Abstract
DoriC was first launched in 2007 as a database of replication origins (oriCs) in bacterial genomes and has since been constantly updated to integrate the latest research progress in this field. The database was subsequently extended to include the oriCs in archaeal genomes as well as those in plasmids. This latest release, DoriC 12.0, includes the oriCs in both draft and complete prokaryotic genomes. At the same time, the number of oriCs in the database has also increased significantly and currently contains over 200 000 bacterial entries distributed in more than 40 phyla. Among them, a large number are from bacteria in new phyla whose oriCs were not explored before. Additionally, new oriC features and improvements have been introduced, especially in the visualization and analysis of oriCs. Currently, DoriC is considered as an important database in the fields of bioinformatics, microbial genomics, and even synthetic biology, providing a valuable resource as well as a comprehensive platform for the research on oriCs. DoriC 12.0 can be accessed at https://tubic.org/doric/ and http://tubic.tju.edu.cn/doric/.
INTRODUCTION
In general, DNA replication is initiated at replication origins, which encode the information instructions of replication initiation and regulation (1,2). The bacterial replication origin (oriC) usually includes DnaA-binding sites, binding motifs of replication regulatory proteins, and an AT-rich DNA unwinding element (DUE). Similarly, the archaeal oriCs consist of a DUE and a number of conserved repeats known as origin recognition boxes (ORBs), which act as binding sites for the origin recognition proteins (3). With the exception of the conserved core oriC functional elements (e.g. DnaA box and DnaA-trios), the functional elements within oriCs are highly diverse, revealing the complexity of the replication initiation and regulation mechanisms. Thus, the identification of oriCs is essential for understanding their structure and functions, which would provide further insights into the regulatory mechanisms of the initiation step in DNA replication. Compared with experimental means, the in silico prediction of oriCs is more efficient and economical, rendering the method especially suitable for dealing with large-scale genome sequences. However, a variety of factors make the prediction of oriCs more complicated than expected, such as atypical GC skews, species-specific DnaA boxes, and loss of the dnaA gene by genome reduction. Consequently, the oriCs in a number of sequenced genomes could not be identified and some are even annotated incorrectly in the genome reports. To address this problem, we had carried out systematic research on oriCs over the last two decades and developed the Ori-Finder system in 2008 using the Z-curve theory and comparative genomics method (4,5). At the same time, we also launched DoriC, a database of oriCs in prokaryotic genomes based on the prediction of the Ori-Finder system. Initially, the DoriC database only contained predicted or experimentally confirmed oriCs in hundreds of bacterial chromosomes (6). Subsequently, DoriC 5.0 provided more than 1000 oriCs in both bacterial and archaeal chromosomes (7), and DoriC 10.0 included over 10 000 oriCs of chromosomes and plasmids in prokaryotic genomes (8). In addition to the information on oriCs, graphical views of GC, AT, RY and MK disparity curves and relevant information about the species or genome (e.g. organism, lineage, chromosome topology, dnaA gene position) have also been supplied to users. Moreover, the DoriC database provides the basic search and BLAST functions. Currently, the DoriC database is considered as an important database in the fields of bioinformatics, microbial genomics, and even synthetic biology, providing a valuable resource for relevant research on oriCs (9–11). The DoriC database has facilitated research on oriC functional elements (12), the replication mechanism (13), strand-biased analysis (14,15) and large-scale oriC data mining (16). Additionally, the database is also used in research involving metagenomics, such as the study of bacterial growth dynamics (17–19) and oriC prediction (20).
With the rapid development of genome sequencing technology and the continuous reduction in sequencing costs, increasingly more prokaryotic genomes are being sequenced. However, as of 9 April 2022, complete genomes only accounted for ∼8% of all available bacterial genomes deposited in the National Center for Bioinformatics Information (NCBI) databases. To make full use of these genome data and facilitate user access to more comprehensive information about oriCs, DoriC has been updated to the new version, DoriC 12.0, which presents with updates in oriC numbers as well as contents and functions.
DATABASE UPDATES
Significant increase in oriC data
Complete and draft genomes together with their available annotation files were downloaded from the NCBI RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/) on 9 April 2022 (21,22). First, the oriCs in bacterial genomes were predicted using Ori-Finder 2022 (https://tubic.org/Ori-Finder2022/) (23), an updated version of Ori-Finder, which adopts a new scoring system to consider the currently available oriC characteristics comprehensively and shows better prediction performance compared with the original version. Then, the predicted oriCs were added to the DoriC database after manual curation. Consequently, the DoriC database has been substantially expanded by including the oriCs predicted by Ori-Finder 2022. Now, the bacterial chromosomes included in DoriC 12.0 have increased from 7580 to 205,798 distributed in more than 40 phyla.
Updated functions
New search functions
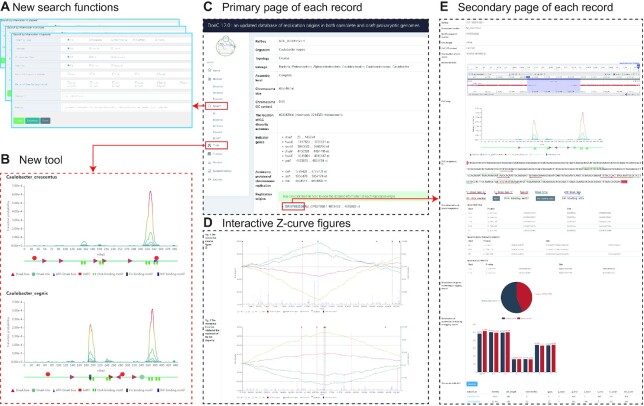
DoriC 10.0 supported the search of RefSeq ID, DoriC ID, organism, lineage and other fields. The extended search functions in DoriC 12.0 make it convenient for users to search and use the database more efficiently. The search interfaces are designed according to the different characteristics of the bacterial, archaeal and plasmid data (Figure 1A), and the search results can be downloaded directly. For example, the search fields provided by the search interface of bacterial data include genome assembly level, chromosome topology, chromosome type, oriC type, regulatory protein, and gene flanking the oriC.
Figure 1.
Overview of the DoriC 12.0 interface. (A) New search functions. Different search interfaces are designed for search of bacterial, archaeal and plasmid data. (B) New tool for comparing the distribution of functional elements in the oriCs. The oriC maps of Caulobactor crescentus and Caulobactor segnis generated by the new tool have been presented here as an example. (C) Primary page of each record. The primary page presents basic information about the genome, genes encoding the regulatory proteins of chromosome replication, and interactive Z-curve figures. (D) Interactive Z-curve figures. (E) Secondary page of each record. The secondary page presents information of each oriC, such as characteristic visualization of oriC sequence, repeat sequences discovered by MEME, strand-biased analysis and homologous oriCs searched by BLAST.
New tool
Although the lengths and nucleotide sequences of different oriCs vary considerably, bacterial oriCs share similar functional elements (24). Therefore, DoriC 12.0 provides a tool for comparative analysis of the distribution of functional elements in bacterial oriCs. Users can enter a comma-separated DoriC ID list, extract the oriC maps recorded in DoriC 12.0 for comparison, and thereby determine if there are any similarities or differences. Additionally, users can also upload one or multiple sequences in a FASTA file to generate the corresponding oriC maps. For example, although the lengths and sequences of the oriCs of Caulobactor crescentus and Caulobactor segnis are not completely consistent, the number and relative position of CtrA-binding motifs seem to be conserved (Figure 1B) (25). These oriC maps may also be helpful in the analysis of oriC sequences acquired by users through experiments, metagenomic data analysis, or other methods.
Updated contents
Redesigned web page
To enhance user experience, the web page has been redesigned to be more intuitive. The home page displays statistical information (e.g. the number of bacterial chromosomes at different assembly levels) through which users can gain a general understanding of the DoriC database. Each record in DoriC 12.0 usually contains two page: the primary page (Figure 1C) showing information about the genome and the secondary page (Figure 1E) showing information about each oriC.
Interactive Z-curve figure
The Z-curve figure shows the four disparity curves, the distribution of DnaA boxes, indicator genes, potential oriCs and replication terminus, making it convenient for users to observe the relative positions of these features (Figure 1D). The redesigned Z-curve figure is interactive, allowing users to zoom into the figure to view more details on the changes in the base composition, select all or only a few sets of data for analysis, and hover over a particular site to see the details of indicator genes, potential oriCs, and replication terminus.
Characteristic visualization of oriC sequences
The characteristic visualization of oriC sequences (‘oriC map’ and ‘oriC sequences’ in Figure 1E) is designed to help users observe the distribution of functional elements within the oriCs and to facilitate exploration of the binding mechanism of the initiator proteins and regulatory proteins. At present, three main types of bacterial functional elements can be visualized in DoriC 12.0: (i) binding motifs of initiator protein DnaA, such as DnaA boxes, DnaA-trios, and the ATP-DnaA boxes (26); (ii) binding motifs of the replication regulatory proteins SeqA, CtrA, Fis and IHF (27–30); and (iii) AT-rich sequences that might serve as DUEs (31). To facilitate potential new discoveries, all possible functional elements have been searched for within the oriCs recorded in DoriC 12.0. However, it should be noted that the predicted binding motifs may not always be functional as real binding sites, since many of these are restricted to specific lineages (e.g. Fis, IHF and CtrA binding sites are usually found in a subset of Proteobacteria) and are not present throughout the bacterial domain (same GATC methylation sites). The default display of these sites is also set according to the lineage of species. Additionally, the characteristic visualization of oriC sequences is interactive, and users can select functional elements for observation by clicking corresponding buttons. For the archaeal oriCs, the annotated functional elements are mainly ORBs and AT-rich sequences.
More information on each record
More information has been added to each record to assist users in analyzing the oriC comprehensively. A list of genes encoding the regulatory proteins of chromosome replication is provided. Combined with the different binding motifs annotated in the ‘characteristic visualization of oriC sequences’, some new insights related to regulatory mechanisms may be gained. The repeat sequences discovered by the Multiple EM for Motif Elicitation (MEME) tool in predicted oriCs may reveal new functional elements (32). The differences in gene distributions and base compositions between the leading and lagging strands are revealed by the strand-biased analysis. Additionally, the homology search results of each oriC are provided in this release, making comparative analyses of the oriC sequences convenient for users (33).
CONCLUDING REMARKS
The recent development of high-throughput sequencing technology has resulted in a rapidly growing number of prokaryotic genome sequences, especially of bacterial genomes. However, a large number of the oriCs among these genomes remain unknown, necessitating the development of bioinformatics algorithms to address this issue. Based on the prediction results of Ori-Finder 2022, over 200 000 bacterial oriCs have been predicted reliably. This latest release of the DoriC database provides a substantial number of novel oriCs of the bacteria belonging to the new phyla, which have not been discovered before. This will help researchers to further reveal the conserved and diverse features of oriCs, such as their DnaA boxes or other functional elements, which will comprehensively enrich our knowledge about these sequences. Artificial intelligence, especially deep learning, has changed the paradigm of scientific research profoundly. However, it requires massive amounts of training data, something which DoriC 12.0 can provide, such as high-quality oriC data that can be used for data mining via deep learning. New discoveries based on these oriCs are expected, leading to a better understanding of the DNA replication mechanisms. In the future, we will carry out further systematic studies of different types of oriCs to extract their respective features and provide more relevant services for their analysis, making the DoriC database a universal knowledge base on oriCs. The elucidation and application of principles of oriCs based on over 200 000 oriC regions in prokaryotic genomes will contribute to the development of oriC prediction algorithms as well as experimental confirmation and functional analysis of oriCs in prokaryotic genomes.
DATA AVAILABILITY
DoriC 12.0 is freely available to the public without registration or login requirements (https://tubic.org/doric/ and http://tubic.tju.edu.cn/doric/).
ACKNOWLEDGEMENTS
We thank Professor Chun-Ting Zhang for the invaluable assistance and inspiring discussions.
Contributor Information
Mei-Jing Dong, Department of Physics, School of Science, Tianjin University, Tianjin 300072, China.
Hao Luo, Department of Physics, School of Science, Tianjin University, Tianjin 300072, China.
Feng Gao, Department of Physics, School of Science, Tianjin University, Tianjin 300072, China; Frontiers Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin 300072, China; SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300072, China.
FUNDING
National Key Research and Development Program of China [2018YFA0903700 to F.G.]; National Natural Science Foundation of China [21621004 to F.G., 31801104 to H.L.]. Funding for open access charge: National Key Research and Development Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hill N.S., Kadoya R., Chattoraj D.K., Levin P.A.. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 2012; 8:e1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolanski M., Donczew R., Zawilak-Pawlik A., Zakrzewska-Czerwinska J.. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2015; 5:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ekundayo B., Bleichert F.. Origins of DNA replication. PLoS Genet. 2019; 15:e1008320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao F., Zhang C.T.. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinf. 2008; 9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo H., Zhang C.T., Gao F.. Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front. Microbiol. 2014; 5:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao F., Zhang C.T.. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007; 23:1866–1867. [DOI] [PubMed] [Google Scholar]
- 7. Gao F., Luo H., Zhang C.T.. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 2013; 41:D90–D93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo H., Gao F.. DoriC 10.0: an updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucleic Acids Res. 2019; 47:D74–D77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pei L., Schmidt M., Wei W.. Synthetic biology: an emerging research field in china. Biotechnol. Adv. 2011; 29:804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhulin I.B. Databases for microbiologists. J. Bacteriol. 2015; 197:2458–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lioy V.S., Junier I., Boccard F.. Multiscale dynamic structuring of bacterial chromosomes. Annu. Rev. Microbiol. 2021; 75:541–561. [DOI] [PubMed] [Google Scholar]
- 12. Pelliciari S., Dong M.J., Gao F., Murray H.. Evidence for a chromosome origin unwinding system broadly conserved in bacteria. Nucleic Acids Res. 2021; 49:7525–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sankar T.S., Wastuwidyaningtyas B.D., Dong Y., Lewis S.A., Wang J.D.. The nature of mutations induced by replication-transcription collisions. Nature. 2016; 535:178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sobetzko P., Travers A., Muskhelishvili G.. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sc. U.S.A. 2012; 109:E42–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen W.H., Lu G., Bork P., Hu S., Lercher M.J.. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat. Commun. 2016; 7:11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo H., Quan C.L., Peng C., Gao F.. Recent development of Ori-Finder system and DoriC database for microbial replication origins. Brief. Bioinf. 2019; 20:1114–1124. [DOI] [PubMed] [Google Scholar]
- 17. Gao Y., Li H.. Quantifying and comparing bacterial growth dynamics in multiple metagenomic samples. Nat. Methods. 2018; 15:1041–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joseph T.A., Chlenski P., Litman A., Korem T., Pe’er I.. Accurate and robust inference of microbial growth dynamics from metagenomic sequencing reveals personalized growth rates. Genome Res. 2022; 32:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korem T., Zeevi D., Suez J., Weinberger A., Avnit-Sagi T., Pompan-Lotan M., Matot E., Jona G., Harmelin A., Cohen N.et al.. Microbiome growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015; 349:1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ionescu D., Bizic-Ionescu M., De Maio N., Cypionka H., Grossart H.-P.. Community-like genome in single cells of the sulfur bacterium Achromatium oxaliferum. Nat. Commun. 2017; 8:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022; 50:D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W., O’Neill K.R., Haft D.H., DiCuccio M., Chetvernin V., Badretdin A., Coulouris G., Chitsaz F., Derbyshire MyraK., Durkin A.S.et al.. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2020; 49:D1020–D1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dong M.J., Luo H., Gao F.. Ori-Finder 2022: a comprehensive web server for prediction and analysis of bacterial replication origins. Genomics, Proteomics Bioinf. 2022; 10.1101/2022.08.09.503306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leonard A.C., Méchali M.. DNA replication origins. Cold Spring Harbor Perspect. Biol. 2013; 5:a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marczynski G.T., Shapiro L.. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 2002; 56:625–656. [DOI] [PubMed] [Google Scholar]
- 26. Speck C., Messer W.. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001; 20:1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chung Y.S., Brendler T., Austin S., Guarne A.. Structural insights into the cooperative binding of SeqA to a tandem GATC repeat. Nucleic Acids Res. 2009; 37:3143–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brassinga A.K.C., Siam R., McSween W., Winkler H., Wood D., Marczynski G.T.. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J. Bacteriol. 2002; 184:5789–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shao Y., Feldman-Cohen L.S., Osuna R.. Functional characterization of the Escherichia coli Fis–DNA binding sequence. J. Mol. Biol. 2008; 376:771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hales L.M., Gumport R.I., Gardner J.F.. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J. Bacteriol. 1994; 176:2999–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhabinskaya D., Madden S., Benham C.J.. SIST: stress-induced structural transitions in superhelical DNA. Bioinformatics. 2014; 31:421–422. [DOI] [PubMed] [Google Scholar]
- 32. Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME suite. Nucleic Acids Res. 2015; 43:W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y.et al.. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DoriC 12.0 is freely available to the public without registration or login requirements (https://tubic.org/doric/ and http://tubic.tju.edu.cn/doric/).