Abstract
Introduction:
Rhesus macaques are natural hosts to multiple viruses including rhesus cytomegalovirus (RhCMV), rhesus rhadinovirus (RRV), and Simian Foamy Virus (SFV). While viral infections are ubiquitous, viral transmissions to uninfected animals are incompletely defined. Management procedures of macaque colonies include cohorts that are Specific Pathogen Free (SPF). Greater understanding of viral transmission would augment SPF protocols. Moreover, vaccine/challenge studies of human viruses would be enhanced by leveraging transmission of macaque viruses to recapitulate expected challenges of human vaccine trials.
Materials and methods:
This study characterizes viral transmissions to uninfected animals following inadvertent introduction of RhCMV/RRV/SFV-infected adults to a cohort of uninfected juveniles. Following co-housing with virus-positive adults, juveniles were serially evaluated for viral infection.
Results:
Horizontal viral transmission was rapid and absolute, reaching 100% penetrance between 19 – 78 weeks.
Conclusions:
This study provides insights into viral natural histories with implications for colony management and modeling vaccine-mediated immune protection studies.
Keywords: Cytomegalovirus, Herpesvirus, Spumavirus, Nonhuman primates, Macaque
INTRODUCTION
Nonhuman primates (NHP) are invaluable for evaluating conceptual, treatment, and prevention strategies for many aspects of human development and disease.(1–3) As NHP can be hosts for multiple endemic microbial agents, breeding cohorts of animals that are Specific Pathogen Free (SPF) for particular viruses that can act as confounding elements in disease pathogenesis under certain physiological conditions have been developed.(4–12) In particular, SPF Level 1 (SPF1) animals are confirmed to be uninfected with Herpes B virus, Simian Immunodeficiency Virus (SIV), the Type D Simian Retrovirus (SRV), and Simian T-lymphotropic Virus (STLV), whereas SPF2 animals are SPF1 animals that are also uninfected with SFV, RhCMV, and RRV. Establishing and maintaining SPF colonies requires the capability to accurately identify infected animals, both in primary screening and in follow-up surveillance, in addition to a management system that prevents contact (direct or indirect) between SPF animals and infected or untested animals (barrier maintenance). Since their initiation by the National Center for Research Resources (NCRR) in 1989, SPF macaque colonies have successfully improved animal health and reproduction by elimination of potential animal pathogens, improved the quality of NHP used in biomedical research by providing animals free of potentially confounding intercurrent infections (13), and reduced potential sources of human occupational exposure to selected NHP viruses. The initial SPF program at the California National Primate Research Center (CNPRC) focused on development of a sustainable breeding population of SPF1 animals. Following the successful establishment and maintenance of an SPF1 population, subsequent development of SPF2 was initiated to further restrict the repertoire of commensal viruses in NHP.(9)
As a standard part of the derivation process to generate SPF populations, adults are routinely introduced to groups of juveniles to increase socialization and breeding competence.(4, 14) The inadvertent but brief introduction of three SPF1 adult rhesus macaques to 31 SPF2 juveniles, all co-housed within a self-contained, corn crib housing unit presented an opportunity to collect prospective samples from the juveniles to monitor transmission and dissemination of SFV, RhCMV, and RRV within this group of naïve (uninfected) juveniles. Observations of the natural history and analysis of the patterns of spread by mucosal transmission of these viruses in NHP provide valuable insights for colony management and for their use as animal models for vaccine and therapeutic research.
Nonhuman primates (NHP) are natural hosts to a variety of exogenous retroviruses and herpes viruses including SPF, RRV, and RhCMV. SFV is a member of the Spumavirus genus of retroviruses and is an adventitious viral outgrowth of cultured cells exposed to bodily fluids of NHP. SFV elicits a notable syncytial cytopathic effect (CPE) in infected cell cultures resulting in a pathognomonic foamy-like appearance within infected cells.(15) A wide range of cell types from a variety of species are susceptible to both in vivo and in vitro infection with SFV. In some cell types, such as human fibroblasts, SFV produces a highly lytic infection, whereas in other cell types such as transformed lymphoid, myeloid, and erythroid cells, SFV can sustain a chronic infection producing virus particles in the absence of significant cell death. A phenotype of SFV infection in vivo, in both acute and chronic infections is the absence of any association with clinical outcomes, even in immunodeficient animals coinfected with SIV or SRV.(15) Horizontal transmission is thought to occur via exchange of salivary fluid.(16–18)
RRV is a member of the RV2 subgroup of gamma-2-herpesviruses, which naturally infect rhesus macaques and other NHP species.(19–23) Since it is closely related to the RV1 subgroup Kaposi sarcoma herpesvirus in humans and Retroperitoneal Fibromatosis Herpesvirus in macaques, RRV has been investigated for mechanisms of pathogenesis as well as use as a vaccine vector for heterologous antigens.(24–26) RRV manifests the characteristic ability of herpesviruses to establish lifelong persistence in immune competent hosts.(27) Infections are usually asymptomatic in immunocompetent natural hosts, but associations with lymphoproliferative disease resembling multicentric Castlemans disease, non-Hodgkins lymphoma, and retroperitoneal fibromatosis in immune compromised SIV-infected hosts have been reported.(28–30) There are no published prevalence data for RRV in wild monkeys, and reports from breeding colonies indicate endemic prevalence rates of 6 to 75%.(31) RRV appears to be readily transmissible with infection occurring at an early age, likely through virus shed in saliva.(27, 32)
RhCMV is a member of the Betaherpesvidae subfamily of herpesviruses, and it is an endemic component of the virome in both breeding cohorts and wild populations.(33–35) Seroprevalence rates approach 100% by one year of age in animals maintained within their natal group.(33) Viral DNA can be detected (via qPCR) in multiple bodily fluids for decades after primary infection (33, 36–40), including in saliva, urine, and breast milk of lactating dams. As with SFV and RRV, the social propensity of NHP in intermixing breeding cohorts and the frequent asymptomatic shedding of virus in bodily fluids enables multiple exposures between infected and uninfected animals and horizontal transmission to other animals. Since the breeding age of rhesus macaques is 4 – 5 years, primary, non-iatrogenic infection with RhCMV during pregnancy is likely to be exceedingly rare in non-SPF animals, and there have been no reports of naturally occurring congenital RhCMV infection and/or sequelae consistent with intrauterine RhCMV exposure.(41) Leveraging the development of SPF2 breeding-age animals, a model of transplacental transmission of RhCMV has been developed following primary exposure to RhCMV early in the second trimester.(42) While RhCMV establishes a lifelong asymptomatic infection in immune competent hosts, RhCMV is a significant cause of morbidity and mortality in immunodeficient animals coinfected with either SRV (43) or SIV (44), and in immunologically immature fetal macaques of seropositive dams following direct fetal inoculation.(45) Studies of the biology, viral mechanisms of persistence, and pathogenesis of RhCMV have provided insights to better understanding of and prevention/treatment modalities for human CMV.(46–51)
RESULTS
This study was prompted by the inadvertent co-housing of three breeding age rhesus macaques (5 years, unknown at the time to be infected with RhCMV, RRV, and SFV) for 3 – 24 days with 31 uninfected juvenile macaques (9 – 16 months) that had been repeatedly confirmed to be uninfected with all three endemic viruses (SPF2) (Fig. 1A). The juveniles had been raised since birth apart from animals known to be infected with viruses defining SPF1 (Herpes B, SIV, SRV, STLV) and SPF2 (SPF1 plus RhCMV, RRV, SFV) status. As a normal process for expanding SPF cohorts, juveniles reared with other juveniles are cohoused with adult animals for socialization purposes.
Figure 1.
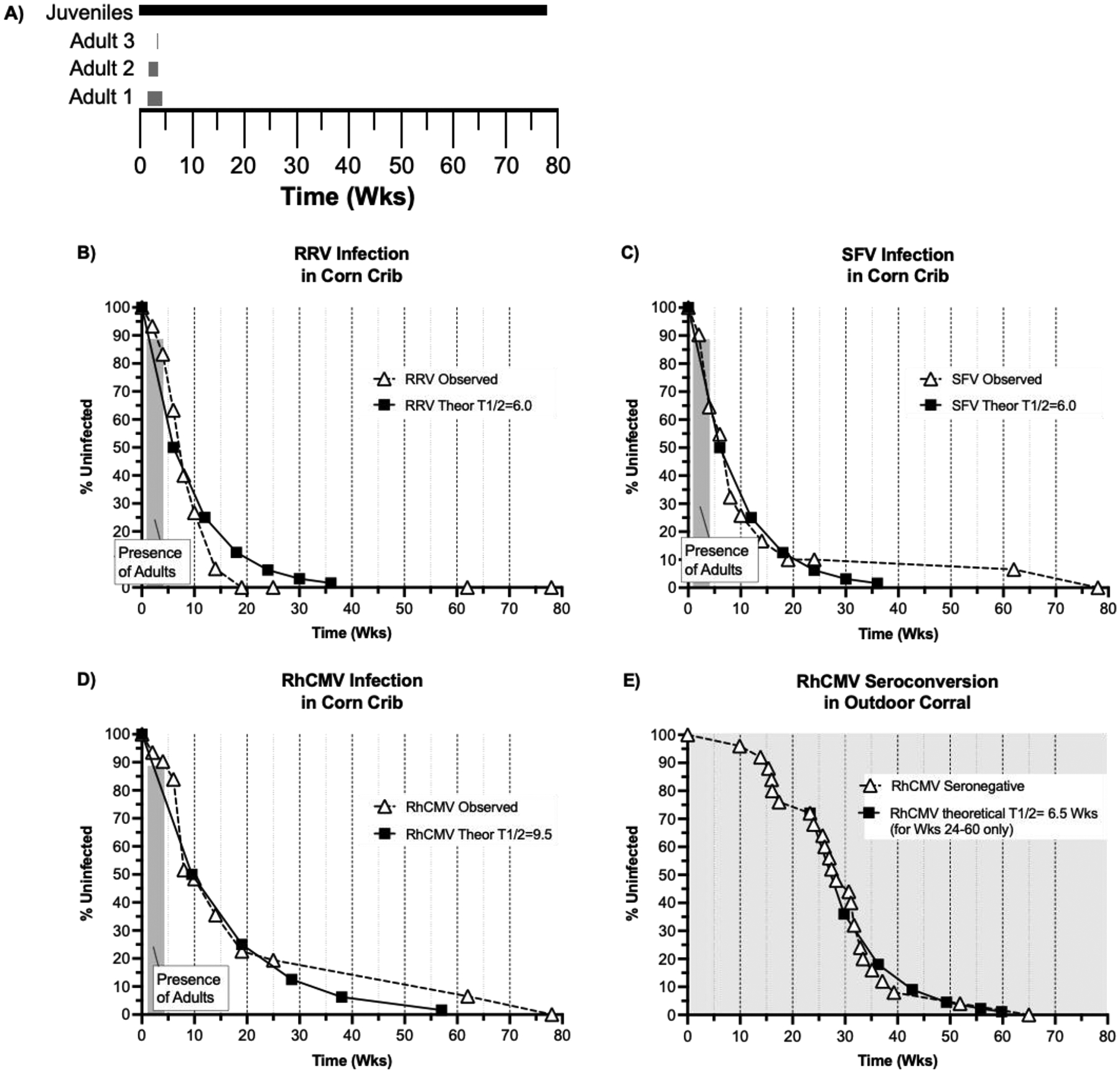
(A) Co-housing timeline (X-axis in Weeks-Wks) for the SPF2 juveniles (black box) and the 3 adults infected with RRV, SFV, and RhCMV (gray boxes). Infection rates for (B) RRV, (C) SFV, and (D) RhCMV in corn crib housing (dashed lines and triangles). The graphs plot the % uninfected frequency (Y-axis) versus time (in weeks, X-axis). The shaded box indicates the period of co-housing with virus-infected adults (Shown in part A). Theoretical infection curves (T1/2 = half-life of uninfected status) for each virus are plotted (solid line and squares). (E) The seroconversion rate for neonates born within an outdoor breeding corral are presented (adapted from a previous publication (33)). The T1/2 curve is only plotted from Wk 24 to Wk 60 (post weaning only). The shading indicates that neonates were continuously co-housed with RhCMV-infected animals after birth.
In this study early during SPF2 colony development, 31 juveniles that had been housed in pairs since birth and repeatedly confirmed to be SPF2 were group-housed on Day 0 in an outdoor housing structure (corn crib). Per normal protocols for socializing juvenile macaques to adulthood (4, 14), two adults were introduced into the corn crib: 1 female on Day 9 and 1 male on Day 10 (Fig. 1A). At the time of introduction into the corn crib of SPF2 juveniles, both adults were thought to be SPF2. The adult male was removed on Day 23 due to social incompatibility and replaced with a second male, which was co-housed in the corn crib from Days 23 – 26, at which time it was removed also because of social incompatibility, leaving the adult female as the sole adult in the corn crib. During the time of co-housing, all animals were free to interact, enabling frequent exposure to bodily fluids either directly (saliva) or indirectly via fomite, such as partially eaten food pellets. The co-housing of adults and juveniles was terminated on Day +33 when it was recognized that the adults had never been evaluated for RhCMV, RRV, and SPF infection status, and the remaining adult female was removed. The three adults were subsequently confirmed to be infected with all three viruses (data not shown). Blood had been drawn from the juveniles on Day 14, and prospective blood and saliva samples were drawn upon recognition of the potential introduction of RRV, RhCMV, and SPF into the juvenile cohort. Longitudinal samples from the juveniles (blood, saliva) were analyzed for immunological (development of IgG antibodies – RhCMV, RRV, SFV) and/or molecular (virus-specific PCR – RRV, SFV) to detect for evidence of horizontal transmission from the adults to the juveniles. The results are presented in Tables 1A – 1C and Figures 1B – 1D.
TABLE 1A:
Detection of RRV in Juvenile Macaques
| Weeks post Exposure |
RRV Ab Positive % (# positive/# susceptible) |
RRV DNA Positive (PBMC) # Pos/# tested † | Total Infected (%) ¥ (# positive/# susceptible) |
|---|---|---|---|
| 2 | 0 (0/31) | 2/29 | 6.5 |
| 4 | 12.9 (4/31) | 3/28 | 16.7 |
| 6 | 16.1 (5/31) | 9/26 | 36.7 |
| 8 | 35.5 (11/31) | 17/30 | 60.0 |
| 10 | 48.4 (15/31) | 19/26 | 73.3 |
| 14* | 60.0 (18/30)* | 28/30* | 93.3* |
| 19* | 76.7 (23/30)* | 26/29* | 100* |
| 25* | 80.0 (24/30)* | 24/26* | 100* |
| 62* | 96.7 (29/30)* | 9/21* | 100* |
| 78* | 96.7 (29/30)* | 13/27* | 100* |
One juvenile was lost to the study between weeks 10 and 14, prior to infection with RRV and post infection with RhCMV and SFV. The infection rate for RRV after week 10 reflects that there were 30 juveniles in toto susceptible to RRV infection.
A juvenile was considered infected with RRV if either antibody and/or DNA was detected.
Not all PBMC samples available for RRV PCR.
TABLE 1C:
Detection of RhCMV in Juvenile Macaques
| Weeks post Exposure | RhCMV Ab Positive (%) (# positive/# susceptible) |
|---|---|
| 2 | 6.5 (2/31) |
| 4 | 9.7 (3/31) |
| 6 | 16.1 (5/31) |
| 8 | 48.4 (15/31) |
| 10 | 51.6 (17/31) |
| 14 | 64.5 (20/31) |
| 19 | 77.4 (24/31) |
| 25 | 80.6 (25/31) |
| 62 | 93.5 (29/31) |
| 78 | 100 (31/31) |
Analyses of immune and molecular parameters of viral infection demonstrated that horizontal transmission of each virus was rapid, occurring within 2 weeks of the introduction of virus-infected adults into a cohort of uninfected juvenile monkeys. De novo development of RhCMV-specific IgG was detected in 2 juveniles at 2-weeks, and viral nucleic acid for RRV and SFV was detected in 2 and 3 juveniles, respectively. Infection was progressive and eventually absolute. The rates of primary infection were similar between the three viruses, but distinctions were noted. To estimate the rate of primary infection, a theoretical half-life (T1/2) for remaining uninfected was plotted for each virus (Fig. 1B–1D). For both RRV and SFV, a T1/2 = 6 weeks for both viruses tracked the observed infection rate for the first 10 and 25 weeks, respectively, under this co-housing arrangement. For RRV after 10 weeks (Fig. 1B), the observed infection rate continued a linear decline such that 100% penetrance of infection was observed at 19 weeks (Table 1A and Fig. 1B). For SFV after 25 weeks, infection of the remaining 4 animals was slower than the initial T1/2 of 6 weeks, and 100% infection was not observed until 78 weeks (Table 1B and Fig. 1C). For RhCMV, an initial T1/2 = 9 weeks tracked closely with the observed infection rate through 25 weeks post introduction of the virus-infected adults (Fig. 1D). Thereafter, the infection of the remaining 7 animals was slower than the initial T1/2 of 9 weeks, and 100% infection was not observed until 78 weeks (Table 1C and Fig. 1D). The last 7 animals to become infected with RhCMV were distinct from the last 4 animals to become infected with SFV. It is worth noting that, since the last virus-infected adult was removed from the cohort by Day 33, progressive primary infections thereafter resulted from juvenile-to-juvenile horizontal transmission versus the initial adult-to-juvenile the transmission.
TABLE 1B:
Detection of SFV in Juvenile Macaques
| Weeks post Exposure |
SFV Ab Positive % (# positive/# susceptible) |
SFV DNA (PBMC) or RNA # Pos/# tested † | Total Infected (%) ¥ (# positive/# susceptible) |
|---|---|---|---|
| 2 | 0 (0/31) | 3/31 | 3/31 (9.7) |
| 4 | 16.1 (5/31) | 11/31 | 11/31 (35.5) |
| 6 | 38.7 (12/31) | 12/31 | 14/31 (45.2) |
| 8 | 58.1 (18/31) | 19/31 | 21/31 (71.0) |
| 10 | 64.5 (20/31) | 19/31 | 23/31 (74.2) |
| 14 | 77.4 (24/31) | 21/30 | 25/31 (83.3) |
| 19 | 83.9 (26/31) | 23/30 | 27/31 (90.0) |
| 25 | 83.9 (26/31) | 23/30 | 27/31 (90.0) |
| 62 | 93.5 (29/31) | 11/27 | 29/31 (93.5) |
| 78 | 100 (31/31) | 15/23 | 31/31 (100) |
A juvenile was considered infected with SFV if either antibody and/or nucleic acid was detected.
Not all PBMC/saliva samples available for SFV PCR.
A previous study quantified seroconversion to RhCMV in 25 neonates born into an outdoor breeding corral housing 142 animals (0 – 14 years) in which 131 of the corral-housed animals were RhCMV seropositive.(33) Differences in this previous study of horizontal transmission in a breeding corral and the corn crib study described herein include differences in the (1) the number of susceptible animals (25 versus 31, respectively), (2) the number of co-housed animals potentially secreting RhCMV in bodily fluids (131 versus 3, respectively), and (3) the length of time of susceptibility to mucosal exposure to infectious virus from sentinel, virus-positive cohorts (455 versus 33 days, respectively). There were several prominent distinctions between the rates of primary infection in these two cohorts of animals (Fig. 1E).
For neonates born into a breeding corral, the observed rate of RhCMV seroconversion appeared to be bi-phasic with the two phases occurring corresponding to 0 – 17.5 weeks and 23 – 65 weeks (Fig. 1E). Secondly, the observed rate of seroconversion in the second phase closely tracks a theoretical T1/2 = 6.5 weeks, faster than the theoretical T1/2 (9 weeks) in the corn crib (Fig. 1D). Lastly, the time of 100% penetrance of RhCMV infection occurred sooner in the breeding corral compared to the corn crib (65 versus 78 weeks, respectively).
The apparent two phases of transmission (Fig. 1E) are coincident with time of weaning of neonates in rhesus macaques, which generally occurs between 4 – 7.5 months of age (~17 – 32 weeks).(52, 53) Based on this developmental landmark in mother/infant bonding in macaques, the first 7 seroconversion events in the neonates within the breeding corral most likely represented maternal-to-infant transmission events. Thereafter in the second phase, when infants are increasingly separated from their dams and interacting with other animals, seroconversion probably represented transmission from co-housed animals. The apparent increased rate of theoretical transmission in the second phase of transmission within the breeding corral (Fig. 1E), compared to the theoretical rate of transmission in the corn crib (Fig. 1D), suggests that the force of infection of RhCMV is a function of the number of close interactions of uninfected animals with virus-shedders.
Discussion
The key take-home lesson for colony management of macaques is that the introduction of a single non-SPF animal into an SPF cohort will rapidly end virus-free status due to rapid horizontal transmission to and primary infections of non-infected animals. Put another way, once a single animal is confirmed as positive for viral infection, the cohort can be considered as compromised for SPF status, as far as these three viruses are concerned. Since macaques can have detectable viral RRV or SFV nucleic acid in saliva and/or PBMC prior to development of virus-specific IgG (Table 1), rigorous screening by methodologies beyond RRV and SPF seroconversion should be implemented whenever there is uncertainty about the potential for viral infection. All three viruses are exceedingly efficient at spreading with complete penetrance in the population, mimicking natural dissemination of HCMV and non-human CMVs in their natural (non-breeding facilities) host species including humans (54), rhesus macaques (35, 55), and mice.(56–61) As members of the Herpesvirinae family of viruses, 400 million years of evolution (62) has enabled herpesviruses like RRV and RhCMV to be exceedingly fit for infection throughout their host species. Whereas Foamy viruses appear to be widespread in NHP, prevalence in humans of Human Foamy Virus is quite limited.(63, 64) Beyond colony management concerns, these results shed important light on RhCMV natural history in particular. In combination with other studies, greater understanding of horizontal transmission of RhCMV by non-iatrogenic interventions can be leveraged to recapitulate the challenges facing human HCMV vaccine trials. In particular, quantifying the protective efficacy of HCMV vaccines can be confounded by differential repeated mucosal exposures to varying titers of challenge virus that may be antigenically divergent from the epitope specificities of the vaccine antigen(s).(65)
One underlying precept of optimal vaccine design is that the vaccine approach should account for salient aspects of the natural history of the targeted pathogen, such as rates of shedding and transmission, titers of the pathogen relevant for transmission, frequency and length of exposure of the vaccinee to infectious pathogen, frequency and magnitude of shedding of the pathogen by an infected vaccinee, and resistance/susceptibility to reinfection following primary infection. While the former statement is applicable to any pathogen, studies of RhCMV infection in rhesus macaques serve as precedent for development of HCMV vaccines by factoring in salient aspects of RhCMV natural history.
The mechanistic basis for horizontal transmission in this study highlights the role of shedding in the natural history of RhCMV in particular, and by extension the natural history of HCMV. Once the last adult was removed from the cohort of juveniles, subsequent infection of the remaining juveniles occurred by shedding of virus from juveniles and subsequent horizontal transmission to mucosal surfaces of uninfected animals. Based on prior studies, it is likely that most infections resulted in prolonged shedding in, at least, saliva and urine, and subsequent horizontal transmission. Cross-sectional studies of the saliva of naturally infected animals demonstrated that ~80% of infected animals persistently secrete high copy numbers of viral genomes (104 – 106 genomes/ml of saliva) for years after primary infection, long past the time when 100% seroprevalence is achieved in the population (1.5 years).(38, 39) It should be noted that the 100% penetrance of RhCMV is not an artifact of breeding facilities. In rhesus macaque groups living in their natural settings (close proximity to humans), RhCMV infection within a “wild” population strongly mirrors the seroprevalence in macaque breeding facilities.(35) Moreover, complete dissemination of RhCMV within a cohort, as well as with SFV and RRV, occurs long before the animals reach sexual maturity (4–5 years). A viral phenotype of persistent RhCMV shedding beyond the requisite time necessary to achieve infection in all cohorts at-risk for primary infection begs the following question. What selective advantage is conferred to the virus by prolonged virus shedding by immune animals within a fully immune population? The clinical ramification of prolonged shedding is that an HCMV-infected individual shedding virus in bodily fluids, particularly young children, represents an infectious threat to those at-risk for primary and non-primary HCMV infection, notably pregnant women.(65) Recent studies involving juvenile macaques vaccinated against RhCMV antigens and then exposed to virus by a mucosal route of challenge offers clues.
Juvenile macaques vaccinated against RhCMV glycoprotein B (gB), phosphoprotein 65, and the gH-anchored pentameric complex (PC) essential for epithelial/endothelial cell tropism develop levels of antigen-specific immune responses comparable in levels to those detected in naturally infected animals.(66) However, the vaccinees exhibited no protection against viral acquisition, viremia, or shedding in saliva and urine compared to placebo controls following an oral mucosa challenge model designed to recapitulate natural infection. Notably, the absence of vaccine-mediated protection was observed with a homologous RhCMV challenge using a strain of RhCMV (UCD52) from which the vaccine antigens were designed. The homologous vaccine/challenge method was used because another study demonstrated that plasma from monkeys inoculated with either of two RhCMV strains exhibiting genetic drift in the coding regions for gB and some of those forming the PC (UCD52 and UCD56) do not cross-neutralize the other non-homologous strain.(67) The simplest interpretation of the absence of protective efficacy in the vaccine/oral challenge results in this study (66) is that despite high levels of antiviral immunity in the periphery circulating antiviral immunity is not operative at the oral mucosa, the site of RhCMV challenge. This putative scenario leads directly to two intertwined hypothetical scenarios. (1) The apparent absence of protective immunity at the mucosal surface is mediated by specific viral functions that suppress recall and/or signaling of the presence of infected cells expressing viral antigens to memory effector functions. (2) Viral functions that suppress circulating immunity to the site of mucosal entry of virus also suppress the trafficking of peripheral antiviral immunity to mucosal sites of persistent viral shedding. Studies of HCMV infection in children and adults led to the hypothesis that development of HCMV-specific CD8 T cells was primarily an immune-mediated defense against systemic disease, as opposed to immune-mediated restriction against local viral replication at mucosal surfaces.(68)
Taken together, the results of these studies strongly suggest that the prolonged period of shedding of virus in bodily fluids for years presumably confers a selective advantage to the virus by enabling reinfection of immune cohorts and further horizontal spread of the virus.(69) If so, then the concept of ‘herd immunity’ through natural spread of the virus has no relevance for RhCMV. In addition, calculation of the basic reproductive number (R0) has no relevance for RhCMV, and probably HCMV, because it is virtually impossible to calculate the number of transmission events, both primary and non-primary, from an infected host shedding virus for years.
Because of prolonged shedding beyond the 1.5 years necessary to achieve 100% seroprevalence, sexually mature female macaques (≥4–5 years) would undoubtedly be repeatedly exposed to RhCMV. There are multiple genetic/antigenic variants circulating in NHP at large breeding facilities.(66, 67, 70, 71) Further, prior infection with endemic strains of RhCMV does not appear to restrict iatrogenic reinfections with RhCMV variants.(72–77) While RhCMV has been demonstrated to be fully capable of transplacental transmission following iatrogenic inoculation (intravenous) (42), there have been no reports of non-iatrogenic instances of congenital sequelae in neonatal or spontaneously aborted macaques. While the reasons for this remain to be identified, one theoretical but unproven possibility is that repeated reinfections with antigenic variants may broaden antiviral immune responses such that the potential for maternal-to-fetal transmission is greatly reduced. One clinical ramification of such a scenario is that current vaccine approaches to development of a protective HCMV vaccine to reduce the incidence of congenital HCMV infection only target women who have never been previously infected with HCMV.(65) However, epidemiological studies indicate that only 14% of women who are of childbearing age are seronegative for HCMV infection.(54) Since all pregnant women are at-risk for either primary or non-primary HCMV infection and subsequent transplacental transmission(78), vaccine approaches targeting only seronegative women leave 86% of women of childbearing age without any benefit from such approaches. Accordingly, there is an urgent clinical imperative to investigate whether the apparent absence of reports describing natural (i.e., non-iatrogenic) instances of congenital RHCMV sequelae has an immunologic basis, particularly related to repeated reinfections with antigenic variants endemic in rhesus macaque breeding facilities. Such a hypothesis may explain another aspect of RhCMV shedding.
It has also been observed that there is an age-associated decline in the frequency and titer of virus from infected macaques. Whereas 78% of 3 – 5-year animals in co-housed breeding facilities shed virus, only 28% of animals 12 – 20 years of age shed virus in addition to demonstrating a significant reduction of RhCMV genomes in the saliva of older animals that had detectable viral DNA, compared to the younger animals.(38) While the basis of the decline in the frequency and genome copies of viral DNA by shedding animals is unknown, this phenotype of shedding has potential vaccine implications. As with the absence of reports regarding breeding age female macaques giving birth to infants with evidence of congenital RhCMV disease, there could also be a potential immunological basis for the decline in shedding, such as broadening of antiviral immune specificities following multiple reinfections. A fundamental issue to be resolved then is what immune specificities associate with an absence of non-iatrogenic transplacental transmission and/or the age-related decline in RhCMV shedding. Answers to such unresolved aspects of RhCMV natural history could yield clinically relevant paradigms for HCMV vaccine development.
From an immunological perspective, significant differences between juvenile and adult macaques have been observed in lymphoid cell subset numbers, B cell subsets (79), T cell activation markers, and cytokine responses in T cells in response to non-specific or RhCMV antigen stimulation.(38) Notably, there are no changes in neutralizing antibody responses over time. Conceivably, repeated reinfections of immune animals with antigenic variants in circulation at National Primate Research Centers could theoretically broaden or initiate de novo immune responses that restrict shedding in bodily fluids over time and/or prevent congenital transmission. Further, does the mechanistic basis for the reduction in shedding correlate with an increased resistance to reinfection across a mucosal surface?(66)
However, while reduction in shedding could conceivably be immunological in nature, the age-related decline in shedding could also be due solely or inter-dependently with viral and/or physiologic determinants. Age-related declines in shedding may reflect that there is a progressively smaller selective benefit to shedding as animals age. As animals age and most likely have fewer direct interactions with other animals, any selective advantage to the virus gained from a high shedding profile earlier in life may diminish and no longer confer any selective advantage. Finally, diminished shedding may simply reflect the process of aging in the infected animal. It is very possible that mucosal epithelial cells that give rise to the cells supporting the replication and shedding of virus at mucosal surfaces likely exhibit age-related declines in their cell division frequency and differentiation processes, progressively reducing the level of viral replication over time. The long-term resolution of RhCMV shedding and reinfection in RhCMV natural history have critical vaccine-related implications. While no population-based studies of reinfection in NHP have yet been done, prolonged shedding of virus supports the hypothesis that prolonged shedding and presumed reinfection is integral to the natural history of RhCMV. If so, the ability of RhCMV to overcome prior immunity generated from prior infection leads directly to the supposition that RhCMV could overcome prior immunity generated through prior vaccination if vaccine approaches recapitulate the type of immune responses generated during natural infection.(80)
In sum, the natural history of CMV in NHP offer an unparalleled opportunity to rigorously evaluate the protective efficacy of vaccine approaches that precisely recapitulate the challenges HCMV vaccinees would be expected to face, namely repeated mucosal exposure to differing titers of potential antigenic variants. The results presented herein for corn crib-housed cohorts or previously with larger corral housing (33) enable statistically powered group sizes in a relatively short timeframe (1 – 1.5 years) to quantify the protective efficacy of vaccination against (1) acquisition of virus, (2) shedding of virus, and (3) protection against reinfection. Results from such studies could lead to vaccine strategies that block both primary and non-primary HCMV infection as well as evaluate whether vaccination of young children to prevent shedding would protect close contacts of the vaccine recipients (pregnant women).
MATERIALS
Note:
This study was performed in 2002, and the methodologies described below were standard operating procedures at that time. Procedures and companies listed below may have changed in the intervening time.
Study Population:
Animals were maintained in a fully Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility in accordance with the Animal Welfare Act, and the Guide for the Care and Use of Laboratory Animals.(81–83) All procedures involving animals used in this study were approved by the University of California, Davis Institutional Animal Care and Use Committee (IACUC). A group of 31 rhesus macaques (11 female, 20 male) aged 9 – 16 months, were group-housed together (Day 0) in a corn crib, a structure facilitating frequent interactions amongst animals. The juveniles had previously been repeatedly determined to be negative for B Virus, SRV, SIV, STLV, SFV, RhCMV, and RRV infection (data not shown). For purposes of socialization, three B Virus, SRV, SIV, STLV negative (SPF1) adults (were housed with the group on a short-term basis. These adults had inadvertently been omitted from testing for SFV, RhCMV, and RRV infection at the time, but were later found to be infected with all three viruses. The adults were (1) 5 years/4 months old female (co-housed from Days +9 to +33), (2) 5 years old male (co-housed from Days +10 to +23), and (3) 5 year/1 month old male (co-housed from Days +23 to +26). The co-housing timeline is presented in Figure 1A.
Upon recognition that adults had not been pre-screened for SFV, RhCMV, and RRV, thereby potentially exposing the juveniles to all three viruses by contact with virus-laden bodily fluids of these 3 adults over a 24-day period, heparinized whole blood and saliva samples were collected bi-weekly for 3 months from week +2 to week +25, and then monthly out to 6 months for virus-specific IgG antibody and nucleic acid testing (SFV and RRV). Monkeys with no initial evidence of infection continued to be tested sporadically for 78 weeks total.
SFV:
The assays used were those validated and in routine use for SFV surveillance at the time of testing.(18)
SFV Antibody Enzyme Immunoassay (EIA):
Plasma from heparinized blood was tested for the presence of IgG antibodies to SFV using an EIA formatted using a viral lysate prepared from Cf2th cells infected with SFV (exhibiting >70% CPE) as the target antigen. Uninfected Cf2th cells served as a negative control antigen. Plasma samples were evaluated at a 1:100 dilution. Antibody binding was detected with horseradish peroxidase-conjugated goat anti-human IgG diluted 1:1000 (MP Biomedicals, Irvine, California) followed by ABTS (Kirkegaard & Perry, Gaithersburg, MD). Absorbency at 450 nm (A405) was read using a Tecan Sunrise Reader (Durham, NC). Any plasma sample with an A405 value for SFV antigen that was ≥0.1 units greater than its control antigen well was considered positive. This EIA approach was successfully tested on 804 samples using this assay and Endpoint PCR in our routine SPF testing algorithm (described below).
SFV DNA PCR:
At each collection time point, DNA was extracted from peripheral blood mononuclear cells by solid phase capture column purification and elution (Gentra, Minneapolis, Minnesota) for qualitative provirus detection by nested end point DNA PCR. The assay used degenerate primers GTG-GIA-AGG-TGG-AAA-GGA-AAA-ATA-GTG-AIA; ITA-IAG-AII-IIC-IAA-TTT-CCT-GTA-AAA-GAG-A FOR THE FIRST REACTION AND IGT-IGG-IIG-ICC-TIC-IAA-GTG-GTA-TGA; IAA-ITC-AAG-TGT-ATC-III-ITT-TGC-AAA-IGG for the nested PCR.(84) Inosine bases were included to provide more flexibility in annealing small nucleotide variances, while targeting the highly conserved polymerase gene.(85) The 153kb PCR amplicon was visualized on an agarose gel with ethidium bromide staining. Beta-globin detection was performed simultaneously to serve as an internal control for the presence of amplifiable DNA using a standard thermocycler profile and reagent cocktail (85) (limit of detection: 100 copies/reaction - unpublished data).
SFV RNA PCR:
Saliva was collected using a sterile, absorbent paper pad attached to a plastic wand (Saliva Diagnostic Systems, Framingham, MA; now available from Thermo Fisher). Viral RNA was purified from saliva using a solid phase capture spin column kit (Zymo Research, Orange, California). cDNA transcription was performed using commercial VILO cDNA reagents (Life Technologies / Invitrogen, Grand Island, NY) followed by quantitative Real Time q-RT PCR (Life Technologies/Applied Biosystems, Grand Island, NY) to the SFV-1 polymerase gene (GenBank Accession# X54482) (forward primer: 5’-CTT CAG GTC AAA ATG GAT CCT-3’, reverse primer 5’-ATC CCA GTG GGC TTT TAA TTT AGT TC-3’, probe 6FAM-5’-CCT CCA GCC TCT GGA AGC GGA AAT-3’ –TAMRA). The primer/probe set was formatted with Primer Express software (Life Technologies / Applied Biosystems, Grand Island, NY) was performed. A parallel GAPDH assay was also performed on each sample to ensure the presence of reverse-transcribable RNA under standard q-RT PCR conditions.(85)
RRV:
RRV DNA PCR:
RRV DNA sequences were detected using the Applied Biosystems 7900 real time PCR TaqMan platform. DNA from both the buffy coat and saliva samples were tested for the presence of the RRV DNA polymerase gene using forward primer TTTAACCGGCTATAACATCTCAAACT, reverse primer CCGGTTTTTATTTTTGTGTATTCGT and probe FAM-CGATCTCCCGTACCTAAT-MGBNFQ. Based on serial dilutions of plasmid, the limit of detection of the real time PCR assay was between 10 – 100 copies in a 50ul reaction. The single copy cellular Oncostatin M gene was included as a control housekeeping gene.(86)
RRV Antibody Indirect Immunofluorescence Assay (IFA):
A previously described IFA was also used to detect antibody in select plasma samples.(27) Telomerized rhesus fibroblasts (87) were plated and grown and infected on 16-well chamber slides and exposed to either infectious RRV supernatant or control media. Following acetone fixation, reaction with sample plasma, and washing, the cells were incubated with Fluorescein isothiocyanate (FITC) conjugated goat anti-monkey IgG (MP BioMedical, Solon, OH) to visualize specific antibody binding to RRV infected as compared to uninfected cells.
RRV and RhCMV:
Multiplex Microbead Assays:
A multiplex microbead immunoassay was used to facilitate detection of IgG antibodies to multiple viruses.(88–90) The remaining available archived plasmas from this group of monkeys were tested for the presence of antibodies to a panel of viruses including SFV, RRV, RhCMV using a commercially available panel of simian antigen coated beads (Charles River Laboratories, Wilmington, MA). In this assay the plasma samples were individually incubated with microbeads of defined spectral properties to coat bead with a distinct antigen (viral or control), and then collectively combined together in a liquid suspension microarray. After washing, the beads were reacted with biotinylated detector antibodies and then, after additional washing, Streptavidin, R-Phycoerythrin conjugate (SAPE). After a final washing to remove all unbound material, the beads were analyzed in a Bioplex reader (BioRad, Hercules, CA). By monitoring the spectral properties of the beads and amount of SAPE fluorescence associated with each, the presence or absence of antibody to the specific virus antigens was quantified.
Acknowledgements
We thank the staff of the California National Primate Research Center for their expert care of our study animals and facilitation of sample collection. We also acknowledge the valuable scientific and technical contributions of our past and present laboratory staff, including Nicholas Lerche, Matthew Deane, Patricia Todd, and Amanda Carpenter.
Funding information
This work was supported by the NIH Office of Research Infrastructure Program, Office of the Director grants CNPRC P51OD011107 and U42 OD010990.
Footnotes
Ethics statement The stated ethical policies of the journal were strictly followed. All animal housing and protocols involving animals were in accordance with the recommendations of the U.S. Department of Agriculture Animal Welfare Act regulations, the Guide for the Care and Use of Laboratory Animals, and regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All experiments were approved by the University of California, Davis (UCD) Animal Care and Use Committee (IACUC), prior to the start of the study.
Conflicts of interest All of the authors declare that there are no conflicts of interest related to this study
Literature Cited
- 1.Bliss-Moreau E, Amara RR, Buffalo EA, Colman RJ, Embers ME, Morrison JH, Quillen EE, Sacha JB, Roberts CT. 2021. Improving rigor and reproducibility in nonhuman primate research. Am J Primatol 83:e23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarantal AF, Noctor SC, Hartigan-O’Connor DJ. 2022. Nonhuman Primates in Translational Research. Annu Rev Anim Biosci 10:441–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren WC, Harris RA, Haukness M, Fiddes IT, Murali SC, Fernandes J, Dishuck PC, Storer JM, Raveendran M, Hillier LW, Porubsky D, Mao Y, Gordon D, Vollger MR, Lewis AP, Munson KM, DeVogelaere E, Armstrong J, Diekhans M, Walker JA, Tomlinson C, Graves-Lindsay TA, Kremitzki M, Salama SR, Audano PA, Escalona M, Maurer NW, Antonacci F, Mercuri L, Maggiolini FAM, Catacchio CR, Underwood JG, O’Connor DH, Sanders AD, Korbel JO, Ferguson B, Kubisch HM, Picker L, Kalin NH, Rosene D, Levine J, Abbott DH, Gray SB, Sanchez MM, Kovacs-Balint ZA, Kemnitz JW, Thomasy SM, Roberts JA, Kinnally EL, Capitanio JP, et al. 2020. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry PA, Strelow L. 2008. Development of breeding populations of rhesus macaques (Macaca mulatta) that are specific pathogen-free for rhesus cytomegalovirus. Comparative medicine 58:43–46. [PMC free article] [PubMed] [Google Scholar]
- 5.Lerche NW, Yee JL, Jennings MB. 1994. Establishing specific retrovirus-free breeding colonies of macaques: an approach to primary screening and surveillance. Lab Anim Sci 44:217–221. [PubMed] [Google Scholar]
- 6.Morton WR, Agy MB, Capuano SV, Grant RF. 2008. Specific pathogen-free macaques: definition, history, and current production. Ilar j 49:137–144. [DOI] [PubMed] [Google Scholar]
- 7.Fujiomto K, Takano J, Narita T, Hanari K, Shimozawa N, Sankai T, Yosida T, Terao K, Kurata T, Yasutomi Y. 2010. Simian betaretrovirus infection in a colony of cynomolgus monkeys (Macaca fascicularis). Comp Med 60:51–53. [PMC free article] [PubMed] [Google Scholar]
- 8.Kanthaswamy S, Kou A, Satkoski J, Penedo MC, Ward T, Ng J, Gill L, Lerche NW, Erickson BJ, Smith DG. 2010. Genetic characterization of specific pathogen-free rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC). Am J Primatol 72:587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerche NW, Simmons JH. 2008. Beyond specific pathogen-free: biology and effect of common viruses in macaques. Comp Med 58:8–10. [PMC free article] [PubMed] [Google Scholar]
- 10.Yee JL, Grant R, Van Rompay KK, Kuller L, Carpenter A, Watanabe R, Huebner R, Agricola B, Smedley J, Roberts JA. 2017. Emerging diagnostic challenges and characteristics of simian betaretrovirus infections in captive macaque colonies. J Med Primatol 46:149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yee JL, Grant RF, Van Rompay KKA, Roberts JA, Kuller L, Cunningham JL, Simmons JH, Papin JF. 2020. In vitro and In vivo Susceptibility of Baboons (Papio sp.) to Infection with and Apparent Antibody Reactivity to Simian Betaretrovirus (SRV). Comp Med 70:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yee JL, Vanderford TH, Didier ES, Gray S, Lewis A, Roberts J, Taylor K, Bohm RP. 2016. Specific pathogen free macaque colonies: a review of principles and recent advances for viral testing and colony management. J Med Primatol 45:55–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerche NW. 2010. Simian retroviruses: infection and disease--implications for immunotoxicology research in primates. J Immunotoxicol 7:93–101. [DOI] [PubMed] [Google Scholar]
- 14.Schapiro SJ, Lee-Parritz DE, Taylor LL, Watson L, Bloomsmith MA, Petto A. 1994. Behavioral management of specific pathogen-free rhesus macaques: group formation, reproduction, and parental competence. Lab Anim Sci 44:229–234. [PubMed] [Google Scholar]
- 15.Voevodin A, Marx P. 2009. Simian Virology. Wiley-Blackwell. [Google Scholar]
- 16.Murray SM, Picker LJ, Axthelm MK, Linial ML. 2006. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J Virol 80:663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenröder O, Spatz W, Volk B, Böhm N, Toniolo A, Neumann-Haefelin D, Schweizer M. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7–14. [DOI] [PubMed] [Google Scholar]
- 18.Blewett EL, Black DH, Lerche NW, White G, Eberle R. 2000. Simian foamy virus infections in a baboon breeding colony. Virology 278:183–193. [DOI] [PubMed] [Google Scholar]
- 19.Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. Journal of Virology 71:9764–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strand K, Harper E, Thormahlen S, Thouless ME, Tsai C, Rose T, Bosch ML. 2000. Two distinct lineages of macaque gamma herpesviruses related to the Kaposi’s sarcoma associated herpesvirus. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 16:253–269. [DOI] [PubMed] [Google Scholar]
- 21.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol 74:3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosch ML, Strand KB, Rose TM. 1998. Gammaherpesvirus sequence comparisons. J Virol 72:8458–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruce AG, Thouless ME, Haines AS, Pallen MJ, Grundhoff A, Rose TM. 2015. Complete genome sequence of Pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirus. J Virol 89:3888–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro IM, Ricciardi MJ, Gonzalez-Nieto L, Rakasz EG, Lifson JD, Desrosiers RC, Watkins DI, Martins MA. 2021. Recombinant Herpesvirus Vectors: Durable Immune Responses and Durable Protection against Simian Immunodeficiency Virus SIVmac239 Acquisition. J Virol 95:e0033021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estep RD, Govindan AN, Manoharan M, Li H, Fei SS, Park BS, Axthelm MK, Wong SW. 2020. Molecular analysis of lymphoid tissue from rhesus macaque rhadinovirus-infected monkeys identifies alterations in host genes associated with oncogenesis. PLoS One 15:e0228484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn AS, Bischof GF, Großkopf AK, Shin YC, Domingues A, Gonzalez-Nieto L, Rakasz EG, Watkins DI, Ensser A, Martins MA, Desrosiers RC. 2020. A Recombinant Rhesus Monkey Rhadinovirus Deleted of Glycoprotein L Establishes Persistent Infection of Rhesus Macaques and Elicits Conventional T Cell Responses. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White JA, Todd PA, Yee JL, Kalman-Bowlus A, Rodgers KS, Yang X, Wong SW, Barry P, Lerche NW. 2009. Prevalence of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesvirus in large age-structured breeding groups of rhesus macaques (Macaca mulatta). Comp Med 59:383–390. [PMC free article] [PubMed] [Google Scholar]
- 28.Wong SW, Bergquam EP, Swanson RM, Lee FW, Shiigi SM, Avery NA, Fanton JW, Axthelm MK. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi’s sarcoma-associated herpesvirus. J Exp Med 190:827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orzechowska BU, Powers MF, Sprague J, Li H, Yen B, Searles RP, Axthelm MK, Wong SW. 2008. Rhesus macaque rhadinovirus-associated non-Hodgkin lymphoma: animal model for KSHV-associated malignancies. Blood 112:4227–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansfield KG, Westmoreland SV, DeBakker CD, Czajak S, Lackner AA, Desrosiers RC. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol 73:10320–10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voevodin AF, Marx PA. 2009. Simian virology, p xi, 511 p., 512 p. of plates. Wiley-Blackwell, Ames, Iowa. [Google Scholar]
- 32.White JA, Yang X, Todd PA, Lerche NW. 2011. Longitudinal patterns of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesviruses in age-structured captive breeding populations of rhesus Macaques (Macaca mulatta). Comp Med 61:60–70. [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel P, Weigler BJ, Kerr H, Hendrickx AG, Barry PA. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci 44:25–30. [PubMed] [Google Scholar]
- 34.Andrade MR, Yee J, Barry PA, Spinner A, Roberts JA, Cabello H, Leite JP, Lerche NW. 2003. Prevalence of Antibodies to Selected Viruses in a Long-term Closed Breeding Colony of Rhesus Macaques (Macaca mulatta) in Brazil. Amer J Primatol 59:123–128. [DOI] [PubMed] [Google Scholar]
- 35.Jones-Engel L, Engel GA, Heidrich J, Chalise M, Poudel N, Viscidi R, Barry PA, Allan JS, Grant R, Kyes R. 2006. Temple monkeys and health implications of commensalism, Kathmandu, Nepal. Emerg Infect Dis 12:900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asher DM, Gibbs CJ Jr., Lang DJ. 1969. Rhesus monkey cytomegaloviruses: persistent asymptomatic viruses. Bacteriol Proc 69:191. [Google Scholar]
- 37.Asher DM, Gibbs CJ Jr., Lang DJ, Gajdusek DC, Chanock RM. 1974. Persistent shedding of cytomegalovirus in the urine of healthy Rhesus monkeys. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine 145:794–801. [DOI] [PubMed] [Google Scholar]
- 38.Oxford KL, Dela Pena-Ponce MGA, Jensen K, Eberhardt MK, Spinner A, Van Rompay KK, Rigdon J, Mollan KR, Krishnan VV, Hudgens MG, Barry PA, De Paris K. 2015. The interplay between immune maturation, age, chronic viral infection and environment. Immun Ageing 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberhardt MK, Deshpande A, Fike J, Short R, Schmidt KA, Blozis SA, Walter MR, Barry PA. 2016. Exploitation of Interleukin-10 (IL-10) Signaling Pathways: Alternate Roles of Viral and Cellular IL-10 in Rhesus Cytomegalovirus Infection. J Virol 90:9920–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur A, Itell HL, Ehlinger EP, Varner V, Gantt S, Permar SR. 2018. Natural history of postnatal rhesus cytomegalovirus shedding by dams and acquisition by infant rhesus monkeys. PLoS One 13:e0206330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barry PA, Lockridge KM, Salamat S, Tinling SP, Yue Y, Zhou SS, Gospe SM Jr., Britt WJ, Tarantal AF. 2006. Nonhuman primate models of intrauterine cytomegalovirus infection. ILAR J 47:49–64. [DOI] [PubMed] [Google Scholar]
- 42.Bialas KM, Tanaka T, Tran D, Varner V, Cisneros De La Rosa E, Chiuppesi F, Wussow F, Kattenhorn L, Macri S, Kunz EL, Estroff JA, Kirchherr J, Yue Y, Fan Q, Lauck M, O’Connor DH, Hall AH, Xavier A, Diamond DJ, Barry PA, Kaur A, Permar SR. 2015. Maternal CD4+ T cells protect against severe congenital cytomegalovirus disease in a novel nonhuman primate model of placental cytomegalovirus transmission. Proc Natl Acad Sci U S A 112:13645–13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lerche NW, Osborn KG, Marx PA, Prahalada S, Maul DH, Lowenstine LJ, Munn RJ, Bryant ML, Henrickson RV, Arthur LO, et al. 1986. Inapparent carriers of simian acquired immune deficiency syndrome type D retrovirus and disease transmission with saliva. J Natl Cancer Inst 77:489–496. [PubMed] [Google Scholar]
- 44.Sequar G, Britt WJ, Lakeman FD, Lockridge KM, Tarara RP, Canfield DR, Zhou SS, Gardner MB, Barry PA. 2002. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J Virol 76:7661–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarantal AF, Salamat MS, Britt WJ, Luciw PA, Hendrickx AG, Barry PA. 1998. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta). The Journal of infectious diseases 177:446–450. [DOI] [PubMed] [Google Scholar]
- 46.Barry P, Chang W-L. 2007. Primate Betaherpesviruses, p 1051–1075. In Arvin A, Campadielli G, Moore P, Mocarski E, Roizman B, Whitley R, Yamanishi K (ed), Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge University Press, Cambridge. [PubMed] [Google Scholar]
- 47.Barry PA. 2015. Exploiting viral natural history for vaccine development. Med Microbiol Immunol 204:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deere JD, Barry PA. 2014. Using the nonhuman primate model of HCMV to guide vaccine development. Viruses 6:1483–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.dela Pena MG, Strelow L, Barry PA, Abel K. 2012. Use of specific-pathogen-free (SPF) rhesus macaques to better model oral pediatric cytomegalovirus infection. J Med Primatol 41:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eberhardt MK, Barry PA. 2014. Pathogen Manipulation of cIL-10 Signaling Pathways: Opportunities for Vaccine Development? Current Topics in Microbiology and Immunology 380:93–128. [DOI] [PubMed] [Google Scholar]
- 51.Yue Y, Barry PA. 2008. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Advances in virus research 72:207–226. [DOI] [PubMed] [Google Scholar]
- 52.Reitsema LJ, Partrick KA, Muir AB. 2016. Inter-individual variation in weaning among rhesus macaques (Macaca mulatta): Serum stable isotope indicators of suckling duration and lactation. Am J Primatol 78:1113–1134. [DOI] [PubMed] [Google Scholar]
- 53.Vandeleest JJ, Capitanio JP. 2012. Birth timing and behavioral responsiveness predict individual differences in the mother-infant relationship and infant behavior during weaning and maternal breeding. Am J Primatol 74:734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuhair M, Smit GSA, Wallis G, Jabbar F, Smith C, Devleesschauwer B, Griffiths P. 2019. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev Med Virol doi: 10.1002/rmv.2034:e2034. [DOI] [PubMed] [Google Scholar]
- 55.Kessler MJ, London WT, Madden DL, Dambrosia JM, Hilliard JK, Soike KF, Rawlins RG. 1989. Serological survey for viral diseases in the Cayo Santiago rhesus macaque population. Puerto Rican Health Sci J 8:95–97. [PubMed] [Google Scholar]
- 56.Becker SD, Bennett M, Stewart JP, Hurst JL. 2007. Serological survey of virus infection among wild house mice (Mus domesticus) in the UK. Lab Anim 41:229–238. [DOI] [PubMed] [Google Scholar]
- 57.Goüy de Bellocq J, Baird SJ, Albrechtová J, Sobeková K, Piálek J. 2015. Murine cytomegalovirus is not restricted to the house mouse Mus musculus domesticus: prevalence and genetic diversity in the European house mouse hybrid zone. J Virol 89:406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moro D, Lloyd ML, Smith AL, Shellam GR, Lawson MA. 1999. Murine viruses in an island population of introduced house mice and endemic short-tailed mice in Western Australia. J Wildl Dis 35:301–310. [DOI] [PubMed] [Google Scholar]
- 59.Parker SE, Malone S, Bunte RM, Smith AL. 2009. Infectious diseases in wild mice (Mus musculus) collected on and around the University of Pennsylvania (Philadelphia) Campus. Comp Med 59:424–430. [PMC free article] [PubMed] [Google Scholar]
- 60.Singleton GR, Smith AL, Krebs CJ. 2000. The prevalence of viral antibodies during a large population fluctuation of house mice in Australia. Epidemiol Infect 125:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singleton GR, Smith AL, Shellam GR, Fitzgerald N, Müller WJ. 1993. Prevalence of viral antibodies and helminths in field populations of house mice (Mus domesticus) in southeastern Australia. Epidemiol Infect 110:399–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGeoch DJ, Rixon FJ, Davison AJ. 2006. Topics in herpesvirus genomics and evolution. Virus Res 117:90–104. [DOI] [PubMed] [Google Scholar]
- 63.Ali M, Taylor GP, Pitman RJ, Parker D, Rethwilm A, Cheingsong-Popov R, Weber JN, Bieniasz PD, Bradley J, McClure MO. 1996. No evidence of antibody to human foamy virus in widespread human populations. AIDS Res Hum Retroviruses 12:1473–1483. [DOI] [PubMed] [Google Scholar]
- 64.Ghersi BM, Jia H, Aiewsakun P, Katzourakis A, Mendoza P, Bausch DG, Kasper MR, Montgomery JM, Switzer WM. 2015. Wide distribution and ancient evolutionary history of simian foamy viruses in New World primates. Retrovirology 12:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plotkin SA, Boppana S. 2019. Vaccination against the human cytomegalovirus. Vaccine 37:7437–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J, Wellnitz S, Chi XS, Yue Y, Schmidt KA, Nguyen N, Chen W, Yurgelonis I, Rojas E, Liu Y, Loschko J, Pollozi E, Matsuka YV, Needle E, Vidunas E, Donald RGK, Moran J, Jansen KU, Dormitzer PR, Barry PA, Yang X. 2022. Horizontal transmission of cytomegalovirus in a rhesus model despite high-level, vaccine-elicited neutralizing antibody and T cell responses. J Infect Dis doi: 10.1093/infdis/jiac129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yue Y, Chang WLW, Li J, Nguyen N, Schmidt KA, Dormitzer PR, Yang X, Barry PA. 2022. Pathogenesis of Wild-Type-Like Rhesus Cytomegalovirus Strains following Oral Exposure of Immune-Competent Rhesus Macaques. J Virol 96:e0165321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen SF, Tu WW, Sharp MA, Tongson EC, He XS, Greenberg HB, Holmes TH, Wang Z, Kemble G, Manganello AM, Adler SP, Dekker CL, Lewis DB, Arvin AM. 2004. Antiviral CD8 T cells in the control of primary human cytomegalovirus infection in early childhood. J Infect Dis 189:1619–1627. [DOI] [PubMed] [Google Scholar]
- 69.Lazar K, Rabe T, Goelz R, Hamprecht K. 2020. Human Cytomegalovirus Reactivation During Lactation: Impact of Antibody Kinetics and Neutralization in Blood and Breast Milk. Nutrients 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taher H, Mahyari E, Kreklywich C, Uebelhoer LS, McArdle MR, Moström MJ, Bhusari A, Nekorchuk M, E X, Whitmer T, Scheef EA, Sprehe LM, Roberts DL, Hughes CM, Jackson KA, Selseth AN, Ventura AB, Cleveland-Rubeor HC, Yue Y, Schmidt KA, Shao J, Edlefsen PT, Smedley J, Kowalik TF, Stanton RJ, Axthelm MK, Estes JD, Hansen SG, Kaur A, Barry PA, Bimber BN, Picker LJ, Streblow DN, Früh K, Malouli D. 2020. In vitro and in vivo characterization of a recombinant rhesus cytomegalovirus containing a complete genome. PLoS Pathog 16:e1008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burwitz BJ, Malouli D, Bimber BN, Reed JS, Ventura AB, Hancock MH, Uebelhoer LS, Bhusari A, Hammond KB, Espinosa Trethewy RG, Klug A, Legasse AW, Axthelm MK, Nelson JA, Park BS, Streblow DN, Hansen SG, Picker LJ, Fruh K, Sacha JB. 2016. Cross-Species Rhesus Cytomegalovirus Infection of Cynomolgus Macaques. PLoS Pathog 12:e1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr., Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15:293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, Axthelm MK, Nelson JA, Jarvis MA, Picker LJ, Fruh K. 2010. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr., Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, Malouli D, Xu G, Richards R, Whizin N, Reed JS, Hammond KB, Fischer M, Turner JM, Legasse AW, Axthelm MK, Edlefsen PT, Nelson JA, Lifson JD, Fruh K, Picker LJ. 2013. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340:1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hansen SG, Piatak M Jr., Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Fruh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. 2013. Immune clearance of highly pathogenic SIV infection. Nature 502:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, Lopez CA, Fruh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ. 2016. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science 351:714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Britt WJ. 2018. Maternal Immunity and the Natural History of Congenital Human Cytomegalovirus Infection. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang WL, Gonzalez DF, Kieu HT, Castillo LD, Messaoudi I, Shen X, Tomaras GD, Shacklett BL, Barry PA, Sparger EE. 2017. Changes in Circulating B Cell Subsets Associated with Aging and Acute SIV Infection in Rhesus Macaques. PLoS One 12:e0170154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.eCFR. 02/15/1991 2022. Subpart D - Specifications for the Humane Handling, Care, Treatment, and Transportation of Nonhuman Primates. https://www.federalregister.gov/citation/56-FR-6495; https://www.ecfr.gov/current/title-9/chapter-I/subchapter-A. . Accessed
- 82.National Research Council (U.S.). Committee for the Update of the Guide for the Care and Use of Laboratory Animals., Institute for Laboratory Animal Research (U.S.), National Academies Press (U.S.). 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, D.C. [Google Scholar]
- 83.ORI. 2013. 7 USC, 2131–2156: Animal Welfare Act as Amended. https://ori.hhs.gov/content/Chapter-4-The-Welfare-of-Laboratory-Animals-7-usc-2131-2156-animal-welfare-act-amended. Accessed
- 84.Heneine W, Switzer WM, Sandstrom P, Brown J, Vedapuri S, Schable CA, Khan AS, Lerche NW, Schweizer M, Neumann-Haefelin D, Chapman LE, Folks TM. 1998. Identification of a human population infected with simian foamy viruses. Nat Med 4:403–407. [DOI] [PubMed] [Google Scholar]
- 85.Lerche NW, Cotterman RF, Dobson MD, Yee JL, Rosenthal AN, Heneine WM. 1997. Screening for simian type-D retrovirus infection in macaques, using nested polymerase chain reaction. Lab Anim Sci 47:263–268. [PubMed] [Google Scholar]
- 86.White JA, Todd PA, Rosenthal AN, Yee JL, Grant R, Lerche NW. 2009. Development of a generic real-time PCR assay for simultaneous detection of proviral DNA of simian Betaretrovirus serotypes 1, 2, 3, 4 and 5 and secondary uniplex assays for specific serotype identification. J Virol Methods 162:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang WL, Kirchoff V, Pari GS, Barry PA. 2002. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J Virol Methods 104:135–146. [DOI] [PubMed] [Google Scholar]
- 88.Khan IH, Mendoza S, Yee J, Deane M, Venkateswaran K, Zhou SS, Barry PA, Lerche NW, Luciw PA. 2006. Simultaneous detection of antibodies to six nonhuman-primate viruses by multiplex microbead immunoassay. Clin Vaccine Immunol 13:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuller L, Watanabe R, Anderson D, Grant R. 2005. Development of a whole-virus multiplex flow cytometric assay for antibody screening of a specific pathogen-free primate colony. Diagn Microbiol Infect Dis 53:185–193. [DOI] [PubMed] [Google Scholar]
- 90.Liao Q, Guo H, Tang M, Touzjian N, Lerche NW, Lu Y, Yee JL. 2011. Simultaneous detection of antibodies to five simian viruses in nonhuman primates using recombinant viral protein based multiplex microbead immunoassays. J Virol Methods 178:143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]


