results are available for users to identify the driver regulator of different cell types. The enriched TFs were identified using LISA based on the marker genes of each cluster. TISCH2 displayed the top enriched TFs for malignant cell clusters and other cell clusters using heatmaps separately (Figure 3A). In addition, TISCH2 provides a dot plot representing the top 50 enriched TFs for each cluster, as well as a table listing the information of top-enriched TFs for each cluster (Figure 3B).
Figure 3.
The novel functions of TISCH2. (A and B) The TF enrichment result of HNSC_GSE103322. (A) The heatmap shows the TF enrichment score from LISA of each cluster. The color indicates the scaled enrichment P-value. (B) The dot plot shows the 50 top ranked enriched TFs of cluster 18, which is a subset of Mono/Macro. The color of the circles represents the average expression level of corresponding genes. The names of the top 10 TFs are labeled on the graph. (C and D) The overview of the cell communication result of HNSC_GSE103322. The heatmap displays the number of interactions between any two clusters, and the circus plot exhibits detailed interaction probability for cluster 18 with other clusters. (E) The significant communicated ligand–receptor pairs for the source and target cluster is shown as a bubble plot. (F) The lollipop plot represents the hazard ratio of SPI1. The HR equals 1 as an intercept point, a HR >1 means an increased risk and HR <1 suggests decreased risk. The color represents the significance, where red and blue are significant and light red and light blue are non-significant. (G) A real-time updated heatmap showing the gene–gene correlations of SPI1 across cancer type datasets. Users can select the different cell lineage levels to explore the gene–gene correlation in detail. The title of the heatmap is consistent with the selected gene and cell lineage.
In the CCI tab, TISCH2 provides the cell–cell communication analysis result to explore the interaction between cell types in the current dataset. The number of significantly interacted L–R pairs for all clusters will be shown using a heatmap for a global overview (Figure 3C). For each cell type cluster, TISCH2 will use a circus plot to display the interaction probability of the selected cluster with other cell types (Figure 3D). Finally, TISCH2 allows users to explore the detailed L–R interaction pairs with two bubble plots separately, with the selected cell type cluster both as the source cluster and as the target cluster (Figure 3E).
Here, we use an example to demonstrate the usage of the TF and CCI tabs. It has been reported that SPI1 and STAT1 play critical roles during monocyte to macrophage maturation (32,33). We observed the consistent conclusion that SPI1 and STAT1 with the largest TF score as highly expressed in cluster 18, which are monocyte and macrophage cells in HNSC_GSE103322 (Figure 3A, B). In addition, for the above cluster in the CCI result, we noticed that the monocyte and macrophages could directly interact with CD8T through CD80–CTLA4 to suppress the immune response of CD8T cells (Figure 3D). Hence, the TF and CCI functions enable users to identify the drive regulator and cell communications of interesting cell type subsets.
Gene module functions
The Gene module supports an advanced search for a single gene to explore its expression level across different datasets or cancer types, co-expressed genes and the clinical effect. The average gene expression tab displays the gene expression in different cell types and datasets for all the selected datasets using a heatmap or violin plot.
The survival and gene correlation tabs were new functions in the TISCH2. In the survival tab, a lollipop plot displays the HR of the selected gene across 33 TCGA cancer types, for which a HR >1 means an increased mortality risk, and a HR <1 suggests a decreased mortality risk (Figure 3F). In the gene correlation tab, the top 500 correlated expressed genes for the input gene in all the selected datasets are listed for users to discover the co-expression pattern of genes. By default, TISCH2 will display the correlated genes using all cells by a heatmap; however, to gain an insight into cell type-specific co-expression, TISCH2 also allows the identification of the correlated genes for a certain lineage (Figure 3G). All of the correlation information was also listed in the table at the bottom of the gene correlation tab.
To explore the co-expression patterns and clinical effects of the SPI1 gene, we queried it in the Gene module. We observed that high expression of SPI1 tends to increase risk in THYM and UVM cancer but to decrease the risk in SKCM cancer, indicating that macrophages might have diverse roles in different cancer types (Figure 3F). In addition, the gene correlation result showed CTSZ and CTSH co-expressed with SPI1, especially in liver cancer, such as LIHC_GSE140228_10X and LIHC_GSE166635. CTSZ and CTSH are lysosomal cysteine proteinases important for the phagocytosis function of macrophages, our analyses suggest that the expression of these genes might be regulated by SPI1 in macrophages.
DISCUSSION
We present TISCH2, an updated version of TISCH that contains expanded data volume and new functionalities. TISCH2 shows several advantages compared with the previous version. First, TISCH2 is a more comprehensive TME single-cell data portal, which almost doubled the covered cancer types and tripled the number of cells and datasets of the previous version. Second, TISCH2 provides more functions for users to explore the datasets, including driver TF identification, cell–cell communication analyses, co-expressed genes across datasets and the clinical benefit of the input gene. In the future, the TISCH team will continue to collect newly generated tumor scRNA-seq data and update TISCH regularly. In addition, we will also make an effort to provide new functions such as trajectory analysis, including multiome information such as epigenetic landscapes, and spatial positions of the cell types in the TME. With these new datasets and functions available, TISCH2 and its updated version will greatly benefit a wide range of biomedical users, especially in the oncology and immunology field.
DATA AVAILABILITY
The expression matrix, sample meta information, differential expression gene list, transcription factors and cell–cell interactions displayed in the TISCH database can be directly downloaded from http://tisch.comp-genomics.org/.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Bioinformatics Supercomputer Center of Tongji University for offering computing resources. The authors acknowledge the authors of published studies for sharing their scRNA-seq data on human clinical samples.
Contributor Information
Ya Han, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Yuting Wang, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Xin Dong, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Dongqing Sun, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Zhaoyang Liu, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Jiali Yue, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Haiyun Wang, Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Taiwen Li, State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, Research Unit of Oral Carcinogenesis and Management, Chinese Academy of Medical Sciences, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Chenfei Wang, Shanghai Putuo District People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; Frontier Science Center for Stem Cells, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Natural Science Foundation of China [32222026 and 32170660 to C.W., 81972551 to T.L.]; the Shanghai Rising Star Program [21QA1408200 to C.W.]; the Natural Science Foundation of Shanghai [21ZR1467600 to C.W.]; the Natural Science Foundation of Sichuan Province [2022NSFSC0054 to T.L.]; and the Young Elite Scientist Sponsorship Program by CAST [2021QNRC001 to T.L.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCE
- 1. Rozenblatt-Rosen O., Regev A., Oberdoerffer P., Nawy T., Hupalowska A., Rood J.E., Ashenberg O., Cerami E., Coffey R.J., Demir E.et al.. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell. 2020; 181:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D.et al.. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019; 179:829–845. [DOI] [PubMed] [Google Scholar]
- 3. Chen B., Scurrah C.R., McKinley E.T., Simmons A.J., Ramirez-Solano M.A., Zhu X., Markham N.O., Heiser C.N., Vega P.N., Rolong A.et al.. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell. 2021; 184:6262–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012; 12:252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.S., Nayar S., Greenstein A.J.et al.. Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019; 178:1493–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vasan N., Baselga J., Hyman D.M.. A view on drug resistance in cancer. Nature. 2019; 575:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fairfax B.P., Taylor C.A., Watson R.A., Nassiri I., Danielli S., Fang H., Mahe E.A., Cooper R., Woodcock V., Traill Z.et al.. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 2020; 26:193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giladi A., Amit I.. Single-cell genomics: a stepping stone for future immunology discoveries. Cell. 2018; 172:14–21. [DOI] [PubMed] [Google Scholar]
- 9. Marangoni F., Zhakyp A., Corsini M., Geels S.N., Carrizosa E., Thelen M., Mani V., Prussmann J.N., Warner R.D., Ozga A.J.et al.. Expansion of tumor-associated Treg cells upon disruption of a CTLA-4-dependent feedback loop. Cell. 2021; 184:3998–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun Y., Wu L., Zhong Y., Zhou K., Hou Y., Wang Z., Zhang Z., Xie J., Wang C., Chen D.et al.. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021; 184:404–421. [DOI] [PubMed] [Google Scholar]
- 11. Zeng J., Zhang Y., Shang Y., Mai J., Shi S., Lu M., Bu C., Zhang Z., Zhang Z., Li Y.et al.. CancerSCEM: a database of single-cell expression map across various human cancers. Nucleic Acids Res. 2022; 50:D1147–D1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M., Zhang X., Ang K.S., Ling J., Sethi R., Lee N.Y.S., Ginhoux F., Chen J.. DISCO: a database of deeply integrated human single-cell omics data. Nucleic Acids Res. 2022; 50:D596–D602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z.et al.. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; 47:D900–D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun D., Wang J., Han Y., Dong X., Ge J., Zheng R., Shi X., Wang B., Li Z., Ren P.et al.. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021; 49:D1420–D1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang C., Sun D., Huang X., Wan C., Li Z., Han Y., Qin Q., Fan J., Qiu X., Xie Y.et al.. Integrative analyses of single-cell transcriptome and regulome using MAESTRO. Genome Biol. 2020; 21:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K.. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014; 344,:1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of single-cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanehisa M., Goto A.S.. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W.. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018; 37:38–44. [DOI] [PubMed] [Google Scholar]
- 22. Pont F., Tosolini M., Fournie J.J.. Single-cell signature explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res. 2019; 47:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu C., Su Z.. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics. 2015; 31:1974–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vu R., Jin S., Sun P., Haensel D., Nguyen Q.H., Dragan M., Kessenbrock K., Nie Q., Dai X.. Wound healing in aged skin exhibits systems-level alterations in cellular composition and cell–cell communication. Cell Rep. 2022; 40:111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin Q., Fan J., Zheng R., Wan C., Mei S., Wu Q., Sun H., Brown M., Zhang J., Meyer C.A.et al.. Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 2020; 21:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu Z., Eils R., Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas Research Network Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baran Y., Bercovich A., Sebe-Pedros A., Lubling Y., Giladi A., Chomsky E., Meir Z., Hoichman M., Lifshitz A., Tanay A.. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019; 20:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng L., Qin S., Si W., Wang A., Xing B., Gao R., Ren X., Wang L., Wu X., Zhang J.et al.. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021; 374:abe6474. [DOI] [PubMed] [Google Scholar]
- 31. Caushi J.X., Zhang J., Ji Z., Vaghasia A., Zhang B., Hsiue E.H., Mog B.J., Hou W., Justesen S., Blosser R.et al.. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature. 2021; 596:126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coccia EM, Del Russo N, Stellacci E, Testa U, Marziali G, Battistini A. STAT1 activation during monocyte to macrophage maturation: role of adhesion molecules. Int. Immunol. 1999; 11:1075–1083. [DOI] [PubMed] [Google Scholar]
- 33. Kurotaki D., Sasaki H., Tamura T.. Transcriptional control of monocyte and macrophage development. Int. Immunol. 2017; 29:97–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The expression matrix, sample meta information, differential expression gene list, transcription factors and cell–cell interactions displayed in the TISCH database can be directly downloaded from http://tisch.comp-genomics.org/.