Abstract
Fungal species have undergone and continue to undergo significant nomenclatural change, primarily due to the abandonment of dual species nomenclature in 2013 and the widespread application of molecular technologies in taxonomy allowing correction of past classification errors. These have effected numerous name changes concerning medically important species, but by far the group causing most concern are the Candida yeasts. Among common species, Candida krusei, Candida glabrata, Candida guilliermondii, Candida lusitaniae, and Candida rugosa have been changed to Pichia kudriavzevii, Nakaseomyces glabrata, Meyerozyma guilliermondii, Clavispora lusitaniae, and Diutina rugosa, respectively. There are currently no guidelines for microbiology laboratories on implementing changes, and there is ongoing concern that clinicians will dismiss or misinterpret laboratory reports using unfamiliar species names. Here, we have outlined the rationale for name changes across the major groups of clinically important fungi and have provided practical recommendations for managing change.
Keywords: Candida, clinical fungi, nomenclature, taxonomy
If we accept that the only constant in life is change, we can begin to understand that fungal name changes always have and always will occur. Fungal nomenclature has been undergoing extensive change for more than a decade. This can largely be attributed to the now commonplace role of molecular-based technologies in taxonomy, diagnostics, and epidemiology. Molecular studies have improved the way in which fungal species are defined and identified, permitting refinement of inter- and intraspecies phylogenetic relationships and correction of taxonomical errors arising from the phenotypic classification and identification methods used in the past. For this reason, the long-held convention of fungal species having 2 or more valid names for their teleomorph (sexual) and anamorph (asexual) states was abandoned in 2013 [1]. The subsequent need to rationalize existing names meant that some names in common use have been retained, whereas in other cases they have been replaced by the less commonly used name. Additional impacts of molecular studies include revealing extensive genetic variation within species that were originally ascribed by their morphology, leading to the description of additional species within them. Molecular analyses have shone a light on whether taxonomic groups that have been classified and named on the basis of shared morphological or phenotypic features actually share a single common ancestor (monophyletic) or whether the species have mixed ancestry such that not all species within the group are related (polyphyletic). In the case of polyphyletic genera, transfer of those species that do not share common ancestry into a more appropriate genus is warranted.
These changes form a critical part of an ongoing process of refinement in the way that we understand organisms to have evolved, to interact, and to behave. Changes in fungal species names have been occurring at a rapid pace over the past decade [2–4], and this has led to some heated debate in the arena of social media [5, 6] on the benefits and difficulties caused by such changes in clinical practice. Commonly the name change affects the genus, but the species epithet remains recognizable (eg, Scedosporium prolificans became Lomentospora prolificans), but this is not always the case (eg, Candida krusei became Pichia kudriavzevii); anecdotally, it seems to be the latter situation causing most concern. It is important to note that fungal nomenclature changes must strictly follow the International Code of Nomenclature for algae, fungi, and plants [7], and any wish to preserve certain names or parts thereof, is overridden by the nomenclatural priority of previous legitimate names for the species. However, nomenclatural changes are not new or unique to fungi, and numerous species name changes in the past have been accepted and embedded into clinical practice. Here we review nomenclature changes in clinically important fungi over the past 20 years and make recommendations on incorporating nomenclature change into laboratory reporting and clinical practice.
YEASTS AND YEAST-LIKE FUNGI
Candida
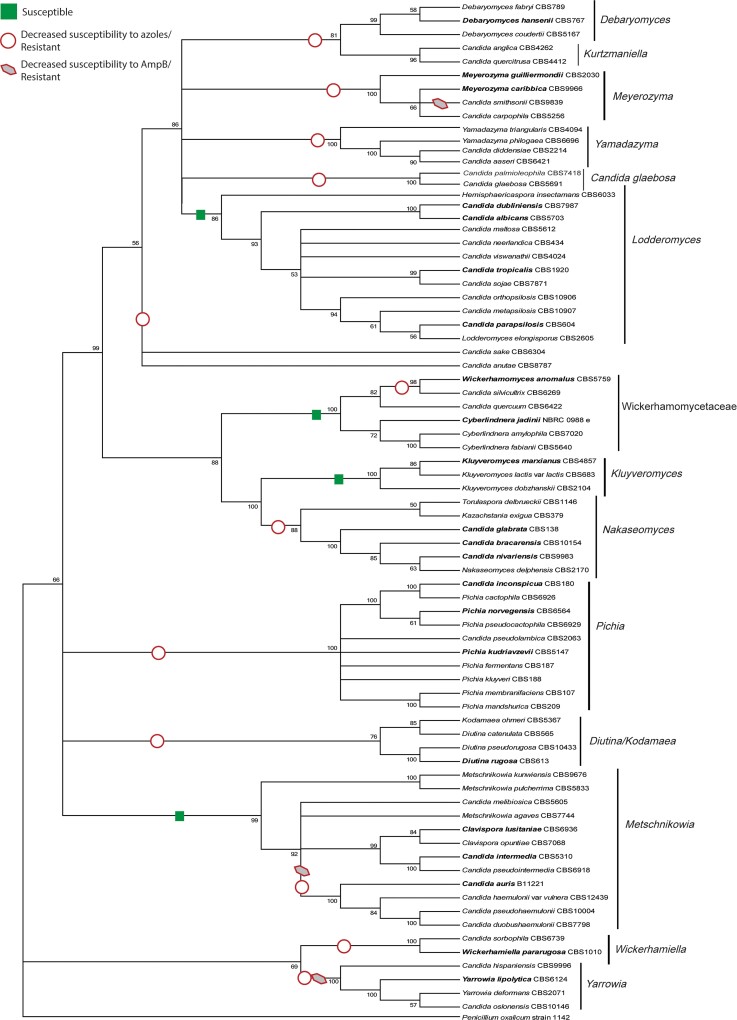
Arguably the group of fungi undergoing the most reclassification in recent times and causing most concern among clinicians and medical laboratorians is the ascomycetous yeasts, and particularly Candida, likely because these are a common cause of invasive and superficial infections encountered in both specialized and nonspecialized microbiology laboratories worldwide. The problem with Candida is that it represents a large, highly polyphyletic group of budding, white colony-forming yeasts in the subphylum Saccharomycotina, originally grouped together because of their similar morphology and lack of a defined teleomorph [8–10]. It does not meet the 3 generally accepted criteria of a genus: (1) monophyly, that is, all species within it evolving from a common ancestor; (2) reasonable compactness in terms of the number of species it encompasses; and (3) members of the genus share evolutionarily derived characteristics [11]. Extensive phylogenetic study of species within the Candida group has revealed a number of well-supported clades that better fit the definition of a genus [8–10]. Figure 1 provides an overview of the relationship between clades within the Candida group. Three of the most common Candida pathogens are Candida albicans, Candida parapsilosis, and Candida tropicalis, which fall into the Lodderomyces clade; this clade contains generally antifungal-susceptible Candida species [10]. Being among the largest clades with demonstrated monophyly, this clade has retained the name Candida. However, Candida glabrata, along with the closely related species Candida bracarensis and Candida nivariensis, form part of the Nakaseomyces clade, and hence have been transferred to a new genus, Nakaseomyces, as Nakaseomyces glabrata, Nakaseomyces bracarensis, and Nakaseomyces nivariensis, respectively, although formal description is still pending [4]. Candida krusei, at one point also being known concurrently by Issatchenckia orientalis, Candida glycerinogenes, and Pichia kudriavzevii [12], belongs to the Pichia clade and was formally described as P kudriavzevii due to the nomenclatural priority of this name over others. Candida norvegensis also forms part of the Pichia clade, and has been transferred to Pichia norvegensis [13]. Both the Nakaseomyces and Pichia clades include species characterized by decreased susceptibility or intrinsic resistance to azole antifungal drugs [10], such that these reclassified genera now represent specific evolutionary traits, the third criterion for a genus (Figure 1).
Figure 1.
Phylogenetic analysis showing the genetic and antifungal susceptibility relationships between 76 Saccharomycotina yeasts within the 14 recognized clades. The tree was based on ribosomal DNA data (18S, ITS1, 5.8S, ITS2, and D1/D2) and constructed using maximum likelihood analysis. Species names in bold indicate those commonly reported in a clinical setting. General antifungal susceptibility properties have been indicated on the tree. Reproduced from Stavrou et al, FEMS Yeast Research 19(4):foz037 [10], with permission from Oxford University Press.
Analyses of 18S and internal transcribed spacer ribosomal DNA (rDNA) have determined that Candida rugosa represents a complex of highly similar species, including C rugosa, Candida pararugosa, Candida neorugosa, and Candida pseudorugosa [14, 15]; these species, along with Candida catenulata and Candida scorzettiae, form a well-separated clade and were transferred to a new genus as Diutina [14]. Other new genera containing former Candida species include Debaryomyces, Clavispora, Kluyveromyces, Meyerozyma, Wickerhamomyces, and Yarrowia. Table 1 summarizes nomenclature changes to date in clinically important yeasts.
Table 1.
Summary of Nomenclature Changes in Clinically Important Yeast-like Fungi
| Previous Name(s) | Current Name | Commonly Associated Infections | Reference |
|---|---|---|---|
| Candida bracarensis | Nakaseomyces bracarensis a | Invasive infections including fungemia | [8] |
| Candida catenulata | Diutina catenulata | Invasive infections including fungemia | [14] |
| Candida colliculosa | Torulaspora delbrueckii | Invasive infections including fungemia | [16] |
| Candida fabianii | Cyberlindnera fabianii | Invasive infections including fungemia | [16] |
| Candida famata | Debaryomyces hansenii | Invasive infections including fungemia | [16] |
| Candida glabrata | Nakaseomyces glabrata a | Invasive infections including fungemia | [8] |
| Candida guilliermondii | Meyerozyma guilliermondii | Invasive infections including fungemia | [16] |
| Candida krusei | Pichia kudriavzevii | Invasive infections including fungemia | [16] |
|
Candida kefyr, Candida pseudotropicalis |
Kluyveromyces marxianus | Invasive infections including fungemia | [16] |
| Candida lipolytica | Yarrowia lipolytica | Invasive infections including fungemia | [16] |
| Candida lusitaniae | Clavispora lusitaniae | Invasive infections including fungemia | [16] |
| Candida nivariensis | Nakaseomyces nivariensis a | Invasive infections including fungemia | [8] |
| Candida neorugosa | Diutina neorugosa | Invasive infections including fungemia | [14] |
| Candida norvegensis | Pichia norvegensis | Invasive infections including fungemia | [16] |
| Candida pararugosa | Diutina pararugosa | Invasive infections including fungemia | [14] |
|
Candida pelliculosa, Pichia anomala |
Wickerhamomyces anomalus | Invasive infections including fungemia | [17] |
| Candida pseudorugosa | Diutina pseudorugosa | Invasive infections including fungemia | [14] |
| Candida rugosa | Diutina rugosa | Invasive infections including fungemia | [14] |
| Cryptococcus albidus | Naganishia albida | Invasive infections including fungemia | [18] |
| Cryptococcus curvatus | Cutaneotrichosporon curvatus | Invasive infections | [18] |
| Cryptococcus cyanovorans | Cutaneotrichosporon cyanovorans | Respiratory infections, especially in cystic fibrosis | [18] |
| Cryptococcus laurentii | Papiliotrema laurentii | Invasive infections including fungemia | [18] |
| Pseudozyma antarctica | Moesziomyces antarticus | Fungemia | [19] |
| Pseudozyma aphidis | Moesziomyces aphidis | Fungemia | [19] |
| Pseudozyma churashimaensis | Dirkmeia churashimaensis | Fungemia | [19] |
| Pseudozyma crassa | Triodiomyces crassus | Fungemia | [19] |
| Pseudozyma parantarctica | Moesziomyces parantarcticus | Fungemia | [19] |
| Pseudozyma siamensis | Ustilago siamensis | Fungemia | [19] |
| Geotrichum capitatum | Magnusiomyces capitatus | Invasive infections including fungemia | [20] |
|
Geotrichum clavatum, Saprochaete clavata |
Magnusiomyces clavatus | Invasive infections including fungemia | [20] |
| Pichia ohmeri | Kodamaea ohmeri | Invasive infections including fungemia | [21] |
| Trichosporon cutaneum | Cutaneotrichosporon cutaneum | Cutaneous/superficial infections | [18] |
| Trichosporon dermatis | Cutaneotrichosporon dermatis | Cutaneous infections, allergic conditions | [18] |
| Trichosporon domesticum | Apiotricum domesticum | Uncertain pathogenicity | [18] |
| Trichosporon loubieri | Apiotrichum loubieri | Invasive infections including fungemia | [18] |
| Trichosporon mucoides | Cutaneotrichosporon mucoides | Cutaneous/superficial infections | [18] |
| Trichosporon montevideense | Apiotrichum montevideense | Invasive infections including fungemia | [18] |
| Trichosporon mycotoxinivorans | Apiotrichum mycotoxinivorans | Invasive infections including fungemia | [18] |
Species is pending formal description.
Several pathogenic Candida species have been described in recent years. Without a doubt, Candida auris, described in 2009 as part of the Candida haemulonii complex, has become the most notorious of these [22]. Candida auris has been associated with large healthcare-related outbreaks globally, and comprises 4 major lineages, each having their own antifungal susceptibility characteristics [23, 24]. Other members of this species complex are Candida duobushaemulonii and Candida vulturna [25, 26]. The latter was indicated as C vulturna pro tempore, indicating that “Candida” is a temporary solution. In fact, these species all cluster within the Clavispora clade [8], suggesting that a name change may be warranted. Candida blankii was described in 1968 but has only recently been recognized as a multidrug-resistant human pathogen [27–31]. It does not group in any of the Candida clades and may, therefore, be the sole representative of an as yet undescribed genus [9].
Cryptococcus
The basidiomycetous yeasts have also undergone substantial taxonomic change based on large-scale phylogenetic evidence [18, 32]. The revision of the genus Cryptococcus coincided with the proposal to elevate the 7 lineages within the Cryptococcus neoformans and Cryptococcus gattii complexes to species [33], which, while now largely accepted, has not been without robust debate [34, 35]. Besides 3 nonpathogenic Cryptococcus species, the genus now contains the major cause of cryptococcosis: C neoformans sensu stricto (previously C neoformans var grubii) and Cryptococcus deneoformans (previously C neoformans var neoformans). Two of 5 pathogenic species within the C gattii complex were renamed to a previously published synonym: C gattii sensu stricto (genotype AFLP4/VGI) and Cryptococcus bacillisporus (AFLP5/VGIII), and Cryptococcus deuterogattii (AFLP6/VGII), Cryptococcus tetragattii (AFLP7/VGIV), and Cryptococcus decagattii (AFLP10/VGVI) were named for their molecular type [33]. Epidemiological studies indicate that various Cryptococcus species have a predilection for certain hosts and exhibit differences in antifungal susceptibility [33]. While identification platforms such as matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS) have the capability to differentiate and identify these species using in-house databases, this may not be accessible to many laboratories on a routine basis; in such cases the organism could be reported as C gattii complex or C neoformans complex as appropriate.
Other clinically relevant Cryptococcus species transferred to other genera were Filobasidium magnum (formerly Cryptococcus magnus), Naganishia adeliensis (formerly Cryptococcus adeliensis), Naganishia albida (formerly Cryptococcus albidus), Naganishia diffluens (formerly Cryptococcus diffluens), Naganishia liquefaciens (formerly Cryptococcus liquefaciens), and Papiliotrema laurentii (formerly Cryptococcus laurentii) [32].
Pseudozyma
Pseudozyma species, which are closely related to smut fungi in the Ustilaginaceae, are emerging as a cause of human fungemia. While reported cases are few, most commonly Pseudozyma aphidis has been identified as the cause of infection, but also Pseudozyma antarctica, Pseudozyma parantarctica, Pseudozyma alboarmeniaca, Pseudozyma churashimaensis, Pseudozyma crassa, Pseudozyma siamensis, and Pseudozyma thailandica [36, 37]. This genus has been demonstrated as polyphyletic, with many species clustering with other genera within the Ustilaginaceae [19]. Pseudozyma aphidis, P antarctica, and P parantarctica clustered with Moesziomyces bullatus and were therefore transferred to this genus as Moesziomyces aphidis, Moesziomyces antarcticus, and Moesziomyces parantarcticus, respectively; a new genus was created for P churashimaensis, now known as Dirkmeia churashimaensis; P crassa was transferred to Triodiomyces as Triodiomyces crassus; P siamensis was transferred to Ustilago as Ustilago siamensis; and the taxonomic status of P alboarmeniaca and P thailandica remains to be resolved [19].
Trichosporon
Trichosporon was greatly expanded by the addition of novel species prior to the taxonomic revision by Liu and colleagues [18, 32]. Currently, Trichosporon includes the clinically relevant species Trichosporon asahii, Trichosporon asteroides, Trichosporon coremiiforme, Trichosporon dohaense, Trichosporon faecale, Trichosporon inkin, Trichosporon japonicum, and Trichosporon ovoides [18, 32]. Trichosporon montevideense and Trichosporon mycotoxinivorans were transferred to Apiotrichum as Apiotrichum montevideense and Apiotrichum mycotoxinivorans, respectively. Trichosporon cutaneum, Trichosporon jirovecii, Trichosporon dermatis, Trichosporon mucoides, Cryptococcus curvatus, and Cryptococcus cyanovorans have been accommodated in the new genus Cutaneotrichosporon, all retaining their species epithets [18].
Geotrichum
Geotrichum is a genus of arthroconidial yeast-like fungi and an emerging cause of fungemia in immunocompromised patients [38]. Originally species were assigned based upon morphological differences only but have since undergone extensive taxonomic revision [38–42]. Examination of 18S rDNA sequences discerned 2 major groups, the first containing Geotrichum species with Galactomyces and Dipodascus teleomorphs, and the second comprising Saprochaete species with Magnusiomyces teleomorphs [39]. Geotrichum clavatum fell into the second group and was thus renamed as Saprochaete clavata, whereas Geotrichum capitatum was renamed as Magnusiomyces capitatus; more recently a multigene phylogenetic analysis supported transferring S clavata to Magnusiomyces as Magnusiomyces clavatus [20]. Thus, Geotrichum candidum remains the only clinically relevant species in this genus.
HYALINE HYPHOMYCETE MOLDS
Aspergillus
Aspergillus species, including the 9 teleomorphic genera associated with them, are among the most common causes of invasive or allergic disease in humans and animals [43, 44], particularly the immunosuppressed, in addition to their devastating impact on agriculture due to mycotoxin production as well as biodiversity and ecological health [45, 46]. The application of “one fungus: one name” to the taxonomy of this group was an area of concern, given the potential for many clinically important Aspergillus species to be renamed according to their teleomorphs [47, 48]. However, multigene phylogenetic studies found that Aspergillus is broadly monophyletic, without overlapping with its sibling genus Penicillium [49, 50]. The monophyly of Aspergillus allowed this name to be maintained for most species in the genus, and the clinical importance of its name to be preserved. Those species commonly known by their teleomorphs were renamed within Aspergillus (eg, Neosartorya fischeri was renamed as Aspergillus fischeri).
Many new Aspergillus species have been described in the past 2 decades, with molecular studies finding numerous genetically distinct species within those which were originally described based on their morphological characteristics. At least 50 genetically distinct species have been identified within the morphologically circumscribed Aspergillus fumigatus, including the pathogenic and antifungal resistant Aspergillus lentulus, A fischeri, and Aspergillus udagawae [51–53]. Molecular investigation of other “morphological species” of Aspergillus have also identified “cryptic species” within [54–57]. Table 2 summarizes nomenclature changes in Aspergillus and other hyaline hyphomycetes.
Table 2.
Summary of Nomenclature Changes in Clinically Important Hyaline Hyphomycete Molds
| Previous Name(s) | Current Name | Commonly Associated Infections | Reference |
|---|---|---|---|
| Acremonium kiliense | Sarocladium kiliense | Fungemia, subcutaneous infections | [58] |
| Acremonium roseogriseum | Gliomastix roseogrisea | Not associated with infection | [58] |
| Acremonium strictum | Sarocladium strictum | Cutaneous, invasive infections | [58] |
| Arthroderma benhamiae | Trichophyton benhamiae | Cutaneous infections | [59] |
|
Cerinosterus cyanescens, Sporothrix cyanescens |
Quambalaria cyanescens | Peritonitis, pneumonia, postsurgical complications | [60] |
| Fusarium dimerum | Bisifusarium dimerum | Keratitis, invasive infections | [61] |
|
Fusarium falciforme, Acremonium falciforme |
Neocosmospora falciformis | Keratitis, invasive infections | [61] |
| Fusarium keratoplasticum | Neocosmospora keratoplastica | Keratitis, invasive infections | [61] |
| Fusarium lichenicola | Neocosmospora lichenicola | Keratitis, invasive infections | [61] |
| Fusarium petroliphilum | Neocosmospora petroliphila | Keratitis, invasive infections | [61] |
| Fusarium solani | Neocosmospora solani | Keratitis, invasive infections | [61] |
|
Geosmithia argillacea, Penicillium argillaceum |
Rasamsonia argillacea | Respiratory infections, especially in cystic fibrosis | [62] |
| Gibberella fujikuroi | Fusarium fujikuroi | Keratitis, invasive infections | [63] |
|
Lecythophora hoffmannii, Phialophora hoffmannii |
Coniochaeta hoffmannii | Subcutaneous infections | [64] |
| Microsporum cookei | Paraphyton cookei | Cutaneous infections | [59] |
| Microsporum fulvum | Nannizzia fulva | Cutaneous infections | [59] |
| Microsporum gallinae | Lophophyton gallinae | Cutaneous infections | [59] |
| Microsporum gypseum | Nannizzia gypsea | Cutaneous infections | [59] |
| Microsporum nanum | Nannizzia nana | Cutaneous infections | [59] |
| Microsporum persicolor | Nannizzia persicolor | Cutaneous infections | [59] |
|
Neosartorya fischeri, Neosartorya pseudofischeri, Aspergillus thermomutatus |
Aspergillus fischeri | Respiratory, invasive infections, allergic conditions | [50] |
| Neosartorya udagawae | Aspergillus udagawae | Respiratory, invasive infections, allergic conditions | [50] |
| Paecilomyces lilacinus | Purpureocillium lilacinum | Keratitis, cutaneous infections | [65] |
| Paecilomyces marquandii | Marquandomyces marquandii | Cutaneous infections (rare) | [66] |
| Penicillium marneffei | Talaromyces marneffei | Systemic infections | [67] |
| Penicillium purpureogenum | Talaromyces purpureogenus | Pulmonary infections (rare) | [67] |
| Trichophyton terrestre | Arthroderma terrestre | Doubtful pathogenicity | [59] |
| Trichophyton ajelloi | Arthroderma uncinatum | Cutaneous infections | [59] |
| Trichophyton mentagrophytes | |||
| var interdigitale | Trichophyton interdigitale | Cutaneous infections | [68] |
| var mentagrophytes | Trichophyton mentagrophytes | Cutaneous infections | [68] |
| genotype VIII | Trichophyton indotineae | Cutaneous infections | [69] |
Penicillium
A 2011 multigene analysis of Penicillium and Talaromyces species found the Biverticillium subgenus of the former to be monophyletic with the latter; thus, species in the subgenus Biverticillium group were transferred to Talaromyces [67]. This included the clinically important Talaromyces marneffei, the only thermally dimorphic species of Penicillum/Talaromyces, which is endemic to tropical areas of Southeast and South Asian countries, predominantly seen as systemic infection in human immunodeficiency virus (HIV)–positive individuals [70]. The red diffusible pigment released into semi-solid media is regarded as a typical T marneffei phenotype; however, several Talaromyces species exhibit this phenotype, including Talaromyces atroroseus and Talaromyces purpureogenus, both described as industrially relevant pigment producers [71, 72]. Both species have been reported as the cause of infection in patients with and without HIV, or with other underlying conditions [73–76].
Paecilomyces
Paecilomyces, a genus of cosmopolitan fungi largely known for their biological control applications against bacteria, phytopathogenic fungi, and nematodes [77], are occasional causes of keratitis and onychomycosis, as well as hyalohyphomycosis in immunocompromised patients [78]. A multilocus phylogenetic study of Paecilomyces found significant variation [65], and the major pathogenic species Paecilomyces variotii, Paecilomyces lilacinus, and Paecilomyces marquandii were each found to group with different families (the Trichocomaceae, Ophiocordycipitaceae, and Clavicipitaceae, respectively). On this basis, P lilacinus and P marquandii were each transferred to a new genus as Purpureocillium lilacinum and Marquandomyces marquandii, respectively [65, 66].
Rasamsonia
Rasamsonia argillacea, often recovered from the airways of patients with cystic fibrosis [79], and a cause of disseminated infections in those with chronic granulomatous disease and immunosuppression [80], bears morphological similarities to Penicillium and Paecilomyces species. Originally classified as Penicillium argillaceum and noted for its thermotolerance, it was transferred to a new genus in 1979, Geosmithia (as Geosmithia argillacea) with teleomorph Talaromyces eburneus [81]. Geosmithia was later found to be polyphyletic [82], paving the way to the eventual creation of a new genus of thermotolerant pathogens, Rasamsonia, for Rasamsonia argillacea, Rasamsonia aegroticola, Rasamsonia eburnea, and Rasamsonia piperina, often referred to as the R argillacea complex [62, 83].
Fusarium and Fusarioid Genera
Modern taxonomy of Fusarium and related genera is based on multilocus phylogenies, accompanied by genomic data, morphological descriptions, and physiological and ecological data. This caused a significant but necessary revision in classification and nomenclature of these fungi. Fusarium and allied fusarioid genera, Bisifusarium (formerly the Fusarium dimerum species complex), and Neocosmospora (formerly the Fusarium solani species complex), contain a genetically diverse group of hyaline fungi with global distribution. They are mainly known as ubiquitous soil saprobes, plant pathogens, and mycotoxin producers; however invasive human infections in immunocompromised patients have high mortality despite antifungal therapy. They are also major causes of fungal keratitis and nondermatophyte onychomycosis. Application of phylogenetic species recognition revealed that there are nearly 500 species in Fusarium. Members of Fusarium species complexes are different in morphology, host association, and molecular characteristics [63] (www.fusarium.org). The majority of human infections are caused by the F solani species complex (FSSC), which contains numerous phylogenetically distinct species. New formal names within Neocosmospora have been proposed for several F solani lineages [61]. The most commonly reported species, under recent revised nomenclature, correspond to Neocosmospora keratoplastica (formerly Fusarium keratoplasticum [FSSC2]), Neocosmospora petroliphila (formerly Fusarium petroliphilum [FSSC1]), Neocosmospora falciformis (formerly Fusarium falciforme [FSSC3 + 4]), Neocosmospora lichenicola (formerly Fusarium lichenicola), and Neocosmospora solani (formerly Fusarium solani [FSSC5]). Notably, morphological species recognition is unable to distinguish Fusarium-like taxa that have been described based on genealogical concordance of phylogenetic species recognition. Thus, the term “fusarioid” was suggested when phenotypic methods are solely used to identify Fusarium-like members of Nectriaceae. Accurate species-level identification of Fusarium and related genera from clinical specimens requires multigene sequencing with comparison to well-curated databases, which is often beyond the capacity of routine diagnostic mycology laboratories. Thus, there is currently no standard approach in reporting of these fungi in clinical practice.
Dermatophytes
Dermatophytes, a group of keratinophilic hyaline hyphomycetes, have traditionally been classified within 3 asexual genera Trichophyton, Microsporum, and Epidermophyton, whereas species with sexual reproduction were placed in within Arthroderma and Nannizzia. While this morphological classification is useful in dermatology clinics and routine diagnostic mycology laboratories, it does not capture the true diversity of this group. A recent multilocus phylogenetic analysis of type and reference strains [59] showed that Trichophyton is polyphyletic and proposed a generic classification scheme for all dermatophytes containing 7 genera—namely, Trichophyton, Epidermophyton, Nannizzia, Microsporum, Lophophyton, Paraphyton, and Arthroderma. Most of the anthropophilic and some zoophilic species remained in 3 older groups of Trichophyton, Microsporum, and Epidermophyton. In contrast, geophilic and some rare zoophilic dermatophytes are now classified in the remaining 4 genera (summarized in Table 2). Under this new scheme, novel geophilic species such as Arthroderma eboreum and Nannizzia aenigmatica have been described. Some older names used to describe distinct phenotypic variants of dermatophytes are no longer in use (eg, Trichophyton megninii, Trichophyton gourvilii, Trichophyton yaoundei, Microsporum boullardii, and Microsporum equinum).
Recent additions to the revised classification include 3 novel species causing tinea corporis, Arthroderma chiloniense [84], Nannizzia perplicata [85], and Trichophyton indotineae [69], the latter being of major clinical significance. Trichophyton indotineae exhibits a high level of terbinafine resistance due to missense mutations is the squalene epoxidase gene, causing extensive recalcitrant infections, mainly in the Indian subcontinent [86], but also reported from Europe [87] and Canada [88].
THERMALLY DIMORPHIC FUNGI
The thermally dimorphic fungal genera Blastomyces, Emergomyces, Histoplasma, Paracoccidioides, and Sporothrix have all significant taxonomic changes. The exception is the genus Coccidioides that 2 decades ago was expanded from a single representative to 2 species, Coccidioides immitis and Coccidioides posadasii, and has been stable ever since [89]. Changes and additions for the other genera are described below and summarized in Table 3.
Table 3.
Summary of Nomenclature Changes in Clinically Important Dimorphic Fungi
| Previous Name(s) | Current Name | Commonly Associated Infections | Reference |
|---|---|---|---|
| Emmonsia crescens | Emergomyces crescens | Adiaspiromycosis | [90] |
| Emmonsia helica | Blastomyces helicus | Systemic infections | [90] |
| Emmonsia parva | Blastomyces parvus | Systemic infections | [90] |
| Emmonsia soli | Emergomyces soli | Not associated with infection | [90] |
| Emmonsia “species 3” | Blastomyces percursus | Systemic infections | [91] |
| Emmonsia “species 5” | Emergomyces africanus | Systemic infections | [91] |
| Emmonsia pasteuriana | Emergomyces pasteurianus | Systemic infections | [91] |
|
Histoplasma capsulatum
var capsulatum |
Histoplasma capsulatum sensu stricto Histoplasma mississippiense Histoplasma ohiense Histoplasma suramericanum |
Systemic infections | [92] |
| Lacazia loboi | Paracoccidioides loboi | Subcutaneous “lobomycosis” | [93] |
| Paracoccidioides brasiliensis |
Paracoccidioides brasiliensis sensu stricto Paracoccidioides americana Paracoccidioides restrepoanaa Paracoccidioides venezuelensis Paracoccidioides lutzii |
Pulmonary, cutaneous infections | [94, 95] |
| Penicillium marneffei | Talaromyces marneffei b | Systemic infections | [67] |
| Sporothrix schenckii |
Sporothrix schenckii sensu stricto Sporothrix brasiliensis Sporothrix globosa Sporothrix luriei |
Subcutaneous infections | [96] |
| Sporothrix pallida |
Sporothrix pallida sensu stricto Sporothrix chilensis Sporothrix humicola Sporothrix mexicana Sporothrix stylites |
Doubtful pathogenicity in humans | [96, 97] |
Published as Paracoccidioides restrepiensis.
See also section on hyaline hyphomycete molds and Table 2.
Histoplasma
Histoplasma capsulatum was until recently represented by 3 varieties: H capsulatum var capsulatum, var duboisii, and var farciminosum. Multiple large phylogenetic studies observed extensive genetic diversity and potentially several new species within the variety capsulatum [98–101]. Using whole genome sequencing, Sepúlveda and colleagues took a first step in revising the genus Histoplasma, splitting H capsulatum var capsulatum into 4 species named H capsulatum sensu stricto (known as the Panama or H81 lineage), Histoplasma mississippiense (NAm1 lineage), Histoplasma ohiense (NAm2 lineage), and Histoplasma suramericanum (LAmA lineage) [92].
Blastomyces and Emmonsia
A phylogenetic analysis revealed 2 evolutionarily distinct lineages within Blastomyces dermatitidis, prompting the recognition of a second species, Blastomyces gilchristii [102]. Changes to the genus Emmonsia led to it being merged with other genera, including Blastomyces; Emmonsia helica, Emmonsia parva, and Emmonsia “species 3” were transferred to Blastomyces as Blastomyces helicus, Blastomyces parvus, and Blastomyces percursus, respectively [90, 103, 104]. This was followed by the description of 2 novel species, Blastomyces silverae, and Blastomyces emzantsi, which has so far only been reported from Southern Africa [90, 105]. The remaining Emmonsia species were transferred to Emergomyces, with Emmonsia “species 5” being renamed Emergomyces africanus [91], a major outbreak-associated clinical species [106]. Emmonsia pasteuriana, Emmonsia crescens, and Emmonsia soli were also transferred to Emergomyces as Emergomyces pasteurianus [91], Emergomyces crescens, and Emergomyces soli [107], respectively. Novel Emergomyces species are Emergomyces canadensis, Emergomyces europaeus, and Emergomyces orientalis [107, 108].
Paracoccidioides
Paracoccidioides is restricted to endemic areas in South America, and Paracoccidioides brasiliensis was considered the only causative agent for >80 years. However, the failure of serology tests to detect some Paracoccidioides infections was noted, and molecular studies determined that these cases were caused by a different species, Paracoccidioides lutzii, previously known as Pb01-like [94]. In addition, several consistently observed lineages led to the diversification of the P brasiliensis species complex into 4 species—Paracoccidioides americana, P brasiliensis sensu stricto, Paracoccidioides restrepoana (as P restrepiensis), and Paracoccidioides venezuelensis—which are more closely related to each other than to P lutzii [95, 109]. Two recently described members of Paracoccidioides have to date been unculturable; Paracoccidioides loboi (previously Lacazia loboi) has been associated with human infections, and Paracoccidioides ceti is linked to disease in marine animals [93].
Sporothrix
Sporothrichosis typically presents as subcutaneous infection caused by traumatic implantation of Sporothrix species. More than 50 species have been described within this genus, but only a small number are proven causes of infections in humans and animals. Until 2007, Sporothrix schenckii was the main causative agent, but molecular investigations showed that this species was highly diverse and 3 new species, Sporothrix brasiliensis, Sporothrix globosa, and Sporothrix mexicana, were subsequently described [96]. Sporothrix brasiliensis has been identified as the cause of large-scale and expanding outbreaks of sporothrichosis among cats with transmission from cats to humans in Brazil and sporadic cases in neighboring countries [110]. The clinically relevant S schenckii, S brasiliensis, S globosa, and Sporothrix luriei are now considered to form the S schenckii complex, while species rarely associated with infection form the Sporothrix pallida complex (S pallida sensu stricto, Sporothrix chilensis, Sporothrix humicola, and S mexicana) [97, 110].
DEMATIACEOUS (MELANIZED) HYPHOMYCETE MOLDS
Scedosporium
Members of the genera Scedosporium and Pseudallescheria have undergone extensive review and reclassification [111] based upon evidence of extensive diversity within the Pseudallescheria boydii complex [112–114]. Two findings are of particular importance in clinical mycology. First, that P boydii and Scedosporium apiospermum were found to represent distinct species on the basis of significant molecular and phenotypic differences and were not in fact anamorph and teleomorph states of a single species [114]. Since Scedosporium has nomenclatural priority, P boydii was transferred into Scedosporium as Scedosporium boydii. Second, Scedosporium prolificans was found to be phylogenetically distinct from all other Scedosporium species and was returned to its original name of 1974, as Lomentospora prolificans [111]. This transfer to a different genus accounts not only for the taxonomic differences between L prolificans and other Scedosporium species, but also the pan-antifungal-resistant nature of this species and differences in its clinical management, compared to other Scedosporium infections. Other clinically relevant species recognized within this group include Scedosporium aurantiacum, Scedosporium dehoogii, and Pseudallescheria angusta [112, 114]. Table 4 summarizes nomenclature changes in these and other dematiaceous hyphomycetes.
Table 4.
Summary of Nomenclature Changes in Clinically Important Dematiaceous Hyphomycetes
| Previous Name(s) | Current Name | Commonly Associated Infections | Reference |
|---|---|---|---|
| Bipolaris australiensis | Curvularia australiensis | Keratitis, cutaneous infections | [115] |
| Bipolaris hawaiiensis | Curvularia hawaiiensis | Keratitis, cutaneous infections | [115] |
| Bipolaris spicifera | Curvularia spicifera | Keratitis, cutaneous infections | [115] |
| Ochroconis gallopava | Verruconis gallopava | Brain and pulmonary infections | [116] |
| Phialophora richardsiae | Pleurostoma richardsiae | Subcutaneous infections | [117] |
| Pseudallescheria boydii | Scedosporium boydii | Osteoarticular, invasive infections | [114] |
| Ramichloridium mackenziei | Rhinocladiella mackenziei | Brain infections | [118] |
| Ramichloridium schulzeri | Myrmecridium schulzeri | Tongue (“golden tongue” syndrome) | [118] |
| Scedosporium prolificans | Lomentospora prolificans | Osteoarticular, invasive infections | [111] |
Bipolaris and Curvularia
Species within the genera Bipolaris and Curvularia represent anamorphic forms of the Cochliobolus teleomorph, and share many morphological similarities, notwithstanding the characteristic curved conidia of some Curvularia species. The taxonomy of this group has long been controversial, and neither genus is monophyletic [119, 120]. Several studies proposed that Bipolaris and Curvularia were in fact synonymous [121, 122], as there is little more than conidial morphology to differentiate them, and many species have conidia that are intermediate between the 2 [122]. Manamgoda and colleagues [115] resolved the conflict with a multigene phylogenetic analysis of a wide range of species including ex-type cultures, which showed 2 distinct clades. On this basis, several common Bipolaris species, including Bipolaris australiensis, Bipolaris hawaiiensis, and Bipolaris spicifera, were transferred into Curvularia as Curvularia australiensis, Curvularia hawaiiensis, and Curvularia spicifera, respectively [115].
Ochroconis
The genus Verruconis was established to accommodate thermophilic species of Ochroconis, which have been isolated from hot springs, thermal soils, sewage from nuclear power plants, and coal waste piles. Ochroconis gallopava, Ochroconis calidifluminalis, and Ochroconis verruculosum were transferred to Verruconis as Verruconis gallopava, Verruconis calidifluminalis, and Verruconis verruculosum, respectively [116], supported by a phylogenetic analysis [123]. The type species V gallopava is a neurotropic pathogen of humans and other warm-blooded animals, mainly birds [124]. Ochroconis species are mesophilic and generally nonpathogenic in mammals, although subcutaneous human infections have been noted by Ochroconis mirabilis [125].
Ramichloridium
Ramichloridium was found to be polyphyletic, forming 8 distinct clades across several orders and families of dematiaceous fungi [118]. Ramichloridium mackenziei, a pathogen associated with high-mortality cerebral infections and prevalent in the Middle East [126], grouped with the Rhinocladiella type species Rhinocladiella atrovirens and was therefore transferred to Rhinocladiella as Rhinocladiella mackenziei. Isolates of Ramichloridium schulzeri formed a distinct cluster away from other genera, leading to the creation of a new genus, Myrmecridium, among which Myrmecridium schulzeri is the only mammalian pathogen. Rhinocladiella aquaspersa and Rhinocladiella similis are known causes of chromoblastomycosis [127, 128].
COELOMYCETES
Significant nomenclature change has also occurred within coelomycetous fungi, those that produce nonsexual conidia within fruiting bodies (summarized in Table 5). Neoscytalidium dimidiatum, formerly Scytalidium dimidiatum, is a plant pathogen associated with a broad spectrum of infections in humans, affecting skin and nails in tropical and subtropical continents and invasive diseases mostly in immunocompromised hosts. Nonmelanized mutants, with white colonies and reduced conidiation, were referred to as Neoscytalidium hyalinum (Scytalidium hyalinum) but have since been synonymized with N dimidiatum [130]. Furthermore, molecular studies revealed that N dimidiatum and its teleomorph, Nattrassia mangiferae (formerly Hendersonula toruloidea), are 2 distinct species and not closely related [131].
Table 5.
Summary of Nomenclature Changes in Clinically Important Coelomycetes
| Previous Name(s) | Current Name | Commonly Associated Infections | Reference |
|---|---|---|---|
| Leptosphaeria senegalensis | Falciformispora senegalensis | Mycetoma | [129] |
| Leptosphaeria tompkinsii | Falciformispora tompkinsii | Mycetoma | [129] |
|
Scytalidium dimidiatum, Scytalidium hyalinum |
Neoscytalidium dimidiatum | Onychomycosis | [130] |
| Hendersonula toruloidea | Nattrassia mangiferae | Onychomycosis | [131] |
| Pyrenochaeta romeroi | Medicopsis romeroi | Mycetoma | [132] |
| Pyrenochaeta mackinnonii | Nigrograna mackinnonii | Mycetoma | [133] |
| Madurella grisea | Trematospheria grisea | Mycetoma | [129] |
Reclassification of species belonging to Pleosporales led to renaming of some of the main etiologic agents of black-grain eumycetoma. Based on the combined DNA sequence data set of the 18S, 28S, RPB2, and TEF1 genes, Leptosphaeria senegalensis was renamed Falciformispora senegalensis and Leptosphaeria tompkinsii as Falciformispora tompkinsii [129]. Likewise, based on the rDNA sequence, the taxonomic position of Madurella grisea was changed to the order Pleosporales, and in 2013 was officially renamed as Trematosphaeria grisea [129]. However, the classification of Madurella mycetomatis, the most common fungal causative agent of mycetoma, remained unchanged. The main taxonomic change in the genus Pyrenochaeta was the reclassification of Pyrenochaeta romeroi to Medicopsis romeroi [132] and Pyrenochaeta mackinnonii as Nigrograna mackinnonii [133]. Based on molecular analysis of the rRNA genes, both fungi were distant from the type species of Pyrenochaeta and from each other; hence, they have been allocated into different genera. It is important to highlight that most members of Pleosporales remain sterile even in prolonged cultures, making their phenotypic identification troublesome. Application of molecular and sequence-based methods are necessary for accurate identification of these fungi.
MUCORALES
The zygomycetes and their classification within Kingdom Fungi has undergone significant change over the past 15 years. The establishment of a phylogenetic system of fungal classification [134] revealed the polyphyletic nature of what was then recognized as the phylum Zygomycota and has since been abolished. It has been replaced by 2 phyla, the Mucoromycotina and the Zoopagomycota, which include the clinically important subphyla Mucoromycotina and Entomophthoromycotina, respectively [135]. As a flow-on effect, the term “zygomycosis,” which described any invasive fungal infection caused by species of the former phylum Zygomycota [136], was replaced by either “mucormycosis” or “entomophthoromycosis.” This was supported by their significant clinical, ecological, and epidemiological differences between the diseases caused by these groups.
Mucor
Within the Mucoromycotina, Mucor is the largest genus, with close to 80 accepted species [137]. Mucor circinelloides is the most clinically important representative of the genus, and a recent in-depth phenotypic and molecular characterization revealed it to be a complex of 16 species [138]. The clinically relevant species in this complex are M circinelloides, Mucor lusitanicus, Mucor griseocyanus, Mucor velutinosus, and Mucor janssenii [138]. Additionally, Rhizomucor variabilis was transferred to Mucor as Mucor irregularis on the basis of rDNA phylogeny [139], becoming somewhat unique among Mucor species due to having rhizoids. Table 6 summarizes nomenclature changes among the clinically important Mucoromycotina.
Table 6.
Summary of Nomenclature Changes in Clinically Important Mucoromycotina
| Previous Name(s) | Current Name | Commonly Associated Infections | Reference |
|---|---|---|---|
|
Absidia corymbifera, Mycocladus corymbifera |
Lichtheimia corymbifera | Sinonasal, subcutaneous, systemic infections | [140] |
| Rhizopus azygosporus | Rhizopus microsporus | Sinonasal, subcutaneous, systemic infections | [141] |
| Rhizopus delemar | Rhizopus arrhizus var delemar | Sinonasal, subcutaneous, systemic infections | [142] |
|
Rhizopus microsporus
var chinensis var oligosporus var rhizopodiformis |
Rhizopus microsporus
(varieties no longer recognized) |
Sinonasal, subcutaneous, systemic infections | [141] |
| Rhizopus oryzae | Rhizopus arrhizus | Sinonasal, subcutaneous, systemic infections | [142] |
| Rhizomucor variabilis | Mucor irregularis | Sinonasal, subcutaneous, systemic infections | [139] |
| Saksenaea vasiformis |
Saksenaea vasiformis sensu stricto Saksenaea erythrospora Saksenaea oblongispora |
Sinonasal, subcutaneous, systemic infections | [143] |
Absidia and Lichtheimia
The genus Absidia was investigated on the basis of multiple gene phylogenies, finding that the species Absidia corymbifera and 4 other species formed a distinct clade that could also be characterized by thermotolerance; these species were reclassified within the genus Lichtheimia, comprising the clinically relevant species Lichtheimia corymbifera, Lichtheimia ramosa, Lichtheimia hyalospora, and Lichtheimia ornata [140, 144].
Rhizopus
Rhizopus oryzae and Rhizopus arrhizus are known to be synonyms of the same species, representing the most common cause of mucormycosis [145, 146]. There has been long-standing debate over which of these is the valid name, well reviewed by Dolatabadi et al [142], with both names in use for many years. This is now settled, with R arrhizus found to be the first valid name [142]. Additional changes in this genus include recognition of Rhizopus delemar as a variety of R arrhizus (ie, R arrhizus var arrhizus and R arrhizus var delemar) and collapse of the varieties within Rhizopus microsporus [141, 142].
Saksenaea
Members of the genus Saksenaea are rarely seen in the clinic, but the majority of reported cases are due to Saksenaea vasiformis and Saksenaea erythrospora [143, 147]. Five additional species have been described during the past decade, although not all have been associated with infection: Saksenaea dorisiae [148], Saksenaea longicolla [149], Saksenaea loutrophoriformis [150], Saksenaea oblongispora [143], and Saksenaea trapizispora [151].
MANAGING CHANGES IN FUNGAL NOMENCLATURE
Nomenclature changes are not new or unique to fungi [152–157]. However, in recent years changes in fungal nomenclature have been numerous, and in the age of social media, criticism has been swift [5]. Concerns include pathology reports containing unfamiliar species names that might be dismissed as nonpathogens (ie, colonizers, laboratory or environmental contaminants) and disruption of molecular and literature databases, as well as interruption of local epidemiology and antifungal susceptibility profiles. Although such concerns are valid, there is little evidence to support them. Recent surveys of Australasian laboratory staff and clinicians found a high level of support (71/92 [77%] laboratories and 204/217 [94%] clinicians) for nomenclature change, providing the previous clinically familiar names are included on reports alongside updated names [158]. This support is further demonstrated by the inclusion of updated nomenclature in the recently published global guidelines for diagnosis and management of rare yeast infections [159] and updated Australasian antifungal guidelines [160].
In fact, common fungal pathogens have undergone numerous name changes in the past; Candida albicans was known by several names including Monilia albicans until 1923, and its associated infection is still sometimes referred to as moniliasis; Candida glabrata was known as Torulopsis glabrata until 1978, a name that was still in common use until the late 1990s and was included in a clinical case report as recently as 2005 [161]. It is unclear to what extent these changes caused concern at the time; however, safe adaptation to the changes was evidently possible.
Concerns that literature and molecular databases will be disrupted by name changes and flooded by redundant “First case of …” reports, are unwarranted. All National Center for Biotechnology Information (NCBI) databases, which include PubMed and GenBank, are underpinned by a standardized taxonomy database, ensuring that any organism-based search term will retrieve all relevant material, regardless of whether the name is current or obsolete [162, 163]. This permits extraction of all relevant literature to guide management. Publicly accessible resources such as Index Fungorum (http://www.indexfungorum.org) and MycoBank (www.mycobank.org) that serve as repositories for nomenclatural information, including whether names are current or obsolete, can assist those unsure of the status of a fungal species name.
Nomenclature updates in proprietary databases, such as those for MALDI-TOF mass spectrometry, will be critical to the successful adaptation to new species names by clinical microbiology laboratories. Unfortunately, this is hindered by the need for manufacturers to meet the requirements of regulatory bodies, such as the US Food and Drug Administration. At this time, the Vitek MS Expanded V3.2 database (bioMérieux, Marcy l’Étoile, France) uses some updated nomenclature (eg, Purpureocillium lilacinum, Lichtheimia corymbifera, Sarocladium kiliense) but also obsolete nomenclature (eg, various Candida species, Scedosporium prolificans). In contrast, the recently released MBT Compass Library Revision G (2021) and MBT Filamentous Fungi Library (2021) (Bruker Daltonics, Bremen, Germany, 2021) accommodates the reclassification of many yeasts and molds. As further database updates are rolled out to laboratories, it can be expected that resistance to nomenclature changes will be reduced. Access to database updates may be dependent upon service contracts and regulatory approval in different regions, but broadly speaking, most laboratories utilizing Vitek MS or MALDI Biotyper systems receive annual database updates without cost. However, in-house databases would require laboratory input to update nomenclature.
Critical to the success of adapting to new nomenclature is education of laboratory staff and of clinicians. The experience in Australia, New Zealand, and the United Kingdom has demonstrated a clear role for external quality assurance programs and reference laboratories in education. Revised names should be incorporated into formal teaching and training programs as well as examinations; this will be greatly supported by the incorporation of updated names in medical reference texts such as the Manual of Clinical Microbiology. The Clinical and Laboratory Standards Institute (CLSI) recently recognized but stopped short of adopting new names in the M27M44S document [164]. There is enormous potential for organizations such as the CLSI, the European Committee on Antimicrobial Susceptibility Testing, the College of American Pathologists, the International Society for Human and Animal Mycoses, the Mycoses Study Group Education and Research Consortium, the European Confederation of Medical Mycology, and the Australia and New Zealand Mycoses Interest Group to play an important role, perhaps through joint working groups, in the education of laboratory staff through workshops, newsletters, and the development of guidelines.
In the absence of endorsed guidelines on adapting to nomenclature change, we make the following recommendations for clinical microbiology laboratories:
It is recommended that all microbiology laboratories, regardless of size, geographic location, degree of mycology expertise, or supervision structure, should take steps toward utilization of updated fungal nomenclature as soon as is practical. Ultimately, this will provide consistency in reporting between laboratories nationally and internationally, reducing the potential for confusion and support the education of laboratory staff and clinicians.
When reporting an organism using new/updated nomenclature, the previous name must also be included on the report; for example, “Growth of Pichia kudriavzevii (Candida krusei)” or “Growth of Pichia kudriavzevii. This species was formerly known as Candida krusei.” Depending on the laboratory information system, the above comments may be coded to occur automatically and ensure consistency in the approach.
It may be necessary to include the previous names on reports for 5 years, or longer, depending on the range of specimens and requesting clinicians and the perceived level of acceptance of the nomenclature into common use.
For taxa where the identification method used is insufficiently sensitive or robust to identify reliably to species level, reporting to species complex level is useful, along with clinically pertinent comments as appropriate; for example, “Growth of Cryptococcus gattii complex” or “Growth of Aspergillus fumigatus complex. This species complex includes a number of pathogenic species that may have reduced susceptibility to one or more antifungal drugs.” In these situations, it may also be important to indicate the method used for identification.
The reporting strategy must be consistent, requiring all laboratory staff to be educated. New names should be updated in the laboratory information system and document-controlled procedure manuals and as soon as practicable.
CONCLUSIONS
Change in the nomenclature for any pathogen is an inevitable and necessary part of the scientific process, a result of refinement and correction of past taxonomic errors, and offers possibilities for improved recognition of clinically relevant biological characteristics, such as antifungal resistance or thermotolerance. The real issue is managing change to best serve those who work with these organisms.
Contributor Information
Sarah E Kidd, National Mycology Reference Centre, SA Pathology, Adelaide, South Australia, Australia; School of Biological Sciences, Faculty of Sciences, University of Adelaide, Adelaide, South Australia, Australia.
Alireza Abdolrasouli, Department of Medical Microbiology, King's College Hospital, London, United Kingdom; Department of Infectious Diseases, Imperial College London, London, United Kingdom.
Ferry Hagen, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; Institute of Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, The Netherlands; Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, The Netherlands.
Notes
Author contributions . S. E. K. developed the concept for the manuscript. All authors contributed equally to reviewing the literature, writing, and editing of the manuscript.
Acknowledgments . The authors acknowledge the efforts of Index Fungorum (http://www.indexfungorum.org) and Mycobank (www.mycobank.org) as nomenclatural repositories for fungi.
Potential conflicts of interest . The authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hawksworth DL, Crous PW, Redhead SA, et al. . The Amsterdam declaration on fungal nomenclature. IMA Fungus 2011; 2:105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Warnock DW. Name changes for fungi of medical importance, 2012 to 2015. J Clin Microbiol 2017; 55:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warnock DW. Name changes for fungi of medical importance, 2016–2017. J Clin Microbiol 2019; 57:e01183–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borman AM, Johnson EM. Name changes for fungi of medical importance, 2018 to 2019. J Clin Microbiol 2021; 59:e01811–20. Erratum in: J Clin Microbiol 2021; 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kidd SE, Halliday CL, McMullan B, Chen SC, Elvy J. New names for fungi of medical importance: can we have our cake and eat it too? J Clin Microbiol 2021; 59:e02730–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borman AM, Johnson EM. Reply to Kidd, et al, “New names for fungi of medical importance: can we have our cake and eat it too?” J Clin Microbiol 2021; 59:e02896–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turland NJ, Wiersema JH, Barrie FR, et al. , eds. 2018: International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China. Regnum Vegetabile 159.2017. Glashütten, Germany: Koeltz Botanical Books. 10.12705/Code.2018 [DOI]
- 8. Daniel HM, Lachance MA, Kurtzman CP. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek 2014; 106:67–84. [DOI] [PubMed] [Google Scholar]
- 9. Kurtzman CP, Robnett CJ. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res 2013; 13:23–33. [DOI] [PubMed] [Google Scholar]
- 10. Stavrou AA, Lackner M, Lass-Flörl C, Boekhout T. The changing spectrum of Saccharomycotina yeasts causing candidemia: phylogeny mirrors antifungal susceptibility patterns for azole drugs and amphotericin B. FEMS Yeast Res 2019; 19:foz037. [DOI] [PubMed] [Google Scholar]
- 11. Gill FB, Slikas B, Sheldon FH. Phylogeny of titmice (Paridae): II. Species relationships based on sequences of the mitochondrial cytochrome-B gene. Auk 2005; 122:121–43. [Google Scholar]
- 12. Douglass AP, Offei B, Braun-Galleani S, et al. . Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PLoS Pathog 2018; 14:e1007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leask BG, Yarrow D. Pichia norvegensis sp. nov. Sabouraudia 1976; 14:61–3. [PubMed] [Google Scholar]
- 14. Khunnamwong P, Lertwattanasakul N, Jindamorakot S, Limtong S, Lachance MA. Description of Diutina gen. nov., Diutina siamensis, f.a. sp. nov., and reassignment of Candida catenulata, Candida mesorugosa, Candida neorugosa, Candida pseudorugosa, Candida ranongensis, Candida rugosa and Candida scorzettiae to the genus Diutina. Int J Syst Evol Microbiol 2015; 65:4701–9. [DOI] [PubMed] [Google Scholar]
- 15. Ming C, Huang J, Wang Y, et al. . Revision of the medically relevant species of the yeast genus Diutina. Med Mycol 2019; 57:226–33. [DOI] [PubMed] [Google Scholar]
- 16. Kurtzman CP, Fell JW, Boekhout T. The yeasts: a taxonomic study. 5th ed. Amsterdam, The Netherlands: Elsevier Science, 2011. [Google Scholar]
- 17. Kurtzman CP. Phylogeny of the ascomycetous yeasts and the renaming of Pichia anomala to Wickerhamomyces anomalus. Antonie Van Leeuwenhoek 2011; 99:13–23. [DOI] [PubMed] [Google Scholar]
- 18. Liu XZ, Wang QM, Göker M, et al. . Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol 2015; 81:85–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang QM, Begerow D, Groenewald M, et al. . Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud Mycol 2015; 81:55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaplan E, Al-Hatmi AMS, Ilkit M, et al. . Molecular diagnostics of arthroconidial yeasts, frequent pulmonary opportunists. J Clin Microbiol 2018; 56:e01427–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamada Y, Suzuki T, Matsuda M, Mikata K. The phylogeny of Yamadazyma ohmeri (Etchells et Bell) Billon-Grand based on the partial sequences of 18S and 26S ribosomal RNAs: the proposal of Kodamaea gen. nov. Saccharomycetaceae). Biosci Biotechnol Biochem 1995; 59:1172–4. [DOI] [PubMed] [Google Scholar]
- 22. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009; 53:41–4. [DOI] [PubMed] [Google Scholar]
- 23. Chow NA, Muñoz JF, Gade L, et al. . Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 2020; 11:e03364–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Jong AW, Hagen F. Attack, defend and persist: how the fungal pathogen Candida auris was able to emerge globally in healthcare environments. Mycopathologia 2019; 184:353–65. [DOI] [PubMed] [Google Scholar]
- 25. Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, et al. . Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol 2012; 50:3641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sipiczki M, Tap RM. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. Int J Syst Evol Microbiol 2016; 66:4009–15. [DOI] [PubMed] [Google Scholar]
- 27. Nobrega de Almeida J Jr, Campos SV, Thomaz DY, et al. . Candida blankii: an emergent opportunistic yeast with reduced susceptibility to antifungals. Emerg Microbes Infect 2018; 7:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Al-Haqqan A, Al-Sweih N, Ahmad S, et al. . Azole-resistant Candida blankii as a newly recognized cause of bloodstream infection. New Microbes New Infect 2018; 26:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chowdhary A, Stielow JB, Upadhyaya G, Singh PK, Singh A, Meis JF. Candida blankii: an emerging yeast in an outbreak of fungaemia in neonates in Delhi, India. Clin Microbiol Infect 2020; 26:648.e5–8. [DOI] [PubMed] [Google Scholar]
- 30. Kollu VS, Kalagara PK, Islam S, Gupte A. A report of Candida blankii fungemia and possible endocarditis in an immunocompetent individual and the review of literature. Cureus 2021; 13:e14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mirchin R, Czeresnia JM, Orner EP, Chaturvedi S, Murphy K, Nosanchuk JD. The continuing emergence of Candida blankii as a pathogenic fungus: a new case of fungemia in a patient infected with SARS-CoV-2. J Fungi (Basel). 2022; 8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu XZ, Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud Mycol 2015; 81:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hagen F, Khayhan K, Theelen B, et al. . Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 2015; 78:16–48. [DOI] [PubMed] [Google Scholar]
- 34. Kwon-Chung KJ, Bennett JE, Wickes BL, et al. . The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2017; 2:e00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hagen F, Lumbsch HT, Arsic Arsenijevic V, et al. . Importance of resolving fungal nomenclature: the case of multiple pathogenic species in the Cryptococcus genus. mSphere 2017; 2:e00238-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Telles JP, Ribeiro VST, Kraft L, Tuon FF. Pseudozyma spp. human infections: a systematic review. Med Mycol 2021; 59:1–6. [DOI] [PubMed] [Google Scholar]
- 37. Chowdhary A, Sharada K, Singh PK, et al. . Outbreak of Dirkmeia churashimaensis fungemia in a neonatal intensive care unit, India. Emerg Infect Dis 2020; 26:764–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graeff L Durán, Seidel D, Vehreschild MJ, et al. ; FungiScope Group . Invasive infections due to Saprochaete and Geotrichum species: report of 23 cases from the FungiScope Registry. Mycoses 2017; 60:273–9. [DOI] [PubMed] [Google Scholar]
- 39. de Hoog GS, Smith MT. Ribosomal gene phylogeny and species delimitation in Geotrichum and its teleomorphs. Stud Mycol 2004; 50:489–515. [Google Scholar]
- 40. de Hoog GS, Smith MT. Chapter 45, Magnusiomyces Zender (1977). In: Kurtzman CP, Fell JW, Boekhout T, eds. The yeasts, a taxonomic study. 5th ed. Amsterdam, The Netherlands: Elsevier; Science, 2011:565–74. [Google Scholar]
- 41. de Hoog GS, Smith MT. Chapter 91, Geotrichum Link: Fries (1832). In: Kurtzman CP, Fell JW, Boekhout T, eds. The yeasts, a taxonomic study. 5th ed. Amsterdam, The Netherlands: Elsevier Science, 2011:1279–86. [Google Scholar]
- 42. de Hoog GS, Smith MT. Chapter 97, Saprochaete Coker & Shanor ex D.T.S. Wagner & Dawes (1970). In: Kurtzman CP, Fell JW, Boekhout T, eds. The yeasts, a taxonomic study. 5th ed. Amsterdam, The Netherlands: Elsevier; Science, 2011:1317–27. [Google Scholar]
- 43. Thompson GR 3rd, Young JH. Aspergillus infections. N Engl J Med 2021; 385:1496–509. [DOI] [PubMed] [Google Scholar]
- 44. Seyedmousavi S, Guillot J, Arné P, et al. . Aspergillus and aspergilloses in wild and domestic animals: a global health concern with parallels to human disease. Med Mycol 2015; 53:765–97. [DOI] [PubMed] [Google Scholar]
- 45. Shabeer S, Asad S, Jamal A, Ali A. Aflatoxin contamination, its impact and management strategies: an updated review. Toxins (Basel) 2022; 14:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fisher MC, Henk DA, Briggs CJ, et al. . Emerging fungal threats to animal, plant and ecosystem health. Nature 2012; 484:186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pitt JI, Taylor JW. Aspergillus, its sexual states and the new International Code of Nomenclature. Mycologia 2014; 106:1051–62. [DOI] [PubMed] [Google Scholar]
- 48. Taylor JW, Göker M, Pitt JI. Choosing one name for pleomorphic fungi: the example of Aspergillus versus Eurotium, Neosartorya and Emericella. Taxon 2016; 65:593–601. [Google Scholar]
- 49. Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 2011; 70:1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kocsubé S, Perrone G, Magistà D, et al. . Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Stud Mycol 2016; 85:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005; 97:1316–29. [DOI] [PubMed] [Google Scholar]
- 52. Katz ME, Dougall AM, Weeks K, Cheetham BF. Multiple genetically distinct groups revealed among clinical isolates identified as atypical Aspergillus fumigatus. J Clin Microbiol 2005; 43:551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Badali H, Cañete-Gibas C, McCarthy D, et al. . Species distribution and antifungal susceptibilities of Aspergillus section fumigati isolates in clinical samples from the United States. J Clin Microbiol 2022; 60:e0028022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zoran T, Sartori B, Sappl L, et al. . Azole-Resistance in Aspergillus terreus and related Species: an emerging problem or a rare phenomenon? Front Microbiol 2018; 9:516. Erratum in: Front Microbiol 2019; 9:3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takeda K, Suzuki J, Watanabe A, et al. . The accuracy and clinical impact of the morphological identification of Aspergillus species in the age of cryptic species: a single-centre study. Mycoses 2022; 65:164–70. [DOI] [PubMed] [Google Scholar]
- 56. Imbert S, Cassaing S, Bonnal C, et al. . Invasive aspergillosis due to Aspergillus cryptic species: a prospective multicentre study. Mycoses 2021; 64:1346–53. [DOI] [PubMed] [Google Scholar]
- 57. Nargesi S, Jafarzadeh J, Najafzadeh MJ, et al. . Molecular identification and antifungal susceptibility of clinically relevant and cryptic species of Aspergillus sections Flavi and Nigri. J Med Microbiol 2022; 71. doi: 10.1099/jmm.0.001480 [DOI] [PubMed] [Google Scholar]
- 58. Summerbell RC, Gueidan C, Schroers HJ, et al. . Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol 2011; 68:139–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. de Hoog GS, Dukik K, Monod M, et al. . Toward a novel multi-locus phylogenetic taxonomy for the dermatophytes. Mycopathologia 2017; 182:5–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Beer ZW, Begerow D, Bauer R, Pegg GS, Crous PW, Wingfield MJ. Phylogeny of the Quambalariaceae fam. nov., including important Eucalyptus pathogens in South Africa and Australia. Stud Mycol 2006; 55:289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sandoval-Denis M, Crous PW. Removing chaos from confusion: assigning names to common human and animal pathogens in Neocosmospora. Persoonia 2018; 41:109–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Houbraken J, Spierenburg H, Frisvad JC. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek 2012; 101:403–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Crous PW, Lombard L, Sandoval-Denis M, et al. . Fusarium: more than a node or a foot-shaped basal cell. Stud Mycol 2021; 98:100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Khan Z, Gené J, Ahmad S, et al. . Coniochaeta polymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecythophora species to Coniochaeta. Antonie Van Leeuwenhoek 2013; 104:243–52. [DOI] [PubMed] [Google Scholar]
- 65. Luangsa-Ard J, Houbraken J, van Doorn T, et al. . Purpureocillium, a new genus for the medically important Paecilomyces lilacinus. FEMS Microbiol Lett 2011; 321:141–9. [DOI] [PubMed] [Google Scholar]
- 66. Mongkolsamrit S, Khonsanit A, Thanakitpipattana D, et al. . Revisiting Metarhizium and the description of new species from Thailand. Stud Mycol 2020; 95:171–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Samson RA, Yilmaz N, Houbraken J, et al. . Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol 2011; 70:159–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gräser Y, Kuijpers AF, Presber W, De Hoog GS. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol 1999; 37:315–30. [DOI] [PubMed] [Google Scholar]
- 69. Kano R, Kimura U, Kakurai M, et al. . Trichophyton indotineae sp. nov.: a new highly terbinafine-resistant anthropophilic dermatophyte species. Mycopathologia 2020; 185:947–58. [DOI] [PubMed] [Google Scholar]
- 70. Cao C, Xi L, Chaturvedi V. Talaromycosis (penicilliosis) due to Talaromyces (Penicillium) marneffei: insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. Mycopathologia 2019; 184:709–20. [DOI] [PubMed] [Google Scholar]
- 71. Frisvad JC, Yilmaz N, Thrane U, Rasmussen KB, Houbraken J, Samson RA. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS One 2013; 8:e84102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bhatnagar S, Kobori T, Ganesh D, Aoyagi H. Fungal pigment-assisted silver nanoparticle synthesis and their antimicrobial and cytotoxic potential. Methods Mol Biol 2022; 2469:65–78. [DOI] [PubMed] [Google Scholar]
- 73. Atalay A, Koc AN, Akyol G, Cakir N, Kaynar L, Ulu-Kilic A. Pulmonary infection caused by Talaromyces purpurogenus in a patient with multiple myeloma. Infez Med 2016; 24:153–7. [PubMed] [Google Scholar]
- 74. Li L, Chen K, Dhungana N, Jang Y, Chaturvedi V, Desmond E. Characterization of clinical isolates of Talaromyces marneffei and related species, California, USA. Emerg Infect Dis 2019; 25:1765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Surja SS, Adawiyah R, Houbraken J, et al. . Talaromyces atroroseus in HIV and non-HIV patient: a first report from Indonesia. Med Mycol 2020; 58:560–3. [DOI] [PubMed] [Google Scholar]
- 76. Aboutalebian S, Mahmoudi S, Okhovat A, Khodavaisy S, Mirhendi H. Otomycosis due to the rare fungi Talaromyces purpurogenus, Naganishia albida and Filobasidium magnum. Mycopathologia 2020; 185:569–75. [DOI] [PubMed] [Google Scholar]
- 77. Moreno-Gavíra A, Huertas V, Diánez F, Sánchez-Montesinos B, Santos M. Paecilomyces and its importance in the biological control of agricultural pests and diseases. Plants (Basel) 2020; 9:1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Antas PR, Brito MM, Peixoto É, Ponte CG, Borba CM. Neglected and emerging fungal infections: review of hyalohyphomycosis by Paecilomyces lilacinus focusing in disease burden, in vitro antifungal susceptibility and management. Microbes Infect 2012; 14:1–8. [DOI] [PubMed] [Google Scholar]
- 79. Abdolrasouli A, Bercusson AC, Rhodes JL, et al. . Airway persistence by the emerging multi-azole-resistant Rasamsonia argillacea complex in cystic fibrosis. Mycoses 2018; 61:665–73. [DOI] [PubMed] [Google Scholar]
- 80. Stemler J, Salmanton-García J, Seidel D, et al. . Risk factors and mortality in invasive Rasamsonia spp. infection: analysis of cases in the FungiScope registry and from the literature. Mycoses 2020; 63:265–74. [DOI] [PubMed] [Google Scholar]
- 81. Pitt J. Geosmithia gen. nov. for Penicillium lavendulum and related species. Can J Bot 1979; 57:2021–30. [Google Scholar]
- 82. Ogawa H, Yoshimura A, Sugiyama J. Polyphyletic origins of species of the anamorphic genus Geosmithia and the relationships of the cleistothecial genera: evidence from 18S, 5S and 28S rDNA sequence analyses. Mycologia 1997; 89:756–71. [Google Scholar]
- 83. Giraud S, Favennec L, Bougnoux ME, Bouchara JP. Rasamsonia argillacea species complex: taxonomy, pathogenesis and clinical relevance. Future Microbiol 2013; 8:967–78. [DOI] [PubMed] [Google Scholar]
- 84. Brasch J, Beck-Jendroschek V, Voss K, Yurkov A, Gräser Y. Arthroderma chiloniense sp. nov. Isolated from human stratum corneum: description of a new Arthroderma species. Mycoses 2019; 62:73–80. [DOI] [PubMed] [Google Scholar]
- 85. Borman AM, Szekely A, Fraser M, Lovegrove S, Johnson EM. A novel dermatophyte relative, Nannizzia perplicata sp. nov., isolated from a case of tinea corporis in the United Kingdom. Med Mycol 2019; 57:548–56. [DOI] [PubMed] [Google Scholar]
- 86. Tang C, Kong X, Ahmed SA, et al. . Taxonomy of the Trichophyton mentagrophytes/T. interdigitale species complex harboring the highly virulent, multiresistant genotype T. indotineae. Mycopathologia 2021; 186:315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jabet A, Brun S, Normand AC, et al. . Extensive dermatophytosis caused by terbinafine-resistant Trichophyton indotineae, France. Emerg Infect Dis 2022; 28:229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Posso-De Los Rios CJ, Tadros E, Summerbell RC, Scott JA. Terbinafine resistant Trichophyton indotineae isolated in patients with superficial dermatophyte infection in Canadian patients. J Cutan Med Surg 2022; 26:371–6. [DOI] [PubMed] [Google Scholar]
- 89. Fisher MC, Koenig GL, White TJ, Taylor JW. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 2002; 94:73–84. [PubMed] [Google Scholar]
- 90. Jiang Y, Dukik K, Muñoz JF, et al. . Phylogeny, ecology and taxonomy of systemic pathogens and their relatives in Ajellomycetaceae (Onygenales): Blastomyces, Emergomyces, Emmonsia, Emmonsiellopsis. Fung Div 2018; 90:245–91. [Google Scholar]
- 91. Dukik K, Muñoz JF, Jiang Y, et al. . Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Onygenales). Mycoses 2017; 60:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sepúlveda VE, Márquez R, Turissini DA, Goldman WE, Matute DR. Genome sequences reveal cryptic speciation in the human pathogen Histoplasma capsulatum. mBio 2017; 8:e01339–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vilela R, Huebner M, Vilela C, et al. . The taxonomy of two uncultivated fungal mammalian pathogens is revealed through phylogeny and population genetic analyses. Sci Rep 2021; 11:18119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Teixeira MM, Theodoro RC, Oliveira FF, et al. . Paracoccidioides lutzii sp. nov.: biological and clinical implications. Med Mycol 2014; 52:19–28. [DOI] [PubMed] [Google Scholar]
- 95. Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet Biol 2017; 106:9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Marimon R, Cano J, Gené J, Sutton DA, Kawasaki M, Guarro J. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol 2007; 45:3198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rodrigues AM, Della Terra PP, Gremião ID, Pereira SA, Orofino-Costa R, de Camargo ZP. The threat of emerging and re-emerging pathogenic Sporothrix species. Mycopathologia 2020; 185:813–42. [DOI] [PubMed] [Google Scholar]
- 98. Jofre GI, Singh A, Mavengere H, et al. . An Indian lineage of Histoplasma with strong signatures of differentiation and selection. Fungal Genet Biol 2022; 158:103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rodrigues AM, Beale MA, Hagen F, et al. . The global epidemiology of emerging Histoplasma species in recent years. Stud Mycol 2020; 97:100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Teixeira MM, Patané JS, Taylor ML, et al. . Worldwide phylogenetic distributions and population dynamics of the genus Histoplasma. PLoS Negl Trop Dis 2016; 10:e0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Theodoro RC, Scheel CM, Brandt ME, Kasuga T, Bagagli E. PRP8 Intein in cryptic species of Histoplasma capsulatum: evolution and phylogeny. Infect Genet Evol 2013; 18:174–82. [DOI] [PubMed] [Google Scholar]
- 102. Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis. PLoS One 2013; 8:e59237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schwartz IS, Kenyon C, Feng P, et al. . 50 years of Emmonsia disease in humans: the dramatic emergence of a cluster of novel fungal pathogens. PLoS Pathog 2015; 11:e1005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schwartz IS, Wiederhold NP, Hanson KE, Patterson TF, Sigler L. Blastomyces helicus, a new dimorphic fungus causing fatal pulmonary and systemic disease in humans and animals in western Canada and the United States. Clin Infect Dis 2019; 68:188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Maphanga TG, Birkhead M, Muñoz JF, et al. . Human blastomycosis in South Africa caused by Blastomyces percursus and Blastomyces emzantsi sp. nov., 1967 to 2014. J Clin Microbiol 2020; 58:e01661–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kenyon C, Bonorchis K, Corcoran C, et al. . A dimorphic fungus causing disseminated infection in South Africa. N Engl J Med 2013; 369:1416–24. [DOI] [PubMed] [Google Scholar]
- 107. Jiang Y, Tsui CKM, Ahmed SA, et al. . Intraspecific diversity and taxonomy of Emmonsia crescens. Mycopathologia 2020; 185:613–27. [DOI] [PubMed] [Google Scholar]
- 108. Wang P, Kenyon C, de Hoog S, et al. . A novel dimorphic pathogen, Emergomyces orientalis (Onygenales), agent of disseminated infection. Mycoses 2017; 60:310–19. [DOI] [PubMed] [Google Scholar]
- 109. Roberto TN, de Carvalho JA, Beale MA, et al. . Exploring genetic diversity, population structure, and phylogeography in Paracoccidioides species using AFLP markers. Stud Mycol 2021; 100:100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rodrigues AM, de Hoog GS, de Camargo ZP. Sporothrix species causing outbreaks in animals and humans driven by animal-animal transmission. PLoS Pathog 2016; 12:e1005638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lackner M, de Hoog GS, Yang L, et al. . Proposed nomenclature for Pseudallescheria, Scedosporium and related genera. Fungal Divers 2014; 67:1–10. [Google Scholar]
- 112. Gilgado F, Cano J, Gené J, Guarro J. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol 2005; 43:4930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zeng JS, Fukushima K, Takizawa K, et al. . Intraspecific diversity of species of the Pseudallescheria boydii complex. Med Mycol 2007; 45:547–58. [DOI] [PubMed] [Google Scholar]
- 114. Gilgado F, Cano J, Gené J, Sutton DA, Guarro J. Molecular and phenotypic data supporting distinct species statuses for Scedosporium apiospermum and Pseudallescheria boydii and the proposed new species Scedosporium dehoogii. J Clin Microbiol 2008; 46:766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Manamgoda DS, Cai L, McKenzie EHC. A phylogenetic and taxonomic re-evaluation of the Bipolaris–Cochliobolus–Curvularia complex. Fungal Divers 2012; 56:131–44. [Google Scholar]
- 116. Samerpitak K, Van der Linde E, Choi H-J, et al. . Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Divers 2014; 65:89–126. [Google Scholar]
- 117. Réblová M, Miller AN, Rossman AY, et al. . Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 2016; 7:131–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol 2007; 58:57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Berbee ML, Pirseyedi M, Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999; 91:964–77. [Google Scholar]
- 120. Manamgoda DS, Cai L, Bahkali AH. Cochliobolus: an overview and current status of species. Fungal Divers 2011; 51:3–42. [Google Scholar]
- 121. Von Arx JA, Luttrell ES. Whole fungus (Kendrick B, ed.). Ottawa: National Museum of Natural Sciences and the Kananaskis Foundation; 1979; 1:260–1. [Google Scholar]
- 122. Goh KT, Hyde KD, Lee KLD. Generic distinction in Helminthosporium complex based on restriction analysis of the nuclear ribosomal RNA gene. Fungal Divers 1998; 1:85–1. [Google Scholar]
- 123. Giraldo A, Sutton DA, Samerpitak K, et al. . Occurrence of Ochroconis and Verruconis species in clinical specimens from the United States. J Clin Microbiol 2014; 52:4189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Revankar SG, Sutton DA. Melanized fungi in human disease. Clin Microbiol Rev 2010; 23:884–928. Erratum in: Clin Microbiol Rev 2012; 25:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shi D, Lu G, Mei H, et al. . Subcutaneous infection by Ochroconis mirabilis in an immunocompetent patient. Med Mycol Case Rep 2016; 11:44–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Mohammadi R, Mohammadi A, Ashtari F, et al. . Cerebral phaeohyphomycosis due to Rhinocladiella mackenziei in Persian Gulf region: a case and review. Mycoses 2018; 61:261–5. [DOI] [PubMed] [Google Scholar]
- 127. Badali H, Bonifaz A, Barrón-Tapia T, et al. . Rhinocladiella aquaspersa, proven agent of verrucous skin infection and a novel type of chromoblastomycosis. Med Mycol 2010; 48:696–703. [DOI] [PubMed] [Google Scholar]
- 128. Santos DWCL, de Azevedo CMPES, Vicente VA, et al. . The global burden of chromoblastomycosis. PLoS Negl Trop Dis 2021; 15:e0009611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ahmed SA, van de Sande WWJ, Stevens DA, et al. . Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia 2014; 33:141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Crous PW, Slippers B, Wingfield MJ, et al. . Phylogenetic lineages in the Botryosphaeriaceae. Stud Mycol 2006; 55:235–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Huang S-K, Tangthirasunun N, Phillips AJL, et al. . Morphology and phylogeny of Neoscytalidium orchidacearum sp. nov. (Botryosphaeriaceae). Mycobiology 2016; 44:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. de Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW. Redisposition of Phoma-like anamorphs in Pleosporales. Stud Mycol 2013; 75:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shaw JJ, Spakowicz DJ, Dalal RS, et al. . Biosynthesis and genomic analysis of medium-chain hydrocarbon production by the endophytic fungal isolate Nigrograna mackinnonii E5202H. Appl Microbiol Biotechnol 2015; 99:3715–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hibbett DS, Binder M, Bischoff JF, et al. . A higher-level phylogenetic classification of the fungi. Mycol Res 2007; 111:509–47. [DOI] [PubMed] [Google Scholar]
- 135. Spatafora JW, Chang Y, Benny GL, et al. . A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016; 108:1028–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ajello L, Dean DF, Irwin RS. The zygomycete Saksenaea vasiformis as a pathogen of humans with a critical review of the etiology of zygomycosis. Mycologia 1976; 68:52–62. [PubMed] [Google Scholar]
- 137. Walther G, Wagner L, Kurzai O. Updates on the taxonomy of Mucorales with an emphasis on clinically important taxa. J Fungi (Basel) 2019; 5:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wagner L, Stielow JB, de Hoog GS, et al. . A new species concept for the clinically relevant Mucor circinelloides complex. Persoonia 2020; 44:67–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Alvarez E, Cano J, Stchigel AM, et al. . Two new species of Mucor from clinical samples. Med Mycol 2011; 49:62–72. [DOI] [PubMed] [Google Scholar]
- 140. Hoffmann K, Walther G, Voigt K. Mycocladus vs. Lichtheimia, a correction (Lichtheimiaceae fam. nov., Mucorales, Mucoromycotina). Mycol Res 2009; 113:277–8. [Google Scholar]
- 141. Dolatabadi S, Walther G, Gerrits van den Ende AHG, de Hoog GS. Diversity and delimitation of Rhizopus microsporus. Fung Div 2014; 64:145–63. [Google Scholar]
- 142. Dolatabadi S, de Hoog GS, Meis JF, Walther G. Species boundaries and nomenclature of Rhizopus arrhizus (syn. R. oryzae). Mycoses 2014; 57(Suppl 3):108–27. [DOI] [PubMed] [Google Scholar]
- 143. Alvarez E, Garcia-Hermoso D, Sutton DA, et al. . Molecular phylogeny and proposal of two new species of the emerging pathogenic fungus Saksenaea. J Clin Microbiol 2010; 48:4410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Alastruey-Izquierdo A, Hoffmann K, de Hoog GS, et al. . Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus). J Clin Microbiol 2010; 48:2154–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Schipper MAA. A revision of the genus Rhizopus. 1. The Rhizopus stolonifer-group and Rhizopus oryzae. Stud Mycol 1984; 25:20–34. [Google Scholar]
- 146. Ellis JJ. Species and varieties in the Rhizopus arrhizus–Rhizopus oryzae group as indicated by their DNA complementarity. Mycologia 1985; 77:243–7. [Google Scholar]
- 147. Prakash H, Chakrabarti A. Global epidemiology of mucormycosis. J Fungi (Basel) 2019; 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Labuda R, Bernreiter A, Hochenauer D, et al. . Saksenaea dorisiae sp. nov., a new opportunistic pathogenic fungus from Europe. Int J Microbiol 2019; 2019:6253829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Nam B, Lee DJ, Choi YJ. High-temperature-tolerant fungus and Oomycetes in Korea, including Saksenaea longicolla sp. nov. Mycobiology 2021; 49:476–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Crous PW, Wingfield MJ, Burgess TI, et al. . Fungal planet description sheets: 558–624. Persoonia 2017; 38:240–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Crous PW, Wingfield MJ, Burgess TI, et al. . Fungal planet description sheets: 469–557. Persoonia 2016; 37:218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Munson E, Carroll KC. An update on the novel genera and species and revised taxonomic status of bacterial organisms described in 2016 and 2017. J Clin Microbiol 2019; 57:e01181–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Janda JM. Proposed nomenclature or classification changes for bacteria of medical importance: taxonomic update 5. Diagn Microbiol Infect Dis 2020; 97:115047. [DOI] [PubMed] [Google Scholar]
- 154. Mathison BA, Bradbury RS, Pritt BS. Medical parasitology taxonomy update, January 2018 to May 2020. J Clin Microbiol 2021; 59:e01308–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Loeffelholz MJ, Fenwick BW. Taxonomic changes for human and animal viruses, 2018 to 2020. J Clin Microbiol 2021; 59:e01932–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Panda A, Islam ST, Sharma G. Harmonizing prokaryotic nomenclature: fixing the fuss over phylum name flipping. mBio 2022; 13:e0097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Walker PJ, Siddell SG, Lefkowitz EJ, et al. . Changes to virus taxonomy and to the International Code of Virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch Virol 2021; 166:2633–48. [DOI] [PubMed] [Google Scholar]
- 158. Kidd SE, Halliday CL, Haremza E, Gardam DJ, Chen SCA, Elvy JA. Attitudes of Australasian clinicians and laboratory staff to changing fungal nomenclature: has mycological correctness really gone mad? Microbiol Spectr 2022; 10:e0237721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Chen SC, Perfect J, Colombo AL, et al. . Global guideline for the diagnosis and management of rare yeast infections: an initiative of the ECMM in cooperation with ISHAM and ASM. Lancet Infect Dis 2021; 21:e375–86. Erratum in: Lancet Infect Dis 2021; 21:e363. [DOI] [PubMed] [Google Scholar]
- 160. Chang CC, Blyth CC, Chen SC, et al. . Introduction to the updated Australasian consensus guidelines for the management of invasive fungal disease and use of antifungal agents in the haematology/oncology setting, 2021. Intern Med J 2021; 51(Suppl 7):3–17. [DOI] [PubMed] [Google Scholar]
- 161. Brickey TW, Trotter JF, Johnson SP. Torulopsis glabrata fungemia from infected transjugular intrahepatic portosystemic shunt stent. J Vasc Interv Radiol 2005; 16:751–2. [DOI] [PubMed] [Google Scholar]
- 162. Federhen S. The NCBI taxonomy database. Nucleic Acids Res 2012; 40(database issue):D136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Schoch CL, Ciufo S, Domrachev M, et al. . NCBI taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antifungal susceptibility testing of yeasts. 3rd ed. CLSI guideline M27M44SEd3. Wayne PA: CLSI; 2022.