Abstract
Viruses are widely recognized as critical members of all microbiomes. Metagenomics enables large-scale exploration of the global virosphere, progressively revealing the extensive genomic diversity of viruses on Earth and highlighting the myriad of ways by which viruses impact biological processes. IMG/VR provides access to the largest collection of viral sequences obtained from (meta)genomes, along with functional annotation and rich metadata. A web interface enables users to efficiently browse and search viruses based on genome features and/or sequence similarity. Here, we present the fourth version of IMG/VR, composed of >15 million virus genomes and genome fragments, a ≈6-fold increase in size compared to the previous version. These clustered into 8.7 million viral operational taxonomic units, including 231 408 with at least one high-quality representative. Viral sequences in IMG/VR are now systematically identified from genomes, metagenomes, and metatranscriptomes using a new detection approach (geNomad), and IMG standard annotation are complemented with genome quality estimation using CheckV, taxonomic classification reflecting the latest taxonomic standards, and microbial host taxonomy prediction. IMG/VR v4 is available at https://img.jgi.doe.gov/vr, and the underlying data are available to download at https://genome.jgi.doe.gov/portal/IMG_VR.
INTRODUCTION
All life on Earth is seemingly infected by virus(es) (1,2). The virosphere, i.e. the entire diversity of viruses on Earth, likely rivals or surpasses the diversity of all cellular life forms (3). This knowledge of extant viral diversity is based on genomes from all types of nucleic acids from single-stranded RNA to double-stranded DNA, with differences in size and complexity spanning several orders of magnitudes. Over the last decade, metagenomics has become the primary approach by which this extended virosphere is being explored (4,5). While metagenomics does not provide mechanistic biological insights that might be enabled through viral cultivation in a laboratory setting, it facilitates high-throughput exploration of the viral genomic diversity across Earth's biomes. The growing importance of metagenome-derived viral genomes, a.k.a. ‘Uncultivated virus genomes’ or ‘UViGs’, led to the development of standard protocols and QC criteria to better identify, analyze, and share these genomes in 2019 (6).
Several studies have gathered large collections of UViGs, typically either focusing on a single environment (e.g. ‘human gut’), or type of virus (e.g. ‘RNA viruses’). In the human gut microbiome, for instance, 3 UViG datasets were recently released each including between ≈30 000 and ≈200 000 distinct viral genomes (7–9), while for soil ecosystems, the PIGEON database recently compiled 266 125 distinct virus genomes (10). Meanwhile, RNA viruses were a recent focus of multiple studies either across ecosystems (11,12) or centered on a single environment, e.g. the Tara Oceans RNA virus dataset (13). Most of these resources are released as flat files and not associated with a dedicated user interface to explore, browse and search these viral genomes.
IMG/VR was first released in 2016 as a separate resource dedicated to viral genomes within the IMG platform (14,15). It initially provided access to the ‘Earth Virome’ dataset (16), which included DNA virus genomes identified in metagenomes from a broad range of ecosystems. IMG/VR was subsequently updated with new UViGs from additional IMG datasets and new dedicated search and analysis tools, making it the largest database of UViGs currently available (17,18). Here, we present the 4th version of the IMG/VR database, which leverages a new and improved virus detection approach to gather a catalog of >15 millions UViGs now including both DNA and RNA viruses, either identified as viral contigs or integrated proviruses in genomes, metagenomes, or metatranscriptomes, all systematically annotated and available through the IMG/VR user interface (UI) (https://img.jgi.doe.gov/vr/) and for download (https://genome.jgi.doe.gov/portal/IMG_VR/IMG_VR.home.html), along with updated online search and analysis tools.
MATERIALS AND METHODS
Automatic virus identification from IMG datasets
Uncultivated viral genomes (UViGs) were mined from public assemblies retrieved from IMG/M on 2022-04-10 using geNomad (version 1.1.0) (19). A total of 28 865 metagenomes, 7258 metatranscriptomes, 83 858 isolate genomes of Bacteria and Archaea, 4342 single amplified genomes (SAGs), and 10 499 metagenome-assembled genomes (MAGs) were processed. A minimum length of 2 kb (for sequences with direct or inverted terminal repeats) or 4 kb (for the remaining sequences) was required for metagenomes, isolate genomes, SAGs and MAGs. For metatranscriptomes, a length cutoff of 2 kb was required throughout. geNomad's score calibration functionality, which infers sample composition to estimate probabilities for each classification, was employed to determine a cutoff that resulted in an estimated false discovery rate of 2%.
UViG completeness/contamination estimation and provirus quality control
Completeness and contamination of single-scaffold viral genomes was estimated with CheckV (version 1.0.1, database version 1.3) (20). CheckV’s AAI-based estimation of completeness was used if the confidence of the estimation was qualified as medium or high; the HMM-based estimate was used otherwise. For multi-scaffold giant virus MAGs (GVMAGs), completeness and contamination estimates, based on the presence and copy number of marker genes, were retrieved from the original study (21).
As proviruses are found integrated in host genomes and because automatic boundary determination is error-prone, CheckV was used to trim host regions from the edges of geNomad-identified proviruses to reduce the amount of contamination by host genes. Contamination by non-protein-coding genes was removed by trimming regions that encoded ribosomal rRNAs, obtained from IMG/M annotation.
UViG clustering into vOTUs
Single-scaffold UViGs were clustered into viral operational taxonomic units (vOTUs) following MIUViG guidelines (95% ANI—average nucleotide identity; 85% AF—aligned fraction) (6). To that end, sequences were first aligned in an all-versus-all BLAST (version 2.13.0+) (22) search that was executed with the following parameters: ‘-task megablast -evalue 1e-5 -max_target_seqs 20000’. Next, ANI and AF were computed by aggregating all the high-scoring segment pairs between each pair of UViGs (code available at https://bitbucket.org/berkeleylab/checkv/src/master/scripts/anicalc.py) and a graph was built by connecting the pairs that fulfilled the minimum ANI and AF requirements. Finally, the Leiden algorithm (23) (as implemented in the igraph Python library, version 0.9.10) was used to cluster sequences into vOTUs by identifying communities of highly interconnected nodes (resolution parameter = 1.0, code available at https://github.com/apcamargo/bioinformatics-snakemake-pipelines/tree/main/genome-ani-leiden-clustering-pipeline). Multi-scaffold GVMAGs were clustered separately and their vOTUs were directly imported from IMG/VR v3 (18).
Virus prediction confidence and UViG quality assessment
UViGs were separated into ‘high-confidence virus’ and ‘putative virus’ based on the amount of evidence supporting their viral origin. Briefly, points were allocated for each UViG that fulfilled certain criteria (listed below) and the genomes that were awarded at least three points were moved to the high-confidence tier.
3 points: Clustered with a RefSeq r213 virus genome in a vOTU; three or more geNomad virus hallmark markers.
2 points: High-confidence CheckV AAI completeness estimate; two geNomad virus hallmark markers.
1 point: Medium-confidence CheckV AAI completeness estimate; one geNomad virus hallmark marker; direct or inverted terminal repeats; two or more matches to CRISPR spacers.
Sequences with low coding density (open reading frame coverage < 60%), high number of ambiguous nucleotides (number of N > 50, n = 105 469), or containing concatemers (≥1 kb repeat of the 5' end of the sequence or ≥10 kb repeat within the sequence, n = 7079) were never assigned to the high-confidence tier, regardless of the number of points they received. UViGs that were obtained from RefSeq (24) or that were identified in external projects such as Inoviruses (25), GVMAGs (21), and RNA Viruses in Metatranscriptomes (RVMT) (12), were automatically assigned to the high-confidence tier.
Genomes within the high-confidence tier were further categorized into four different groups based on their completeness, estimated using CheckV (database version 1.3): high-quality (≥90%), medium quality (50–90%), low-quality (<50%), and unsure (>120% or no completeness estimate). For GVMAGs, completeness estimates are based on the presence of Nucleocytoviricota marker genes and were retrieved from the original study (21).
Taxonomic assignment
Sequences that were imported from RefSeq were assigned to their NCBI taxonomy. UViGs derived from the RVMT and the GVMAG projects were designated to taxonomic lineages listed in their original studies. The remaining UViGs were tentatively assigned to viral taxa as defined in the International Committee on Taxonomy of Viruses’ (ICTV) Release #37 (26). The following taxonomic classification methods were used in order of priority: (i) clustering with RefSeq virus genomes; (ii) geNomad's marker-based taxonomic assignment; (iii) similarity to viral proteins in NCBI NR; (iv) vOTU consensus. UViGs that clustered together with RefSeq references in vOTUs received the taxonomy of the reference genome. For the marker-based assignment, geNomad (version 1.1.0) was employed to classify sequences using taxonomically informative protein profiles. To assign taxonomy based on similarity to viral proteins from NCBI’s NR (retrieved in 2022-05-19), we used MMseqs2’s (version 13.45111) (27) taxonomy module with the ‘- -start-sens 4 -s 6 - -sens-steps 2’ parameters. Finally, UViGs that were not designated to any taxon using the previously described methods were assigned to the consensus taxon within their vOTU, obtained using the ‘find_majority_vote’ function in taxopy (version 0.10.2) (28). The ICTV taxdump used for all the taxonomic assignment methods was generated using TaxonKit (version 0.11.1) (29).
Host assignment
UViGs identified within isolate whole-genome sequencing assemblies or SAGs were assigned a host taxonomy prediction based on the taxonomy of the source genome. The remaining viruses were assigned to hosts by indirect association through matches to a database of CRISPR spacers or exact k-mer matches to bacterial and archaeal genomes derived from NCBI GenBank (release 242; 15 February 2021) and recent large-scale MAG studies (30–33). We annotated all host genomes using GTDB-Tk (version 2.1.0) (34) and the GTDB database r207 (35) to ensure consistency.
CRISPR spacers were identified from the 1.6 million genomes from NCBI and MAG studies (see above) using a combination of CRT (version 1.1) (36) and PILER-CR (version 1.06) (37). Our database contained 28.6 million spacers after filtering potentially spurious (38) CRISPR arrays and problematic spacers, and 4.8 million unique CRISPR spacers after dereplication with MMseqs2 (version 13.45111, parameters: ‘easy-cluster - -min-seq-id 1.0 -c 1.0’). Viruses were matched to the spacer database using blastn (version 2.9.0+, parameters: ‘-max_target_seqs = 1000 -word_size = 8 -dust = no’) and we only considered alignments with at least 25 bp, a maximum of 1 mismatch, and that covers ≥95% of the spacer length. After these filtering criteria, a total of 1 017 950 (21.2%) CRISPR spacers matched 1 003 550 UViGs.
PHIST (version 1.0.0) (39) was used to perform k-mer matching between viruses and bacterial and archaeal genomes from NCBI and MAG studies. To improve computational throughput and reduce skew by highly represented pathogens, we included a maximum of 10 randomly selected genomes per host lineage, resulting in a database of 230 600 prokaryotic genomes. To reduce spurious matches, we only considered virus-host connections where at least 20% of viral k-mers were found in a prokaryotic genome.
Each virus was then assigned to the host taxon, at the lowest taxonomic rank, having at least two connections and representing >70% of all connections. In cases where a virus had an assigned host via CRISPR spacer and k-mer approaches, we chose the method that was supported by a greater number of host connections. The full, filtered, and clustered datasets of CRISPR spacers can be found at: https://portal.nersc.gov/cfs/m342/crisprDB.
RESULTS
Overview of IMG/VR v4 novel features
Compared to its previous release, IMG/VR v4 has a number of major changes which include:
A new de novo virus discovery using a virus identification pipeline based on geNomad allowing for improved detection of viruses and more quantitative metrics for prediction confidence.
An expanded selection of reference viral genomes.
Estimates of genome quality with an updated version of CheckV, which now includes a 27% larger reference database of complete viral genomes.
An updated taxonomic assignment schema that follows the taxa described in ICTV’s Taxonomy Release #37.
A larger set of host predictions, using information from isolate genomes, an expanded database of CRISPR spacers, and k-mer matches to a large database of prokaryotic genomes.
Updated online tools to search for viral sequences through gene composition, nucleotide similarity, or amino acid similarity.
Tighter integration into the IMG database ecosystem including, e.g. the possibility to connect IMG/VR data to individual genome, scaffold, and gene carts.
Below we provide a more detailed description of each of these new features.
Improved detection of viral sequences and database composition
To perform automatic identification of viral genomes within all IMG sequencing data for IMG/VR v4, we developed geNomad (available at https://github.com/apcamargo/genomad), which replaced the IMG Virus Detection Pipeline (40) used in previous versions of the database. geNomad is a hybrid virus detection tool that uses both gene content and a neural network that takes in nucleotide sequences to estimate the ‘viralness’ of each input sequence. For IMG/VR v4, geNomad presents several advantages compared to the previous IMG/VR prediction pipeline and other existing tools: (i) high-throughput and speed—which allowed us to process billions of scaffolds in IMG/M within a week using the NERSC’s Cori computer cluster; (ii) broad taxonomic coverage—which allowed the identification of multiple viral groups that were underrepresented in the previous version—such as RNA viruses, retroviruses, ssDNA viruses, Megaviricetes, and Tokiviricetes; (iii) automatic identification of proviruses; (iv) increased identification of short viral sequences, achieved through increased marker coverage and the nucleotide-based neural network classifier and (v) better distinction from plasmids, since those are explicitly identified and treated as a separate category by geNomad.
In addition to virus identification, geNomad automated annotation results were leveraged to provide: (i) automatic identification of viruses that likely employ alternative genetic codes (translation tables 4 and 15); (ii) gene-level annotation using a total of 227 897 marker protein profiles (41) that are used to find viruses that have a similar gene composition (now used in the ‘Find similar UViGs’ tool, see ‘Search and browse interface’ section); (iii) taxonomic assignment using a set of taxonomically-informative profiles (see ‘Taxonomic, host, and ecological distribution’ section); (iv) detection of virus hallmarks, which are used for identifying the sequences that were assigned to the ‘high-confidence virus’ tier (see ‘IMG/VR v4 UViGs are stratified using quality metrics and represent a large expansion over the previous release’ section).
Expanded Reference virus genome database
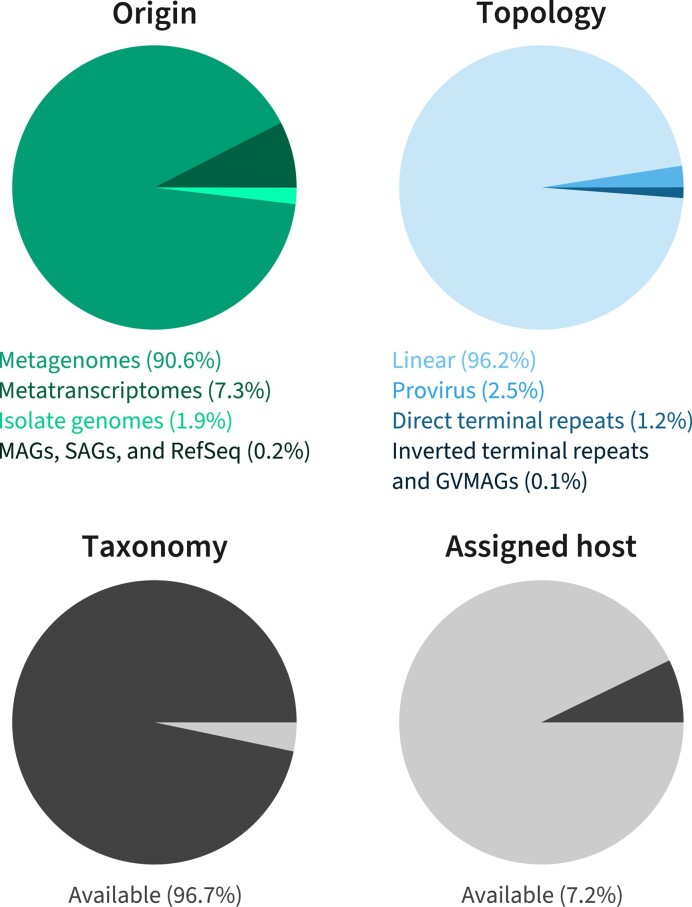
IMG/VR v4 also contains sequences imported from RefSeq (release 213) and viruses identified in independent projects that focused on specific viral groups: Inoviruses (Inoviridae family), giant viruses (Nucleocytoviricota phylum), and RNA viruses (Orthornavirae kingdom) (Table 1). In total, IMG/VR now includes 13 971 references from RefSeq (genomes and segments) and 15 663 652 UViGs identified across 25 603 metagenomes, 6755 metatranscriptomes, 62 342 genomes of isolated prokaryotes, 854 SAGs, and 3514 MAGs (Figure 1)—a 6.7-fold increase over the previous IMG/VR release. The majority of these UViGs are linear sequences, could be assigned a taxonomy using the geNomad taxonomically-informative profiles, but still lack host prediction (Figure 1, and see below).
Table 1.
Data sources used to build the v4 release of IMG/VR, as well as the number of UViGs and vOTUs within each. Numbers for high-confidence viruses and vOTUs containing high-confidence viruses in the geNomad-identified UViGs are shown within parenthesis. For additional statistics: https://portal.nersc.gov/cfs/m342/imgvr_stats/
| Source | Reference | Description | No. of UViGs | No. of vOTUs |
|---|---|---|---|---|
| IMG/M mining with geNomad | (19) | Mining IMG/M’s public sequencing data using geNomad | 15 303 607 (5 202 181) | 8 452 288 (2 744 855) |
| RefSeq r213 | (24) | Viral genomes imported from NCBI | 13 971 | 12 410 |
| Inoviruses | (25) | Custom search of extrachromosomal and integrated Inoviridae genomes using marker genes and gene content features | 228 | 5281 |
| Giant virus MAGs (GVMAGs) | (21) | Custom search for Nucleocytoviricota MAGs in IMG/M | 2055 | 2005 |
| RVMT | (12) | Identification of Orthornavirae in IMG/M metatranscriptomes using a custom set of RNA-dependent RNA polymerase models. | 348 762 | 163 939 |
Figure 1.
IMG/VR v4 composition regarding UViG origin, scaffold topology, taxonomic assignment and availability of host data. Each pie chart displays the number of UViGs from different types of datasets (‘Origin’), with different sequence topologies (‘Topology’), classified into an existing virus taxon at any rank (‘Taxonomy’), and with at least one host prediction available (‘Assigned host’).
IMG/VR v4 UViGs are stratified using quality metrics and represent a large expansion over the previous release.
The automatic identification of viral sequences in sequencing data resulted in an explosion of the known diversity of the virosphere over the last few years. Yet, computational methods for virus genome detection can sometimes retrieve sequences for which the viral origin is uncertain (42). Two notable examples are: (i) sequences for which there is little functional information, but are classified as viruses either because of the presence of hypothetical genes that have been previously observed in viruses or because of nucleotide signatures captured by methods such as neural networks and (i) short sequences that are enriched in functions associated with viruses (e.g. addiction modules and mobility genes) but encode no virus hallmark(s). These putative viral sequences can be very useful if the intent is to identify novel viruses, but can also hinder studies that require clean data, such as the identification of viral-encoded auxiliary metabolic genes, as these putative viral sequences can easily include non-viral sequences such as plasmids and microbial genomic islands (20).
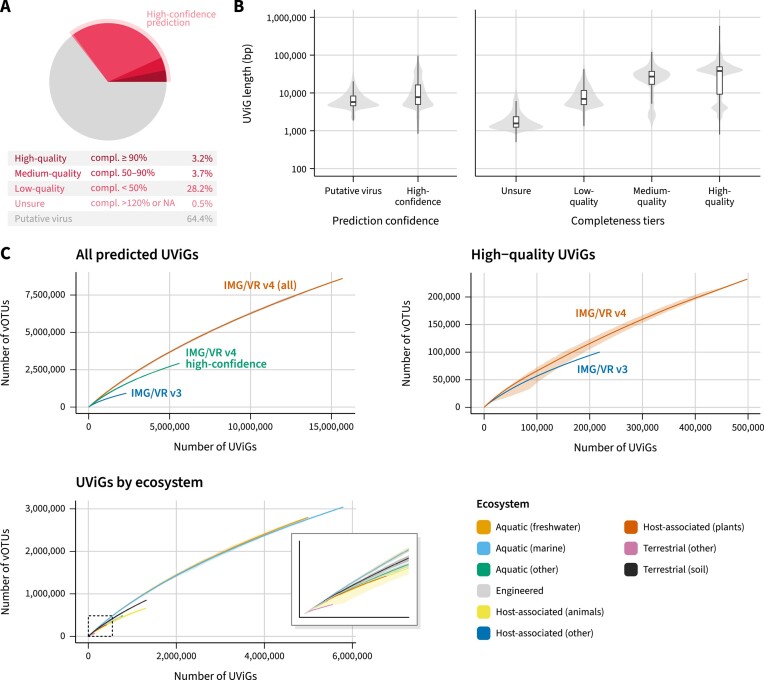
To help users identify the well-supported viral sequences, while also providing the larger set of viral predictions for users interested in these putative viral sequences, the IMG/VR v4 database is stratified into two main confidence tiers: ‘high-confidence virus’ and ‘putative virus’. The former, which represents about 35% of the database (or 5 576 197 UViGs clustered into 2 917 516 vOTUs, Table 1), contains UViGs for which additional evidence of viral origin (Figure 2A) could be gathered, and is enriched with bona fide viral genomes at the cost of reduced sensitivity, i.e. missing some novel viruses and short sequences for which gene signal is scarce. The latter includes sequences that were flagged as viruses by geNomad, but do not fulfill the criteria for inclusion in the ‘high-confidence’ tier (see Methods).
Figure 2.
(A) Fraction of high-confidence UViGs within IMG/VR v4 and the contribution of each of the completeness tiers. (B) Left: length distribution of UViGs classified as putative viruses and high-confidence viruses. Right: length distribution of the different completeness tiers within high-confidence predictions. (C) Accumulation curves of vOTUs across different subsets of IMG/VR. The top left panel shows the accumulation of vOTUs as a function of the number of UViGs for all sequences in IMG/VR v4, only high-confidence sequences, or for the sequences that were already present in IMG/VR v3. The top right panel shows similar accumulation curves considering only high-quality (i.e. high-confidence and ≥90% complete) UViGs. In this panel, for IMG/VR v3 high-quality UViGs, only sequences that were available through the IMG/VR v3 interface and are still available in the IMG/VR v4 database were included. Finally, the bottom panel shows similar accumulation curves separated by ecosystem and considering all sequences in IMG/VR v4. Each curve is the average of 50 random permutations, with the minimum and maximum value at each step indicated with a gray (top two panels) or colored (bottom panel) outline.
Compared to the previous release, an additional 4 164 835 (74.7%) high-confidence UViGs were added to IMG/VR v4. Within those, 65.4% were from new prokaryotic genomes, metagenomes, metatranscriptomes, and external datasets. The remaining new sequences were detected due to the lower minimum sequence length threshold (12.9%), or increased detection sensitivity from the geNomad pipeline (21.7%).
Commonly needed information for the downstream analysis of viral sequences is the completeness estimate of the genomes. Assembly of high-throughput sequencing data often results in fragmented chromosomes, which preclude analyses that require complete or near complete genomes, such as delimitation of novel taxa. In IMG/VR v4 UViGs were evaluated for completeness using an updated version of CheckV’s database, which includes an expanded database of complete genomes (a 27% increase) compared to the version used in IMG/VR v3, that provides more accurate estimates for a number of underrepresented viral lineages including Lavidaviridae, Inoviridae, Nucleocytoviricota, Tectiviridae and RNA phages.
Within the high-confidence tier, UViGs were stratified based on their completeness into high-quality (completeness ≥ 90%; 495 576 UViGs), medium quality (completeness between 50% and 90%; 570 984 UViGs), low-quality (completeness < 50%; 4 427 996 UViGs), or unsure (completeness > 120% or no completeness estimate; 81 641 UViGs; Figure 2B). Most of the high-quality UViGs (314 758, or 62.51%) are newly available in IMG/VR v4 compared to v3. In addition, 78 137 high-quality UViGs (15.8%) from the MGV dataset (9), that were only available for download in IMG/VR v3, are now also available for interactive analysis in the IMG/VR v4 web interface. This version 4 thus represents a substantial expansion in the number of complete and near-complete genomes available in IMG/VR.
IMG/VR v4 UViGs were clustered at the standard 95% ANI 85% AF thresholds into 8 606 551 vOTUs (5 846 590 of which are singletons)—a 9.2-fold increase over the previous version (Figure 2C). Accumulation curves based on the vOTU clustering show no sign of reaching a plateau, either when taken globally, restricted to only high-quality sequence, or evaluated for individual ecosystems. This seemingly exponential growth of viral genome data in IMG/VR is consistent with previous observations (18), and suggests that many more viruses remain to be discovered.
Taxonomic, host, and ecological distribution
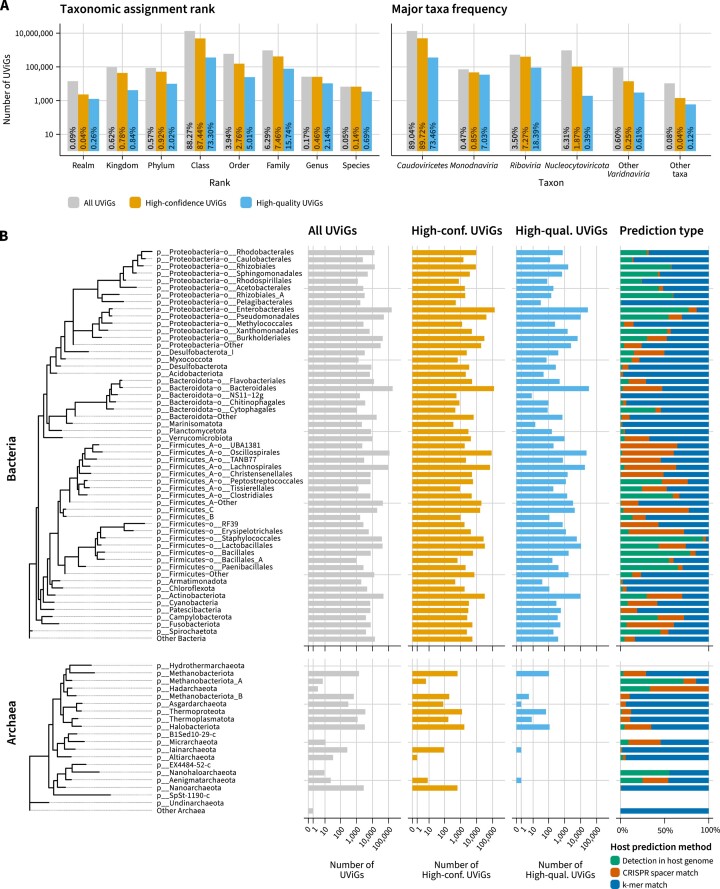
In the v4 release, IMG/VR taxonomy is based on the taxa described in ICTV Taxonomy Release #37. Notably, this release introduced binomial naming for viral species and extinguished the Myoviridae, Siphoviridae, and Podoviridae families of tailed phages (43). Additionally, a new automatic taxonomy assignment method, based on a set of 85 158 taxonomically informative geNomad marker profiles, was introduced to improve the taxonomic coverage of the database.
In total, 15 161 577 UViGs—or 96.7%—were assigned to viral taxa at different ranks (Figure 3A), up from 61.4% in IMG/VR v3. Among the high-confidence viruses the proportion is higher, as 5 557 099 sequences—or 99.7%—are taxonomically annotated. Across the entire dataset, the majority of the sequences (86.1%) were assigned to the Caudoviricetes class (Figure 3A), which encompasses tailed phages that are highly abundant across ecosystems (16). Of note, IMG/VR v4 includes 404 245 high-confidence UViGs (7.3% of the high-confidence set) assigned to the Riboviria realm, which includes all the RNA viruses. This represents a substantial increase over IMG/VR v3 (8622 UViGs) and it is due to the inclusion of scaffolds identified in the RVMT project and the usage of geNomad. Other important taxa, such as the Monodnaviria (47 146 high-confidence UViGs, 0.85%), Nucleocytoviricota (104,220 high-confidence UViGs, 1.9%), and other Varidnaviria (14 063 high-confidence UViGs, 0.25%) comprise the majority of the taxonomically-assigned UViGs (Figure 3A).
Figure 3.
(A) Left: number of UViGs assigned to each taxonomic rank. For each UViG, only the most specific rank was considered. Right: Number of UViGs assigned to major taxa. Percentages over the bars represent the fraction of UViGs within each group (all UViGs, high-confidence viruses, high-quality viruses) that is represented by that bar. (B) Frequency of bacterial (top tree) and archaeal (bottom tree) hosts to which UViGs were assigned to. Counts for all UViGs (gray), high-confidence UViGs (orange), and high-quality UViGs (blue) are shown in separate columns. Relative frequencies for each of the host assignment methods are shown in the fourth column. Trees were retrieved from GTDB (release 207).
UViGs in IMG/VR v4 were tentatively assigned to bacterial and archaeal hosts using three different approaches. Connections between viral genomes and hosts were made directly, via guilt-by-association of viruses identified within host genomes, or indirectly, by performing CRISPR spacer and k-mer matches in a large database of prokaryotic genomes. Host taxa were harmonized to reflect GTDB r207 taxonomy. Across the entire dataset, 1 125 839 UViGs (7.2%) were assigned to a host lineage. The fraction was higher for the high-confidence UViGs, where 764 426 genomes (13.8%) were linked to putative hosts. Within this set, 40.4% of the virus-host connections were made using CRISPR spacer matches, 30.9% by direct detection in the host genome, and 28.7% using k-mer matches. High confidence UViGs were mostly assigned to Bacteria (99.4%), especially to the Proteobacteria (34.2%), ‘Firmicutes_A’ (25.7%), Bacteroidota (17.7%), Firmicutes (10.4%), and Actinobacteriota (4.3%) phyla. Within Archaea, most UViGs were assigned to Halobacteriota (35.9%), Thermoproteota (25.8%), Nanoarchaeota (13.6%) and Methanobacteriota (13%) (Figure 3B).
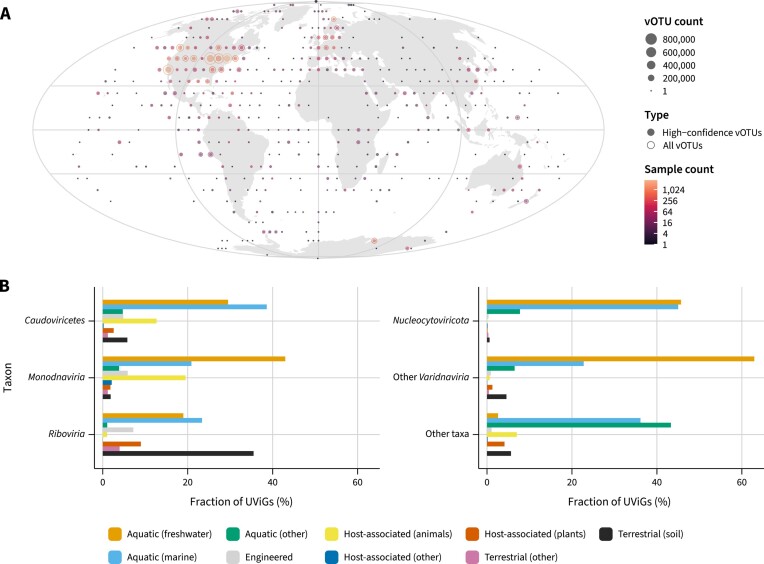
IMG/VR v4 leverages metadata from the Genomes OnLine Database (GOLD) (44) to provide geographical (Figure 4A) and ecosystem (Figure 4B) data to all the UViGs identified in IMG/M metagenomes and metatranscriptomes. Geographical coordinates of the samples show that IMG/VR v4 encompasses UViGs identified across all continents and oceans, and that most of these viral genomes come from North America and Europe. Regarding ecosystem distribution, high-confidence UViGs were mostly detected in samples classified as marine (2 006 477 UViGs, 37.6%), freshwater (1 565 841 UViGs, 29.3%), and animal-associated (609 925 UViGs, 11.4%) (Figure 2C). Taxonomy data revealed that there is strong association between some taxa and ecosystems, as Nucleocytoviricota and other Varidnaviria are more common in aquatic environments and rarely found in animal-associated samples, and Riboviria are relatively much more common in soils than other taxa.
Figure 4.
(A) Geographical distribution of the IMG/VR v4 viruses at the vOTU level based on the coordinates of IMG/M metagenomes and metatranscriptomes. Rings represent the total number of vOTUs within an area and filled circles represent the number of vOTUs with at least one high-confidence prediction. Data points are colored according to the number of samples where UViGs were detected. Samples were binned in fixed intervals across the longitude and latitude. UViGs identified in microbial genomes or imported from RefSeq are not represented. (B) Environmental distribution of major virus taxa. Bars represent the fraction of UViGs that were found within metagenomes and metatranscriptomes assigned to each of the major ecosystem classes.
Search and browse interface
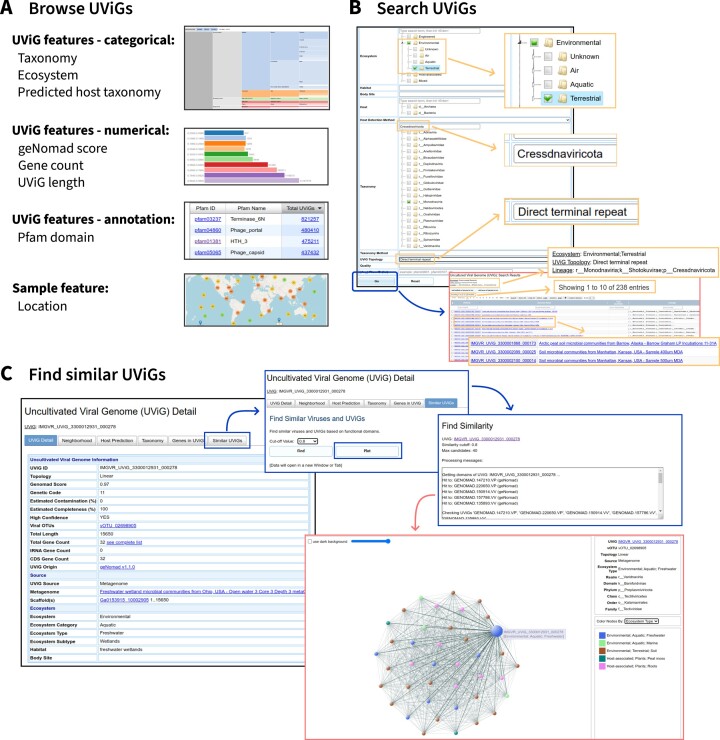
The search and browse interfaces from IMG/VR v3 were enhanced to provide optimal display of the new metrics and features now associated with IMG/VR v4 sequences. The browse menu allows users to explore UViGs based on categorical features through treemaps, numerical features through interactive bar charts, Pfam annotations through a searchable table, and sample location via an interactive map (Figure 5A). In particular, browsable numerical features now include the geNomad confidence score, a new metric in IMG/VR v4 reflecting the confidence in the viral nature of the sequence.
Figure 5.
(A) IMG/VR’s web interface allows users to browse UViGs according to multiple features related to the viral genome and the sample where it was identified. (B) Diverse metadata queries can be combined to search for UViGs. (C) The ‘Find similar UViGs’ tool allows users to find viruses with similar gene composition to a given query by identifying UViGs with similar sets of geNomad markers.
As far as UViG searches, in addition to the features already available in IMG/VR v3, UViGs can now be selected based on their geNomad classification score, their type of source dataset, e.g. metagenome or metatranscriptome, or their confidence category (Figure 5B). This enables users to select a subset of the >15 million IMG/VR v4 UViGs based on multiple criteria, and obtain an UViG list that can be further filtered interactively and/or downloaded for offline analysis.
Finally, the ‘Similar UViGs’ functionality, which is available on a UViG detail page and enables searching of the entire UViG database based on gene content using the current UViG as ‘hook’, was also modified in IMG/VR v4. This search now uses geNomad's markers (see below), instead of the Pfam domain annotation in IMG/VR v3, to identify similar UViGs. This means that this search now also leverages conserved hypothetical proteins lacking a Pfam domain match but detected across enough viral genomes to be included in geNomad protein clusters. As in IMG/VR v3, the UViGs detected as similar to the ‘hook’ can be displayed in a table or through an interactive network (Figure 5C).
Complementary download and user sequence analysis
Next to the browse and search interfaces, IMG/VR also provides online analysis tools for users to apply to their own UViGs and download bundles. In terms of analysis, IMG/VR v4 still provides online blastn to the UViG database, blastp to the predicted cds from these UViGs, along with blastn to the IMG CRISPR spacer database. All these are available through the ‘Viral/Spacer Blast’ button on the home page and provide a straightforward way for users to compare individual UViGs to the IMG/VR database. In addition, a dedicated tool for giant virus taxonomic assignment of user MAGs and contigs, named ‘GVClass’ (https://github.com/NeLLi-team/gvclass), is available through IMG/VR v4 home page.
For larger scale analyses, we recommend users download the IMG/VR v4 database and perform sequence similarity offline. To facilitate this, we provide file bundles to download, available through the ‘Download IMG/VR database’ link on the home page. Specifically, two folders are provided for IMG/VR v4, each containing a FASTA file of genomes, FASTA file of predicted proteins, a single table of UViG metadata, a table summarizing UViG host predictions, and a readme file. The first folder includes data from the entire IMG/VR v4 database, i.e. >15 million UViGs, while the other, identified with an ‘-hc’ suffix, provides the same file for the 5 576 197 UViGs identified as high-confidence. Lastly, the geNomad markers used for virus sequence identification and taxonomic classification are provided for download through a link on the IMG/VR home page.
CONCLUSION
Genomes from uncultivated viruses (‘UViGs’), obtained without cultivation of the corresponding virus in the laboratory but instead identified in shotgun sequencing datasets, are reshaping our understanding of the global virosphere. However, because the vast majority of this data derived solely from computational prediction, some specific cautions and challenges are important to note. First and foremost, while the performance and accuracy of virus sequence prediction tools greatly improved over the last few years, these predictions remain imperfect, especially for short sequences and for viruses closely related to other types of mobile genetic elements, such as plasmids. Since different types of UViGs analyses can accommodate different levels of non-viral sequences, and to enable users to apply their own preferred stringency level, IMG/VR v4 now features the most confident predictions in a ‘high-confidence’ category, and provides the raw geNomad score associated with each UViG for further refinement.
The other major limitation of UViGs compared to virus isolates is the lack of host association. Computational host prediction methods are still under very active development, with new and improved approaches published seemingly every year. Meanwhile, current prediction methods typically suffer from either low recall or low accuracy. For IMG/VR v4, a conservative set of methods and cutoffs was selected, which should yield accurate predictions but only for a minority of UViGs. Nevertheless, these host predictions should be interpreted carefully and not considered as definitive virus-host associations. Moving forward, we expect host prediction methods will keep improving in the years to come and will be complemented by a broader application of novel experimental assays for virus-host association including proximity ligation sequencing (45).
Since 2016, IMG/VR has been a flagship database in the field of viral ecology by providing a large collection of virus genomes from a broad range of environments and virus types. The new IMG/VR v4 version presented here holds the largest UViGs collection yet, and will undoubtedly yield many novel insights into the ecological parameters driving viral diversity, virus evolutionary history, and viral-encoded functional diversity. A number of the approaches and data used to build IMG/VR are also relevant for other types of mobile genetic elements, such as plasmids. In particular, geNomad automatically identifies both virus-like and plasmid-like elements separately. Given the potential importance of plasmids in microbiomes, a separate IMG system based on a similar architecture as IMG/VR but dedicated to plasmid-like sequences, is currently being developed to enable large-scale exploration of plasmid diversity.
DATA AVAILABILITY
IMG/VR data is freely available through the online UI (https://img.jgi.doe.gov/vr/) and for download (https://genome.jgi.doe.gov/portal/IMG_VR/IMG_VR.home.html).
Contributor Information
Antonio Pedro Camargo, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Stephen Nayfach, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
I-Min A Chen, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Krishnaveni Palaniappan, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Anna Ratner, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Ken Chu, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Stephan J Ritter, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
T B K Reddy, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Supratim Mukherjee, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Frederik Schulz, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Lee Call, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Russell Y Neches, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Tanja Woyke, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Natalia N Ivanova, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Emiley A Eloe-Fadrosh, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Nikos C Kyrpides, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Simon Roux, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
FUNDING
U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]; National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy. Funding for open access charge: Office of Science of the U.S. Department of Energy (operated under Contract No. DE-AC02-05CH11231).
Conflict of interest statement. None declared.
REFERENCES
- 1. Breitbart M., Rohwer F.. Here a virus, there a virus, everywhere the same virus?. Trends Microbiol. 2005; 13:278–284. [DOI] [PubMed] [Google Scholar]
- 2. Koonin E.V., Dolja V.V., Krupovic M., Kuhn J.H.. Viruses defined by the position of the virosphere within the replicator space. Microbiol. Mol. Biol. Rev. 2021; 85:e00193-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koonin E.V., Dolja V.V., Krupovic M., Varsani A., Wolf Y.I., Yutin N., Zerbini F.M., Kuhn J.H.. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020; 84:e00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sommers P., Chatterjee A., Varsani A., Trubl G.. Integrating viral metagenomics into an ecological framework. Annu. Rev. Virol. 2021; 8:133–158. [DOI] [PubMed] [Google Scholar]
- 5. Greninger A.L. A decade of RNA virus metagenomics is (not) enough. Virus Res. 2018; 244:218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roux S., Adriaenssens E.M., Dutilh B.E., Koonin E.V., Kropinski A.M., Krupovic M., Kuhn J.H., Lavigne R., Brister J.R., Varsani A.et al.. Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol. 2019; 37:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tisza M.J., Buck C.B.. A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2023202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camarillo-Guerrero L.F., Almeida A., Rangel-Pineros G., Finn R.D., Lawley T.D.. Massive expansion of human gut bacteriophage diversity. Cell. 2021; 184:1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nayfach S., Páez-Espino D., Call L., Low S.J., Sberro H., Ivanova N.N., Proal A.D., Fischbach M.A., Bhatt A.S., Hugenholtz P.et al.. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 2021; 6:960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. ter Horst A.M., Santos-Medellín C., Sorensen J.W., Zinke L.A., Wilson R.M., Johnston E.R., Trubl G., Pett-Ridge J., Blazewicz S.J., Hanson P.J.et al.. Minnesota peat viromes reveal terrestrial and aquatic niche partitioning for local and global viral populations. Microbiome. 2021; 9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar R.C., Taylor J., Lin V., Altman T., Barbera P., Meleshko D., Lohr D., Novakovsky G., Buchfink B., Al-Shayeb B.et al.. Petabase-scale sequence alignment catalyses viral discovery. Nature. 2022; 602:142–147. [DOI] [PubMed] [Google Scholar]
- 12. Neri U., Wolf Y.I., Roux S., Camargo A.P., Lee B., Kazlauskas D., Chen I.M., Ivanova N., Zeigler Allen L., Paez-Espino D.et al.. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell. 2022; 185:4023–4037. [DOI] [PubMed] [Google Scholar]
- 13. Zayed A.A., Wainaina J.M., Dominguez-Huerta G., Pelletier E., Guo J., Mohssen M., Tian F., Pratama A.A., Bolduc B., Zablocki O.et al.. Cryptic and abundant marine viruses at the evolutionary origins of earth's RNA virome. Science. 2022; 376:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paez-Espino D., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Szeto E., Pillay M., Huang J., Markowitz V.M., Nielsen T.et al.. IMG/VR: a database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res. 2016; 45:gkw1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen I.-M.A., Chu K., Palaniappan K., Ratner A., Huang J., Huntemann M., Hajek P., Ritter S., Varghese N., Seshadri R.et al.. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 2021; 49:D751–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C.. Uncovering earth's virome. Nature. 2016; 536:425–430. [DOI] [PubMed] [Google Scholar]
- 17. Paez-Espino D., Roux S., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Huntemann M., Reddy T.B.K., Pons J.C., Llabrés M.et al.. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2019; 47:D678–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roux S., Páez-Espino D., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Reddy T.B.K., Nayfach S., Schulz F., Call L.et al.. IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021; 49:D764–D775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Camargo A. apcamargo/genomad: geNomad v1.1.0 (v1.1.0). Zenodo. 2022; 10.5281/zenodo.7015982. [DOI] [Google Scholar]
- 20. Nayfach S., Camargo A.P., Schulz F., Eloe-Fadrosh E., Roux S., Kyrpides N.C.. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021; 39:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schulz F., Roux S., Paez-Espino D., Jungbluth S., Walsh D.A., Denef V.J., McMahon K.D., Konstantinidis K.T., Eloe-Fadrosh E.A., Kyrpides N.C.et al.. Giant virus diversity and host interactions through global metagenomics. Nature. 2020; 578:432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y., Ye W., Zhang Y., Xu Y.. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015; 43:7762–7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Traag V.A., Waltman L., van Eck N.J.. From louvain to leiden: guaranteeing well-connected communities. Sci. Rep. 2019; 9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roux S., Krupovic M., Daly R.A., Borges A.L., Nayfach S., Schulz F., Sharrar A., Matheus Carnevali P.B., Cheng J.-F., Ivanova N.N.et al.. Cryptic inoviruses revealed as pervasive in bacteria and archaea across earth's biomes. Nat. Microbiol. 2019; 4:1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B.. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 2018; 46:D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steinegger M., Söding J.. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017; 35:1026–1028. [DOI] [PubMed] [Google Scholar]
- 28. Camargo A., Borry M.. apcamargo/taxopy: v0.10.2 (v0.10.2). Zenodo. 2022; 10.5281/zenodo.7010602. [DOI] [Google Scholar]
- 29. Shen W., Ren H.. TaxonKit: a practical and efficient NCBI taxonomy toolkit. J. Genet. Genomics. 2021; 48:844–850. [DOI] [PubMed] [Google Scholar]
- 30. Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., Wu D., Paez-Espino D., Chen I.-M., Huntemann M.et al.. A genomic catalog of earth's microbiomes. Nat. Biotechnol. 2021; 39:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Almeida A., Nayfach S., Boland M., Strozzi F., Beracochea M., Shi Z.J., Pollard K.S., Sakharova E., Parks D.H., Hugenholtz P.et al.. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021; 39:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P.et al.. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019; 176:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Merrill B.D., Carter M.M., Olm M.R., Dahan D., Tripathi S., Spencer S.P., Yu B., Jain S., Neff N., Jha A.R.et al.. Ultra-deep sequencing of hadza hunter-gatherers recovers vanishing microbes. 2022; biorRxiv doi:31 March 2022, preprint: not peer reviewed 10.1101/2022.03.30.486478. [DOI] [PMC free article] [PubMed]
- 34. Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H.. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics. 2022; btac672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parks D.H., Chuvochina M., Rinke C., Mussig A.J., Chaumeil P.-A., Hugenholtz P.. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022; 50:D785–D794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bland C., Ramsey T.L., Sabree F., Lowe M., Brown K., Kyrpides N.C., Hugenholtz P.. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinf. 2007; 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar R.C. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinf. 2007; 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitrofanov A., Alkhnbashi O.S., Shmakov S.A., Makarova K.S., Koonin E.V., Backofen R.. CRISPRidentify: identification of CRISPR arrays using machine learning approach. Nucleic Acids Res. 2021; 49:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zielezinski A., Deorowicz S., Gudyś A.. PHIST: fast and accurate prediction of prokaryotic hosts from metagenomic viral sequences. Bioinformatics. 2022; 38:1447–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paez-Espino D., Pavlopoulos G.A., Ivanova N.N., Kyrpides N.C.. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat. Protoc. 2017; 12:1673–1682. [DOI] [PubMed] [Google Scholar]
- 41. Camargo A. geNomad database (1.1) [Data set]. Zenodo. 2022; 10.5281/zenodo.7084650. [DOI] [Google Scholar]
- 42. Ponsero A.J., Hurwitz B.L.. The promises and pitfalls of machine learning for detecting viruses in aquatic metagenomes. Front. Microbiol. 2019; 10:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., Davison A.J., Dempsey D.M., Dutilh B.E., García M.L.et al.. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2021). Arch. Virol. 2021; 166:2633–2648. [DOI] [PubMed] [Google Scholar]
- 44. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Sundaramurthi J.C., Lee J., Kandimalla M., Chen I.-M.A., Kyrpides N.C., Reddy T.B.K.. Genomes online database (GOLD) v.8: overview and updates. Nucleic Acids Res. 2021; 49:D723–D733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marbouty M., Baudry L., Cournac A., Koszul R.. Scaffolding bacterial genomes and probing host-virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Sci. Adv. 2017; 3:e1602105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
IMG/VR data is freely available through the online UI (https://img.jgi.doe.gov/vr/) and for download (https://genome.jgi.doe.gov/portal/IMG_VR/IMG_VR.home.html).