Abstract
Amyotrophic lateral sclerosis is a rapidly progressing neurodegenerative disease characterized by the degeneration of motor neurons and loss of various muscular functions. Dyslipidaemia is prevalent in amyotrophic lateral sclerosis with aberrant changes mainly in cholesterol ester and triglyceride. Despite this, little is known about global lipid changes in amyotrophic lateral sclerosis or in relation to disease progression. The present study incorporated a longitudinal lipidomic analysis of amyotrophic lateral sclerosis serum with a comparison with healthy controls using advanced liquid chromatography-mass spectrometry. The results established that diglyceride, the precursor of triglyceride, was enriched the most, while ceramide was depleted the most in amyotrophic lateral sclerosis compared with controls, with the diglyceride species (18:1/18:1) correlating significantly to neurofilament light levels. The prenol lipid CoQ8 was also decreased in amyotrophic lateral sclerosis and correlated to neurofilament light levels. Most interestingly, the phospholipid phosphatidylethanolamine and its three derivatives decreased with disease progression, in contrast to changes with normal ageing. Unsaturated lipids that are prone to lipid peroxidation were elevated with disease progression with increases in the formation of toxic lipid products. Furthermore, in vitro studies revealed that phosphatidylethanolamine synthesis modulated TARDBP expression in SH-SY5Y neuronal cells. Finally, diglyceride, cholesterol ester and ceramide were identified as potential lipid biomarkers for amyotrophic lateral sclerosis diagnosis and monitoring disease progression. In summary, this study represents a longitudinal lipidomics analysis of amyotrophic lateral sclerosis serum and has provided new insights into multiple pathways of lipid dysregulation in amyotrophic lateral sclerosis.
Keywords: amyotrophic lateral sclerosis, lipidomics, biomarkers
Phan et al. report a longitudinal lipidomics analysis of amyotrophic lateral sclerosis serum and have provided new insights into multiple pathways of lipid dysregulation in amyotrophic lateral sclerosis.
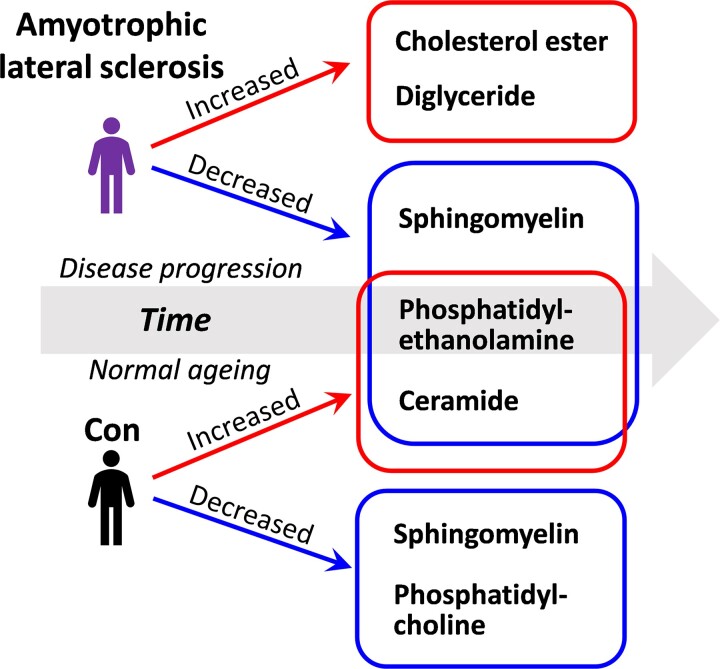
Graphical Abstract
Graphical abstract.
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by progressive degeneration of motor neurons that results in muscle atrophy, dysarthria, gradual paralysis and death usually within 2–5 years from diagnosis.1,2 ALS overlaps with another neurodegenerative disease, frontotemporal dementia (FTD), with TDP-43 as the predominant pathology in both diseases.3 The overwhelming majority, ∼90%, of ALS cases are sporadic. In familial cases, mutations in over 20 genes have been associated with ALS, including TAR DNA binding protein (TARDBP) that encode TDP-43.4 Currently, there is no cure for ALS and the FDA-approved drug Riluzole extends survival, albeit only by ∼3 months.5 Although the exact mechanism by which Riluzole improves survival is unclear, it appears to reduce glutamate release, attenuate persistent sodium and inward calcium currents,6,7 whilst at the same time leading to an improvement in functional capacity of mitochondria.8
Mitochondrial dysfunction is a predominant feature of ALS and has been associated with multiple pathogenic pathways in ALS, with a number of ALS candidate genes implicated in mitochondrial dysfunction, e.g. TDP-43, which forms aggregates in mitochondria and causes disruption to mtDNA transcription, ATP production and calcium homeostasis.9 The function of mitochondria is utmost essential in cell survival and metabolism, which includes ATP production via oxidative phosphorylation, calcium homeostasis and lipid biosynthesis. The role of mitochondria in neurons is of great importance by the fact that neurons consume high levels of ATP. In calcium homeostasis, mitochondria play a crucial role in regulating calcium concentrations that impact neurotransmitter release.
In terms of lipid biosynthesis, mitochondria are a major producer of the lipids phosphatidylethanolamine (PE)10 and coenzyme Q10 (CoQ10),11 which are important in the mitochondrial respiratory chain. PE is generated by four independent pathways in eukaryotic cells, including the PISD pathway in the mitochondria.12 Lipid dysregulation is recognized as a common feature of ALS, although most studies have focused mainly on cholesterol and triglycerides (TGs).13,14 Presently, very little is known about PE in the context of ALS. In contrast, the importance of CoQ10 in ALS has been well documented and CoQ10 has been trialled as a therapeutic supplement to improve mitochondrial function and survival.15
In respect to cholesterol and TG, studies produced inconsistent results with contradictory changes in the level of total cholesterol and TG.13,16,17 The one consistent result, however, has been the strong association between elevated serum cholesterol and TG levels with prolonged survival of ALS patients.18-20 The increase in cholesterol ester (ChE) has been shown to be associated with ALS risk in a retrospective metabolic study.21 ChE is also known to mediate oxidative stress-induced death in ALS motor neurons.22
Early studies were based on lipid measurements using indirect enzymatic/colorimetric assays. These assays are, however, limited to measuring only a few lipid classes, e.g. cholesterol and TG that are present in moderate-to-high abundance in serum. Furthermore, they cannot measure individual lipid species, of which there are thousands in the serum. Recently, analysis of lipids using mass spectrometry-based lipidomics has come to the fore in the field of neurodegenerative diseases, including ALS.23-25 It has enabled the measurement of all lipid classes and lipid species present in the serum with greater accuracy and specificity. Importantly, it has allowed the detection of small, yet significant, changes in lipids that are critical to disease processes. Recent lipidomics analyses of FTD serum have revealed that lipid dysregulation underlies the FTD pathophysiology linked to neurodegeneration.26,27
The present study incorporated a longitudinal analysis of lipidomics in the serum of ALS patients. Our primary aim was to identify lipid pathways that were altered in ALS, to understand lipid dysregulation that may contribute to ALS pathogenesis.28 Our secondary aim was to identify lipids for the purpose of developing biomarkers for ALS diagnosis and for monitoring disease progression.
Materials and methods
Amyotrophic lateral sclerosis patient cohort
Individuals diagnosed with sporadic ALS29,30 (n = 28; Supplementary Table 1) were recruited from the forefront multidisciplinary ALS clinic at the University of Sydney Brain and Mind Centre. Results were compared with those of healthy control volunteers31 (n = 22; Supplementary Table 1) without neurological or psychiatric disorders or cognitive impairment. Blood samples were collected at two-time points (i.e. T1 and T2) for each individual. The mean number of months between the two visits was 12 months for ALS and 23 months for controls. The study was approved by the University of New South Wales (approval number: HC12573) and the University of Sydney (approval numbers: 2012/160, 2014/539 and 2017/928) human research ethics committees. All methods were carried out in accordance with the relevant guidelines and regulations. Blood samples were obtained following written informed consent from the participant and/or primary career as legal representatives. All participants underwent a neurological examination, a comprehensive cognitive assessment and a structural brain MRI and met the current consensus diagnostic criteria for ALS32 or no neurological disease. Blood samples (9 mL) were collected in tubes (BD Vacutainer SST II Advance Tube #367958), and serum was prepared by centrifugation at 3500 rpm for 10 min at 4°C, which was then aliquoted and stored at −80°C until use.
Neurofilament assays
Frozen serum samples were sent to Quanterix (Billerica, MA, USA) and the concentration of neurofilament light (NFL) was measured using the Simoa HD-1 analyzer and software. Samples were randomized, blinded and measured in duplicates using NFL standards. The concentration of phosphorylated neurofilament heavy (pNFH) was measured using a pNFH ELISA kit (RD191138300R, Assay Matrix) and CLARIOstar plate reader (BMG Labtech).
Lipid extraction and liquid chromatography/mass spectrometry
Lipid extraction and liquid chromatography/mass spectrometry were carried out as per our previously published methods.26,27,33 Relative abundance of lipids was obtained from peak areas normalized to internal standards and data were analysed using LipidSearch software 4.1.16.
Thin-layer chromatography
Thin-layer chromatography was conducted on ALS serum to determine lipid concentrations using the Sulfo-Phospho-Vanillin method, as previously described.34
Calcium assay
Calcium assay was carried out on ALS and control serum using a calcium assay kit (Abcam, ab102505) and by following the manufacturer’s protocol as previously described.26
Malondialdehyde assay
Malondialdehyde (MDA) was carried out on ALS and control serum using a MDA assay kit (Abcam, ab118970) and by following the manufacturer’s protocol as previously described.26
Cell studies
SH-SY5Y neuronal cells were cultured in 12-well plates in Dulbecco’s modified Eagle’s medium containing 10% foetal calf serum, 1% glutamax, 0.5% glucose, 100 IU/mL penicillin and 100 μg/mL streptomycin at 37°C in humidified air containing 5% CO2. Cells were cultured in six-well plates and transfected with a vector containing PISD cDNA or TARDBP cDNA or vector-only control using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s protocol. After 48 h, the cells were harvested and total RNA was prepared for gene expression studies.
RNA extraction and quantitative PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen) following the manufacturer’s protocol. All procedures were carried out using RNase-free reagents and consumables. The 4 µg of RNA was reverse transcribed into cDNA using Moloney-murine leukaemia virus reverse transcriptase and random primers (Promega, Madison, WI, USA) in a 20 μL reaction volume. Gene expression was measured by quantitative polymerase chain reaction (qPCR) using the BioRad CFX Connect (BioRad, Australia) and the fluorescent dye SYBR Green (BioRad), following the manufacturer’s protocol. Briefly, each reaction (20 μL) contained 1× mastermix, 5 pmoles of primers and 1 μL of cDNA template. Amplification was carried out with 40 cycles at 94°C for 15 s and at 60°C for 1 min. Gene expression was normalized to the geometric mean of three housekeeping genes, GAPDH, β-ACTIN and PPIA. A no-template control was included for each PCR amplification assay.
Statistical analyses
Statistical analyses of the lipidomics data set were conducted in R version 4.0.3, using the lipidr package35 for differential expression and discriminant analysis. Lipid expression intensities were normalized with probabilistic quotient normalization36 and log2 transformed before being analysed for differential expression37 in each of the three comparisons. The first compared cases with ALS to healthy controls and the last two compared T1 to T2 within either of the ALS or the control cohorts, to look at the progression over time. Subsequent lipid set enrichment analyses38 were performed for each class of lipids to determine whether the class overall was differentially expressed. TGs were additionally analysed in lipid sets according to chain length and unsaturation and a linear model of the form [log2(FC) = β0 + β1 × unsaturation + β2 × chain length], so as to extract the statistically significant trend in this lipid class. Multivariate analyses (general linear model) with age and sex as covariates were used to determine differences in lipids and proteins using SPSS Statistics 26 (IBM, Chicago, IL, USA) and a significance set at P < 0.05. Association analyses were performed using Pearson’s correlation and statistical significance set at P < 0.05. In vitro cell studies were conducted in n = 6 replicates and significance was determined using the Student’s t-test with P < 0.05 considered significant. Individual data plots were generated using GraphPad Prism 9.
Results
Lipidomics analysis of amyotrophic lateral sclerosis serum
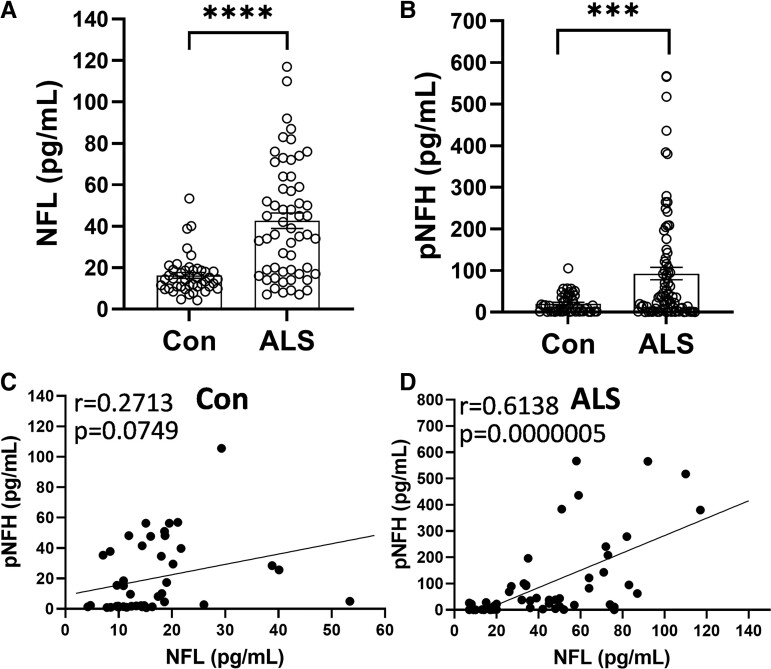
Given that lipid dysregulation is increasingly recognized as a key feature in a number of neurodegenerative diseases, a comprehensive lipidomics analysis was undertaken of ALS serum with the aim of understanding lipid dysregulation in ALS and identifying potential lipid biomarkers for ALS. Firstly, we measured NFL and pNFH chains to verify the manifestation of neurodegeneration in our ALS cohort. Both NFL and pNFH levels were significantly elevated in ALS compared with controls (Fig. 1A and B) and the two proteins correlated strongly with one another in the ALS cohort (Fig. 1C and D).
Figure 1.
Evidence of neurodegeneration in the ALS cohort. (A) NFL t-test, ****P < 0.00005, t = 6.039 and (B) pNFH t-test, ***P < 0.0005, t = 3.698 chains were elevated in ALS (n = 56) compared with controls (n = 44). Pearson’s correlation between NFL and pNFH in controls (C) and in ALS (D).
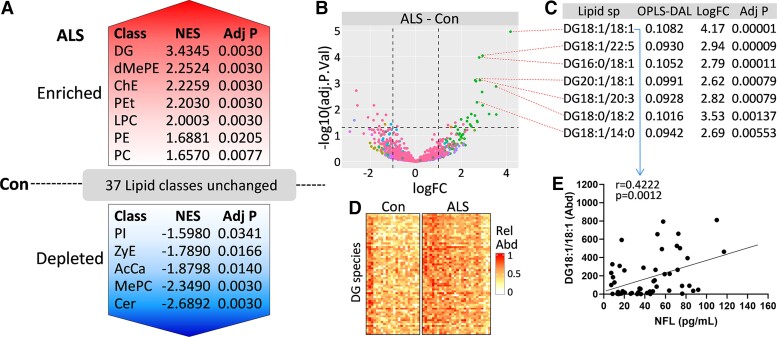
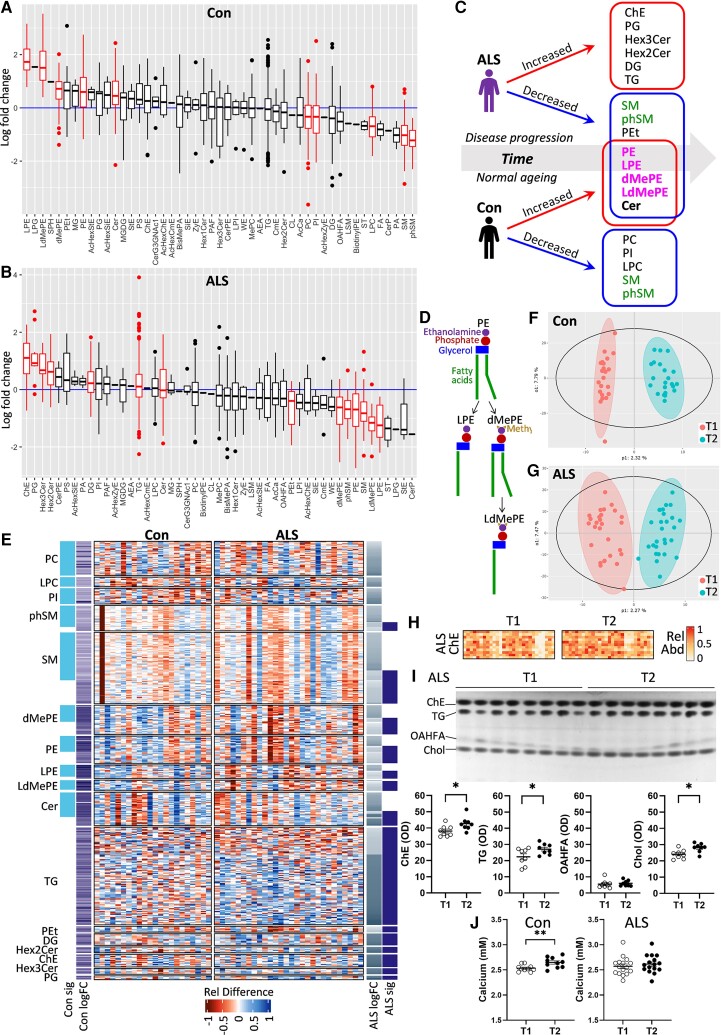
We then extracted total lipids from the same serum samples and the abundance of lipids was analysed using liquid chromatography-mass spectrometry and the LipidSearch software. Statistical analyses on the lipid data set were carried out using the R statistics package lipidr, covarying for age and sex. To detect preferential regulation of lipid classes in ALS serum compared with controls, we applied the lipid set enrichment analysis (LSEA) method to the data set. Enriched lipids were ranked by normalized enrichment score (NES) and significance. We found that seven lipid classes [diglyceride (DG), dimethylphosphatidylethanolamine (dMePE), ChE, phosphatidylethanol (PEt), lysophosphatidylcholine (LPC), PE, phosphatidylcholine (PC)] were enriched in ALS compared with controls with DG having the highest NES (Fig. 2A). The five lipid classes [ceramide (Cer), methylphosphatidylcholine (MePC), acylcarnitine (AcCa), ZyE, phytosphingosine (PI)] were depleted in ALS compared with controls with Cer having the lowest NES (Fig. 2A). These lipids belong to different functional groups with AcCa linked closely to mitochondrial function. The 37 other lipid classes were not altered in ALS compared with controls (Fig. 2A). A volcano plot shows that the most significantly altered lipid species was DG (18:1/18:1; Fig. 2B and C). A heatmap of DG species shows an overall increase in the relative abundance of DG in ALS compared with controls (Fig. 2D). Also, DG (18:1/18:1) correlated significantly with NFL levels in ALS (Fig. 2E). DG (18:1/18:1) could therefore be considered as a candidate biomarker for ALS diagnosis.
Figure 2.
Lipidomics analysis of ALS serum. (A) LSEA of lipid classes altered in ALS (n = 56) serum compared with controls (n = 44) was performed using the lipidr package for differential expression and discriminant analysis. Lipid expression intensities were normalized with probabilistic quotient normalization and log2 transformed before being analysed for differential expression and significance set at adjusted P < 0.05. The significant lipids were ranked by NES and significance. A volcano plot shows altered lipid species (B) with DG species being the most significantly altered species (C). (D) A heatmap showing DG species in ALS compared with controls. (E) DG (18:1/18:1) species correlated significantly with NFL levels in ALS.
Detection of coenzyme Q lipids
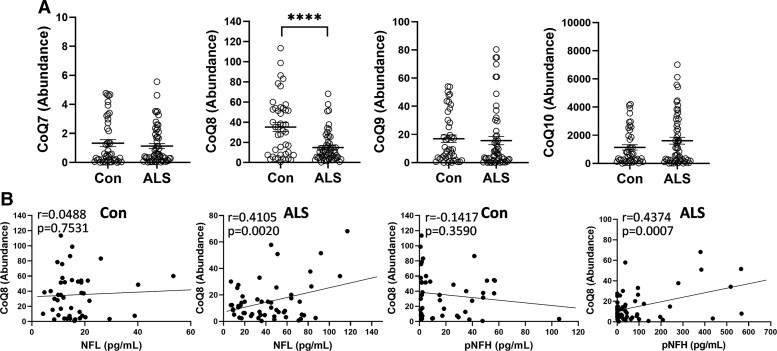
Another group of lipids that was detected by the lipidomics analysis was CoQ (also known as prenol lipids). These lipids are components of the electron transport chain that produces ATP in the mitochondria. We detected CoQ with 7, 8, 9 and 10 isoprene units, i.e. CoQ7, CoQ8, CoQ9 and CoQ10, respectively, with CoQ10 being the most abundant (Fig. 3A). Only CoQ8 was altered, i.e. decreased, in ALS serum compared with controls (Fig. 3A). CoQ8 also correlated significantly to NFL and pNFH in ALS (Fig. 3B). Other CoQ levels were not correlated to either NFL or pNFH (data not shown). CoQ8 could also be considered as a candidate biomarker for ALS diagnosis.
Figure 3.
Measurement of coenzyme Q lipids in ALS serum. (A) A comparison of coenzyme Q lipids CoQ7, CoQ8, CoQ9 and CoQ10 in ALS (n = 56) serum compared with controls (n = 44), t-test, ****P < 0.0001, t = 4.504. (B) Pearson’s correlation between CoQ8 and NFL and pNFH in controls and in ALS.
Changes in lipids with disease progression
Little is known about the global lipid changes in ALS with respect to disease progression. Here, we analysed lipids in ASL and control serum at two-time points (i.e. T1 and T2). We used the LSEA enrichment analysis to detect preferential regulation of lipids in T2 compared with T1 and lipids were ranked by their fold changes with time. We found that 10 lipid classes were significantly altered with time in controls (Fig. 4A), whereas 14 were significantly altered with time in ALS (Fig. 4B). Most interestingly, five [PE, lysophosphatidylethanolamine (LPE), dMePE, lysodimethylphosphatidylethanolamine (LdMePE) and Cer] of the eight lipids that increased with time in controls decreased with time in ALS (Fig. 4C). These were PE and PE derivatives (LPE, dMePE and LdMePE; Fig. 4D). PE is predominantly produced in the mitochondria. Sphingomyelin (SM) and phSM were the only lipids that decreased with time in both ALS and controls (Fig. 4C). In contrast, no same lipids were increased with time in ALS and controls (Fig. 4C). A heatmap shows the relative difference between T2 and T1 for each species for all of the lipid classes that were significantly altered with time (Fig. 4E), e.g. the abundance of TG species, overall, increased with time in ALS (Fig. 4E). Effects of time on the serum lipidome were demonstrated with the supervised orthogonal partial least squares discriminant analysis (OPLS-DA) method and a clear separation between the two-time points was evident for both the control (Fig. 4F) and ALS (Fig. 4G) cohorts. ChE was the most enriched lipid with time in ALS serum (Fig. 4B) and this was further illustrated by a heatmap of ChE species that shows an overall increase in the relative abundance of ChE in T2 compared with T1 (Fig. 4H). This result was validated using an alternative method of measuring lipids—thin layer chromatography—which showed a significant increase in ChE, along with validation of other lipids (Fig. 4I). Finally, we measured calcium levels and showed that they are increased with time in controls, but not in ALS (Fig. 4J).
Figure 4.
A longitudinal lipidomics assessment of ALS serum. LSEA of lipids altered in T2 compared with T1 in controls (A) and in ALS (B) was performed using the lipidr package for differential expression and discriminant analysis. Lipid expression intensities were normalized with probabilistic quotient normalization and log2 transformed before being analysed for differential expression and significance set at adjusted P < 0.05; significantly altered lipids are in red. (C) A summary of changes in lipids with time. Red boxes contain significantly increased lipids and blue boxes contain significantly decreased lipids. Green text indicates lipids decreased in both ALS and controls. Pink text indicates PE or PE derivatives. (D) Derivation of LPE, dMePE and LdMePE from PE. (E) A heatmap showing the relative difference between T2 and T1 for each species for all of the lipid classes that were significantly altered with time. (F) OPLS-DA of lipidome in T1 and T2 in controls (F) and in ALS (G). (H) A heatmap of ChE species in T2 compared with T1 in ALS. (I) Validation of mass spectrometry results using thin layer chromatography of ALS T1 (n = 8) and T2 (n = 8), t-test, *P < 0.05, t = 2.383 (ChE), t = 2.151 (TG), t = 2.913 (Chol). (J) Calcium levels of control T1 (n = 10) and T2 (n = 10), t-test, **P < 0.01, t = 2.931 and ALS T1 (n = 16) and T2 (n = 16).
To identify and formulate a lipid signature for ALS for disease diagnosis and disease progression, the three LSEA analyses (Con versus ALS, Con T1 versus T2, ALS T1 versus T2) were examined together. Of all the lipids present in the serum, only DG and ChE were enriched in ALS compared with controls and further enriched with disease progression, whereas Cer was depleted in ALS compared with controls and further depleted with disease progression. Therefore, DG, ChE and Cer and/or a combination of these three could be used for developing biomarkers for monitoring the disease progression of ALS.
Analysis of lipid unsaturation
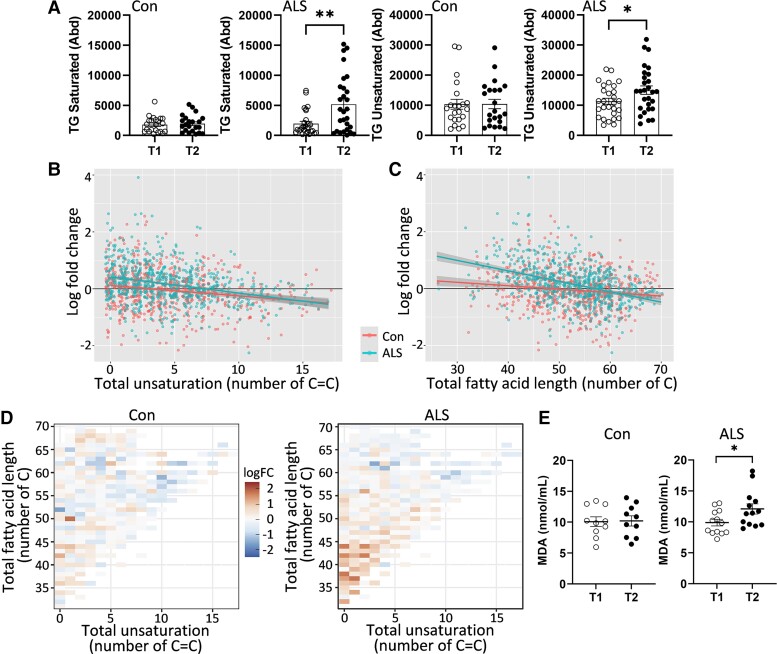
Apart from changes to lipid abundance, changes to lipid chemistry also impact on cell pathophysiology. We were particularly interested in the changes to lipid unsaturation with disease progression. Lipids become unsaturated when the hydrogen atoms are replaced by carbon–carbon double bonds (C=C) in the fatty acid chains. Importantly, unsaturated lipids are susceptible to lipid peroxidation and the greater the number of C=C the greater the susceptibility to lipid peroxidation. Lipid peroxidation results in the formation of toxic lipid products, i.e. lipid aldehydes, that cause oxidative stress and neurodegeneration.39 We analysed the changes in TG unsaturation with time in ALS serum and controls. TG has three fatty acid chains and is the most abundant lipid in the serum with the highest number of carbon atoms in the fatty acid chains and therefore most relevant to lipid peroxidation. We found that both saturated and unsaturated TG levels were elevated with time in ALS, whereas both unaltered with time in controls (Fig. 5A). This was further demonstrated by a plot of individual TG species that shows, overall, a greater fold-change in ALS compared with controls (Fig. 5B). The overall change in terms of chain length was also greater in ALS compared with controls (Fig. 5C). These changes were more pronounced in TG species with fewer C=C and shorter chain lengths (Fig. 5D).
Figure 5.
Analysis of lipid unsaturation. (A) Abundance of saturated t-test, *P < 0.05 t = 2.118 and unsaturated t-test, **P < 0.005, t = 3.288 TG in ALS (n = 28) and controls (n = 22). (B) A fold-change plot of individual TG species in terms of total unsaturation. (C) A fold-change plot of individual TG species in terms of total fatty acid length. (D) Heat maps show that changes were more pronounced in TG species with fewer C=C and shorter chain length in ALS. (E) Verification of increased lipid peroxidation product, MDA, with time in ALS (n = 13) t-test, *P < 0.05, t = 2.248 and control (n = 10).
To investigate the effect of the increased unsaturation in ALS on lipid peroxidation, we then assessed the level of MDA, a major lipid aldehyde, in the same ALS samples at two time points. MDA is highly reactive and toxic to cells.39 We found that MDA levels were significantly increased with time (Fig. 5E). These results indicate that lipid peroxidation increases with disease progression in ALS.
Effect of changes to PE synthesis on TARDBP expression
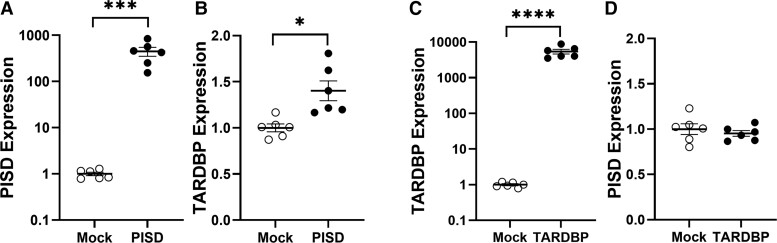
Since TDP-43 (encoded by the TARDBP gene) is the major pathology in ALS with ∼95% ALS cases bearing this phenotype,40 we were interested in the effect of changes to PE synthesis on TARDBP expression. PE is predominantly synthesized in the mitochondria by the key synthesis gene PISD. We altered the expression of PISD in SH-SY5Y neuronal cells and measured the effect on TARDBP expression and found that overexpression of PISD (Fig. 6A) upregulated TARDBP transcription (Fig. 6B). In a separate experiment, we altered the expression of TARDBP (Fig. 6C) and found no changes to PISD transcription (Fig. 6D), indicating that changes in TARDBP expression is downstream of PE synthesis.
Figure 6.
Effect of changes to PE synthesis on TARDBP expression. (A) SH-SY5Y neuronal cells (n = 6) were transfected with PISD cDNA t-test, ***P < 0.005, t = 4.513 and (B) its impact on TARDBP mRNA expression (n = 6) t-test, **P < 0.01, t = 3.483 was measured by qPCR. (C) In a separate experiment, SH-SY5Y neuronal cells (n = 6) were transfected with TARDBP cDNA t-test, ****P < 0.0001, t = 6.680 and (D) its impact on PISD mRNA expression (n = 6) was measured by qPCR.
Discussion
Increasing evidence shows that lipids are intrinsically linked to neurodegenerative processes with recent advances in mass spectrometry-based lipidomics having substantially improved our understanding of the importance of lipid dysregulation in neurodegenerative diseases. Comprehensive lipidomics analysis of blood serum from ALS patients established that DG, dMePE, ChE, PEt, LPC, PE and PC were increased, while Cer, MePC, AcCa, ZyE and PI were decreased in ALS compared with controls. DG was increased the most with the highest enrichment score and significance. Of all the DG species, DG (18:1/18:1) had the highest OPLS-DA loading score and significance, correlated significantly with NFL and therefore is a strong candidate for ALS biomarker development. Consistent with our results, an earlier lipidomics analysis of plasma showed that the DG class was significantly enriched in ALS compared with controls.25 We also detected a group of prenol lipids and found that CoQ8 levels were significantly decreased in ALS compared with controls. CoQ8 levels were also correlated to NFL levels and thus CoQ8 could also be considered as a biomarker for ALS.
We were also interested in understanding the lipid dysregulation associated with disease progression and analysed ALS and control serum in longitudinal samples. We observed that a greater number of lipid classes (i.e. 14) were altered with time in ALS compared with 10 in controls. The six lipid classes were increased with time in ALS, including the glycerolipids DG and TG, which is consistent with other lipidomics analysis of ALS plasma.25 We also observed that ChE levels increased with time in ALS. Similarly, ChE (18:1), ChE (18:2) and ChE (22:5) were shown to be significant markers of disease progression of ALS.24 Most interestingly, five lipid classes (PE, LPE, dMePE, LdMePE and Cer) that increased with time in controls decreased with time in ALS. In other words, the levels of these lipids were altered conversely with time in ALS compared with controls. LPE, dMePE and LdMePE are all derived from PE. In a mouse model, decreases in PE resulted in a greater availability of DG and increased formation of TG.41 Consistent with this, we observed increases in DG and TG with decreases in PE in ALS. These observations suggest that PE is particularly associated with disease progression of ALS. However, virtually nothing is known about PE in the context of ALS.
PE is a major membrane phospholipid that is particularly enriched in the brain, nerves and spinal cord, making up ∼45% of total phospholipid. It is synthesized in the mitochondria via the phosphatidylserine decarboxylase pathway by the PISD gene and is highly enriched in the organelle.42 It is also synthesized via a pathway steps located in the endoplasmic reticulum. PE has a cone shape because of its relatively small head group (see Fig. 4D) and exerts a lateral pressure that maintains membrane curvature and it is thought that PE regulates mitochondrial structure and morphology.43 The importance of PE in mitochondria is demonstrated by the fact that deletion of PISD in mice causes extensive mitochondrial fragmentation and lethality in embryonic mice.44 We also provided in vitro evidence that changes in PISD alter TARDBP expression in SH-SY5Y neuronal cells. The role of PE is also related to modulating Ca2+ ion levels in the function of the ATPase enzyme. Dysregulation of Ca2+ homeostasis in the mitochondria is evident at disease onset and progression of ALS that results in loss of mitochondrial membrane potential and subsequent dysfunction.45 Furthermore, as a major constituent of membranes, PE mediates membrane movement that allows successful cell growth and division,46 which would be important in neurogenesis. A large number of cytosolic proteins bind non-covalently to PE. These so-called PE-binding proteins are highly conserved in evolution and are involved in a number of processes, including neuronal development and the regulation of several signalling pathways.
A number of studies have shown that PE also plays a critical role in the oxidative phosphorylation of the electron transport chain that is relevant to ALS pathogenesis. In one study, knockdown of PISD in PSB-2 cells impaired the activities of electron transport chain complex and ATP production.43 In another study, PE was shown to be associated with the ubiquinol:cytochrome c reductase complex47 and that inclusion of PE in proteoliposomes reconstituted with the electron transport chain complex enhanced oxidative phosphorylation.48 Furthermore, PE was shown to modulate NADH:ubiquinone oxidoreductase complex activity in bovine mitochondria.49 These and other studies indicate that PE is essential in the regulation of mitochondrial function and structure, although the precise mechanisms by which PE regulates these processes are unclear.
LPE was also decreased with disease progression in ALS. It is derived from PE by the enzymatic removal of one of the two fatty acids (see Fig. 4D). Unlike PE, LPE is a minor constituent of cell membranes and has a different role to PE. It partakes in cell signalling and enzymatic activation. As a lysophospholipid, LPE typically binds to protein targets, such as receptors, kinases and phosphatases, activating specific cellular responses in a broad range of biological processes. Emerging evidence suggests that LPE acts like a neurotrophic factor that promotes neuronal differentiation and migration and that suppresses apoptosis via the MAPK signal cascade.50
Another group of lipids that partake in the mitochondrial respiratory chain is CoQ molecules. In our lipidomics analysis, we detected CoQ7, CoQ8, CoQ9 and CoQ10 but only CoQ8 levels were reduced in ALS compared with controls. Moreover, only CoQ8 levels were correlated to NFL levels in ALS. CoQ8 levels were, however, minor compared with CoQ10, which is regarded as the most important CoQ since it plays a pivotal role in the production of cellular energy in the mitochondria. Much research focus has been on the role of CoQ10 in ALS pathogenesis and on CoQ10 as a therapeutic supplement. In mouse studies, supplementation of CoQ10 reduced the decline in mitochondrial function associated with ageing.15 Supplementation of CoQ10 in a mouse model of ALS produced promising results of prolonged survival.51 However, a Phase II clinical trial of CoQ10 supplementation for 9 months had no significant effect on the disease progression of 185 ALS patients.52 In fact, most, if not all, clinical trials of the CoQ10 supplementation in patients with other neurodegenerative diseases produced, overall, no positive results. A likely reason for these failures could be due to, as a lipid molecule, the extreme hydrophobic nature of CoQ10, which renders it low bioavailability when orally administered. It is likely that only a fraction of CoQ10 ingested is absorbed from the digestive tract into the bloodstream.
Apart from CoQ8, AcCa and Cer were also decreased in ALS compared with controls. AcCa is a key mitochondrial lipid that transports long-chain fatty acids into the mitochondria, where the fatty acids are oxidized to produce ATP energy.53 Some Cer species modulate mitochondrial function and oxidative phosphorylation in the production of ATP.54,55 Cardiolipin is another lipid that is closely associated with mitochondria and although we detected this lipid, there was no significant change in ALS compared with controls.
Another aspect of lipid chemistry that could contribute to ALS pathogenesis is lipid peroxidation caused by increases in fatty acid unsaturation. We analysed the degree of unsaturation in TG, which is the most abundant lipid in the serum with the highest number of carbon atoms in the fatty acid chains. We revealed that unsaturated TG levels were significantly increased with time in ALS. Unsaturated lipids are chemically unstable and are prone to peroxidation (a process of oxidative degradation) that results in the formation of lipid aldehydes that are highly toxic to cells. We showed that the lipid aldehyde, MDA, was increased with time in ALS serum. Lipid peroxidation is particularly relevant to neurodegenerative diseases, because the brain is highly enriched in lipids and has a high oxygen consumption. Lipid aldehydes have been shown to contribute to the pathogenesis of a number of neurodegenerative diseases, including FTD.39,56,57 We also observed that increases in unsaturation were more prominent in TG species with shorter fatty acid chains. Interestingly, TG species with short-to-medium fatty acid chains have been shown to be associated with increases in resting energy expenditure58 as demonstrated by the hypermetabolism and resultant malnutrition common among ALS patients.59,60 Overall, these results suggest increased susceptibility to lipid peroxidation in ALS with disease progression and also support the use of lipid measurements to detect changes in lipid peroxidation in ALS serum.
Finally, in order to formulate a lipid signature for ALS, we examined the three LSEA analyses (Con versus ALS, Con T1 versus T2, ALS T1 versus T2). Of all the lipids present in the serum, only DG and ChE were increased in ALS compared with controls and further increased with disease progression and therefore could be considered as biomarkers for ALS diagnosis and for monitoring disease progression. Cer could also be considered as a potential biomarker for ALS as it was decreased in ALS compared with controls and further decreased with disease progression. However, these lipid changes are not entirely specific to ALS but could be associated with other neurodegenerative diseases and therefore further work is required to evaluate lipid changes across multiple neurodegenerative diseases. DG (18:1/18:1) and CoQ8 were identified as strong candidates for biomarker development for ALS. In a recent lipidomics study of ALS plasma, TG (68:12) was found to be correlated with NFL.24 SM and phSM were decreased with time in both ALS and controls and therefore could be considered as markers for normal ageing. Future work in biomarker development could include synthesizing specific lipid species and carrying out targeted quantification.
In conclusion, the present study utilized mass spectrometry-based lipidomics to identify multiple pathways of lipid dysregulation in ALS. Our findings provide new evidence that supports the concepts that aberrations in cellular structure and function contribute to ALS pathology, via specific effects in individual lipidomic pathways, potentially guiding more precise therapeutic approaches.61
Supplementary Material
Acknowledgement
The authors are grateful to the ForeFront research participants in the ALS and Frontier research clinics at the University of Sydney Brain and Mind Centre and to Nicole Mueller for coordinating the Frontier donor programme.
Glossary
Abbreviations
- AcCa =
acylcarnitine
- ALS =
amyotrophic lateral sclerosis
- Cer =
ceramide
- ChE =
cholesterol ester
- CoQ =
coenzyme Q
- DG =
diglyceride
- dMePE =
dimethylphosphatidylethanolamine
- FTD =
frontotemporal dementia
- LdMePE =
lysodimethylphosphatidylethanolamine
- LPC =
lysophosphatidylcholine
- LPE =
lysophosphatidylethanolamine
- MePC =
methylphosphatidylcholine
- NFL =
neurofilament light
- PC =
phosphatidylcholine
- PE =
phosphatidylethanolamine
- PEt =
phosphatidylethanol
- PG =
phosphatidylglycerol
- phSM =
sphingomyelin(phytosphingosine)
- PI =
phosphatidylinositol
- pNHL =
neurofilament heavy
- SM =
sphingomyelin
- TARDBP =
TAR DNA binding protein
- TG =
triglyceride
- ZyE =
zymosterol ester.
Contributor Information
Katherine Phan, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Medical Sciences, Sydney, NSW, Australia.
Ying He, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Medical Sciences, Sydney, NSW, Australia.
Surabhi Bhatia, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Medical Sciences, Sydney, NSW, Australia.
Russell Pickford, Bioanalytical Mass Spectrometry Facility, University of New South Wales, Sydney, NSW, Australia.
Gordon McDonald, The University of Sydney, Sydney Informatics Hub, Sydney, NSW, Australia.
Srestha Mazumder, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia.
Hannah C Timmins, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia.
John R Hodges, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia.
Olivier Piguet, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Psychology, Sydney, NSW, Australia.
Nicolas Dzamko, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Medical Sciences, Sydney, NSW, Australia.
Glenda M Halliday, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Medical Sciences, Sydney, NSW, Australia.
Matthew C Kiernan, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; Institute of Clinical Neurosciences, Royal Prince Alfred Hospital, Sydney, NSW, Australia.
Woojin Scott Kim, The University of Sydney, Brain and Mind Centre, Sydney, NSW, Australia; The University of Sydney, School of Medical Sciences, Sydney, NSW, Australia.
Funding
This work was supported by funding to ForeFront, a collaborative research group dedicated to the study of FTD and ALS, from the National Health and Medical Research Council of Australia (NHMRC) grants (#1132524, #1095127). M.C.K. is a NHMRC Practitioner Fellow (#1156093). G.M.H. is a NHMRC Senior Leadership Fellow (#1176607). O.P. is supported by a NHMRC Senior Research Fellowship (GNT1103258).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain Communications online.
Data availability
Lipidomics raw data were generated at Bioanalytical Mass Spectrometry Facility, University of New South Wales. Derived data supporting the findings of this study are available from the corresponding author, upon reasonable request. Other patient data cannot be made publicly available because the ethical approval and the informed consent from the patients included in this study did not cover placing the data into publicly open repositories.
References
- 1. Goutman SA, Hardiman O, Al-Chalabi A, et al. Recent advances in the diagnosis and prognosis of amyotrophic lateral sclerosis. Lancet Neurol. 2022;21(5):480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7(11):639–649. [DOI] [PubMed] [Google Scholar]
- 3. Tan RH, Kril JJ, Fatima M, et al. TDP-43 proteinopathies: Pathological identification of brain regions differentiating clinical phenotypes. Brain. 2015;138(10):3110–3122. [DOI] [PubMed] [Google Scholar]
- 4. Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet (London, England). 2011;377(9769):942–955. [DOI] [PubMed] [Google Scholar]
- 5. Kiernan MC, Vucic S, Talbot K, et al. Improving clinical trial outcomes in amyotrophic lateral sclerosis. Nat Rev Neurol. 2021;17(2):104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dharmadasa T, Kiernan MC. Riluzole, disease stage and survival in ALS. Lancet Neurol. 2018;17(5):385–386. [DOI] [PubMed] [Google Scholar]
- 7. Vucic S, Lin CS, Cheah BC, et al. Riluzole exerts central and peripheral modulating effects in amyotrophic lateral sclerosis. Brain. 2013;136(5):1361–1370. [DOI] [PubMed] [Google Scholar]
- 8. Siniscalchi A, Bonci A, Mercuri NB, Bernardi G. Effects of riluzole on rat cortical neurones: An in vitro electrophysiological study. Br J Pharmacol. 1997;120(2):225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benson BC, Shaw PJ, Azzouz M, Highley JR, Hautbergue GM. Proteinopathies as hallmarks of impaired gene expression, proteostasis and mitochondrial function in amyotrophic lateral sclerosis. Front Neurosci. 2021;15:783624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831(3):543–554. [DOI] [PubMed] [Google Scholar]
- 11. Mancuso M, Orsucci D, Calsolaro V, Choub A, Siciliano G. Coenzyme Q10 and neurological diseases. Pharmaceuticals (Basel). 2009;2(3):134–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calzada E, Onguka O, Claypool SM. Phosphatidylethanolamine metabolism in health and disease. Int Rev Cell Mol Biol. 2016;321:29–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahmed RM, Highton-Williamson E, Caga J, et al. Lipid metabolism and survival across the frontotemporal dementia-amyotrophic lateral sclerosis spectrum: Relationships to eating behavior and cognition. J Alzheimers Dis. 2017;61(2):773–783. [DOI] [PubMed] [Google Scholar]
- 14. Dupuis L, Corcia P, Fergani A, et al. Dyslipidemia is a protective factor in amyotrophic lateral sclerosis. Neurology. 2008;70(13):1004–1009. [DOI] [PubMed] [Google Scholar]
- 15. Takahashi M, Takahashi K. Water-soluble CoQ10 as a promising anti-aging agent for neurological dysfunction in brain mitochondria. Antioxidants (Basel). 2019;8(3):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang JW, Kim S-M, Kim H-J, et al. Hypolipidemia in patients with amyotrophic lateral sclerosis: A possible gender difference? J Clin Neurol. 2013;9(2):125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiò A, Calvo A, Ilardi A, et al. Lower serum lipid levels are related to respiratory impairment in patients with ALS. Neurology. 2009;73(20):1681–1685. [DOI] [PubMed] [Google Scholar]
- 18. Dorst J, Kühnlein P, Hendrich C, Kassubek J, Sperfeld AD, Ludolph AC. Patients with elevated triglyceride and cholesterol serum levels have a prolonged survival in amyotrophic lateral sclerosis. J Neurol. 2011;258(4):613–617. [DOI] [PubMed] [Google Scholar]
- 19. Huang R, Guo X, Chen X, et al. The serum lipid profiles of amyotrophic lateral sclerosis patients: A study from south-west China and a meta-analysis. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16(5-6):359–365. [DOI] [PubMed] [Google Scholar]
- 20. Liu J, Luo X, Chen X, Shang H. Lipid profile in patients with amyotrophic lateral sclerosis: A systematic review and meta-analysis. Front Neurol. 2020;11:567753–567753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bjornevik K, Zhang Z, O'Reilly ÉJ, et al. Prediagnostic plasma metabolomics and the risk of amyotrophic lateral sclerosis. Neurology. 2019;92(18):e2089–e2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cutler RG, Pedersen WA, Camandola S, Rothstein JD, Mattson MP. Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann Neurol. 2002;52(4):448–457. [DOI] [PubMed] [Google Scholar]
- 23. Goutman SA, Guo K, Savelieff MG, et al. Metabolomics identifies shared lipid pathways in independent amyotrophic lateral sclerosis cohorts. Brain. 2022;145(12):4425–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sol J, Jove M, Povedano M, et al. Lipidomic traits of plasma and cerebrospinal fluid in amyotrophic lateral sclerosis correlate with disease progression. Brain Commun. 2021;3(3):fcab143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Area-Gomez E, Larrea D, Yun T, et al. Lipidomics study of plasma from patients suggest that ALS and PLS are part of a continuum of motor neuron disorders. Sci Rep. 2021;11(1):13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phan K, He Y, Pickford R, et al. Uncovering pathophysiological changes in frontotemporal dementia using serum lipids. Sci Rep. 2020;10(1):3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He Y, Phan K, Bhatia S, et al. Increased VLCFA-lipids and ELOVL4 underlie neurodegeneration in frontotemporal dementia. Sci Rep. 2021;11(1):21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katzeff JS, Bright F, Phan K, et al. Biomarker discovery and development for frontotemporal dementia and amyotrophic lateral sclerosis. Brain. 2022;145(5):1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shefner JM, Al-Chalabi A, Baker MR, et al. A proposal for new diagnostic criteria for ALS. Clin Neurophysiol. 2020;131(8):1975–1978. [DOI] [PubMed] [Google Scholar]
- 30. Kiernan MC, Shefner JM, Kaji R, Burke D. Amyotrophic lateral sclerosis: A new diagnostic paradigm. J Neurol Neurosurg Psychiatry. 2020;91(9):903–904. [DOI] [PubMed] [Google Scholar]
- 31. Ahmed RM, MacMillan M, Bartley L, et al. Systemic metabolism in frontotemporal dementia. Neurology. 2014;83(20):1812–1818. [DOI] [PubMed] [Google Scholar]
- 32. Al-Chalabi A, Hardiman O, Kiernan MC, Chio A, Rix-Brooks B, van den Berg LH. Amyotrophic lateral sclerosis: Moving towards a new classification system. Lancet Neurol. 2016;15(11):1182–1194. [DOI] [PubMed] [Google Scholar]
- 33. Kim WS, Jary E, Pickford R, et al. Lipidomics analysis of behavioral variant frontotemporal dementia: A scope for biomarker development. Front Neurol. 2018;9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng YS, Zheng Y, VanderGheynst JS. Rapid quantitative analysis of lipids using a colorimetric method in a microplate format. Lipids. 2011;46(1):95–103. [DOI] [PubMed] [Google Scholar]
- 35. Mohamed A, Molendijk J, Hill MM. Lipidr: A software tool for data mining and analysis of lipidomics datasets. J Proteome Res. 2020;19(7):2890–2897. [DOI] [PubMed] [Google Scholar]
- 36. Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78(13):4281–4290. [DOI] [PubMed] [Google Scholar]
- 37. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021;060012:1-40. [Google Scholar]
- 39. Reed TT. Lipid peroxidation and neurodegenerative disease. Free Radic Biol Med. 2011;51(7):1302–1319. [DOI] [PubMed] [Google Scholar]
- 40. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- 41. Fullerton MD, Hakimuddin F, Bonen A, Bakovic M. The development of a metabolic disease phenotype in CTP:Phosphoethanolamine cytidylyltransferase-deficient mice. J Biol Chem. 2009;284(38):25704–25713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Percy AK, Moore JF, Carson MA, Waechter CJ. Characterization of brain phosphatidylserine decarboxylase: Localization in the mitochondrial inner membrane. Arch Biochem Biophys. 1983;223(2):484–494. [DOI] [PubMed] [Google Scholar]
- 43. Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem. 2013;288(6):4158–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steenbergen R, Nanowski TS, Beigneux A, Kulinski A, Young SG, Vance JE. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J Biol Chem. 2005;280(48):40032–40040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tradewell ML, Cooper LA, Minotti S, Durham HD. Calcium dysregulation, mitochondrial pathology and protein aggregation in a culture model of amyotrophic lateral sclerosis: Mechanistic relationship and differential sensitivity to intervention. Neurobiol Dis. 2011;42(3):265–275. [DOI] [PubMed] [Google Scholar]
- 46. Emoto K, Kobayashi T, Yamaji A, et al. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc Natl Acad Sci U S A. 1996;93(23):12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pfeiffer K, Gohil V, Stuart RA, et al. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278(52):52873–52880. [DOI] [PubMed] [Google Scholar]
- 48. Schagger H, Hagen T, Roth B, Brandt U, Link TA, von Jagow G. Phospholipid specificity of bovine heart bc1 complex. Eur J Biochem. 1990;190(1):123–130. [DOI] [PubMed] [Google Scholar]
- 49. Sharpley MS, Shannon RJ, Draghi F, Hirst J. Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry. 2006;45(1):241–248. [DOI] [PubMed] [Google Scholar]
- 50. Nishina A, Kimura H, Sekiguchi A, Fukumoto RH, Nakajima S, Furukawa S. Lysophosphatidylethanolamine in Grifola frondosa as a neurotrophic activator via activation of MAPK. J Lipid Res. 2006;47(7):1434–1443. [DOI] [PubMed] [Google Scholar]
- 51. Matthews RT, Yang L, Browne S, Baik M, Beal MF. Coenzyme Q10 administration increases brain mitochondrial concentrations and exerts neuroprotective effects. Proc Natl Acad Sci U S A. 1998;95(15):8892–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaufmann P, Thompson JL, Levy G, et al. Phase II trial of CoQ10 for ALS finds insufficient evidence to justify phase III. Ann Neurol. 2009;66(2):235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem J. 2002;361(3):417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Di Paola M, Cocco T, Lorusso M. Ceramide interaction with the respiratory chain of heart mitochondria. Biochemistry. 2000;39(22):6660–6668. [DOI] [PubMed] [Google Scholar]
- 55. Gudz TI, Tserng KY, Hoppel CL. Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J Biol Chem. 1997;272(39):24154–8. [DOI] [PubMed] [Google Scholar]
- 56. Martinez A, Carmona M, Portero-Otin M, Naudi A, Pamplona R, Ferrer I. Type-dependent oxidative damage in frontotemporal lobar degeneration: Cortical astrocytes are targets of oxidative damage. J Neuropathol Exp Neurol. 2008;67(12):1122–1136. [DOI] [PubMed] [Google Scholar]
- 57. Liou CJ, Tong M, Vonsattel JP, de la Monte SM. Altered brain expression of insulin and insulin-like growth factors in frontotemporal lobar degeneration: Another degenerative disease linked to dysregulation of insulin metabolic pathways. ASN Neuro. 2019;11:1759091419839515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. St-Onge M-P, Jones PJH. Physiological effects of medium-chain triglycerides: Potential agents in the prevention of obesity. J Nutr. 2002;132(3):329–332. [DOI] [PubMed] [Google Scholar]
- 59. Bouteloup C, Desport JC, Clavelou P, et al. Hypermetabolism in ALS patients: An early and persistent phenomenon. J Neurol. 2009;256(8):1236–1242. [DOI] [PubMed] [Google Scholar]
- 60. Desport JC, Torny F, Lacoste M, Preux PM, Couratier P. Hypermetabolism in ALS: Correlations with clinical and paraclinical parameters. Neurodegener Dis. 2005;2(3-4):202–207. [DOI] [PubMed] [Google Scholar]
- 61. Shibuya K, Otani R, Suzuki YI, Kuwabara S, Kiernan MC. Neuronal hyperexcitability and free radical toxicity in amyotrophic lateral sclerosis: Established and future targets. Pharmaceuticals (Basel). 2022;15(4):433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Lipidomics raw data were generated at Bioanalytical Mass Spectrometry Facility, University of New South Wales. Derived data supporting the findings of this study are available from the corresponding author, upon reasonable request. Other patient data cannot be made publicly available because the ethical approval and the informed consent from the patients included in this study did not cover placing the data into publicly open repositories.