Abstract
T cells are critical orchestrators of the adaptive immune response that optimally eliminates a specific pathogen. Aberrant T cell development and function are implicated in a broad range of human disease including immunodeficiencies, autoimmune diseases, and allergic diseases. Accordingly, therapies targeting T cells and their effector cytokines have drastically improved the care of patients with immune dysregulatory diseases. Newer discoveries concerning T cell mediated antitumor immunity and T cell exhaustion have further prompted development of highly effective and novel treatment modalities for malignancies, including checkpoint inhibitors and antigen-reactive T cells. Recent discoveries are also uncovering the depth and variability of T cell phenotypes: while T cells have long been described using a subset-based classification system, next-generation sequencing technologies suggest an astounding degree of complexity and heterogeneity at the single cell level.
Introduction
T cells are critical orchestrators that set the tone of an adaptive immune response to optimally eliminate a specific pathogen. Inappropriately activated T cells and their products are major drivers of autoimmune and allergic disease, and therapies targeting T cells and their effector cytokines have revolutionized the treatment of these diseases. More recently, efforts are being made to propagate and engineer antigen-reactive T cells to treat malignancies and other diseases 1-3. In a brief review such as this, it is impossible to do justice to all facets of T cell biology; instead, we will try to focus on some of the more topical issues and touch on salient clinical issues.
T cell development
Conventional αβ T cells are categorized as CD4+ helper T cells, which provide help to other immune cells, or CD8+ cytotoxic T cells, which kill their target cells. T cells express a unique T cell receptor (TCR), which recognizes small peptide fragments presented on major histocompatibility complex (MHC) molecules. CD4+ helper cells are MHC class II (MHC-II) restricted, while CD8+ cytotoxic T cells are MHC-I restricted. So called nonconventional T cells are discussed below.
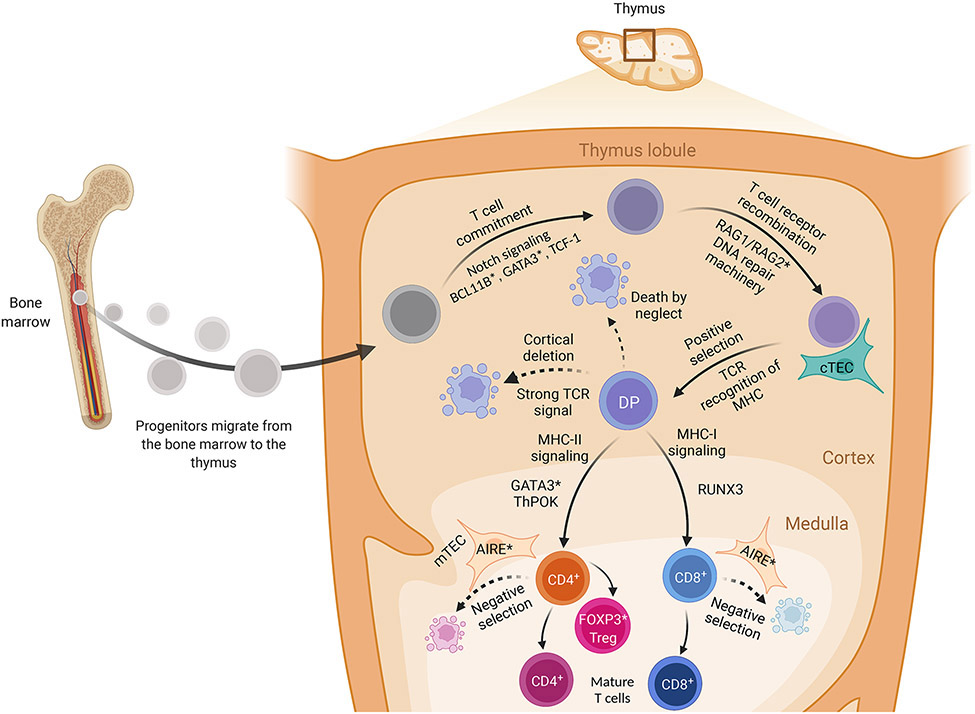
During T cell development (Figure 1), precursor cells arrive in the thymus from the bone marrow. In the thymus, T cells undergo commitment to the T cell lineage, TCR rearrangement, positive and negative selection, and lineage differentiation. T cell commitment involves the loss of multipotent potential and relies on Notch signaling and on Bcl11b, Gata3, and TCF-14-12.
Figure 1: T cell development.
T cell progenitors, or precursor cells, arrive in the thymus from the bone marrow. They lose multipotent potential and commit to the T cell lineage under control of Notch signaling and Bcl11b, Gata3, and TCF-1. These committed thymocytes subsequently recombine their T cell receptors (TCRs) under the control of RAG enzymes and DNA repair machinery to generate a diverse TCR repertoire. TCR recombination is followed by positive selection of T cells that recognize self-MHC:peptide complexes; this occurs in the thymic cortex and is mediated by thymic cortical epithelial cells. The thymocytes then migrate to the medulla where they are tested for reactivity against self-peptides-by medullary thymic epithelial cells; self-reactive T cells are negatively selected and deleted. Next, developing thymocytes differentiate into CD4+ vs. CD8+ T cells under the control of Runx3 (CD8+) or Gata3 and ThPOK (CD4+). A subset of CD4+ T cells develop in to FOXP3+ regulatory T cells (Treg), whereas the others mature into naïve T cells.
TCR recombination is achieved by RAG-mediated rearrangement of V and J segments of the TCRα gene, and V, D, and J segments of the TCRβ gene, followed by DNA repair of the resultant breaks13-16. This rearrangement allows for the generation of a hugely diverse repertoire of TCRs, capable of recognizing essentially any peptide fragment. Patients with inactivating RAG mutations are unable to rearrange their T and B cell receptors; this leads to immunodeficiency and - in some cases - autoimmunity mediated by expanded oligoclonal autologous T cells17.
Positive selection of T cells that recognize self-MHC:peptide complexes is mediated by cortical thymic epithelial cells18-20. TCR engagement by self-MHC:peptide complex also downregulates RAG expression21. The transcription factor RORγ promotes thymocyte survival during this process via BCL-XL expression22. Positively selected thymocytes migrate to the medulla where they are tested for reactivity against self-peptides-by medullary thymic epithelial cells. These cells ectopically express various tissue-restricted antigens, a process-mediated by the transcription factor AIRE. Loss-of-function (LOF) AIRE mutations cause the autoimmune disorder APECED (APS1), where autoreactive T cells escape negative selection and mediate tissue damage23, 24 Clonal deletion can also occur in the cortex, presumably through different mechanisms25, 26.
Next, developing thymocytes differentiate into CD4+ vs. CD8+ cells depending on their MHC-restriction. MHC class II restricted thymocytes express Gata3 and ThPOK, which promote CD4+ commitment; MHC-I-restricted thymocytes express Runx3 and become CD8+27-30. During terminal maturation, CD4+ and CD8+ T cells start expressing sphingosine 1 phosphate receptor 1 (S1PR1), which promotes entry into the circulation31, 32. S1P also mediates egress of T cells from secondary lymphoid organs; the S1PR inhibitors fingolimod, siponimod and ozanimod are approved for multiple sclerosis33.
Naïve T cells
After thymic development, T cells exist as naïve pluripotent cells, remaining in the G0 phase of the cell cycle due to quiescence factors like Foxp1 and Klf2 until they encounter their cognate antigen34, 35 Naïve cells are heterogeneous and include recent thymic emigrants as well as several subgroups of mature naïve T cells34. Although T cell activation takes place in secondary lymphoid organs; naïve T cells are widely distributed in lymphoid and nonlymphoid tissues34, 36.
T cell activation
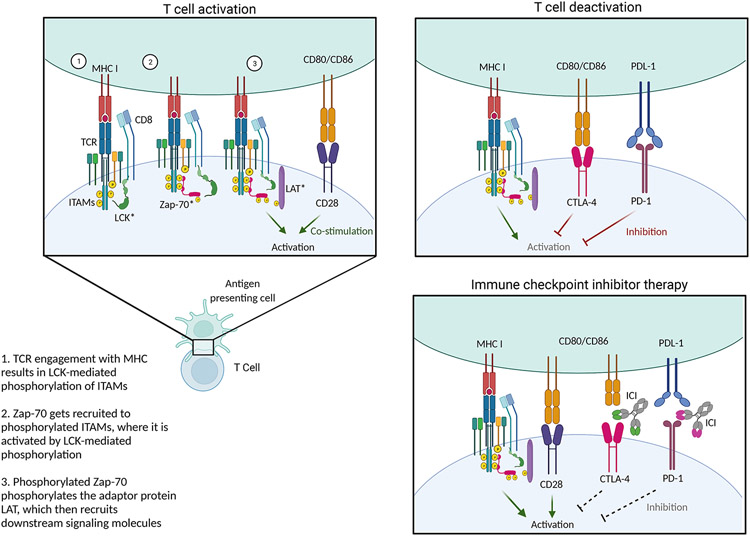
There are approximately 2x1011 naïve T cells in the human body, expressing about 1010 different TCRs capable of recognizing various antigenic peptides 37 Peptides derived from pathogens and transformed cells are presented by antigen presenting cells (APCs) in the context of MHC-II to activate CD4+ T cells. By contrast, all nucleated cells express MHC-I and can present antigen to activate cytotoxic CD8+ T cells. The TCR is a multi-protein complex consisting of αβ chains noncovalently attached to multidimeric CD3 proteins (Figure 2). CD3 molecules have cytoplasmic tails with immunoreceptor tyrosine activation motifs (ITAMs)38. TCR activation by peptide:MHC initiates a complex series of phosphorylation events39. CD4 and CD8 molecules recruit Lck, which phosphorylates the ITAMs on CD3 and TCRζ, causing them to recruit Zap70 kinase, which is also phosphorylated by Lck 40, 41. Zap70 then phosphorylates the adaptor protein LAT, which recruits downstream signaling molecules42. Mutation of LAT and ZAP70 can underlie severe combined immunodeficiency (SCID), whereas LCK mutations cause T cell immunodeficiency (Figure 2) 42-44.
Figure 2. T cell activation.
The T cell receptor (TCR) is a multi-protein complex in which αβ chains are noncovalently attached to CD3 proteins, whose cytoplasmic tails contain immunoreceptor tyrosine activation motifs (ITAMs). When TCRs are activated by peptide:MHC, a complex series of phosphorylation events ensues. First, CD4 and CD8 molecules recruit Lck, a kinase that phosphorylates the ITAMs on CD3 and TCRζ. This allows the ITAMs to recruit Zap70 kinase, which is also phosphorylated by Lck. Phosphorylated Zap70 then phosphorylates the adaptor protein LAT, which recruits various downstream signaling molecules.
TCR stimulation is insufficient to activate a T cell and by itself causes the T cell to become hyporesponsive, or anergic. A second signal required for T cell activation is provided costimulatory receptor binding. CD28 is expressed on most CD4+ and CD8+ T cells and binds to the B7 family of molecules, primarily B7-1 (CD80) and B7-2 (CD86). CD28 promotes cell survival, proliferation, metabolism, and cytokine production through diverse mediators including PI3 kinase (PI3K), Akt, Itk, NF-κB, and p38 MAPK45. Other costimulatory molecules include ICOS, 4-1BB, OX-40, CD2, CD5, and LFA-145, 46. Costimulatory domains, including intracellular signaling domains of CD28 and 4-1BB, have been used to enhance the function of chimeric antigen receptor (CAR) T cells specific for tumor antigens2. The CTLA4-fusion protein abatacept blocks costimulation and is FDA-approved for the treatment of multiple autoimmune and immune-mediated conditions.
Integrins are cell surface receptors that mediate interactions between cells and other cells, or between cells and the extracellular matrix; they are important for T cell trafficking47. The α4β7 integrin inhibitor vedolizumab is approved for inflammatory bowel disease (IBD), and integrin-targeted therapies are under development for fibrosis, cardiac disease, and malignancy48. Upon activation, T cells upregulate surface expression of integrins. This helps to stabilize T cell interactions with APCs, thereby enhancing signal transduction. TCR signaling then feeds back on integrins to increase their avidity, a process termed "inside-out" signaling49.
Activation induces a complex series of transcriptional and epigenetic changes that are partly shaped by the environment in which the T cell is activated 50-53. TCR-dependent transcription factors (TFs) including NF-κB, NFAT, and AP-1 induce IL-2 production and upregulate the high affinity IL-2 receptor subunit CD25, increasing T cell responsiveness to IL-254. Binding of IL-2 to its high-affinity receptor causes T cells to proliferate about 1,000-fold in the secondary lymphoid organs, producing a large population of lymphoblasts, also termed effector cells.
Studying antigen-specific responses
Despite the central role of antigen recognition to T cell effector function, several barriers have historically limited the identification of antigen-specific T cells in vivo. These include the low frequency of peripheral antigen-specific T cells, weak or polyspecific MHC-TCR binding, and the wide array of potential T cell epitopes55. Advances in computational biology, combined with comprehensive peptide libraries and novel computational methods can now allow identification of novel antigen-TCR combinations56, 57. While computational assays do not provide information about the function and phenotypes of the expanded T cell clones, they can be a powerful tool in combination with other methods. For example, TCR sequencing can be paired with single cell transcriptomic profiling of the expanded T cell clones. To functionally evaluate T cell responses to putative cognate antigens, directed methods like peptide-induced cytokine production and tagged peptide-MHC tetramers can specifically profile antigen-specific T cell abundance, location, function, and phenotype, focusing on one antigen at a time55. Such techniques confirm antigen-reactivity in vivo and can distinguish between functional vs. exhausted T cells, active vs. latent infection, or T helper subset phenotype.
Antigen-specific T cells have perhaps been best-studied in the context of infection. In tuberculosis infection, for example, directed functional assays that identify antigen-specific T cells outperform tuberculin skin testing in distinguishing patients with active vs. latent infection58, 59. Using newer methodologies can also be helpful in the context of infection: TCR sequencing has been used to identify antigenic signals of active vs. latent cytomegalovirus (CMV) infection60. This is important because CMV is highly prevalent but causes active disease in only a small minority of infected subjects; hence, identifying TCR-antigen signals associated with active infection could help target patients for antiviral treatment. During the COVID-19 pandemic, antigen-specific T cell studies have been used to define associations with disease severity, track vaccine responses, predict recognition of novel variants, and identify a mechanism of superantigen recognition – or non-antigen-specific/polyclonal T cell activation – that leads to multisystemic inflammatory syndrome in children61-67.
In other human diseases, computational methods have enabled a broad range of discoveries. In monogenic primary immunodeficiencies like RAG1 deficiency and common variable immunodeficiency, specific TCR repertoire deficiencies are associated with distinct clinical phenotypes43, 68-70 In malignant tumors, TCR profiling of circulating and tumor-resident T cells has revealed intratumoral T cell dysfunction that improved with checkpoint inhibitors71. In complex autoimmune diseases like scleroderma, and SLE, TCR sequencing has revealed clonal expansion, and targeted functional assays have identified autoimmunity-specific self-reactive TCRs 71, 72. Tetramer studies have uncovered a population of Th17 cells specific to the autoantigen U1-snRNP73. In patients with cat allergies, Tetramer-Guided Epitope Mapping has identified novel cat allergen epitopes that are potential targets for desensitization74. These findings help to identify the antigens that allow dysregulated T cell mediated immune responses to develop and persist, which could represent novel therapeutic targets.
CD4+ T subsets
During CD4+ T cell activation, different microenvironmental signals including pathogens and cytokines induce specific lineage-determining TFs (LDTF). LDTFs bind to target DNA regulatory regions and induce the transition of target gene loci from an inactive state to a poised state75. This enables the differentiation into CD4+ effector T cells subsets, including T helper 1 (Th), Th2, Th9, Th17, Th22, and follicular helper (Tfh) cells. Each subset promotes an immune response to a different family of pathogens by activating CD8+ T cells, NK cells, B cells, myeloid cells, and non-professional immune cells. For this reason, CD4+ T cells are designated “T helper cells”. It is now appreciated that selective cytokine production is not unique to T cells but is rather paralleled by innate lymphoid cells (ILCs) that employ that same LDTFs in a non-antigen-specific manner.
Historically, T helper subsets were viewed as fixed states: once a T cell committed to a specific subset, it selectively produced a hallmark effector cytokine and could not easily acquire a different phenotype (Figure 3). For example, T helper 1 (Th1) cells produce interferon gamma (IFN-γ) whereas Th2 cells produce IL-4, IL-5, and IL-13. While these concepts have utility in explaining some of the behaviors of CD4+ T cells, newer immunophenotyping techniques reveal substantial heterogeneity, complexity, and plasticity of T effector subsets.
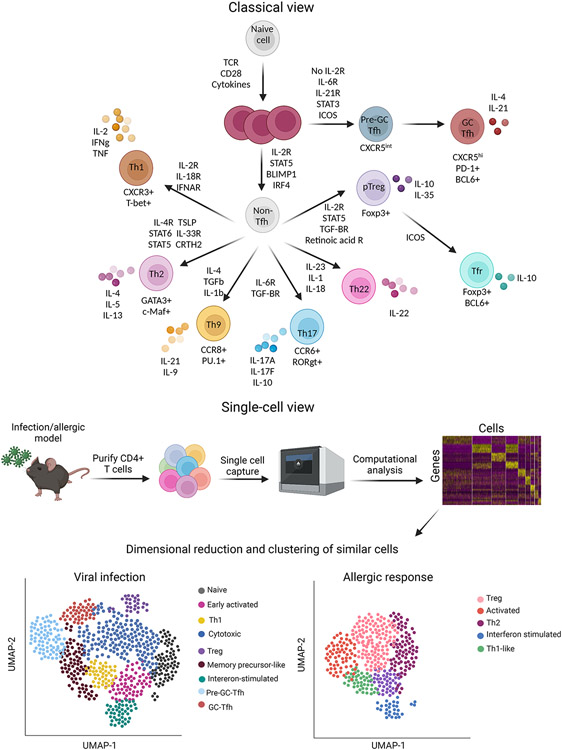
Figure 3: CD4+ subsets and heterogeneity.
The classical view of CD4+ differentiation holds that naïve T cells differentiate into distinct subsets under the control of subset-specific environmental signals and downstream transcription factors. In this view, T helper 1 (Th1) cells express T-bet and produce interferon gamma (IFN-γ) whereas Th2 cells express Gata3 produce IL-4, IL-5, and IL-13. Th17 cells express RORγt and produce IL-17-family cytokines, Th9 cells express PU.1 and produce IL-9, and Th22 cells produce IL-22. Peripherally derived regulatory T cells promote immune tolerance, whereas T follicular helper (Tfh) cells are critical for B cell responses. In many cases subsets can by identified by the expression of specific cell surface markers; for example, CXCR3 marks Th1 cells while CCR6 marks Th17 cells. However, single cell technologies reveal that there is substantial overlap and heterogeneity between these different subsets in vivo.
Th1 cells
Along with CD8+ T cells and NK cells, Th1 cells produce IFN-γ in response to signals elicited by intracellular microbes, including macrophage-derived IL-12 and macrophage/NK-derived IFN-γ (Figure 3)76. IL-12 and IFN-γ activate STAT4 and STAT1, which induce the T-bet (encoded by Tbx21) to promote Th1 differentiation76. Loss of function (LOF) mutations of STAT1 are linked to disseminated infection with intracellular bacteria and viruses, whereas gain of function (GOF) mutations cause autoimmunity and mucocutaneous candidiasis77. Similarly, inactivating mutations in IL12, IFNG and TBX21 increase susceptibility to mycobacterial disease; STAT4 LOF mutations are linked to fungal disease77-80. IFN-γ enhances antigen presentation via upregulation of MHC-I and MHC-II, promotes B cell class switching to IgG1 and IgG3, and inhibits class switching to IgE76. Th1 cells also express the chemokine receptor CXCR3, which senses CXCL9 and CXCL10, causing Th1 to accumulate in microbe-containing granulomas81. Ustekinumab targets IL-12 and IL-23 via the shared subunit IL-12B (p40) and is approved for the treatment of plaque psoriasis and inflammatory bowel disease. The IFN-γ monoclonal antibody emapalumab is approved for hemophagocytic lymphohistiocytosis.
Th2 cells
Th2 cells produce IL-4, IL-13, and IL-5, which are part of a family termed type 2 cytokines (Figure 3). Th2 cells gain full effector capacity in tissues by interacting with various cells including ILCs and neurons, which produce Th2-promoting factors in response to allergens, helminths, and other stimuli. Th2-promoting cytokines include IL- 4, IL-25, IL-33, and thymic stromal lymphopoietin; neuropeptides like substance P, neuromedin U, and CGRP also promote Th2 differentiation82, 83. Other factors like Prostaglandin D2 induce Th2 differentiation through chemoattractant receptor-homologous molecule (CRTH2)84. GATA3, the LDTF driving Th2 differentiation, has many additional functions; GATA3 haploinsufficiency therefore causes a complex disease that includes defective Th2 specification82. Other Th2-promoting TFs include STAT6, Notch, IRF-4, Growth factor independent-1 (Gfi-1), and Bcl11b82. Conversely, repressor of GATA3 (encoded by Zbtb32) limits Th2 differentiation82.
Th2-derived IL-4, IL-13 and IL-5 mobilize eosinophils, basophils, and mast cells. IL-4 also promotes B cell growth, immunoglobulin class switching to IgG4 and IgE, and additional Th2 differentiation; IL-13 increases mucus secretion from airway and gut epithelial cells76. Dupilumab targets the shared receptor for IL-4 and IL-13 and is approved for treatment of atopic dermatitis and asthma. The IL-5 and IL-5R blocking antibodies mepolizumab, reslizumab, and benralizumab are approved for eosinophilic asthma; mepolizumab is also approved for hypereosinophilic syndrome, eosinophilic granulomatosis with polyangiitis, and chronic rhinosinusitis with nasal polyps77.
Th17 cells
Along with gamma-delta T cells, invariant natural killer T (iNKT) cells, and ILC3s, Th17 cells produce the signature cytokine IL-17A, as well as IL-17F and IL-21 (Figure 3)85. Extracellular bacteria and fungi promote Th17 differentiation by eliciting production of IL-6, IL-21, IL-23, IL-1β, and TGF-β85. IL-6, IL-23, IL-1β, and TGF-β promote expression of Rorγt, whereas IL-6, IL-21 and IL-23 activate STAT3. IL17F and RORC inactivating mutations lead to chronic mucocutaneous candidiasis86. Dominant negative mutations of STAT3 underlie hyperimmunoglobulin E syndrome (HIES), which is characterized by mucocutaneous candidiasis and susceptibility to staphylococcal infection77. Other factors that promote Th17 differentiation include RORα, BATF, IRF4, aryl hydrocarbon receptor, IκBζ, HIF-1, and RUNX187. Th17 cells highly express the chemokine receptor CCR688. CCR6 binds to CCL20, which is expressed by epithelial cells, synoviocytes, and Th17 cells within inflamed tissues88. After migrating to inflamed tissues, Th17 cells indirectly recruit myeloid cells by inducing G-CSF, CCL2, and chemokines; thus, IL-17 represents an important link between innate and adaptive immunity88. As will be discussed, Th17 cells are heterogenous and can co-produce IFN-γ; they can also shift their phenotype to become Th1 cells over time85. Th17 cells are implicated in the pathogenesis of autoimmune diseases including psoriasis and spondyloarthritis. Accordingly, biologics targeting IL-17 and its receptor (secukinumab, ixekizumab, brodalimumab) are effective for this group of conditions, as are antibodies that block Th17 differentiation by targeting IL-23 (guselkumab, risenkizumab, ustekinumab).
Tfh and Tph cells
Unlike other T helper effector subsets defined by selective cytokine production, Tfh cells are recognized by expression of CXCR5 (Figure 3). CXCR5 allows Tfh cells to localize in B cell-rich follicles and germinal centers, where they promote B cell responses. The signals that promote Tfh differentiation differ from those that induce other effector subsets because Tfh cells must be activated twice89, 90. Both activation events involve ICOS-ICOSL signaling, but they are mediated by two different types of APC: usually dendritic cells (DCs) first, followed by B cells. Other cytokines and TFs that enhance Tfh differentiation in mice include IL-6 and IL-27 via STAT3; TGF-β, Activin A, IL-12, and IL-23 promote human Tfh differentiation through STAT3 and STAT491, 92. Tfh-promoting factors also include PD-1, the ubiquitin ligase ITCH and TFs BATF, IRF4, TCF1/LEF1, and ASCL289. Most of these stimuli act by inducing BCL6, which limits differentiation of other subsets. By contrast, IL-2 represses BCL6 through STAT5 and BLIMP1 (encoded by Prdm1); BLIMP1 and BCL6 repress each other to fine tune responsiveness to IL-289. Other inhibitors of Tfh differentiation and function include FOXO1, FOXP1, and KLF289.
Tfh cells promote humoral immunity by inducing B cell proliferation, survival, and differentiation to plasmablasts or germinal center (GC) cells. Within the GC, Tfh cells upregulate CXCR5 and drive affinity maturation. IL-21 and CD40L are the most critical Tfh-derived signals, although other factors can also provide B cell help90. Accordingly, Tfh subsets have been described that provide help in different inflammatory milieus: Tfh1 for Th1-associated responses, Tfh2 for type 2 responses, and Tfh17 for Th17-associated responses93. Although there is some evidence for plasticity between Tfh subsets, the extent of Tfh heterogeneity and plasticity is still being delineated93.
Insights from single cell technologies have revealed a Tfh-related CD4 subset designated T peripheral helper cells (Tph), which reside in inflamed peripheral tissues94. Because Tfh and Tph frequencies often correlate, the two subsets are thought to develop under similar environmental signals94 Tph cells express PD-1 and produce IL-21. However, they express lower BCL6, more Blimp1 and more CCR2, CX3CR1, and CCR5 than Tfh cells94. They are less efficient than Tfh at providing help to naïve B cells, but they strongly induce affinity maturation in memory B cells94.
Treg cells
Regulatory T cells (Treg) do not orchestrate responses to specific pathogens, instead promoting peripheral and central tolerance. Most naturally occurring Tregs (nTregs) develop in the thymus when T cells with intermediate-affinity TCR for self-peptide encounter self-antigen and develop into antigen-specific suppressive cells (Figure 1)95. Intermediate-affinity TCR can respond to self-antigen, so nTregs undergo activation and proliferation at lower concentrations of self-antigen than their effector counterparts. A smaller group of Tregs derives from T effector cells that differentiate into peripherally derived Tregs (pTregs) outside of the thymus (Figure 3). In vitro, IL-2 and TGF-β strongly induce Treg development; Tregs are also exquisitely dependent upon IL-2 in vivo. Both nTregs and pTregs are defined by expression of FOXP3; inactivating FOXP3 mutations cause Immunodyregulation, polyendocrinopathy, enteropathy, X-linked (IPEX), which is typified by severe autoimmunity affecting various organs and by food allergy96. Some Tfh cells express FOXP3 and are designated Tfr cells.
Tregs likely exert their suppressive effects through contact-dependent mechanisms, secretion of cytokines, and production of other factors. Ectopic Foxp3 expression confers Treg-mediated suppressive capacity, although Foxp3-deficient cells generated under Treg-promoting conditions in vitro are also capable of suppression95, 97. Exogenous IL-2 and CD25 are critical for Treg survival and suppression. The checkpoint molecules CTLA4 and PD-1 are also key modulators of Treg function; CTLA4 inhibition is highly toxic to Tregs, allowing for enhanced antitumor responses. PD-1 inhibition, by contrast, enhances the suppressive activity of tumor-resident Tregs: Treg-specific PD-1 deletion causes tumor hyperproliferation; this may underlie paradoxical hyperproliferation in some patients who receive PD-1 blockade for cancer98.
Th9 cells and Th22 cells
Th9 cells are identified by their production of IL-9 (Figure 3). In vitro, Th9 cells are induced by a combination of IL-4 and TGF-β and express PU.1; accordingly, they have a role in allergic diseases and antihelminth responses97. However, Th9 cells are also found in patients with autoimmunity and promote antitumor immunity99. Other major Th9 inducers include IL-2 acting via STAT5, TSLP, IL-25 (IL-17RB), TNF family members, epithelial growth factor, and hypoxia. Th9 cells are phenotypically unstable compared to other subsets, and it has been proposed that they are an early activated Th2 subset; however, in vivo-generated Th9 cells appear to be a mature effector population with a distinct identity99-101.
Th22 cells are defined by production of IL-22 (Figure 3), which promotes mucosal immunity by reducing epithelial permeability, inducing chemokine expression in epithelial cells, promoting hepatic antimicrobial responses, and directly regulating commensal microorganisms102. IL-22 is induced by IL-23, IL-1, and IL-18; microbiota can also promote IL-22 production by activating AHR102. Like the relationship between Th9 and Th2 cells, that between Th22 and Th17 cells is unclear. Th17 cells and ILC3s can produce IL-22, yet a distinct population of IL-22-expressing T cells has also been described, suggesting that Th22 cells may represent a distinct T helper subset103, 104 However, there are no LDTFs linked to Th22 cells, and many IL-22-modulating TFs are expressed in Th17 cells, including SMADs, BATF, IRF4, and RORγt.
Subsets vs. states: the changing view of T helper specification
The discovery of distinct T helper subsets and elucidation of underlying mechanisms and transcriptomic programs was a crucial insight into the mechanisms by which CD4+ T cells target discrete immune responses against specific pathogens. However, improved methods for assessing the complex biology of T cells and ILCs – including transcriptomic and genomic accessibility profiling – suggest more a dynamic and plastic view of T helper subset specification (Figure 3). This comprises various states of activation, proliferation, differentiation, and memory that are shaped by local tissue environments, including metabolic and neuronal inputs. In this respect, lymphocyte programs are better conceptualized as multidimensional continua with a range of outcomes shaped by myriad intrinsic and extrinsic factors76.
Particularly in recent years, cutting edge assays like single cell RNAseq (scRNAseq) single cell ATACseq (scATACseq), and spatial transcriptomics have been extensively utilized to study CD4+ T cell states at the steady state and in various diseases including infection, cancer, and airway inflammation105-110. Single cell RNAseq enables the measurement of the transcriptomes of individual cells, offering unprecedented insight into the heterogeneity of cell subsets. Similarly, single cell ATACseq measures chromatin accessibility of individual cells. More recent technologies measure gene expression and chromatin accessibility in the same cell, termed ‘multiomics.’ Briefly, cells are purified from a tissue and ‘captured’ and processed. The result is a matrix of cells and genes (or chromosomal coordinates for ATACseq). Computational analyses then group cells together into clusters based on their transcriptomic or epigenomic similarity.
Single cell studies suggest that CD4+T cell transcriptional programs are heterogeneous, with populations that do not fit the classical view of subset differentiation. These populations are often classified as "ambiguous" or "unknown" and have features of several different effector states. This includes co-expression of hallmark effector cytokines like Ifng (Th1) and Il17a (Th17), or Il13 (Th2) and Il17a (Th17)106, 108 These phenotypes are largely influenced by environmental cues including microbiota and metabolic factors like free fatty acids 106, 108, 111. Multiple LDTFs can also be co-expressed in a given CD4+T cell. Tregs are a particularly good example: in addition to Foxp3, they can also express T-bet, Gata3, Rorγt, and Bcl6. T-bet promotes Treg trafficking to sites of Th1-associated inflammation, Gata3 is critical for Treg function and trafficking, and Rorγt is important for GI-resident Treg function112-114. This is an evolving field that could itself be the subject of a separate review.
CD8+ T cells: general concepts
In the 1970s, CD8+ T cells were identified as the primary cytotoxic T lymphocyte (CTL) subset115. Since then, CD8+ T cells have been implicated in a host of additional functions. In addition to promoting the cytotoxicity of infected or malignant target cells, CTL-induced cell death is important in the elimination of intracellular infections and tumor surveillance. Hence, tumor-specific CTLs are being engineered to treat various malignancies116. Activated CTLs induce apoptosis via several mechanisms. Perforin-containing granules released from CTL lysosomes create pores in target cell membranes, causing direct damage and permitting influx of other cytotoxic material. CTL granules also contain granzymes, which trigger target cell apoptosis by activating a caspase cascade, directly damaging DNA, and inducing pro-apoptotic cytokine release117. CTLs also produce cytokines like IFN-γ and TNF-α that enhance cytotoxicity and activate macrophage to promote target cell death.
CD8+ Distribution, subsets, and heterogeneity
Like CD4+ T cells, CD8+ cells receive various inputs after TCR activation that promote their proliferation and function. IL-7 and IL-15 promote constant self-renewal to maintain stable CD8+ populations (Figure 4)118. These include effector memory CD8+ T cells, which promote early immune responses, and long-lived central memory CD8+ cells, which prevent reinfection118. Resident memory CD8+ T cells reside in mucosal tissues that interface with the external environment and help prevent influx of pathogens; ICOS is important for their development118, 119. Re-activation of these memory CTLs is CD4-dependent and results in a robust cytotoxic response. In vivo, CD8+ T cells comprise a heterogeneous effector-to-memory gradient (Figure 4) that depends on environmental cues and genetic regulators like Tcf1, AP-1, Tox, Foxo1, STAT3, Zeb1, Bach2, and Bcl6120-126.
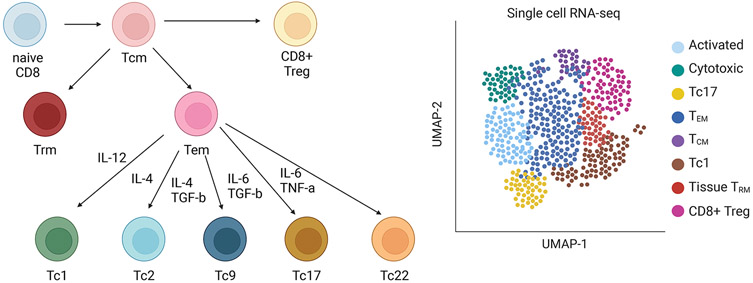
Figure 4. CD8+ subsets and heterogeneity.
CD8+ cells comprise several subsets. These include effector memory CD8+ T cells, which promote early immune responses; long-lived central memory CD8+ cells, which prevent reinfection; and resident memory CD8+ T cells, which reside in mucosal tissues and prevent influx of pathogens. Effector memory T cells are described to have several of their own subgroups, analogous to CD4+ helper subsets. Tc1 are analogous to Th1 and produce IFN-γ, Tc2 are analogous to Th2 and produce type 2 cytokines, Tc9 are analogous to Th9 and produce IL-9, Tc17 are analogous to Th17 and produce IL-17A, and Tc22 are analogous to Th22 and produce IL-22. In vivo, CD8+ T cells are characterized by a large amount of heterogeneity and overlap between these various subsets, with specific clones potentially co-expressing elements of multiple different subsets. This complexity is illustrated by single cell transcriptomic analyses.
Like CD4+ T cells, CD8+ effector memory T cells can also differentiate into different subsets under the influence of factors like IL-6, AHR, and SLAMF7 (Figure 4)127 These are similar to CD4+ subsets: Tc1 (analogous to Th1), Tc2 (analogous to Th2), Tc17 (analogous to Th17), and so forth127. CD8+ Tregs that promote immune tolerance have also been identified; some of these express Foxp3 like their CD4+ counterparts, but many CD8+ T cells with regulatory phenotypes do not express FoxP3128, 129. Regulatory CD8+ T cells can inhibit CTLs and CD4+ T cells via cytokine production or direct cytotoxic action and appear protect against autoimmune diseases129, 130.
CD8+ T cell exhaustion
Although exposure of a memory CD8+ to its cognate antigen generally induces re-activation and cytotoxicity, sustained antigen exposure during chronic infection and cancer can cause exhaustion131. Exhausted CD8+ T cells lose effector function, fail to self-renew, and have defective memory responses. They are characterized by high expression of inhibitory receptors, expression of specific transcriptional regulators, and metabolic dysfunction131, 132. While T cell exhaustion is undesirable in the setting of infection and malignancy, it can protect from autoimmunity133.
Both soluble and cellular mediators can regulate exhaustion in CD8+ T cells. IL-10 and TGF-β induce exhaustion, whereas IL-21 prevents it131. Type 1 interferons and IL-2 can promote or repress exhaustion depending on the timing and duration of exposure131, 134, 135 Mitochondrial stress and reduced mitochondrial fitness strongly induce T cell exhaustion, which may relate to the distinct metabolic profile of exhausted cells 131, 136, 137. Tregs and exhausted APCs also promote CD8+ T cell exhaustion, whereas effector CD4+ T cells antagonize exhaustion131.
Mechanistically, these soluble and cellular mediators act by inducing transcriptional networks that promote an exhausted phenotype131. Many TFs expressed by exhausted T cells are also expressed by conventional CD8+ T cells, including T-bet, Eomes, Tcf-1, and NFAT. NFAT seems particularly important, as increased NFAT:AP-1 ratios promote exhaustion138. TOX also induces an exhausted phenotype downstream of NFAT; the N4A family of TFs has a similar role138. Together, these TFs induce a series of epigenetic changes that is partially reversible but also comprises durable epigenetic scars that prevent the formerly exhausted cells from completely regaining function139.
Checkpoint molecules and immune checkpoint inhibitors
One of the hallmark features of exhausted T cells is expression of inhibitory checkpoint molecules: programmed cell death protein 1 (PD-1) and cytotoxic T lymphocyte antigen 4 (CTLA-4)131. PD-1 is expressed transiently in all T cells after TCR activation, where it binds PD-L1; in exhausted T cells, expression is sustained due to chronic antigen exposure131. Sustained expression of the inhibitory receptor CTLA4, which binds CD80/86 and prevents costimulatory engagement of CD28, also induces CD8+ T cell exhaustion and impaired cellular immunity140. Antagonists of PD1, PD-L1, and CTLA4 are termed immune checkpoint inhibitors (ICI) and prevent or reverse CD8 T cell exhaustion141. Re-activation of these cells promotes a robust cytotoxic immune response to tumor antigens, resulting in tumor clearance131. ICIs are highly effective for numerous malignancies, yet immune system reactivation can cause side effects resembling systemic autoimmune conditions141. These immune related adverse events (IRAEs) have been described in the skin, thyroid, GI tract, joints, and central nervous system141. Treatment with systemic immunomodulators is effective but can restore the T cell exhaustion phenotype, impairing the anti-tumor immune response141. Some newer strategies attempt to promote immune responses within the tumor microenvironment while preventing off-target IRAEs141.
Engineered T cells
Chimeric antigen receptor T cells have revolutionized the field of cancer immunotherapy since they were first used in the early 2010s 142, 143 These T cells express engineered antibodies specific to a target antigen (e.g., CD19 or CD20) as their receptors, which are coupled to costimulatory and signal transduction domains. While they have had tremendous success against hematologic malignancies, they are less efficacious against solid tumors144. Furthermore, some cancers can mutate to escape CAR-T cell recognition, and CAR-T cells themselves can cause toxicity144. Potential approaches to resolve these challenges include engineered safety switches that prevent toxicity and tri-specific CAR-T cells that target multiple epitopes to overcome immune evasion 145-147 Combining CAR-T therapy with mRNA lipid nanoparticle technology can generate CAR-T cells that are effective in vivo and are short-lived, thus preventing unforeseen off-target effects 148
CAR-T cells have additional applications beyond cancer immunotherapy. Recent studies have demonstrated their efficacy in the treatment of cardiac fibrosis, where they can restore heart function149. In autoimmune skin diseases, CAR-T cells have been engineered that target and kill autoreactive B cells, preventing autoimmunity1. Thus, CAR-T cells are a promising therapy that could be used to target a broad range of diseases, representing a major advance in targeted therapeutics.
Non-conventional T cells
In addition to conventional CD4+ and CD8+ T cells, several rare non-conventional populations have substantial immunological roles. γδT cells develop in the thymus, where their TCR is generated by RAG-mediated recombination of Tcrd and Tcrg and recognize antigen independent of MHC presentation150. γδT cells acquire the ability to secrete cytokines during development, allowing for a quick, innate-like response to tissue insult. After development, they preferentially home to the skin, lung, and intestines, where they have an important role in tissue repair 151. γδT cells also have a role in short-term memory, thermogenesis, and neuronal synaptic plasticity; dysregulation is linked to psoriasis and atopic dermatitis150, 152, 153. A major class of γδ T cells is activated by nonpeptide, phosphorylated antigens and are dependent upon butyrophilin, which is similar to co-stimulatory B7 molecules154.
NKT cells have properties of both NK and T cells and recognize lipid antigens presented by the MHC-like CD1d molecule. They develop in the thymus and depend on CD1d+ DP thymocytes rather than thymic epithelial cells155. Like γδT cells, they can quickly produce cytokines upon activation and have roles in infection, autoimmunity, graft-versus-host disease, and allergy155. Invariant NKT cells (iNKT), also referred to as type I NKT cells, recognize the lipid antigen α-galactosylceramide (αGalCer) and are sometimes subdivided into Type 1, Type 2, and Type 17 iNKT cells based on TF expression156. Type II NKT cells are not to be confused with Type 2 iNKT cells – which are a subset of Type I NKT (iNKT) cells and therefore react to αGalCer. Type II NKT cells, which are less well explored than iNKT cells, express a different type of antigen receptor that is not αGalCer-reactive but rather has a larger range of lipid specificities157.
Mucosal associated invariant T (MAIT) cells recognize microbial riboflavin-derivatives presented on MHC I-related MR1 molecules and are more abundant in humans than in mice. They are rapidly activated through TCR-dependent and independent stimuli, have innate-like effector properties, and are important for defense against pathogens, tissue repair and wound healing158. Reduced numbers of MAIT cells are seen in patients with allergic and autoimmune diseases, and MAIT cells may also have a role in antitumor immunity158.
CD4+CD8αα+ intraepithelial lymphocytes are thought to arise from CD4+ T cells in the gut and thus have both helper and cytotoxic functions. CD4+ T cells can extinguish ThPOK expression and can reactivate CD8 expression. CD4+ cytotoxic T cells have also been identified in the setting of viral infections.
T cell trafficking
To mediate immune responses, T cells must migrate to the lymph nodes for activation and then to peripheral tissue to execute their effector functions. T cell trafficking is regulated by a host of different receptors and signals. Naïve T cells enter lymph nodes when L-selectin on their cell surface recognizes its ligands on the node’s postcapillary venule. This causes the T cells to adhere to the venules, roll, Afterwards, they primarily engage in non-informed motion through lymphoid organs to maximize their likelihood of encountering their cognate antigen159. Recent advances in intravital lymph node imaging reveal that naïve CD4+ T cells are localized close to the medulla while CD8+ T cells are distributed within the cortex, in part because CD4+ velocity is depth-dependent while CD8+ T cells maintain a constant velocity regardless of depth160.
After T cells are activated, they are guided by various chemokines, integrins, and other tissue-homing receptors to migrate to various peripheral organs. For example, CLA+ T cells home to the skin, while α4β7+ cells migrate to the gut161. Chemokines are G protein coupled receptors that can function as monomers, homodimers, heterodimers, or oligomers. This variety allows for complex and nuanced regulation of T cell migration and function. In cancers like melanoma, chemokines can promote immune cell trafficking and antitumor immunity; reduced chemokine expression impairs antitumor immunity, and this may become a therapeutic target for tumor immunotherapy161, 162.
Once reaching target organs, cell adhesion molecules like selectins and integrins permit entry into peripheral tissue161. T cells move through peripheral tissue quickly, moving through natural channels in the extracellular matrix 163, 164 Local chemokines and adhesion molecules can influence migration, but recent advances in intravital microscopy have revealed that T cell locomotion depends more on the topography of the extracellular matrix165. The balance of inputs from chemokines, adhesion molecules, and environmental topography is organ-specific and varies between different tissues163.
Aside from potential effects on antitumor immunity, dysfunctional T cell migration can cause monogenic immune dysregulatory disorders. Gain of function mutations CXCR4 prevent leukocyte migration out of bone marrow and primary lymphoid organs. This causes WHIM syndrome, which is typified by HPV infection, hypogammaglobulinemia, immunodeficiency, and myelokathexis166 Conversely, blocking T cell trafficking is a successful therapeutic strategy for inflammatory bowel disease (IBD) and multiple sclerosis (MS). Three FDA-approved monoclonal antibodies target T cell trafficking: vedolizumab (anti-α4/β7 integrin), natalizumab (anti-α4 integrin), along with fingolimod and related small molecules. Although natalizumab increases the risk of progressive multifocal leukoencephalopathy, limiting its clinical use, vedolizumab and fingolimod are widely used for IBD and MS, respectively.
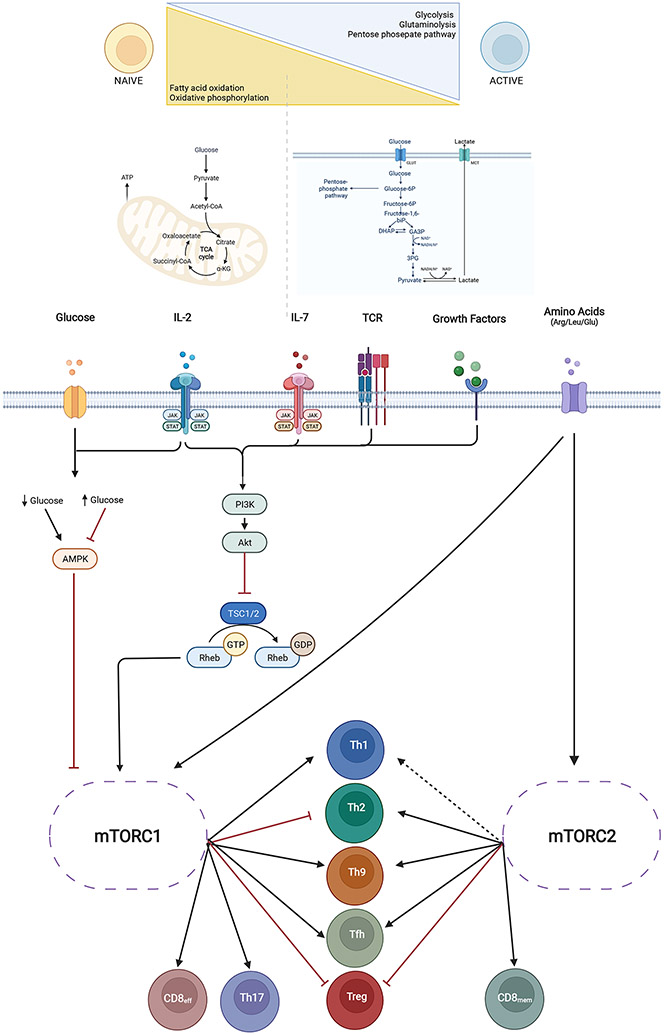
T cell immunometabolism
The energy requirements of T cells vary based on cell identity, activation, and function (Figure 5)167 Naïve T cells have relatively low metabolic demands and primarily rely on fatty acid oxidation (FAO) and oxidative phosphorylation (OXPHOS)167. Activated T cells undergo rapid proliferation, during which OXPHOS is replaced by glycolysis, the pentose phosphate pathway, and glutaminolysis 167, 168 The shift in energy production is accompanied by mitochondrial ultrastructural modifications that facilitate the metabolic transition169. Memory T cells also undergo mitochondrial remodeling that promotes a quiescent metabolic state and increases longevity169. Hypoxia-induced mitochondrial remodeling can also promote T cell exhaustion, reducing antitumor immunity137.
Figure 5. T cell metabolism.
Naïve T cells have relatively low metabolic demands and primarily rely on oxidative phosphorylation, whereas activated and proliferating T cells depend upon glycolysis and glutaminolysis. These metabolic shifts are accompanied by mitochondrial remodeling, which also plays a role in T cell exhaustion and antitumor immunity. Metabolic pathways are extensively targeted by disease-modifying drugs to treat immune-dysregulatory disease. Rapamycin (sirolimus) targets mechanistic Target of Rapamycin (mTOR), which is activated ty T cell receptor (TCR) stimulation and proliferation-inducing cytokines like IL (interleukin) −2 and IL-7. In environments with limited glucose availability, AMP protein kinase (AMPK) inhibits mTOR, pushing T cells towards quiescence and a memory phenotype
Since before the advent of targeted immunomodulation, metabolic pathways have been manipulated to treat immune-dysregulatory diseases. One of the oldest such therapies is rapamycin (sirolimus), which targets mechanistic Target of Rapamycin (mTOR, Figure 5). mTOR is a serine/threonine protein kinase that is activated by TCR stimulation through Pi3K/Akt and Glut1 (glucose transporter 1)167. IL-2 and IL-7, which are important for T cell proliferation and maintenance, also induce mTOR167. mTOR can also be regulated by local microenvironmental nutrients. In environments with limited glucose availability, AMP protein kinase (AMPK) inhibits mTOR, pushing T cells towards quiescence and a memory phenotype (Figure 5)170. AMPK is targeted by the immunomodulatory agent methotrexate. Leucine, glutamine, and arginine also regulate mTOR expression; in patients with atopy due to CARD11 LOF, glutamine supplementation can promote Th1 differentiation through mTOR, rescuing the atopic T cell phenotype171-173.
Aside from these cell-intrinsic immunomodulatory factors, environmental metabolic cues can also dramatically affect T cell function. One of the best studied such inputs is obesity, which causes increased local and systemic production of adipokines, fatty acids, and cytokines167. In obese patients, adipocyte-derived leptin promotes Th17 differentiation. Obesity also reduces PPARγ levels, enhancing Th17 differentiation and worsening skin inflammation174. The PPARγ agonists pioglitazone and rosiglitazone are FDA-approved for the treatment of Type 2 Diabetes and can reduce Th17 activity that contributes to early onset atherosclerosis in autoimmune diseases 175, 176 For example, Treatment of SLE patients with pioglitazone reduced vascular stiffness and cardiometabolic disease 177.
Amino acids can also have profound effects on T cell phenotype and function, apart from their effects on mTOR. Glutamine has pleiotropic roles in T cell metabolism and function and can promote or suppress T cell immune responses in a context-dependent fashion167. For example, glutamine antagonism can reduce tumor burden while promoting T cell activity, identifying glutamine metabolism as a potential target for cancer immunotherapy178. Methionine is important for CD8+ antitumor immunity, and proline enhances the cytotoxic effect of CAR-T cells179, 180. Hence, metabolic reprogramming is being actively investigated for optimization of CAR-T cells and other adoptive T cell therapies167, 179, 181.
T cell epigenetics
The three-dimensional organization of the genome is dynamic and highly cell-type and state specific. Chromatin remodeling is accomplished in part by the regulation of histone modifiers, including methylation and acetylation. Chromatin remodeling is a highly complex process, and relies on multiple TFs, co-factors, and chromatin modifying enzymes182. The importance of chromatin remodeling to T cells is made evident by the profound effects of deleting chromatin-modifying enzymes on T cell development and function 183-185 Further supporting the importance of 3D chromatin structure to T cells, natural genetic variation in 3D chromatin organization can lead to misfolding of key T cell genes, altering downstream gene expression and causing autoimmunity 186.
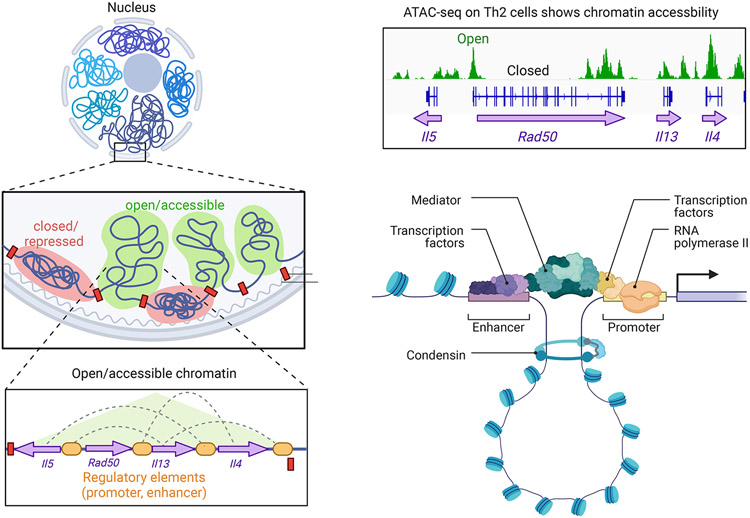
Noncoding DNA regulatory elements (REs) are classified based on their architectural and functional properties. REs that control the expression of nearby genes are termed cis-REs and include promoters, enhancers, and silencers (Figure 6). As their name suggests, enhancers upregulate the expression of many effector molecules, including cytokines, proliferation factors, and apoptosis genes187, 188. In some cases, a block of genes can be co-regulated as an extended locus; one classic example is the IL4-IL5-IL13 locus, which underlies coordinated expression of IL-4, IL-5, and IL-13 in Th2 cells (Figure 6). Enhancers can either strongly induce or fine-tune target gene expression: a subset of enhancers called super-enhancers are particularly important for genes associated with cell identity and function189, 190 Timing is also important: some enhancers become accessible during differentiation or TCR activation, whereas others are already accessible in mature mouse and human thymocytes, suggesting a poised state191, 192. Because many epigenome-wide profiling techniques only capture a “snapshot” in time, identification of functional enhancers can depend upon cell state and timing. Combining bulk sequencing, single cell genomics, and CRISPR-interference assays has partly addressed this problem by looking at multiple T cell states and populations to identify functional enhancers193.
Figure 6. T cell epigenetics.
Chromatin remodeling is a major requirement for regulation of gene expression and involves the architectural transition from an inactive/repressed state to an open/accessible state. Within accessible areas of chromatin, genes can be found in association with noncoding regulatory elements (REs) including promoters – located in proximity to the transcriptional start site (TSS) – and enhancers, which are distal to the TSS. Enhancers can directly interact with target genes by forming transcriptionally active loops, can recruit transcriptional modifiers, or can be transcribed to noncoding regulatory RNAs that have various regulatory roles. As an example, the extended type 2 locus is shown. This includes the Il4, Il13, Rad50, and Il5 genes as well as multiple noncoding regulatory elements that are open/accessible when the genes are open/poised for transcription.
The noncoding portion of the genome can regulate target gene expression through several mechanisms (Figure 6). Noncoding cis-REs can directly interact with target genes, recruit transcriptional modifiers, or can be transcribed to noncoding regulatory RNAs76, 97, 188, 194. Noncoding RNAs include microRNAs (miRs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs)195. miRs are perhaps the best studied of these and have been found to regulate Th1, Th17 and Treg differentiation, as well as CTL-cytotoxicity195-197. In CD8+ T cells, lncRNAs can regulate apoptosis, cytokine production, and cytotoxicity to influence antiviral and antitumor responses198-202. LncRNAs also modulate differentiation and cytokine production in CD4+ T cells, as well as activation and autophagy 194, 198, 203-205. Unlike miRs and lncRNAs, circRNAs can sometimes be translated into proteins or can act as noncoding RNAs. CircRNAs have been shown to regulate cell cycle and apoptosis in T cells, but they are not as well studied as miRs or lncRNAs195. In ways that are largely not understood, epigenomic factors and RNAs likely contribute to immunologic memory.
Conclusions
T cells sit at the center of adaptive immune responses, with CD8+ cells eliminating undesirable cells and CD4+ cells guiding immune responses to protect against specific pathogens. From thymus to circulation to peripheral tissues, T cell development, differentiation, migration, and function are tightly regulated to ensure that T cells have sufficient diversity to respond to a wide host of pathogens but do not contain any self-reactive clones. Upon encountering cognate antigen in the context of MHC and costimulation, T cells become activated, proliferate, and differentiate into effectors that can traffic to various tissues and carry out myriad effector functions. In CD4+ T cells, differentiation classically describes phenotypic skewing towards one of several discrete subsets with an associated lineage-defining transcription factor and one or more hallmark effector cytokines. Recent advances in single cell transcriptomic and epigenomic processing have led to a re-evaluation of this framework, and it is now thought that each T cell exists on a continuum molded by external and cell-intrinsic factors, including immunometabolic factors. In CD8+ cells, differentiation classically results in a similarly distinct commitment to an effector memory, central memory, or resident memory phenotype. As in CD4+ T cells, it is becoming clear in vivo CD8+ T cells are heterogeneous and can co-express genes typical of multiple subsets, due to varied environmental and cell-intrinsic inputs. These signals modulate T cell identity and function through diverse mechanisms including chromatin remodeling – especially of super-enhancers – and modulation of noncoding RNAs. In some settings, T cells – particularly CD8+ T cells – develop exhaustion due to chronic antigen exposure and are unable to optimally function as effectors. Over the last several decades, major advances in the field have identified many disease-causing mechanisms of T cell dysregulation and have sparked the development of multiple therapeutic agents targeting T cells and their effector cytokines. This is a rapidly advancing field, and continued technological advances in intravital imaging, single cell sequencing, and spatial transcriptomics are expected to result in further insights.
Summary Box.
T cells are critical orchestrators of the adaptive immune response whose development is tightly regulated to permit responses to a broad selection of foreign antigen while limiting self-reactivity.
CD4+ T cells provide help to other immune cells and have multiple effector/regulatory subsets with characteristic regulatory transcription factors and effector cytokines.
CD8+ T cells are cytotoxic and also differentiate into different subsets under the influence of various environmental signals and transcription factors
Single cell technologies reveal considerable in vivo plasticity, overlap, and heterogeneity of CD4 and CD8 subsets at a single cell level.
Inhibitory checkpoint molecules limit T cell effector function and can promote T cell exhaustion; pharmacologic inhibitors of checkpoint molecules are used for tumor immunotherapy.
Engineered T cells that react to tumor antigens represent another major type of tumor immunotherapy and are being explored for other clinical indications.
Funding statement:
This work was funded by the intramural research programs of NIAID, NIAMS, and NCI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: J.J.O. and the National Institutes of Health hold patents related to therapeutic targeting of JAKs and have a Collaborative Research Agreement and Development Award with Pfizer. The other authors have no conflicts to disclose.
References
- 1.Ellebrecht CT, Bhoj VG, Nace A, Choi EJ, Mao X, Cho MJ, et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 2016; 353:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science 2018; 359:1361–5. [DOI] [PubMed] [Google Scholar]
- 3.Sadeqi Nezhad M, Seifalian A, Bagheri N, Yaghoubi S, Karimi MH, Adbollahpour-Alitappeh M. Chimeric Antigen Receptor Based Therapy as a Potential Approach in Autoimmune Diseases: How Close Are We to the Treatment? Front Immunol 2020; 11:603237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothenberg EV. T cell lineage commitment: identity and renunciation. J Immunol 2011; 186:6649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou W, Gao F, Romero-Wolf M, Jo S, Rothenberg EV. Single-cell deletion analyses show control of pro-T cell developmental speed and pathways by Tcf7, Spi1, Gata3, Bcl11a, Erg, and Bcl11b. Sci Immunol 2022; 7:eabml920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothenberg EV. Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev 2019; 33:1117–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hozumi K, Negishi N, Tsuchiya I, Abe N, Hirano K, Suzuki D, et al. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur J Immunol 2008; 38:977–85. [DOI] [PubMed] [Google Scholar]
- 8.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med 1996; 184:1137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 1996; 384:474–8. [DOI] [PubMed] [Google Scholar]
- 10.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol 2003; 4:533–9. [DOI] [PubMed] [Google Scholar]
- 11.Hosokawa H, Koizumi M, Masuhara K, Romero-Wolf M, Tanaka T, Nakayama T, et al. Stage-specific action of Runx1 and GATA3 controls silencing of PU.1 expression in mouse pro-T cells. J Exp Med 2021; 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosokawa H, Romero-Wolf M, Yui MA, Ungerbäck J, Quiloan MLG, Matsumoto M, et al. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol 2018; 19:1427–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell 1989; 59:1035–48. [DOI] [PubMed] [Google Scholar]
- 14.Notarangelo LD, Kim MS, Walter JE, Lee YN. Human RAG mutations: biochemistry and clinical implications. Nat Rev Immunol 2016; 16:234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 1990; 248:1517–23. [DOI] [PubMed] [Google Scholar]
- 16.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992; 68:869–77. [DOI] [PubMed] [Google Scholar]
- 17.Delmonte OM, Villa A, Notarangelo LD. Immune dysregulation in patients with RAG deficiency and other forms of combined immune deficiency. Blood 2020; 135:610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol 2003; 21:139–76. [DOI] [PubMed] [Google Scholar]
- 19.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 2014; 14:377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell 1994; 76:17–27. [DOI] [PubMed] [Google Scholar]
- 21.Brändle D, Müller C, Rülicke T, Hengartner H, Pircher H. Engagement of the T-cell receptor during positive selection in the thymus down-regulates RAG-1 expression. Proc Natl Acad Sci U S A 1992; 89:9529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 2000; 288:2369–73. [DOI] [PubMed] [Google Scholar]
- 23.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet 1997; 17:393–8. [DOI] [PubMed] [Google Scholar]
- 24.Finnish-German AC. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet 1997; 17:399–403. [DOI] [PubMed] [Google Scholar]
- 25.McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med 2008; 205:2575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breed ER, Watanabe M, Hogquist KA. Measuring Thymic Clonal Deletion at the Population Level. J Immunol 2019; 202:3226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 2005; 433:826–33. [DOI] [PubMed] [Google Scholar]
- 28.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol 2005; 6:373–81. [DOI] [PubMed] [Google Scholar]
- 29.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002; 111:621–33. [DOI] [PubMed] [Google Scholar]
- 30.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 2003; 100:7731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resop RS, Douaisi M, Craft J, Jachimowski LC, Blom B, Uittenbogaart CH. Sphingosine-1-phosphate/sphingosine-1-phosphate receptor 1 signaling is required for migration of naive human T cells from the thymus to the periphery. J Allergy Clin Immunol 2016; 138:551–7.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004; 427:355–60. [DOI] [PubMed] [Google Scholar]
- 33.Tsai HC, Han MH. Sphingosine-1-Phosphate (S1P) and S1P Signaling Pathway: Therapeutic Targets in Autoimmunity and Inflammation. Drugs 2016; 76:1067–79. [DOI] [PubMed] [Google Scholar]
- 34.van den Broek T, Borghans JAM, van Wijk F. The full spectrum of human naive T cells. Nat Rev Immunol 2018; 18:363–73. [DOI] [PubMed] [Google Scholar]
- 35.Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol 2011; 12:544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friesen TJ, Ji Q, Fink PJ. Recent thymic emigrants are tolerized in the absence of inflammation. J Exp Med 2016; 213:913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lythe G, Callard RE, Hoare RL, Molina-Paris C. How many TCR clonotypes does a body maintain? J Theor Biol 2016; 389:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irving BA, Weiss A. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell 1991; 64:891–901. [DOI] [PubMed] [Google Scholar]
- 39.Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell 1994; 76:263–74. [DOI] [PubMed] [Google Scholar]
- 40.Chan AC, Iwashima M, Turck CW, Weiss A. ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell 1992; 71:649–62. [DOI] [PubMed] [Google Scholar]
- 41.Artyomov MN, Lis M, Devadas S, Davis MM, Chakraborty AK. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc Natl Acad Sci U S A 2010; 107:16916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Au-Yeung BB, Shah NH, Shen L, Weiss A. ZAP-70 in Signaling, Biology, and Disease. Annu Rev Immunol 2018; 36:127–56. [DOI] [PubMed] [Google Scholar]
- 43.Hauck F, Randriamampita C, Martin E, Gerart S, Lambert N, Lim A, et al. Primary T-cell immunodeficiency with immunodysregulation caused by autosomal recessive LCK deficiency. J Allergy Clin Immunol 2012; 130:1144–52 e11. [DOI] [PubMed] [Google Scholar]
- 44.Bacchelli C, Moretti FA, Carmo M, Adams S, Stanescu HC, Pearce K, et al. Mutations in linker for activation of T cells (LAT) lead to a novel form of severe combined immunodeficiency. J Allergy Clin Immunol 2017; 139:634–42 e5. [DOI] [PubMed] [Google Scholar]
- 45.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol 2009; 27:591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.So T, Lee SW, Croft M. Tumor necrosis factor/tumor necrosis factor receptor family members that positively regulate immunity. Int J Hematol 2006; 83:1–11. [DOI] [PubMed] [Google Scholar]
- 47.Pribila JT, Quale AC, Mueller KL, Shimizu Y. Integrins and T cell-mediated immunity. Annu Rev Immunol 2004; 22:157–80. [DOI] [PubMed] [Google Scholar]
- 48.Slack RJ, Macdonald SJF, Roper JA, Jenkins RG, Hatley RJD. Emerging therapeutic opportunities for integrin inhibitors. Nat Rev Drug Discov 2022; 21:60–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, Huth S, Adam D, Selhuber-Unkel C. Reinforcement of integrin-mediated T-Lymphocyte adhesion by TNF-induced Inside-out Signaling. Sci Rep 2016; 6:30452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, et al. STATs shape the active enhancer landscape of T cell populations. Cell 2012; 151:981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 2010; 32:840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 2012; 149:467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 2012; 151:153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 2010; 33:153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joglekar AV, Li G. T cell antigen discovery. Nat Methods 2021; 18:873–80. [DOI] [PubMed] [Google Scholar]
- 56.Zhou C, Zhu C, Liu Q. Toward in silico Identification of Tumor Neoantigens in Immunotherapy. Trends Mol Med 2019; 25:980–92. [DOI] [PubMed] [Google Scholar]
- 57.Lam H, McNeil LK, Starobinets H, DeVault VL, Cohen RB, Twardowski P, et al. An Empirical Antigen Selection Method Identifies Neoantigens That Either Elicit Broad Antitumor T-cell Responses or Drive Tumor Growth. Cancer Discov 2021; 11:696–713. [DOI] [PubMed] [Google Scholar]
- 58.Lalvani A, Pathan AA, McShane H, Wilkinson RJ, Latif M, Conlon CP, et al. Rapid detection of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. Am J Respir Crit Care Med 2001; 163:824–8. [DOI] [PubMed] [Google Scholar]
- 59.Lalvani A, Pathan AA, Durkan H, Wilkinson KA, Whelan A, Deeks JJ, et al. Enhanced contact tracing and spatial tracking of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. The Lancet 2001; 357:2017–21. [DOI] [PubMed] [Google Scholar]
- 60.Huth A, Liang X, Krebs S, Blum H, Moosmann A. Antigen-Specific TCR Signatures of Cytomegalovirus Infection. J Immunol 2019; 202:979–90. [DOI] [PubMed] [Google Scholar]
- 61.Sacco K, Castagnoli R, Vakkilainen S, Liu C, Delmonte OM, Oguz C, et al. Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19. Nat Med 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020; 183:996–1012 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020; 584:457–62. [DOI] [PubMed] [Google Scholar]
- 64.Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol 2022; 23:186–93. [DOI] [PubMed] [Google Scholar]
- 65.Painter MM, Mathew D, Goel RR, Apostolidis SA, Pattekar A, Kuthuru O, et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 2021; 54:2133–42 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022; 603:488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geers D, Shamier MC, Bogers S, den Hartog G, Gommers L, Nieuwkoop NN, et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol 2021; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee YN, Frugoni F, Dobbs K, Tirosh I, Du L, Ververs FA, et al. Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Sci Immunol 2016; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong GK, Millar D, Penny S, Heather JM, Mistry P, Buettner N, et al. Accelerated Loss of TCR Repertoire Diversity in Common Variable Immunodeficiency. J Immunol 2016; 197:1642–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang M, Su Z, Abolhassani H, Zhang W, Jiang C, Cheng B, et al. T Cell Repertoire Abnormality in Immunodeficiency Patients with DNA Repair and Methylation Defects. J Clin Immunol 2022; 42:375–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pauken KE, Lagattuta KA, Lu BY, Lucca LE, Daud AI, Hafler DA, et al. TCR-sequencing in cancer and autoimmunity: barcodes and beyond. Trends Immunol 2022; 43:180–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tiniakou E, Fava A, McMahan ZH, Guhr T, O'Meally RN, Shah AA, et al. Definition of Naturally Processed Peptides Reveals Convergent Presentation of Autoantigenic Topoisomerase I Epitopes in Scleroderma. Arthritis Rheumatol 2020; 72:1375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kattah NH, Newell EW, Jarrell JA, Chu AD, Xie J, Kattah MG, et al. Tetramers reveal IL-17-secreting CD4+ T cells that are specific for U1-70 in lupus and mixed connective tissue disease. Proc Natl Acad Sci U S A 2015; 112:3044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwok WW, Roti M, Delong JH, Tan V, Wambre E, James EA, et al. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol 2010; 125:1407–9 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 2015; 16:144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 2010; 327:1098–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo Y, Alexander M, Gadina M, O'Shea JJ, Meylan F, Schwartz DM. JAK-STAT signaling in human disease: From genetic syndromes to clinical inhibition. J Allergy Clin Immunol 2021; 148:911–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, et al. Human T-bet Governs Innate and Innate-like Adaptive IFN-gamma Immunity against Mycobacteria. Cell 2020; 183:1826–47 e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kerner G, Rosain J, Guerin A, Al-Khabaz A, Oleaga-Quintas C, Rapaport F, et al. Inherited human IFN-gamma deficiency underlies mycobacterial disease. J Clin Invest 2020; 130:3158–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Picard C, Fieschi C, Altare F, Al-Jumaah S, Al-Hajjar S, Feinberg J, et al. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am J Hum Genet 2002; 70:336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldberg MF, Roeske EK, Ward LN, Pengo T, Dileepan T, Kotov DI, et al. Salmonella Persist in Activated Macrophages in T Cell-Sparse Granulomas but Are Contained by Surrounding CXCR3 Ligand-Positioned Th1 Cells. Immunity 2018; 49:1090–102 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakayama T, Hirahara K, Onodera A, Endo Y, Hosokawa H, Shinoda K, et al. Th2 Cells in Health and Disease. Annu Rev Immunol 2017; 35:53–84. [DOI] [PubMed] [Google Scholar]
- 83.Nagashima H, Mahlakoiv T, Shih HY, Davis FP, Meylan F, Huang Y, et al. Neuropeptide CGRP Limits Group 2 Innate Lymphoid Cell Responses and Constrains Type 2 Inflammation. Immunity 2019; 51:682–95 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Honda K, Arima M, Cheng G, Taki S, Hirata H, Eda F, et al. Prostaglandin D2 reinforces Th2 type inflammatory responses of airways to low-dose antigen through bronchial expression of macrophage-derived chemokine. J Exp Med 2003; 198:533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stockinger B, Omenetti S. The dichotomous nature of T helper 17 cells. Nat Rev Immunol 2017; 17:535–44. [DOI] [PubMed] [Google Scholar]
- 86.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science 2015; 349:606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Capone A, Volpe E. Transcriptional Regulators of T Helper 17 Cell Differentiation in Health and Autoimmune Diseases. Front Immunol 2020; 11:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li X, Bechara R, Zhao J, McGeachy MJ, Gaffen SL. IL-17 receptor-based signaling and implications for disease. Nat Immunol 2019; 20:1594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hart AP, Laufer TM. A review of signaling and transcriptional control in T follicular helper cell differentiation. J Leukoc Biol 2022; 111:173–95. [DOI] [PubMed] [Google Scholar]
- 90.Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019; 50:1132–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K, et al. The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol 2014; 15:856–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT, et al. Activin A programs the differentiation of human TFH cells. Nat Immunol 2016; 17:976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olatunde AC, Hale JS, Lamb TJ. Cytokine-skewed Tfh cells: functional consequences for B cell help. Trends Immunol 2021; 42:536–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshitomi H, Ueno H. Shared and distinct roles of T peripheral helper and T follicular helper cells in human diseases. Cell Mol Immunol 2021; 18:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, Ohkura N. Regulatory T Cells and Human Disease. Annu Rev Immunol 2020; 38:541–66. [DOI] [PubMed] [Google Scholar]
- 96.Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest 2000; 106:R75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwartz DM, Farley TK, Richoz N, Yao C, Shih HY, Petermann F, et al. Retinoic Acid Receptor Alpha Represses a Th9 Transcriptional and Epigenomic Program to Reduce Allergic Pathology. Immunity 2019; 50:106–20 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci U S A 2019; 116:9999–10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu Y, Wang Q, Xue G, Bi E, Ma X, Wang A, et al. Th9 Cells Represent a Unique Subset of CD4(+) T Cells Endowed with the Ability to Eradicate Advanced Tumors. Cancer Cell 2018; 33:1048–60 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Micosse C, von Meyenn L, Steck O, Kipfer E, Adam C, Simillion C, et al. Human "TH9" cells are a subpopulation of PPAR-gamma(+) TH2 cells. Sci Immunol 2019; 4. [DOI] [PubMed] [Google Scholar]
- 101.Ulrich BJ, Kharwadkar R, Chu M, Pajulas A, Muralidharan C, Koh B, et al. Allergic airway recall responses require IL-9 from resident memory CD4(+) T cells. Sci Immunol 2022; 7:eabg9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ouyang W, O'Garra A. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 2019; 50:871–91. [DOI] [PubMed] [Google Scholar]
- 103.Roy U, de Oliveira RS, Galvez EJC, Gronow A, Basic M, Perez LG, et al. Induction of IL-22-Producing CD4+ T Cells by Segmented Filamentous Bacteria Independent of Classical Th17 Cells. Front Immunol 2021; 12:671331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barnes JL, Plank MW, Asquith K, Maltby S, Sabino LR, Kaiko GE, et al. T-helper 22 cells develop as a distinct lineage from Th17 cells during bacterial infection and phenotypic stability is regulated by T-bet. Mucosal Immunol 2021; 14:1077–87. [DOI] [PubMed] [Google Scholar]
- 105.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun 2019; 10:4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tibbitt CA, Stark JM, Martens L, Ma J, Mold JE, Deswarte K, et al. Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity 2019; 51:169–84 e5. [DOI] [PubMed] [Google Scholar]
- 107.Seumois G, Ramirez-Suastegui C, Schmiedel BJ, Liang S, Peters B, Sette A, et al. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci Immunol 2020; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kiner E, Willie E, Vijaykumar B, Chowdhary K, Schmutz H, Chandler J, et al. Gut CD4(+) T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat Immunol 2021; 22:216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, et al. The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok. Immunity 2019; 50:91–105.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wen T, Rothenberg ME. Cell-by-cell deciphering of T cells in allergic inflammation. J Allergy Clin Immunol 2019; 144:1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest 2019; 129:2014–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, et al. Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol 2016; 9:444–57. [DOI] [PubMed] [Google Scholar]
- 113.Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest 2011; 121:4503–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 2011; 35:337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shiku H, Kisielow P, Bean MA, Takahashi T, Boyse EA, Oettgen HF, et al. Expression of T-cell differentiation antigens on effector cells in cell-mediated cytotoxicity in vitro. Evidence for functional heterogeneity related to the surface phenotype of T cells. J Exp Med 1975; 141:227–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Raskov H, Orhan A, Christensen JP, Gogenur I. Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br J Cancer 2021; 124:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hay ZLZ, Slansky JE. Granzymes: The Molecular Executors of Immune-Mediated Cytotoxicity. Int J Mol Sci 2022; 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martin MD, Badovinac VP. Defining Memory CD8 T Cell. Front Immunol 2018; 9:2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peng C, Huggins MA, Wanhainen KM, Knutson TP, Lu H, Georgiev H, et al. Engagement of the costimulatory molecule ICOS in tissues promotes establishment of CD8(+) tissue-resident memory T cells. Immunity 2022; 55:98–114 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, McDonald B, et al. Heterogenous Populations of Tissue-Resident CD8(+) T Cells Are Generated in Response to Infection and Malignancy. Immunity 2020; 52:808–24 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yao C, Lou G, Sun HW, Zhu Z, Sun Y, Chen Z, et al. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8(+) T cells. Nat Immunol 2021; 22:370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat Immunol 2019; 20:890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim MV, Ouyang W, Liao W, Zhang MQ, Li MO. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 2013; 39:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10- STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 2011; 35:792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 2016; 17:851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol 2004; 173:883–91. [DOI] [PubMed] [Google Scholar]
- 127.Loyal L, Warth S, Jurchott K, Molder F, Nikolaou C, Babel N, et al. SLAMF7 and IL-6R define distinct cytotoxic versus helper memory CD8(+) T cells. Nat Commun 2020; 11:6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mishra S, Liao W, Liu Y, Yang M, Ma C, Wu H, et al. TGF-beta and Eomes control the homeostasis of CD8+ regulatory T cells. J Exp Med 2021; 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shimokawa C, Kato T, Takeuchi T, Ohshima N, Furuki T, Ohtsu Y, et al. CD8(+) regulatory T cells are critical in prevention of autoimmune-mediated diabetes. Nat Commun 2020; 11:1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jin K, Wen Z, Wu B, Zhang H, Qiu J, Wang Y, et al. NOTCH-induced rerouting of endosomal trafficking disables regulatory T cells in vasculitis. J Clin Invest 2021; 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 2019; 37:457–95. [DOI] [PubMed] [Google Scholar]
- 132.Pritykin Y, van der Veeken J, Pine AR, Zhong Y, Sahin M, Mazutis L, et al. A unified atlas of CD8 T cell dysfunctional states in cancer and infection. Mol Cell 2021; 81:2477–93 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wiedeman AE, Muir VS, Rosasco MG, DeBerg HA, Presnell S, Haas B, et al. Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression. J Clin Invest 2020; 130:480–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kahan SM, Bakshi RK, Ingram JT, Hendrickson RC, Lefkowitz EJ, Crossman DK, et al. Intrinsic IL-2 production by effector CD8 T cells affects IL-2 signaling and promotes fate decisions, stemness, and protection. Sci Immunol 2022; 7:eabl6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu Y, Zhou N, Zhou L, Wang J, Zhou Y, Zhang T, et al. IL-2 regulates tumor-reactive CD8(+) T cell exhaustion by activating the aryl hydrocarbon receptor. Nat Immunol 2021; 22:358–69. [DOI] [PubMed] [Google Scholar]
- 136.Yu YR, Imrichova H, Wang H, Chao T, Xiao Z, Gao M, et al. Disturbed mitochondrial dynamics in CD8(+) TILs reinforce T cell exhaustion. Nat Immunol 2020; 21:1540–51. [DOI] [PubMed] [Google Scholar]
- 137.Scharping NE, Rivadeneira DB, Menk AV, Vignali PDA, Ford BR, Rittenhouse NL, et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat Immunol 2021; 22:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Seo W, Jerin C, Nishikawa H. Transcriptional regulatory network for the establishment of CD8(+) T cell exhaustion. Exp Mol Med 2021; 53:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Abdel-Hakeem MS, Manne S, Beltra JC, Stelekati E, Chen Z, Nzingha K, et al. Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nat Immunol 2021; 22:1008–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 2018; 558:454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: emerging immunological strategies. Nat Rev Drug Discov 2021; 20:899–919. [DOI] [PubMed] [Google Scholar]
- 142.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 2010; 116:4099–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med 2011; 365:725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sterner RC, Sterner RM. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J 2021; 11:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jan M, Scarfo I, Larson RC, Walker A, Schmidts A, Guirguis AA, et al. Reversible ON- and OFF-switch chimeric antigen receptors controlled by lenalidomide. Sci Transl Med 2021; 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu S, Yi M, Qin S, Wu K. Next generation chimeric antigen receptor T cells: safety strategies to overcome toxicity. Mol Cancer 2019; 18:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schneider D, Xiong Y, Wu D, Hu P, Alabanza L, Steimle B, et al. Trispecific CD19- CD20-CD22-targeting duoCAR-T cells eliminate antigen-heterogeneous B cell tumors in preclinical models. Sci Transl Med 2021; 13. [DOI] [PubMed] [Google Scholar]
- 148.Rurik JG, Tombácz I, Yadegari A, Méndez Fernández PO, Shewale SV, Li L, et al. CAR T cells produced in vivo to treat cardiac injury. Science 2022; 375:91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aghajanian H, Kimura T, Rurik JG, Hancock AS, Leibowitz MS, Li L, et al. Targeting cardiac fibrosis with engineered T cells. Nature 2019; 573:430–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ribot JC, Lopes N, Silva-Santos B. γδ T cells in tissue physiology and surveillance. Nat Rev Immunol 2021; 21:221–32. [DOI] [PubMed] [Google Scholar]
- 151.Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, et al. A role for skin gammadelta T cells in wound repair. Science 2002; 296:747–9. [DOI] [PubMed] [Google Scholar]
- 152.Ribeiro M, Brigas HC, Temido-Ferreira M, Pousinha PA, Regen T, Santa C, et al. Meningeal γδ T cell-derived IL-17 controls synaptic plasticity and short-term memory. Sci Immunol 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldberg EL, Shchukina I, Asher JL, Sidorov S, Artyomov MN, Dixit VD. Ketogenesis activates metabolically protective γδ T cells in visceral adipose tissue. Nat Metab 2020; 2:50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rigau M, Ostrouska S, Fulford TS, Johnson DN, Woods K, Ruan Z, et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by gammadelta T cells. Science 2020; 367. [DOI] [PubMed] [Google Scholar]
- 155.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol 2010; 11:197–206. [DOI] [PubMed] [Google Scholar]
- 156.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 2013; 14:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dasgupta S, Kumar V. Type II NKT cells: a distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics 2016; 68:665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol 2019; 20:1110–28. [DOI] [PubMed] [Google Scholar]
- 159.Krummel MF, Bartumeus F, Gerard A. T cell migration, search strategies and mechanisms. Nat Rev Immunol 2016; 16:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choe K, Hontani Y, Wang T, Hebert E, Ouzounov DG, Lai K, et al. Intravital three-photon microscopy allows visualization over the entire depth of mouse lymph nodes. Nat Immunol 2022; 23:330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sackstein R, Schatton T, Barthel SR. T-lymphocyte homing: an underappreciated yet critical hurdle for successful cancer immunotherapy. Lab Invest 2017; 97:669–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hoch T, Schulz D, Eling N, Gomez JM, Levesque MP, Bodenmiller B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci Immunol 2022; 7:eabk1692. [DOI] [PubMed] [Google Scholar]
- 163.Fowell DJ, Kim M. The spatio-temporal control of effector T cell migration. Nat Rev Immunol 2021; 21:582–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Renkawitz J, Kopf A, Stopp J, de Vries I, Driscoll MK, Merrin J, et al. Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature 2019; 568:546–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Reversat A, Gaertner F, Merrin J, Stopp J, Tasciyan S, Aguilera J, et al. Cellular locomotion using environmental topography. Nature 2020; 582:582–5. [DOI] [PubMed] [Google Scholar]
- 166.Hernandez PA, Gorlin RJ, Lukens JN, Taniuchi S, Bohinjec J, Francois F, et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet 2003; 34:70–4. [DOI] [PubMed] [Google Scholar]
- 167.Geltink RIK, Kyle RL, Pearce EL. Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu Rev Immunol 2018; 36:461–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chang CH, Curtis JD, Maggi LB Jr., Faubert B, Villarino AV, O'Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013; 153:1239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 2016; 166:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.He J, Shangguan X, Zhou W, Cao Y, Zheng Q, Tu J, et al. Glucose limitation activates AMPK coupled SENP1-Sirt3 signalling in mitochondria for T cell memory development. Nat Commun 2021; 12:4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012; 149:410–24. [DOI] [PubMed] [Google Scholar]
- 172.Ma CA, Stinson JR, Zhang Y, Abbott JK, Weinreich MA, Hauk PJ, et al. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat Genet 2017; 49:1192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, et al. Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell 2018; 175:1780–95 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bapat SP, Whitty C, Mowery CT, Liang Y, Yoo A, Jiang Z, et al. Obesity alters pathology and treatment response in inflammatory disease. Nature 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miao Y, Zheng Y, Geng Y, Yang L, Cao N, Dai Y, et al. The role of GLS1-mediated glutaminolysis/2-HG/H3K4me3 and GSH/ROS signals in Th17 responses counteracted by PPARgamma agonists. Theranostics 2021; 11:4531–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Castilla C, Jmelnitzky A, Chopita N, Belloni P, Landoni N. [Value of the endoscopic sphinctero-papillotomy by surgical radiocholangiodebitometry]. Acta Gastroenterol Latinoam 1988; 18:173–85. [PubMed] [Google Scholar]
- 177.Hasni S, Temesgen-Oyelakin Y, Davis M, Chu J, Poncio E, Naqi M, et al. Peroxisome proliferator activated receptor-gamma agonist pioglitazone improves vascular and metabolic dysfunction in systemic lupus erythematosus. Ann Rheum Dis 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 2019; 366:1013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ye L, Park JJ, Peng L, Yang Q, Chow RD, Dong MB, et al. A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab 2022; 34:595–614 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 2020; 585:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hao M, Hou S, Li W, Li K, Xue L, Hu Q, et al. Combination of metabolic intervention and T cell therapy enhances solid tumor immunotherapy. Sci Transl Med 2020; 12. [DOI] [PubMed] [Google Scholar]
- 182.Vahedi G Remodeling the chromatin landscape in T lymphocytes by a division of labor among transcription factors. Immunol Rev 2021; 300:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 2004; 20:719–33. [DOI] [PubMed] [Google Scholar]
- 184.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 2001; 15:763–74. [DOI] [PubMed] [Google Scholar]
- 185.Schmidl C, Delacher M, Huehn J, Feuerer M. Epigenetic mechanisms regulating T-cell responses. J Allergy Clin Immunol 2018; 142:728–43. [DOI] [PubMed] [Google Scholar]
- 186.Fasolino M, Goldman N, Wang W, Cattau B, Zhou Y, Petrovic J, et al. Genetic Variation in Type 1 Diabetes Reconfigures the 3D Chromatin Organization of T Cells and Alters Gene Expression. Immunity 2020; 52:257–74.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol 2012; 30:707–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Henning AN, Roychoudhuri R, Restifo NP. Epigenetic control of CD8(+) T cell differentiation. Nat Rev Immunol 2018; 18:340–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sciume G, Mikami Y, Jankovic D, Nagashima H, Villarino AV, Morrison T, et al. Rapid Enhancer Remodeling and Transcription Factor Repurposing Enable High Magnitude Gene Induction upon Acute Activation of NK Cells. Immunity 2020; 53:745–58 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 2015; 520:558–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chopp LB, Gopalan V, Ciucci T, Ruchinskas A, Rae Z, Lagarde M, et al. An Integrated Epigenomic and Transcriptomic Map of Mouse and Human αβ T Cell Development. Immunity 2020; 53:1182–201.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, et al. Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 2016; 165:1120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Giles JR, Manne S, Freilich E, Oldridge DA, Baxter AE, George S, et al. Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 2022; 55:557–74.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Petermann F, Pekowska A, Johnson CA, Jankovic D, Shih HY, Jiang K, et al. The Magnitude of IFN-gamma Responses Is Fine-Tuned by DNA Architecture and the Non-coding Transcript of Ifng-as1. Mol Cell 2019; 75:1229–42 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Taheri M, Barth DA, Kargl J, Rezaei O, Ghafouri-Fard S, Pichler M. Emerging Role of Non-Coding RNAs in Regulation of T-Lymphocyte Function. Front Immunol 2021; 12:756042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mikami Y, Philips RL, Sciume G, Petermann F, Meylan F, Nagashima H, et al. MicroRNA-221 and −222 modulate intestinal inflammatory Th17 cell response as negative feedback regulators downstream of interleukin-23. Immunity 2021; 54:514–25 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol 2014; 15:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Plasek LM, Valadkhan S. lncRNAs in T lymphocytes: RNA regulation at the heart of the immune response. Am J Physiol Cell Physiol 2021; 320:C415–c27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hudson WH, Prokhnevska N, Gensheimer J, Akondy R, McGuire DJ, Ahmed R, et al. Expression of novel long noncoding RNAs defines virus-specific effector and memory CD8(+) T cells. Nat Commun 2019; 10:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A 2015; 112:E3883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kotzin JJ, Iseka F, Wright J, Basavappa MG, Clark ML, Ali MA, et al. The long noncoding RNA Morrbid regulates CD8 T cells in response to viral infection. Proc Natl Acad Sci U S A 2019; 116:11916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ai Y, Wu S, Gao H, Wei H, Tang Z, Li X, et al. Repression of CRNDE enhances the anti-tumour activity of CD8 + T cells against oral squamous cell carcinoma through regulating miR-545–5p and TIM-3. J Cell Mol Med 2021; 25:10857–68. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 203.Fu J, Shi H, Wang B, Zhan T, Shao Y, Ye L, et al. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4(+) T cell activation and Sjögren's syndrome-like autoimmune response. J Autoimmun 2020; 107:102358. [DOI] [PubMed] [Google Scholar]
- 204.Lu J, Wang X, Zhang B, Li P, Du X, Qi F. The lncRNA PVT1 regulates autophagy in regulatory T cells to suppress heart transplant rejection in mice by targeting miR-146a. Cell Immunol 2021; 367:104400. [DOI] [PubMed] [Google Scholar]
- 205.Liu Q, Deng Y, Li C, Xie H, Liu Q, Ming S, et al. LncRNA GAS5 suppresses CD4(+) T cell activation by upregulating E4BP4 via inhibiting miR-92a-3p in systemic lupus erythematosus. Immunol Lett 2020; 227:41–7. [DOI] [PubMed] [Google Scholar]