Abstract
Porphyromonas gingivalis is one of the principal organisms associated with adult periodontitis. Bacterial surface proteins such as fimbriae and gingipain hemagglutinin domains have been implicated as adhesins that actuate colonization of epithelium lining the gingival sulcus. We investigated the genetics of P. gingivalis adhesion to monolayers of epithelial cells using wild-type and gingipain mutant strains. These experiments suggested that arginine-specific gingipain (Rgp) catalytic activity modulated adhesion. From the data obtained with rgp mutants, we constructed a working hypothesis predicting that attachment and detachment of P. gingivalis to epithelial cells were mediated by gingipain adhesin and Rgp catalytic domains, respectively. A membrane-based epithelial cell binding assay, used to locate adhesins in extracellular fractions of wild-type and mutant strains, recognized gingipain peptides as adhesins rather than fimbriae. We developed a capture assay that demonstrated the binding of gingipain adhesin peptides to oral epithelial cells. The adherence of fimbrillin to epithelial cells was detected after heat denaturation of cell fractions. The prediction that Rgp catalytic activities mediated detachment was substantiated when the high level of attachment of an rgp mutant was reduced in the presence of wild-type cell fractions that contained gingipain catalytic activities.
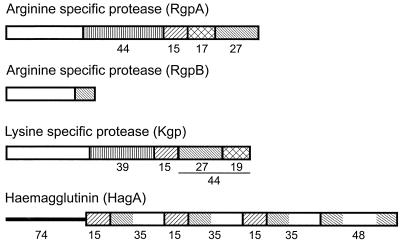
Porphyromonas gingivalis, a gram-negative anaerobe present in subgingival plaque, is one of the bacteria strongly associated with adult periodontitis. The molecular mechanisms leading to colonization of the epithelium that lines the gingival sulcus are poorly understood. Fimbria-deficient mutants of P. gingivalis showed reduced attachment to and invasion of oral epithelial cells (24, 36, 40). In addition, cysteine proteinase (gingipain) hemagglutinin domains have been implicated in tissue colonization either directly through adhesion to extracellular matrix proteins (17, 31) or indirectly by processing the fimbrillin subunit of fimbriae (23). Gingipains are secreted proteins found on the bacterial cell surface, associated with extracellular vesicles, and in culture supernatants. Gingipain genes rgpA and rgpB encode Arg-gingipains (Rgp) A and B, respectively. These enzymes possess arginine-specific amidolytic activity, while a third gene, kgp, encodes an enzyme with lysine-specific amidolytic activity (Lys-gingipain [Kgp]). The Rgp isozymes contain propeptide and catalytic domains, but only RgpA contains a carboxy-terminal extension known as the adhesin domain (Fig. 1). Kgp also contains propeptide, catalytic, and adhesin domains, and the latter shares over 97% homology with the adhesin domain of RgpA (4). Within this family is an additional gene encoding a surface protein, hemagglutinin A. hagA contains three or four copies of a 1.35-kb direct repeat (15) encoding protein sequences that also contain homology to the adhesin domains of RgpA and Kgp. Autoprocessing of the adhesin domains yields a series of peptides (Fig. 1), and it has been determined experimentally that sequences within these peptides possess hemagglutinating and tissue colonization functions (12, 22, 34). Assays developed in this study showed directly that the adhesin peptides, possibly complexed together, bind to epithelial cells, and that gingipain catalytic activities can mediate detachment of bacteria and thus modulate adhesion.
FIG. 1.
Structures and homologies of gingipains RgpA, RgpB, Kgp, and HagA. Catalytic domains are depicted as open boxes, and adhesin domains are shaded to indicate specific peptide regions and their homologies. The three repeat regions within HagA from ATCC 33277 were established by PCR (15). The rgpA and kgp genes from ATCC 33277 have not been sequenced; therefore, the assigned molecular masses (kilodaltons) of the peptides are deduced from published sequences of those from the closely related strain 381.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
P. gingivalis strains ATCC 33277 and 381 and their mutant derivatives were maintained on blood agar as described previously (2). A fimA mutant, from the laboratory of R. Genco (Buffalo, N.Y.), and Rgp mutants were maintained on blood agar, and antibiotics were added to final concentrations of 5 μg/ml (erythromycin) and 1 μg/ml (tetracycline). Unless stated otherwise, all chemicals were from Sigma, St. Louis, Mo.
Hemagglutination assay.
Sheep erythrocytes (BINAX/NEL, Portland, Maine) were washed and resuspended to a final concentration of 2% (vol/vol) in phosphate-buffered saline, pH 7.4 (PBS). Bacteria were harvested after growth for 48 h on blood plates and suspended in PBS with 1 mM l-cysteine to activate hemagglutinin activity. Concentrations of bacteria were adjusted to an A550 of 0.2, and 50-μl aliquots were twofold serially diluted in 96-well plates with PBS, followed by the addition of 50 μl of sheep red blood cells. Plates were incubated at room temperature with shaking for 1 h and then overnight at 4°C without shaking.
Assays for gingipain catalytic activities.
Bacteria were prepared as for the hemagglutination assay and used at an A550 of 0.2. The fluorescent protease substrates αN-benzoyl-l-arginine-7-amido-4-methylcoumarin and t-butyloxycarboyl-Val-Leu-Lys-7-amido-4-methylcoumarin were used to determine Arg-X and Lys-X specific activities, respectively. Arg-X activity was assayed in 0.1 ml of PBS containing 1 mM l-cysteine, 200 μM substrate, and 5 to 50 μl of cell suspension or fraction at room temperature for 10 min. Lys-X activity was assayed in 0.1 ml of PBS containing 1 mM l-cysteine, 10 μM substrate, and 5 to 10 μl of cell suspension or fraction at 40°C for 10 min. Protease reactions were terminated by the addition of Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) to 500 μM. Released 7-amido-4-methylcoumarin was measured with a fluorimeter (model HTS 7000 Plus, Perkin-Elmer, Norwalk, Conn.) with exciting and emitting wavelengths of 365 and 460 nm, respectively. Assays were carried out in duplicate and were repeated at least twice with independent cultures. Protease activities of whole cells were determined as fluorescent counts per min per A550 optical density unit; activities of cell fractions were determined as fluorescent counts per minute per milligram of protein. For comparative purposes, the results were expressed as a percentage of the activity of the parent strain, ATCC 33277.
Attachment of P. gingivalis to oral epithelial cell monolayers.
KB oral epithelial cells (ATCC CCL17) were grown as described previously (7) and prepared for attachment assays by dilution to 5 × 105 cells/ml in complete Dulbeccco's modified Eagle's medium (DMEM) without antibiotics. One milliliter of diluted cells was added per well of a 24-well plate and grown overnight. Immediately before infection with bacteria, KB monolayers were washed three times with PBS (1 ml per well). Bacterial cultures were harvested after 48 h of growth on blood agar plates, washed once, and resuspended in PBS. Bacteria were diluted to approximately 107 cells/ml in DMEM containing only 20 mM HEPES and 2 mM glutamine, and 1-ml aliquots were added to each well containing KB cells. As described previously (2), the number of bacteria attaching to the KB cells after 90 min of incubation was expressed as the percentage of the number of bacteria added per monolayer. The results are from at least three experiments.
Membrane-based epithelial cell binding assay.
The assay was used to measure the adhesion of KB cells to immobilized bacteria and to detect adhesins in cell extracts. Wild-type and mutant P. gingivalis strains varied in plating efficiency; therefore, quantitation of the assay is expressed relative to the number of ATCC 33277 cells at an A550 of 0.2 (ca. 5 × 108 cells/ml), with the assumption that this initial number was equivalent for all strains, and independent of plating efficiency. Thus, at an A550 of 0.2, each 5-μl spot (as shown in Table 2) contained 3 × 106 intact bacteria. Equal volumes (5 μl) of equivalent cell suspensions, or extracts (also 5 μl) prepared from these suspensions, were spotted to Immobilon P membranes (Millipore Corp., Bedford, Mass.). The published protocol (7) was modified in that bacteria were not lysed in situ, and membranes were blocked only overnight in dry milk solution. Subsequent incubation of membranes with KB cells, treatment with glutaraldehyde to fix KB cells bound to bacteria or extracts, and chromogenic detection of bound epithelial cells were as described previously (7).
TABLE 2.
Distribution of gingipain activities in and binding of KB cells to intact bacteria and extracellular fractions from P. gingivalis parent and mutant strainsa
| Strain | Genotype | % Total residual activity in vesicle plus VDS fractionsb
|
Distribution of Rgp and Kgp catalytic activities between extracellular fractionsc and their KB cell binding
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Vesicles
|
VDS
|
||||||||
| Rgp | Kgp | % Activity
|
KB binding | % Activity
|
KB binding | ||||
| Rgp | Kgp | Rgp | Kgp | ||||||
| ATCC 33277 | rgpA+rgpB+ | 100 | 100 | 78 | 65 | + | 22 | 35 | + |
| KDP110 | rgpA | 106 | 102 | 83 | 72 | + | 17 | 28 | + |
| KDP111 | rgpB | 18 | 69 | 82 | 37 | + | 18 | 63 | + |
| KDP112 | rgpA rgpB | 0 | 27 | 0 | 0 | − | 0 | 100 | − |
| DPG3 | fimA | 68 | 51 | 30 | 35 | + | 70 | 65 | + |
All intact bacteria showed the ability to bind KB cells.
Percentage of the level for ATCC 33277.
Percentage of the total activity in the combined vesicle and supernatant fractions.
Fluorometric assay of P. gingivalis attachment to epithelial cells.
KB epithelial cells were seeded in a 96-well tissue culture plate at a density of 104 cells/200 μl/well and incubated in DMEM containing 10% newborn calf serum but without antibiotics. Monolayers were washed twice with PBS before use. Bacterial strains were grown as above, and cells were washed once and resuspended in PBS. Suspensions were twofold serially diluted with DMEM containing 20 mM HEPES and 2 mM glutamine in a separate 96-well plate, and cell densities were measured at A550. Suspensions (100 μl) were transferred to plates containing KB monolayers and incubated for 90 min. P. gingivalis extracellular extracts were added to the concentrations described below, and 500 μM TLCK was added to control assays to inhibit gingipain catalytic activities. Infected monolayers were washed twice with PBS to remove unattached bacteria. Attached bacteria were detected by their ability to hydrolyze the fluorogenic substrate 4-methylumbelliferyl N-acetyl-β-d-glucosaminide (100 μl, 0.13 mM) at 37°C for 60 min. The number of attached bacteria was proportional to the number of umbelliferone fluorescent counts measured with a fluorimeter at exciting and emitting wavelengths of 340 and 440 nm, respectively. In control wells, KB cells were incubated alone and showed no hydrolysis of the fluorogenic substrate.
Preparation of P. gingivalis cell fractions.
Bacteria were suspended at approximately 1010 cells/ml in PBS. Suspensions were vortexed vigorously and then centrifuged at 6,000 × g for 15 min at 4°C. The supernatant wash fractions contained outer membrane vesicles and soluble extracellular proteins. Vesicle-depleted supernatants (VDS) were obtained by a further centrifugation of the wash supernatants at 29,000 × g for 40 min at 4°C to sediment vesicles (8). The supernatant fraction was used in capture assays with KB cells. Vesicles were resuspended in 100 μl of PBS, and gingipain catalytic activities were measured in both vesicle and VDS fractions. Cell extracts were prepared from suspensions of washed bacteria in PBS that were chilled in an ice-water bath and sonicated for 1 min in 15-s bursts alternating with 15 s of cooling. Cell sonicates were also used in capture assays.
Adhesin capture assay.
KB cells were prepared as described previously (7) and resuspended in DMEM–20 mM HEPES–2 mM glutamine. Before their addition to KB cells, VDS were pretreated by either 5 min of boiling to denature proteins or dissociate complexes or the addition of TLCK (500 μM) to inhibit protease activity; untreated supernatants were used as controls. VDS fractions (200 μl containing 300 μg of protein per ml) and KB cells (106 cells in 200 μl of medium) were incubated for 90 min at 37°C in a 5% CO2 incubator. KB cells were washed twice (500 × g, 5 min) to remove loosely binding proteins and then lysed by boiling for 5 min with an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (16). Equal volumes (10 μl) of samples were fractionated by SDS-PAGE (10% polyacrylamide gel; Bio-Rad Inc., Richmond, Calif.), and proteins were either revealed by silver staining or electroblotted (38) to Immobilon P (Millipore). Gingipain-related proteins were detected with rabbit polyclonal primary antibody to recombinant RgpA adhesin domain from G721 to R1262 (a generous gift from M. Curtis, London, United Kingdom). Fimbrillin was detected with rabbit polyclonal primary antibody, kindly provided by A. Sharma, State University of New York at Buffalo. In both cases, antibody binding was detected with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody using the enhanced chemiluminescence Western blot detection system (Amersham-Pharmacia, Piscataway, N.J.).
N-terminal sequencing.
VDS samples were fractionated by SDS-PAGE and electroblotted to an Immobilon P membrane (Millipore), and proteins were located by Ponceau red staining. Automated Edman degradation and sequencing were performed by Midwest Analytical, Inc. (St. Louis, Mo.), with a model 477A protein sequencer (Perkin-Elmer Biosystems, Foster City, Calif.). The 44- and 27-kDa peptides were obtained from the VDS of ATCC 33277. The 39-kDa peptide was obtained from the VDS of DGP3, a fimA mutant of strain 381, to avoid contamination with the similarly sized FimA protein.
RESULTS
Effect of gingipain mutations on P. gingivalis adhesion to epithelial cells.
The attachment level of each single mutant, KDP110 or KDP111, was not statistically different from that of the parent strain, ATCC 33277, in that only 1 to 2% of the input bacteria attached to KB monolayers (Table 1). Enzyme assays showed that their cell-associated Rgp catalytic activities were approximately 50% of the wild type, and Kgp activities were unaffected (Table 1). By contrast, approximately 60% of the input cells of KDP112, the rgpA rgpB double mutant, attached to the monolayers. KDP112 had no detectable Rgp activity and 50% of the parental level of Kgp activity. The results of this series experiments indicated that protease activity, especially that of Rgp, modulated P. gingivalis adherence to epithelial cells. Furthermore, because Rgp catalytic activities are required for processing prefimbrillin, the protein subunit of fimbriae, KDP112 is reported to possess very few fimbriae (23); thus, the results also suggested that proteins other than fimbriae may function as adhesins for epithelial cells.
TABLE 1.
Effects of Arg-gingipain mutations on P. gingivalis attachment to KB oral epithelial cells
| Strain | Genotype | Residual activitya
|
Bacteria attached to epithelial cellsb | |
|---|---|---|---|---|
| Rgp | Kgp | |||
| ATCC 33277 | rgpA+rgpB+ | 100 | 100 | 1.8 ± 1.0 |
| KDP110 | rgpA | 54.5 | 100.4 | 1.1 ± 0.79 |
| KDP111 | rgpB | 52.1 | 92.6 | 2.1 ± 1.6 |
| KDP112 | rgpA rgpB | 0 | 54.3 | 60.5 ± 28.0 |
Percentage of that of the parent strain ATCC 33277.
Percentage of the number of bacteria added to monolayers at 0 min ± standard deviation. The results are the average of at least three experiments in which each strain was assayed in triplicate.
Localization of adhesins.
To identify candidate adhesin proteins in the least complex cell fraction, we first assumed they would be associated with the surface of P. gingivalis cells. Therefore, equivalent suspensions of the parent and mutant bacteria were vortexed vigorously to release loosely attached surface proteins, fimbriae, and extracellular vesicles. Bacteria were removed by centrifugation, and supernatants were further fractionated to yield pellets of extracellular vesicles and VDS containing soluble proteins. Both fractions from parent and mutant strains were assayed for Rgp and Kgp catalytic activities. In addition, equal volumes of each fraction (5 μl containing 2 to 3 μg of protein) were tested for the ability to bind epithelial cells in a membrane-based assay.
As described previously, epithelial cells recognize high-affinity-binding epitopes in this assay and, for example, neither intact nor lysed Escherichia coli cells give a positive reaction (6, 7, 18). As shown in Table 2, at an A550 of 0.2, each 5-μl spot of intact, washed P. gingivalis parent and mutant strains, containing 3 × 106 bacteria, showed a positive KB binding reaction. Variations in binding ability became apparent at higher dilutions; thus, while approximately 8 × 104 cells of ATCC 33277 was the minimum number that still gave a positive reaction, twice as many KDP110 and KDP111, four times as many DPG3, and eight times as many KDP112 cells were required. If a positive reaction depends on a finite number of adhesins, it appears that KDP112 possessed fewer than ATCC 33277 since more mutant cells were required to detect KB binding. Despite this, KDP112 attached to KB monolayers in high numbers, underscoring the importance of Rgp catalytic activities in the modulation of P. gingivalis adhesion to KB cells. By the same argument, DPG3, the fimA mutant, also possessed fewer adhesins than ATCC 33277, but the significant level of Rgp activity associated with DPG3 cells (Table 2) may also account for its low attachment to KB monolayers.
Except for those from KDP112, the rgpA rgpB mutant, vesicle, and VDS fractions from the other strains contained both Rgp and Kgp catalytic activities and were also able to bind epithelial cells (Table 2). In addition, all fractions of strain DPG3, a fimA mutant of P. gingivalis 381 (20), retained the ability to bind KB cells, indicating that fimbriae were not the adhesins recognized in this assay. While intact cells of KDP112 bound epithelial cells, neither the vesicle nor VDS fraction from the mutant contained adhesins, although Kgp catalytic activity was detected in the VDS fraction. This result suggested that in KDP112 adhesins may be restricted to and tightly bound to the outer surface of the cell and that Rgp plays a wider role in the processing or secretion of gingipains.
Identification of gingipain adhesin peptides in VDS fractions of parent and mutant strains.
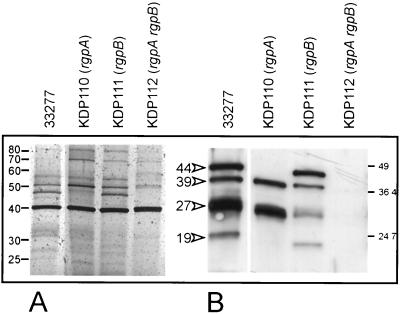
Since the VDS fractions retained epithelial cell binding activity, they were used in further experiments as simpler sources of candidate adhesins, rather than more complex cellular extracts. The VDS fraction from ATCC 33277, the parent strain, contained approximately 10 to 12 proteins by silver staining (Fig. 2A) and retained KB binding activity (Table 2). Western blots of the VDS fractions from the parent and mutant strains were probed with antibody to the adhesin domain of RgpA, which, because of their sequence homologies, also recognizes the adhesin domains of Kgp and HagA (Fig. 1). The antibody reacted with peptides of 44, 39, 27, and 19 kDa in the VDS fraction of ATCC 33277 (Fig. 2B); these were similar in size to the autoprocessing products from the adhesin domains of RgpA and Kgp (Fig. 1). The VDS fraction of the rgpA mutant (KDP110) did not contain the 44-kDa adhesin peptide, consistent with it being derived from RgpA, while the same fraction from the rgpB mutant (KDP111) retained this band. None of these peptides were found in the VDS of the rgpA rgpB double mutant KDP112 (Fig. 2B); therefore, even though Kgp catalytic activity was detected in this fraction (Table 2), apparently it was not associated with the adhesin domain.
FIG. 2.
Gingipain adhesins in VDS fractions from P. gingivalis ATCC 33277 and rgp mutant strains. (A) VDS fractions from parent and mutant strains after SDS-PAGE and silver staining. (B) Western blot of the VDS fractions using a polyclonal antibody to gingipain adhesin domains. The 44-, 39-, 27-, and 19-kDa peptides in the fraction from ATCC 33277 are indicated by arrowheads. The 44-kDa peptide was absent from the rgpA mutant, KDP110. The VDS fraction from the rgpA rgpB mutant, KDP112, did not contain adhesin peptides.
Capture of gingipain adhesin peptides and fimbrillin by KB cells.
To demonstrate that gingipain adhesin peptides bind to epithelial cells, we developed an assay in which KB cells were used to capture adhesins from the VDS fractions. The fractions were native, treated with TLCK to inhibit protease activity, or boiled for 5 min. Following incubation with VDS fractions, KB cells were washed to remove loosely binding proteins and prepared for SDS-PAGE by boiling in electrophoresis sample buffer. Western blots probed with antiadhesin antibody showed that KB cells incubated with native or TLCK-treated VDS fractions from ATCC 33277, KDP111 (rgpB), and DPG3 (fimA) captured peptides of approximately 44, 39, 27, and 19 kDa (Fig. 3A). Extracts of KB cells that had been incubated with the VDS from the rgpA mutant (KDP110) contained only the 39-, 27-, and 19-kDa peptides. We deduced that these peptides were derived from Kgp since the three peptides were not detected in capture assays with a kgp mutant (data not shown). Adhesin peptides were not present in KB cell extracts incubated with the VDS from KDP112 (Fig. 3A) or in KB cells incubated alone (not shown). KB cells that had been incubated with heat-denatured VDS fractions did not contain adhesin peptides (Fig. 3A) but captured a protein that cross-reacted with antifimbrillin antibody (Fig. 3B). This protein was confirmed as fimbrillin based on size (ca. 41 kDa) and its absence in extracts of KB cells incubated with the fimA mutant. Interestingly, the immunoblots also showed that KDP112 produced some protein reactive to antifimbrillin antibody, either as fimbriae or as unassembled fimbrillin subunits, that was released from the cell surface of the mutant.
FIG. 3.
Western blot of KB epithelial cell extracts after capture of adhesins from VDS fractions of P. gingivalis parent and rgp mutant strains. Before incubation with KB cells, VDS fractions were untreated (−), treated with 500 μM TLCK (T), or heated at 100°C for 5 min (triangles). (A) Blots were probed with antibody to gingipain adhesin domains, and cross-reacting adhesin peptides are indicated by arrowheads. (B) Blots were probed with fimbrillin antibody. Fimbrillin (indicated by the arrowhead) showed more intense reactivity in boiled VDS fractions. WT, wild type.
N-terminal sequences of captured adhesins.
The N-terminal sequence of the captured 44-kDa adhesin was SGQAEI (Fig. 4A), identical to that of the 44-kDa adhesin peptide from RgpA (1, 30) and confirming our results and deductions with the capture assay. The sequence of the 39-kDa protein was ANEAKV (Fig. 4A), identical to that of the 39-kDa adhesin peptide from Kgp and the 27-kDa adhesin peptide from RgpA. Since this peptide was also associated with KB cells that had been incubated with the VDS from the rgpA mutant, we inferred that it was derived from Kgp. The N-terminal sequence of the captured 27-kDa protein was PQSVWI, identical to that of the 17-kDa adhesin peptide from RgpA, the 44-kDa C-terminal peptide from Kgp in strain HG66 (28), its 17-kDa autoprocessed product, and the 44-kDa repeat peptides from HagA. Because this peptide was captured by KB cells from the VDS of the rgpA mutant, we reasoned that it was derived from Kgp. Comparison of Kgp 44-kDa adhesin peptide sequences from strains HG66 (28) and 381 (26) showed that the latter did not contain the processing site that would generate component 17- and 27-kDa peptides (Fig. 4B). Sequencing of this region of kgp from ATCC 33277 (data not shown) showed it was identical to that of the closely related strain 381 (5, 21). Alternate downstream processing sites within the Kgp 44-kDa adhesin of ATCC 33277 would yield peptides of approximately 27 and 19 kDa (Fig. 4B).
FIG. 4.
N-terminal amino acid sequences of adhesin peptides and sequence comparison of Kgp 44-kDa adhesin peptides. (A) Cleavage sites and N-terminal sequences of the 44-kDa adhesin from RgpA, the 39-kDa adhesin from Kgp, the putative 27-kDa adhesin from Kgp and alternative cleavage site for the 19-kDa peptide, and the 44-kDa adhesins from HagA. (B) Sequence alignment of Kgp 44-kDa adhesins from strains HG66 and 381. N-terminal sequences of the 17- and 27-kDa peptides from HG66 are underlined, and cleavage sites are indicated by arrows. The N terminus of the proposed 27-kDa peptide from 381 is underlined, and alternative downstream processing sites are indicated by arrows.
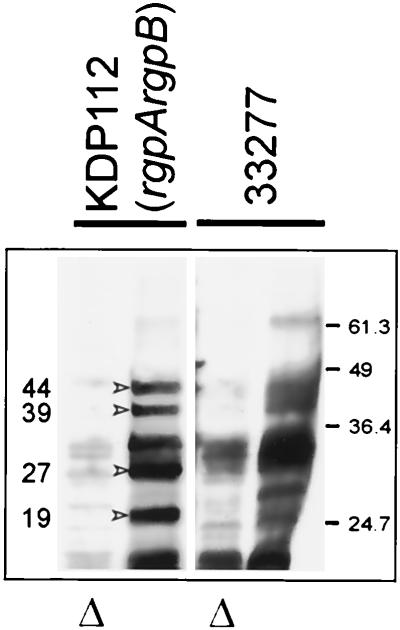
Gingipain adhesins of KDP112 are cell associated.
While intact KDP112 cells bound to KB monolayers in high numbers, adhesins were not detected in their vesicle or VDS fractions (Table 2), implying that they remained cell associated in this strain. Sonicates of washed cells of KDP112 bound KB cells in the membrane assay (data not shown). Furthermore, as shown in Fig. 5, KB cells captured adhesin antibody cross-reacting proteins of 44, 39, 27, and 19 kDa from KDP112 sonicates in amounts at least equivalent to those for the parent strain ATCC 33277. However, the sonicate did not contain Rgp or Kgp proteolytic activity (data not shown), and although we have not ruled out the possibility that a catalytically inactive Kgp precursor was present in this strain, it is also possible that adhesin peptides were derived from cell-associated HagA.
FIG. 5.
Western blot analysis of epithelial cell extracts showing the capture of proteins from cell sonicates of ATCC 33277 and KDP112. Blots were probed with antiadhesin antibody. Triangles denote heat-treated sonicates. The 44-, 39-, 27-, and 19-kDa adhesin peptides (indicated by arrowheads) were present in the cell sonicates of KDP112 and ATCC 33277.
Adhesion of KDP112 to monolayers is modulated by exogenous gingipain catalytic activities.
If gingipain adhesin domains mediate the adhesion of P. gingivalis to epithelial cells and Rgp catalytic activities cause detachment of the organism (Table 1), one prediction is that the addition of these activities back to KDP112 would reduce its high level of attachment to monolayers. To prove the reduced attachment was specifically due to proteolytic activities, their inhibition should restore adhesion of KDP112 to the previous high levels. However, these experiments could not be performed using the conventional viability-based epithelial cell adhesion assay due to loss of P. gingivalis viability upon prolonged incubation with inhibitor. From previous studies we knew that nonviable bacteria could bind epithelial cells (7) and therefore devised a fluorometric assay to measure bacterial attachment to KB monolayers that depended only on detecting the activity of a P. gingivalis surface carbohydrate hydrolase (β-glucosaminidase), not on viable counts. In this assay, adhesion of untreated KDP112 to KB monolayers was approximately 15-fold higher than that of ATCC 33277 (Fig. 6), consistent with results obtained with the viability-based monolayer adhesion assay (Table 1). Increasing amounts of Rgp catalytic activity, provided as increased amounts of the VDS fraction from ATCC 33277, reduced adhesion of KDP112 to monolayers. At the highest concentration of ATCC 33277 VDS (400 μg/ml), there was an approximately 60% decrease in the adhesion of KDP112. Similarly, although adhesion of ATCC 33277 was much lower, its levels were also reduced in the presence of the ATCC VDS fraction. Addition of the VDS fraction of ATCC 33277 together with TLCK restored the adhesion of KDP112 to the original levels and also increased attachment of ATCC 33277. There was a small (10 to 15%) decrease in the attachment of KDP112 to monolayers in the presence of KDP112 VDS (data not shown); and this effect was TLCK sensitive, suggesting it was due to either Kgp or another sensitive proteolytic activity. These results indicated that proteolytic activities, with the genetic data suggesting those of Rgp, mediated detachment of P. gingivalis from epithelial cells.
FIG. 6.
Adhesion of KDP112 to monolayers is modulated by exogenous gingipain catalytic activities. ATCC 33277 (solid bars) or KDP112 (open bars) was added to KB monolayers in the presence of increasing amounts of the gingipain-containing VDS fraction from ATCC 3277, without or with 500 μM TLCK. Adherent bacteria were detected by their ability to hydrolyze 4-methylumbelliferyl N-acetyl-β-d-glusosaminide. Bacterial adhesion levels are proportional to 4-methyl umbelliferone fluorescent counts and are expressed as fluorescent counts ± standard deviations.
DISCUSSION
Failure-proof mechanisms for adhering to epithelial and endothelial cell surfaces are essential for the successful colonization of host tissue by bacteria. Like that of most microorganisms, the natural environment of P. gingivalis is complex; in addition to tissue colonization, it must interact with other bacteria and ward off the challenge of host immune defenses. To control this environment, P. gingivalis has evolved an array of cell surface proteins. Predominant among these are a family of proteinases, the gingipains (3), whose activities have the capacity to degrade host defense proteins (11, 39, 42) and supply peptides and amino acids that are the preferred carbon and nitrogen sources for the organism. In addition to catalytic activity, certain forms of gingipains were known to possess hemagglutinating activity. Catalytic activity alone was associated with purified low-molecular-weight gingipains, while their higher-molecular-weight precursors retained both catalytic and hemagglutinating activities, suggesting that each function may be carried out by different domains within the protein (30). Cloning of the rgpA and kgp genes confirmed that they encoded two functional domains, and their carboxy termini were designated adhesin-hemagglutinins (27, 29). In this study we provide evidence that gingipains play a role in tissue colonization and present a model for their function in adhesion to epithelial cells.
Cognizant of a potential role for gingipains in adherence, we examined the effects of gingipain mutations on P. gingivalis ATCC 33277 adhesion to epithelial cell monolayers. While single rgpA or rgpB mutants showed the same low levels of adherence as the parent strain, an rgpA rgpB double mutant, lacking Rgp catalytic activity, showed a high level of adhesion to epithelial cells. A similar increased level was observed with the parent strain after brief pretreatments with TLCK to inhibit gingipain activities (data not shown). Further, previous experiments (7) that showed higher levels of adhesion of ATCC 33277 to monolayers were carried out in the presence of serum, and substitution with bovine serum albumin produced similar results; therefore, we reasoned that excess, exogenous protein may saturate gingipain catalytic active sites and, as with TLCK, result in the observed higher attachment levels. In the present study, results obtained with mutant strains suggested that gingipain catalytic activities, specifically those of RgpA and RgpB, modulated bacterial attachment to epithelial cells. Using a membrane-based epithelial cell adhesion assay, we determined that adhesins were present in surface protein fractions from wild-type and mutant strains. Also, the results indicated that fimbriae were not the adhesins detected in this assay since intact bacteria and extracellular fractions of a fimA mutant, DPG3, retained the ability to bind KB cells. This is consistent with previous data indicating that the absence of fimbriae did not abrogate the binding of TLCK-treated DPG3 to KB cells (36).
Adhesins in bacterial surface protein fractions were captured by epithelial cells and identified after Western blotting. Four of the captured proteins reacted with polyclonal antibody to gingipain adhesin domains, and from their N-terminal sequences we identified the 44-kDa adhesin peptide from RgpA, the 39-kDa peptide from Kgp, and a 27-kDa peptide derived from either Kgp or HagA. Thus, certain gingipain adhesin peptides have the ability to adhere to epithelial cells. The notion that Rgp catalytic activities modulate adhesion was supported by experiments in which the high level of adhesion of the double mutant, KDP112, was reduced following the addition of increasing amounts of a cell fraction containing these activities.
Several years ago a model was proposed for the secretion, processing, and extracellular localization of RgpA (41). According to the model, the nascent RgpA prepropolypeptide is directed by its signal sequence from the ribosome to the cytoplasmic membrane. After translocation to the periplasm, the propeptide sequence is removed by arginine-specific autolytic processing. The carboxy-terminal adhesin domain contains amphipathic amino acid sequences that direct its incorporation into the outer membrane, where it forms a pore through which the mature proteolytic domain passes to the exterior of the cell. After additional arginine-specific autolytic processing the catalytic domain may be released from the adhesin domain; however, it can also remain noncovalently associated with the membrane-located adhesin. Because of sequence and structural similarities in their adhesin domains, it is likely that Kgp, and possibly HagA, are secreted by a similar mechanism and the adhesin domains also become incorporated into the outer membrane. This pathway is very similar to that described for the secretion of Neiserria gonorrhoeae serine-type immunoglobulin A proteases which were termed autotransporters (32). While RgpA and Kgp have similar general structures and contain amphipathic amino acid sequences within their adhesin domains (27), they do not contain most of the sequence motifs found either within the passenger (N-terminal) or β (C-terminal) domains of the most studied autotransporters, those from the family Enterobacteriaceae (9). However, part of a proposed autotransporter C-terminal signature sequence (19) was detected close to the C termini of Rgp and Kgp. The Enterobacteriaceae are only distantly related to the Bacteroidaceae; therefore, P. gingivalis autotransporters may be similarly evolutionarily distant and retain only the basic structures necessary for their function. Analysis of the P. gingivalis genome sequence may reveal additional proteins of this family from which functional motifs may be identified.
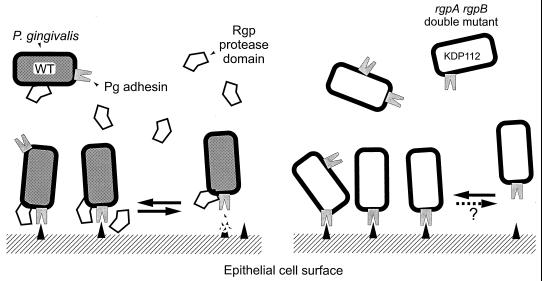
The secretion model is consistent with our results for P. gingivalis adherence to epithelial cells. We hypothesize that adhesion is mediated by gingipain (and possibly HagA) adhesin peptides localized at the surface of the outer membrane or in membrane vesicles, and adhesion is modulated by Rgp catalytic activities that either are noncovalently associated with the adhesin domains or are soluble proteins (Fig. 7). With the parent strain, ATCC 33277, the monolayer adhesion assay measures the net reaction of bacterial attachment, either to an epithelial cell surface receptor or to an extracellular matrix protein, and detachment is caused by the degradation of these receptors by RgpA and RgpB catalytic activities. We propose this as the normal adhesion mode. When catalytic activity is reduced by mutation (or inhibited), the assay measures the large number of bacteria that attach and accumulate on the monolayers but that cannot detach or do so very slowly. The demonstration that gingipain adhesin domains bound to epithelial cells supported our hypothesis regarding the role of gingipains in attachment. In KDP112, the rgpA rgpB double mutant, adhesins were not present in the extracellular fractions but were detected in cell sonicates, suggesting that they were tightly bound to the bacterial surface. To some extent this result supports the model for gingipain secretion in that loss of RgpA and RgpB catalytic activities may reduce the efficiency of processing of Kgp and possibly also HagA. In turn, this might lead to the misincorporation of incorrectly processed adhesins into the outer membrane, where they remain trapped instead of completing their secretion pathway to the cell surface and membrane vesicles. However, epithelial cell binding epitopes within these adhesin domains must still be accessible.
FIG. 7.
Gingipain-mediated adhesion and detachment of P. gingivalis to epithelial cells. We hypothesize that adhesion is mediated by gingipain adhesin peptides localized at the surface of the outer membrane and in membrane vesicles. Rgp catalytic activities, either noncovalently linked with adhesin domains or as soluble proteins, modulate adhesion through digestion of binding substrate. With the parent strain, ATCC 33277, the monolayer adhesion assay measures the net reaction of bacterial attachment to and detachment from receptors or extracellular matrix proteins at the epithelial cell surface. When catalytic activity is reduced by mutation (KDP112), the assay measures the large number of bacteria that attach (via Kgp or HagA adhesin peptides) and accumulate on the monolayers but do not detach or do so very slowly.
It was reported that a large protein complex, dislodged from the surface of strain W50 by mild sonication, contained the catalytic domains of Rgp and Kgp noncovalently linked with their adhesin domains (1). Adhesins identified in the present study may be present in a similar complex dislodged from the surface of ATCC 33277 by our extraction methods. We recognize that there are potentially two classes of adhesins: those that bind directly to epithelial cells (or proteins on their cell surface), and those that may bind to the primary adhesins and thus indirectly to epithelial cells. Our results show that the 39- and 44-kDa peptides from Kgp and RgpA (and possibly HagA) are adhesins for epithelial cells. Evidence from other studies supports this notion. These peptides contain the sequence GVSPKVCKDVTVEGSNEFAPVQNLT, proposed as a colonization epitope and recognized by a monoclonal antibody that conferred passive immunization after topical application, and prevented recolonization by P. gingivalis in periodontitis patients (12). Also within this sequence is the minimum hemagglutinin motif, PVQNLT; a synthetic peptide containing this sequence was shown to inhibit hemagglutination by P. gingivalis vesicles, suggesting that it contained the binding epitope for erythrocytes (34). In a recent study, mice were protectively immunized against P. gingivalis with peptides derived from the catalytic and adhesin domains of RgpA and Kgp (25); interestingly, their sera recognized the RgpA 44-, Kgp 39-, and RgpA 27-kDa antigens, the latter also being present in Kgp. However, until capture assays are repeated with purified peptides, we cannot confirm whether the gingipain adhesin peptides are primary or secondary, or whether they need to be complexed with other proteins. In this context, we reasoned that if fimbriae were primary adhesins, they should also be captured from both nondenatured and TLCK-treated VDS fractions. Instead, heat treatment that dissociated fimbriae to fimbrillin subunits was required to expose epitopes that bound KB cells (Fig. 3B). It is possible that under our experimental conditions, the subunit or partially denatured fimbriae have a higher affinity for epithelial cells than native fimbriae.
The work presented here indicates that adherence to epithelial cells can occur in the absence of fimbriae, although several groups have reported that lack of fimbriae drastically reduced P. gingivalis attachment to and invasion of epithelial cells (24, 36, 40). Those results were obtained using a single mutant, DPG3, derived from strain 381 by inserting an antibiotic resistance gene into the fimA gene. We investigated this mutant and found that it possessed approximately 5% of the Rgp and 13% of the Kgp total catalytic activities of the parent strain. Furthermore, the disposition of these activities at the cell surface of DPG3 was different from that of the parent in that they were easily removed by washing (T. Chen and M. J. Duncan, unpublished data). This suggests that loss of fimbrillin expression may affect gingipain production, just as gingipain mutations appear to affect fimbrillin expression (37) and the formation of fimbriae (10, 23). These findings may also account for the low level of attachment of washed DPG3 mutant cells to epithelial monolayers. Similarly treated cells of strains W50 and W83 also showed negligible attachment to epithelial cells yet possessed high levels of extracellular Rgp and Kgp activities (7; unpublished data). In contrast, the retention of gingipains at the cell surface of strain 381 may result in its consistently higher level of attachment to KB monolayers (36) compared to ATCC 33277 assayed under the same conditions.
Our hypothesis of how gingipains may operate as adhesins does not, however, exclude a role for fimbriae, and cooperative roles have been proposed whereby gingipain catalytic activities cleave extracellular matrix proteins on the surface of gingival epithelial cells to expose cryptic binding epitopes for fimbrial adhesins (13, 14). Consistent with this scenario is the demonstration of RgpA colocalization with fibronectin and α5β1 integrin receptors on the surface of human gingival fibroblasts (33) and the finding that fimbriae bind to KB cells (35). Although heat treatment was required for fimbrial adherence to KB cells under the experimental conditions of the present study, we do not rule out the possibility that a second step in adhesion may involve the binding of fimbriae to modified epithelial cell receptors.
Mutant analyses in this study indicated that gingipain catalytic activities modulated P. gingivalis attachment to epithelial cells, and we presented evidence that gingipain adhesin domains are involved in adherence to epithelial cells. We intend to validate our results with competition studies using purified proteins and domain- and peptide-specific antibodies. In addition, the results suggested that a complex of proteins may be required for binding to epithelial cells, and experiments are under way to define this entity and other potential adhesins. Future studies will also address whether gingipain adhesin peptides bind directly to the epithelial cell surface or to extracellular matrix proteins.
ACKNOWLEDGMENTS
We thank M. Malamy (Tufts University Medical School) for discussions on the mixing experiment.
This work was supported by PHS grant R01 DE10510 (NIDCR).
REFERENCES
- 1.Bhogal P S, Slakeski N, Reynolds E C. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143:2485–2495. doi: 10.1099/00221287-143-7-2485. [DOI] [PubMed] [Google Scholar]
- 2.Chen T, Dong H, Yong R, Duncan M J. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb Pathog. 2000;28:235–247. doi: 10.1006/mpat.1999.0338. [DOI] [PubMed] [Google Scholar]
- 3.Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 4.DeCarlo A A, Paramaesvaran M, Yun P L, Collyer C, Hunter N. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J Bacteriol. 1999;181:3784–3791. doi: 10.1128/jb.181.12.3784-3791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong H, Chen T, Dewhirst F E, Fleischmann R D, Fraser C M, Duncan M J. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect Immun. 1999;67:3416–3423. doi: 10.1128/iai.67.7.3416-3423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan M J, Emory S A, Almira E C. Porphyromonas gingivalis genes isolated by screening for epithelial cell attachment. Infect Immun. 1996;64:3624–3631. doi: 10.1128/iai.64.9.3624-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan M J, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun. 1987;55:111–117. doi: 10.1128/iai.55.1.111-117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 10.Kadowaki T, Nakayama K, Yoshimura F, Okamoto K, Abe N, Yamamoto K. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J Biol Chem. 1998;273:29072–29076. doi: 10.1074/jbc.273.44.29072. [DOI] [PubMed] [Google Scholar]
- 11.Katz J, Sambandam V, Wu J H, Michalek S M, Balkovetz D F. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect Immun. 2000;68:1441–1449. doi: 10.1128/iai.68.3.1441-1449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly C G, Booth V, Kendal H, Slaney J M, Curtis M A, Lehner T. The relationship between colonization and haemagglutination inhibiting and B cell epitopes of Porphyromonas gingivalis. Clin Exp Immunol. 1997;110:285–291. doi: 10.1111/j.1365-2249.1997.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol. 1997;24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 14.Kontani M, Ono H, Shibata H, Okamura Y, Tanaka T, Fujiwara T, Kimura S, Hamada S. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect Immun. 1996;64:756–762. doi: 10.1128/iai.64.3.756-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozarov E, Whitlock J, Dong H, Carrasco E, Progulske-Fox A. The number of direct repeats in hagA is variable among Porphyromonas gingivalis strains. Infect Immun. 1998;66:4721–4725. doi: 10.1128/iai.66.10.4721-4725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lantz M S, Allen R D, Duck L W, Blume J L, Switalski L M, Hook M. Identification of Porphyromonas gingivalis components that mediate its interactions with fibronectin. J Bacteriol. 1991;173:4263–4270. doi: 10.1128/jb.173.14.4263-4270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leong J M, Moitoso de Vargas L, Isberg R R. Binding of cultured mammalian cells to immobilized bacteria. Infect Immun. 1992;60:683–686. doi: 10.1128/iai.60.2.683-686.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loveless B J, Saier M H., Jr A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 20.Malek R, Fisher J G, Caleca A, Stinson M, van Oss C J, Lee J Y, Cho M I, Genco R J, Evans R T, Dyer D W. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Njoroge T, Genco R J, Sojar H T, Hamada N, Genco C A. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect Immun. 1997;65:1980–1984. doi: 10.1128/iai.65.5.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien-Simpson N M, Paolini R A, Reynolds E C. RgpA-Kgp peptide-based immunogens provide protection against Porphyromonas gingivalis challenge in a murine lesion model. Infect Immun. 2000;68:4055–4063. doi: 10.1128/iai.68.7.4055-4063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain) J Biochem (Tokyo) 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto K, Misumi Y, Kadowaki T, Yoneda M, Yamamoto K, Ikehara Y. Structural characterization of argingipain, a novel arginine-specific cysteine proteinase as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch Biochem Biophys. 1995;316:917–925. doi: 10.1006/abbi.1995.1123. [DOI] [PubMed] [Google Scholar]
- 28.Pavloff N, Pemberton P A, Potempa J, Chen W C, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272:1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 29.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 30.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 31.Pike R N, Potempa J, McGraw W, Coetzer T H, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohlner J, Halter R, Beyreuther K, Meyer T F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 33.Scragg M A, Cannon S J, Rangarajan M, Williams D M, Curtis M A. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect Immun. 1999;67:1837–1843. doi: 10.1128/iai.67.4.1837-1843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata Y, Hayakawa M, Takiguchi H, Shiroza T, Abiko Y. Determination and characterization of the hemagglutinin-associated short motifs found in Porphyromonas gingivalis multiple gene products. J Biol Chem. 1999;274:5012–5020. doi: 10.1074/jbc.274.8.5012. [DOI] [PubMed] [Google Scholar]
- 35.Sojar H T, Han Y, Hamada N, Sharma A, Genco R J. Role of the amino-terminal region of Porphyromonas gingivalis fimbriae in adherence to epithelial cells. Infect Immun. 1999;67:6173–6176. doi: 10.1128/iai.67.11.6173-6176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokuda M, Duncan M, Cho M I, Kuramitsu H K. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect Immun. 1996;64:4067–4073. doi: 10.1128/iai.64.10.4067-4073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokuda M, Karunakaran T, Duncan M, Hamada N, Kuramitsu H. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect Immun. 1998;66:1159–1166. doi: 10.1128/iai.66.3.1159-1166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis J, Potempa J. Bacterial proteinases as targets for the development of second-generation antibiotics. Biochim Biophys Acta. 2000;1477:35–50. doi: 10.1016/s0167-4838(99)00278-2. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg A, Belton C M, Park Y, Lamont R J. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto K, Kadowaki T, Okamoto K, Yoneda M, Nakayama K, Misumi Y, Ikehara Y. Intracellular protein catabolism. New York, N.Y: Plenum Press; 1996. [Google Scholar]
- 42.Zhang J, Dong H, Kashket S, Duncan M J. IL-8 degradation by Porphyromonas gingivalis proteases. Microb Pathog. 1999;26:275–280. doi: 10.1006/mpat.1998.0277. [DOI] [PubMed] [Google Scholar]