Abstract
Much of the Ca2+ activity in astrocytes is spatially restricted to microdomains and occurs in fine processes that form a complex anatomical meshwork, the so‐called spongiform domain. A growing body of literature indicates that those astrocytic Ca2+ signals can influence the activity of neuronal synapses and thus tune the flow of information through neuronal circuits. Because of technical difficulties in accessing the small spatial scale involved, the role of astrocyte morphology on Ca2+ microdomain activity remains poorly understood. Here, we use computational tools and idealized 3D geometries of fine processes based on recent super‐resolution microscopy data to investigate the mechanistic link between astrocytic nanoscale morphology and local Ca2+ activity. Simulations demonstrate that the nano‐morphology of astrocytic processes powerfully shapes the spatio‐temporal properties of Ca2+ signals and promotes local Ca2+ activity. The model predicts that this effect is attenuated upon astrocytic swelling, hallmark of brain diseases, which we confirm experimentally in hypo‐osmotic conditions. Upon repeated neurotransmitter release events, the model predicts that swelling hinders astrocytic signal propagation. Overall, this study highlights the influence of the complex morphology of astrocytes at the nanoscale and its remodeling in pathological conditions on neuron‐astrocyte communication at so‐called tripartite synapses, where astrocytic processes come into close contact with pre‐ and postsynaptic structures.
Keywords: calcium microdomains, computational neuroscience, intracellular signaling, nano‐morphology, reaction–diffusion simulations
Main Points
Astrocyte nano‐morphology favors the compartmentalization of biochemical signals.
This compartmentalization promotes local Ca2+ activity and signal propagation robustness.
In contrast, its pathological remodeling upon swelling attenuates Ca2+ activity.
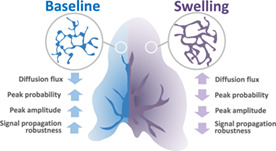
1. INTRODUCTION
Astrocytes are glial cells of the central nervous system that are essential for brain development and function (Verkhratsky and Nedergaard 2018). They notably modulate neuronal communication at synapses. Astrocytic Ca2+ signals are triggered by neurotransmitters released by active neurons, which can trigger the release of neuroactive molecules by the astrocyte, referred to as gliotransmitters. The first type of astrocytic Ca2+ signals that has been observed was Ca2+ waves that propagate through gap junctions in astrocyte networks (Giaume and Venance 1998). Ca2+ waves have also been observed in the branches of single astrocytes, sporadically propagating to the soma (Haustein et al. 2014; Bindocci et al. 2017). The recent development of Ca2+ imaging techniques with improved spatial and temporal resolution has revealed the existence of spatially‐restricted Ca2+ signals in astrocytes, referred to as microdomains or hotspots (Di Castro et al. 2011; Panatier et al. 2011; Stobart, Ferrari, Barrett, Glück, et al. 2018; Srinivasan et al. 2015; Shigetomi et al. 2013; Sherwood et al. 2017; Otsu et al. 2015; Lind et al. 2013; Bindocci et al. 2017; Agarwal et al. 2017; Arizono et al. 2020; Otsu et al. 2015; Grosche et al. 1999). These local Ca2+ signals account for the vast majority ( 80%) of astrocytic Ca2+ activity and occur in fine processes, which occupy 75% of the astrocytic volume (Bindocci et al. 2017), forming the spongiform domain, also referred to as the gliapil. Given that one astrocyte may contact tens of thousands of synapses simultaneously (Bushong et al. 2002) via these fine processes, local and fast Ca2+ signals might enable the astrocyte to powerfully yet precisely control the flow of information through synaptic circuits. Importantly, reactive astrocytes, hallmark of brain diseases, display aberrant amplitude, duration, frequency and spatial spread of Ca2+ signals (Shigetomi et al. 2019; Nedergaard et al. 2010; Lee et al. 2022).
Cellular micro‐morphology lends itself to the compartmentalization of biochemical signals. For example, the anatomical design of dendritic spines restricts the diffusion of Ca2+ to the activated synapse, which reduces cross‐talk between nearby synapses (Santamaria et al. 2011; Tonnesen et al. 2014; Yuste et al. 2000; Noguchi et al. 2005; Yasuda 2017; Holcman and Schuss 2011). The complex shapes of Bergmann glia (Grosche et al. 1999) and perisynaptic “astrocytic compartments” along major branches (Panatier et al. 2011) have been proposed to restrict Ca2+ signals to the vicinity of synapses. Fine processes of the spongiform domain, however, cannot be resolved by diffraction‐limited light microscopy (Rusakov 2015), so that the contribution of their morphology to shaping local Ca2+ signals is poorly understood. Our recent 3D STED study (Arizono et al. 2020) revealed the structural basis of compartmentalized spontaneous Ca2+ signals in fine astrocytic processes. Importantly, pathological changes in astrocytic morphology (Lafrenaye and Simard 2019), such as “astrocytic swelling,” are paired with aberrant Ca2+ signals (Shigetomi et al. 2019). We have recently reported that swelling can also occur at the level of fine astrocytic processes (Arizono, Bancelin, et al. 2021). The effect of such a remodeling of astrocytic nano‐morphology on the local Ca2+ signals involved in regulating synapses remains yet unclear. Together, there is a great interest in understanding the mechanistic link between the nano‐morphology of astrocytic processes and Ca2+ profiles in health and disease. Computational approaches make it possible to simulate different geometrical scenarios, in a much more systematic and controlled way than what could be done experimentally. Computational models can thus help us gain insights into the impact of morphological parameters on Ca2+ activity.
Here, we use computational tools to explore the role of the anatomical design of the gliapil. To do so, we perform simulations in branchlet geometries that we designed based on super‐resolution microscopy data reported in live tissue (Arizono et al. 2020), consisting of nodes, which host Ca2+ microdomains, and their intervening shafts of variable widths. Our simulation results suggest that the nanoscale design of the spongiform domain effectively decreases diffusion flux, which increases Ca2+ peak probability, duration and amplitude in the stimulated and neighboring nodes. To test those predictions, we performed Ca2+ recordings in organotypic hipppocampal cultures in hypo‐osmotic conditions, where the normal node‐shaft arrangement is altered. In line with our model predictions, Ca2+ activity in hypo‐osmotic conditions was decreased compared to Ca2+ activity in normal tissue. We further found that, upon repeated neuronal stimulation, thin shafts allow signal propagation even if some stimuli are omitted, thus allowing for a more robust signal propagation.
Overall, our study sheds light on the influence of the nanoscale morphology of the complex spongiform domain of astrocytes on Ca2+ microdomain activity and indicates that pathological morphological changes may substantially affect their Ca2+ activity.
2. METHODS
2.1. Stochastic spatially‐explicit voxel‐based simulations
In order to model astrocyte Ca2+ signals in astrocyte branchlets, we have used the voxel‐based “GCaMP” implementation of the Inositol 3‐Phosphate (IP3) receptor‐dependent Ca2+ signaling model from Denizot et al. (2019), using the same reaction scheme and parameter values (Figure 1b). Briefly, we model Ca2+ fluxes in and out of the cytosol, mediated by channels and pumps on the endoplasmic reticulum (ER) and on the plasma membrane. signals occur when some IP3R channels are in the open state. IP3 can be synthesized by the Ca2+‐dependent activity of phospholipase Cδ (PLCδ) and the removal of IP3 molecules from the cytosol is expressed as a single decay rate. IP3R kinetics is described by a Markov model, derived from De Young & Keizer's model (De Young and Keizer 1992). Each IP3R molecule contains one IP3 binding site and two binding sites. An IP3R is in the open state when in state (first Ca site and IP3 bound, second Ca site free). Depending on the simulation, other diffusing molecules were added to the model, such as the fluorescent molecule ZsGreen and fluorescent indicators, here 10 μM of GCaMP6s. GCaMPs are genetically‐encoded indicators (GECIs) that are derived from the fluorescent protein GFP and the buffer calmodulin (see Shigetomi et al. (2016) for a review on GECIs). Unless specified otherwise, simulation time T was 50 s. Data collection interval was 1 ms. For further details on the kinetic scheme and model assumptions, please refer to Denizot et al. (2019).
FIGURE 1.
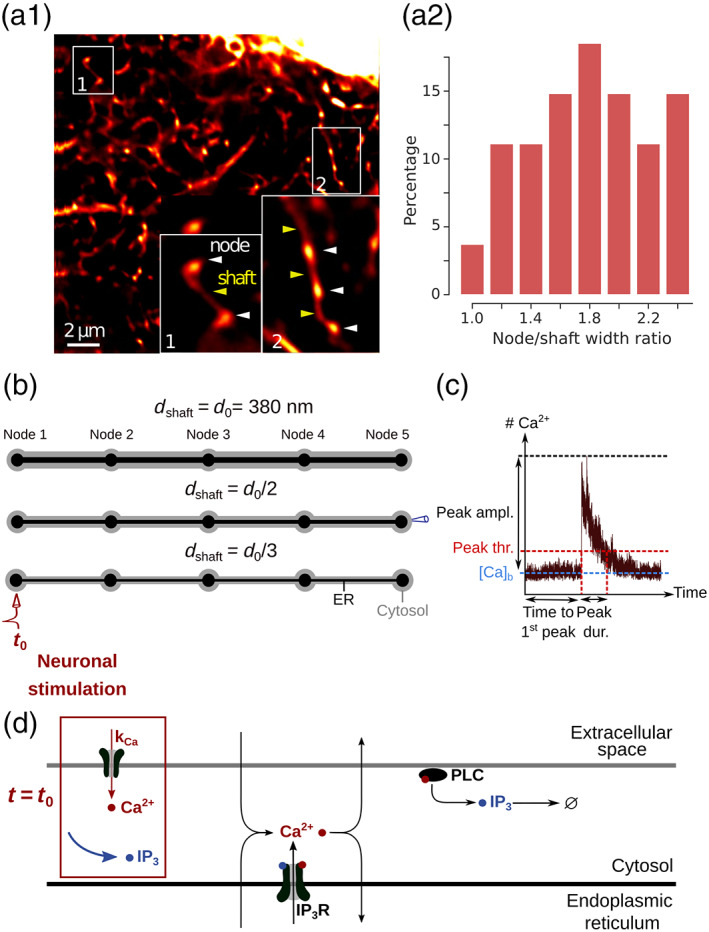
Geometries and kinetic scheme used for simulating dynamics in node/shaft structures of the gliapil. (a1) Representative STED image showing the astrocytic spongiform domain. Zoom‐in images show its anatomical units: Nodes and shafts. (a2) Distribution of the ratio between node width and the width of its neighboring shaft, measured using STED microscopy, n = 28. (b) Idealized geometries designed to reproduce the node/shaft ultrastructure of the spongiform domain. Nodes are approximated as spheres of diameter 380 nm and shafts as 1 μm‐long cylinders, based on their recent characterization in live tissue by Arizono et al. (2020) (Figure 1i–k. Shaft width varied based on experimentally‐observed node‐shaft width ratios reported in panel b: nm, and . The geometries designed in this study, referred to as “5 nodes,” contain 5 identical nodes and 4 identical shafts. Unless specified otherwise, ER geometry (black) also consists in node/shaft successions (see Methods). The associated cytosolic volume, plasma and ER membrane areas are presented in Table 1. (c) Example of a trace of the model, recorded in one node. activity is quantified by measuring time to 1st peak, peak probability, amplitude, duration and frequency. A peak is detected when the number of ions in the recorded node increases above peak threshold (see Methods). (d) Biochemical processes included in the model. can enter/exit the cytosol from/to the extracellular space or the endoplasmic reticulum (ER), resulting from the activity of channels/pumps. and IP3 diffuse in the cytosol following Brownian motion. The kinetics of channels corresponds to the 8‐state Markov model from Denizot et al. (2019), adapted from De Young and Keizer (1992); Bezprozvanny et al. (1991). When both IP3 and are bound to IP3R activating binding sites, the IP3R is in open state and enters the cytosol. can activate phospholipase C δ (PLCδ), which results in the production of IP3. For more details, please refer to Denizot et al. (2019). Neuronal stimulation is simulated as an infusion of IP3 in the cytosol and the opening of channels at the plasma membrane with an influx rate (see Methods).
The model was implemented using the STochastic Engine for Pathway Simulation (STEPS, http://steps.sourceforge.net/), a python package performing exact stochastic simulation of reaction–diffusion systems (Hepburn et al. 2012). More precisely, simulations were performed using the spatial stochastic “Tetexact” solver, based on Gillespie's Stochastic Simulation Algorithm (SSA) (Gillespie 1977), extended for diffusive fluxes across voxels in tetrahedral meshes (Hepburn et al. 2012). Diffusion is treated as a first order reaction in the SSA system, whose rate is determined by the diffusion coefficient of the diffusing species.
Simulations in STEPS can be performed in complex geometries in three spatial dimensions. Space is divided into well‐mixed tetrahedral compartments, referred to as voxels. Reactions between two molecules can only occur if they are located within the same voxel. Diffusion events are modeled as a decrease of the number of molecules in the original voxel and an increase in the number of molecules in its neighboring voxel. Boundary conditions, except when specified otherwise, were reflective. In other words, mobile molecules could not diffuse away from the geometry, as if they were “bouncing” onto the plasma membrane. STEPS enables to compute, in complex 3D geometries, reactions and diffusion in the cytosol as well as reactions between cytosolic molecules and molecules located at the plasma or ER membrane.
2.2. Geometries
Typical astrocyte branchlet geometries were designed from their recent experimental characterization in live tissue at high spatial resolution (50 nm in x–y) (Arizono et al. 2020). Those geometries consist in alternations of bulbous structures, nodes, connected to each other with cylindrical structures, shafts. Geometries with different shaft widths were designed using Trelis software (https://www.csimsoft.com/trelis, Figure 1a). The geometry of a node was approximated as being a sphere of diameter 380 nm. Shaft geometry consisted in a 1 μm long cylinder. Shaft diameter was defined relative to node diameter. For example, shaft diameter was the same as node diameter, that is, = = 380 nm. Similarly, shaft diameter was 190 nm and 127 nm for = and , respectively. Cones were positioned between spheres and cylinders in order to create a smoother transition between nodes and shafts, better approximating the geometry observed experimentally. Cytosolic volume was thus V 1 = 0.620 μm3, V 2 = 0.263 μm3, and V 3 = 0.195 μm3, for = , and , respectively. Please note that molecular concentrations were the same in all simulations, so that the total number of molecules in basal conditions in shafts varied depending on shaft width. A subset of simulations was performed in a geometry with V 1 = 0.258 μm3. This geometry is characterized, similarly to geometries with = , by a node/shaft width ratio of 1. It contains cylinders of length 750 nm, diameter 285 nm and spheres of diameter 285 nm. As a first approximation, ER geometry was considered to be similar to the geometry of the astrocyte branchlet: node/shaft successions. ER nodes were aligned with cytosolic nodes. As no quantification of the ratio between astrocytic ER volume and cellular volume was found in the literature, ER volume was 10% of the total branchlet volume, based on available data in neurons (Spacek and Harris 1997). As the shape and distribution of the ER in fine processes have not been characterized in live tissue but are likely highly variable, additional simulations were performed in meshes with various ER shapes: “No ER,” “Node ER,” and “Cyl ER,” in which there was no ER, discontinuous ER in nodes or cylindrical ER, respectively (Figures S7–S9). The cytosolic volume, plasma and ER membrane surface areas of those 3D geometries are presented in Table 1.
TABLE 1.
Characteristics of the geometries of astrocyte branchlets used in this study. is the cytosolic volume, is the area of the plasma membrane and is the area of the ER membrane. Volumes are expressed in nm3 and areas in nm2. Meshes are available at http://modeldb.yale.edu/266928.
| Geometry | (nm3) | (nm2) | (nm2) | |||
|---|---|---|---|---|---|---|
| “5nodes” |
|
|
|
|||
| “5nodes” |
|
|
|
|||
| “5nodes” |
|
|
|
|||
| “No ER” |
|
|
0.00 | |||
| “No ER” |
|
|
0.00 | |||
| “No ER” |
|
|
0.00 | |||
| “Node ER” |
|
|
|
|||
| “Node ER” |
|
|
|
|||
| “Node ER” |
|
|
|
|||
| “Cyl ER” |
|
|
|
|||
| “Cyl ER” |
|
|
|
|||
| “Cyl ER” |
|
|
|
A sensitivity study was performed to investigate the effect of voxel size on the kinetics of the molecular interactions modeled. Information on the voxel sizes of the different meshes used is presented in Table S1. Results are presented in Figure S1. Meshes that contained voxels that were <50 nm3 were characterized by aberrant kinetics, resulting in aberrant average numbers of molecules in a given state. Those results thus suggest that to prevent errors due to voxel size, meshes should not display voxel sizes that are <50 nm3. We have thus made sure, while meshing the geometries in which simulations were ran, that no voxels were <50 nm3. Minimum voxel size was 443, 1100, and 447 nm3, for = , and geometries, respectively.
In a subset of simulations, ER geometry varied. The shape of the cell was the same as in “5nodes” geometries (Figure 1). ER geometry consisting of node/shaft alternations, described above, is referred to as “Node/shaft ER.” “No ER” geometry contains no ER. “Node ER” is characterized by a discontinuous ER geometry, consisting in spheres of diameter 54 nm, located in cellular nodes. “Cyl ER” corresponds to a cylindrical ER, of length = 6274 nm and a diameter of 108, 54, and 36 nm, for = , and , respectively. The associated cytosolic volume, ER and plasma membrane area are presented in Table 1.
2.3. Protocol for simulating bleaching experiments
In order to test whether the idealized geometries presented in Figure 1 are a good approximation of the spongiform ultrastructure of astrocyte branchlets, we have simulated their fluorescence recovery after photobleaching (FRAP) experiments. Briefly, laser pulses are simulated on a node (region of interest) while the fluorescence level is being recorded. At bleaching time, the fluorescence level in the region of interest decreases to . Then, because of the diffusion of fluorescent molecules into the region of interest, fluorescence increases until it reaches a new steady state, . We characterize node compartmentalization by measuring the time taken by fluorescing molecules to diffuse into the node to reach . In other words, a high node compartmentalization will be associated with a high value of . Thus, three main parameters characterize bleaching traces: , and .
To mimic bleaching experiments in fine branchlets performed by (Arizono et al. 2020), ZsGreen molecules were added to simulation space. After 2 s of simulation, providing the basal level of fluorescence, 60% of ZsGreen molecules were bleached. In order to fit and that were measured experimentally, and as bleaching time lasted 10 ms in experiments and 1 ms in simulations, the bleached volume in simulations was adjusted depending on the geometry (see Figure 2a). Bleaching was simulated as a transition from ZsGreen molecules to ZsGreen‐bleached molecules, the latter being considered as non‐fluorescing molecules. Screenshots of simulations, illustrating the diffusion of ZsGreen and ZsGreen‐bleached molecules, are presented in Figure S2b. The number of ZsGreen molecules in the central node was recorded over simulation time and a fit was performed following Equation (1) to determine the values of , and .
| (1) |
where I(t) is the level of fluorescence measured at time t. The coefficient of diffusion, , and the concentration, [ZsGreen], of ZsGreen were adjusted to fit experimental data. Indeed, the amplitude of [ZsGreen] fluctuations at steady state is inversely proportional to the number of ZsGreen molecules in the geometry. In other words, fluorescence signals are more noisy when [ZsGreen] is low. Moreover, the autocorrelation of those fluctuations depends on the coefficient of diffusion of ZsGreen, . If increases, the autocorrelation of Lag, where Lag is the autocorrelation delay, will decrease faster as Lag increases. Comparing the fluctuations of [ZsGreen] and its autocorrelation in experiments and in simulations thus enabled to find the values of and of [ZsGreen] that allowed for the best fit to experimental data. In the simulations presented here, = 90 and [ZsGreen] = 25 μM.
FIGURE 2.
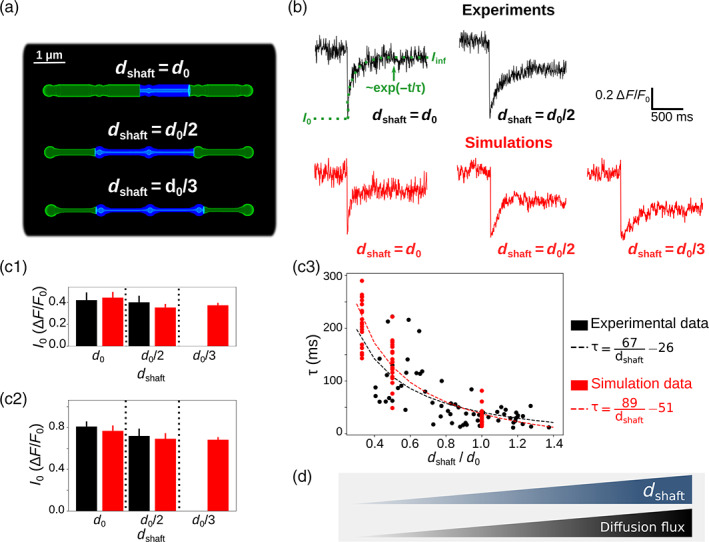
Simulations confirm that thin shafts favor node compartmentalization. (a) Geometries of different shaft widths , , and , used in the bleaching simulations. Blue color represents the bleached volume, which varied depending on the value of in order to fit experimental values of and . (b) Representative experimental (top) and simulation (bottom) traces for different shaft width values. , and were calculated using Equation (1). Note that simulations were also performed for . (c) Quantification of (c1), (c2), and (c3) values in simulations (red) compared to experiments (black). Note that no experimental data was available for . In c1 and c2, n = 5 × 2 and 20 × 3 for experiments and simulations, respectively. Data are presented as mean ± STD. In c3, n = 66 and n = 20 × 3 for experiments and simulations, respectively. is negatively correlated to in experiments (n = 66 from 7 slices; Spearman r = −0.72, p < 0.001***) and simulations (n = 60; Spearman r = −0.89, p < 0.001***). Black and red lines represent curve fit of as a function of of the form for experiments and simulations, respectively. (d) Schematic summarizing the conclusion of this figure: Diffusion flux increases with . In that sense, thin shafts favor node compartmentalization. Data in panels c1 and c2 are represented as mean ± STD, n = 20 for each geometry.
2.4. Protocols for simulating neuronal stimulation
In order to investigate the propagation of signals from nodes that contact neuronal spines, we have developed two different protocols for our simulations, performed in the geometries presented in Figure 1. As nodes were the site of signal initiation (Arizono et al. 2020) and as most spines contacted nodes rather than shafts, we have simulated neuronal stimulation in nodes. To simulate neuronal stimulation, IP3 and were infused in the cytosol at stimulation time. IP3 infusion reflects the production of IP3 by phospholipase C that results from the activation of G q‐G‐protein‐coupled receptors (GPCRs). infusion mimics the influx of in the cytosol through channels at the plasma membrane. The rate of this neuronal activity‐induced influx, , varied within a physiological range of values, from 0 to 1000 s−1 (Wu et al. 2018; Brazhe et al. 2018). Signals were recorded both in the stimulated node, Node 1, and in the neighboring node, Node 2.
In the first protocol, 100 IP3 molecules were infused in Node 1, at t = t 0 = 1 s, while activity was monitored in Node 1 and in the neighboring node, Node 2 (see e.g., Figure 3a). Neuronal activity‐induced influx was mediated by generic channels at the plasma membrane. Twenty‐five of those channels were placed on the plasma membrane of Node 1, corresponding to a similar density to the IP3R density on the ER membrane, and were set to an inactive state. At stimulation time t = t 0, channels were set to an active state, resulting in an influx of within the cytosol at rate = 0–1000 s−1. At t = t 0 + 1, channels were set back to their initial inactive state. Simulations were performed in geometries with varying shaft width .
In the second protocol, we have investigated signal propagation in the node/shaft geometry depending on shaft width when several nodes were successively stimulated. In “5 nodes” geometries, 50 IP3 molecules were infused at t 0 = 5 s, , , in Nodes 1, 2, 3, and 4, respectively. During the whole simulation time, activity was recorded in Node 5 (see Figure 5). In a subset of simulations, stimulation of Nodes 2, 3 and 4 occurred with a probability , with .
FIGURE 3.
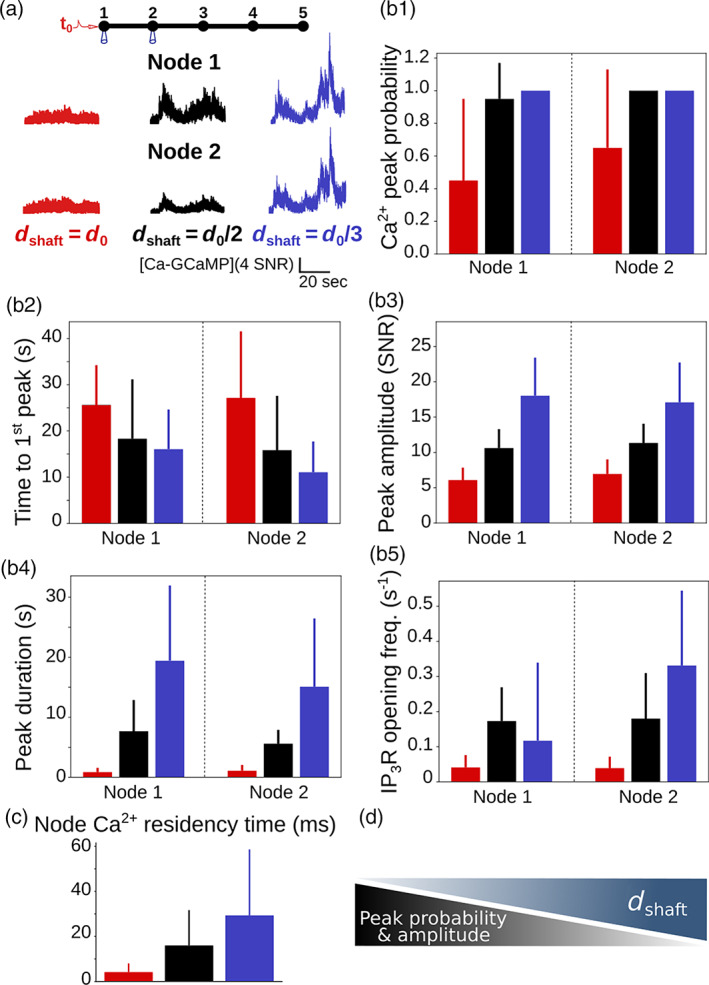
peak probability, amplitude and duration increase when shaft width decreases. (a) (top) Neuronal stimulation protocol simulated for each geometry: Node 1 was stimulated at t = t 0 = 1 s, while activity was monitored in node 2. Representative traces for shaft width (red), (black) and (blue), expressed as SNR (see Methods). (b) Quantification of the effect of on signal characteristics data are represented as mean ± STD, n = 20. peak probability increases (***, b1), time to 1st peak decreases (***, b2), peak amplitude (***, b3) and duration (***, b4) increase when decreases. (c) residency time in node 1 increases when decreases (***, n = 300). (d) Schematic summarizing the main result from this figure: peak probability and amplitude increase when shaft width decreases.
FIGURE 5.
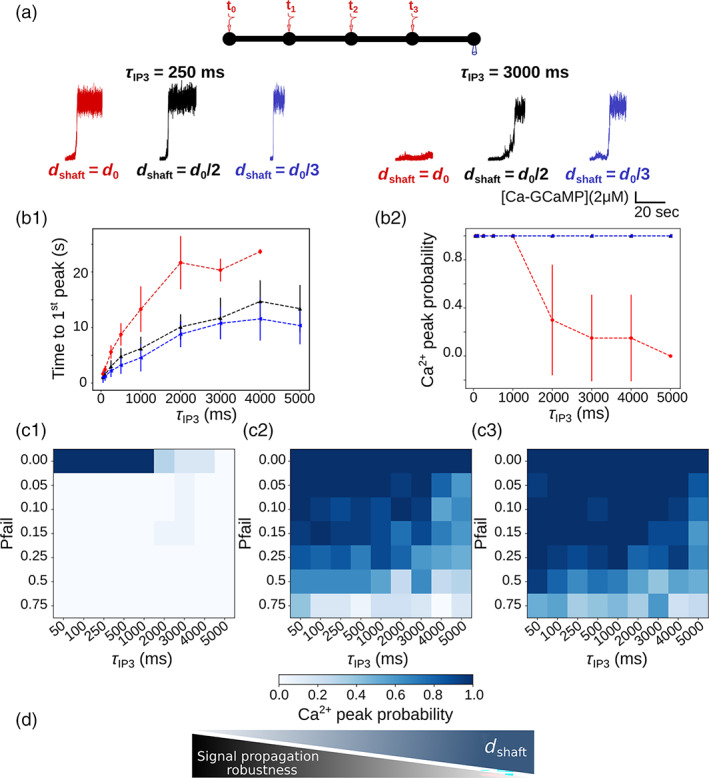
Thin shafts favor a more robust signal propagation upon repeated neurotransmitter release events. (a) (top) Neuronal stimulation protocol: Node 1 is stimulated at t = t 0 = 5 s, node 2 at , node 3 at and node 4 at , = 0 . activity is recorded in node 5. (bottom) representative traces in node 5 for shaft width (red), (black) and (blue), with = 250 ms (left) and 3000 ms (right), expressed as SNR (see Methods). For all values of tested, simulation time was 25 s. (b1) Time to 1st peak increases with for = (***), (***) and (***). T‐tests revealed that for any value of , time to 1st peak is higher for = compared to = and . Time to 1st peak is significantly higher when = compared to = , for most values of (p = 0.032*, 0.0025**, 0.034*, 0.016*, and 0.019* for = 250, 500, 1000, 4000, and 5000 ms, respectively). (b2) peak probability in node 5 is lower for = compared to = and . peak probability decreases as increases for = (***). (c) peak probability in node 5 (colorbar) as a function of and of the probability of failure of node stimulation , for = (c1), = (c2) and = (c3), with . (d) Schematic summarizing the main conclusion of this figure: Decreased shaft width allows signal propagation despite omitted node stimulation, thus favoring more robust signal propagation. Data are represented as mean ± STD, n = 20 for each value of and of . Lines in panel B are guides for the eyes.
2.5. Code accessibility
The simulation code, implemented with STEPS 3.5.0, and the meshes are available on ModelDB (McDougal et al. 2017) at http://modeldb.yale.edu/266928. The original model from Denizot et al. (2019) is available at http://modeldb.yale.edu/247694.
2.6. Peak detection and analysis
The same strategy as developed by Denizot et al. (2019) was used for detecting and analyzing signals. Briefly, basal concentration of , , was defined based on a histogram of the number of ions in the absence of neuronal stimulation. Peak initiation corresponded to the time when [Ca2+] was higher than the following threshold: , where is the standard deviation of [Ca2+] histogram in the absence of neuronal stimulation. The value of n was set by hand depending on signal/noise ratio of the simulation of interest. Peak termination corresponded to the time when [Ca2+] decreased below the peak threshold.
Several parameters were analyzed to characterize signals. Peak duration corresponds to the time between peak initiation and peak termination. Peak amplitude corresponds to the maximum number of ions recorded in the studied node during peak duration. It is expressed as signal to noise ratio . Time to 1st peak corresponds to the delay between the beginning of the simulation and the first peak detection, measured in the cellular compartment of interest. Peak probability corresponds to the fraction of simulations in which at least one peak was detected during simulation time in the region of interest. residency time was measured by performing n = 300 simulations for each value of , in which only 1 ion was added to node 1, without other molecular species. residency time corresponds to the time taken by the ion to diffuse away from node 1.
2.7. Organotypic hippocampal slice cultures
All experiments were conducted as described in Arizono et al. (2020) and were in accordance with the European Union and CNRS UMR5297 institutional guidelines for the care and use of laboratory animals (Council directive 2010/63/EU). Organotypic hippocampal slices (Gähwiler type) were dissected from 5 to 7‐d‐old wild‐type mice and cultured 5 to 8 week in a roller drum at 35°C, as previously described (Gähwiler 1981).
2.8. Viral infection
AAV9‐GFAP‐GCaMP6s (Stobart, Ferrari, Barrett, Stobart, et al. 2018) was injected by brief pressure pulses (40 ms; 15 psi) into the stratum radiatum of 2–3‐week old slices from Thy1‐YFP‐H (JAX:003782) mice 4–6 weeks prior to the experiment.
2.9. Image acquisition
For Ca2+ imaging, we used a custom‐built setup based on an inverted microscope body (Leica DMI6000), as previously described in Tonnesen et al. (2011). We used a 1.3 NA glycerol immersion objective equipped with a correction collar to reduce spherical aberrations and thereby allow imaging deeper inside brain tissue (Urban et al. 2011). The excitation light was provided by a pulsed diode laser (l = 485 nm, PicoQuant, Berlin, Germany). The fluorescence signal was confocally detected by an avalanche photodiode (APD; SPCM‐AQRH‐14‐FC; PerkinElmer). The spatial resolution of the setup was around 200 nm (in x–y) and 600 nm (z). Confocal time‐lapse imaging (12.5 × 25 μm, pixel size 100 nm) was performed at 2 Hz for 2.5 min in artificial cerebrospinal fluid containing 125 mM NaCl, 2.5 mM KCl, 1.3 mM MgCl2, 2 mM CaCl2, 26 mM NaHCO3, 1.25 mM NaH2PO4, 20 mM D‐glucose, 1 mM Trolox; 300 mOsm; pH 7.4. Perfusion rate was 2 ml/min and the temperature 32°C. Hypo‐osmotic stress (300–200 mOsm) was applied by perfusing ACSF with reduced NaCl concentration (119–69 mM NaCl).
2.10. Image analysis
Image analysis was performed as described in Arizono et al. (2020). events in the spongiform structure were detected and analyzed using the ImageJ LC_Pro plugin (Francis et al. 2012), which allows automatic and unbiased detection of events.
2.11. Experimental design and statistical analysis
For each parameter set, 20 simulations, with different seed values, were generated to get a large enough sample size for statistical analysis while keeping the computational cost of the work to a minimum. Each parameter describing dynamics was expressed as mean ± standard deviation. The effect of on each signal characteristic was tested using one‐way ANOVA. Comparison between two different conditions was performed using unpaired Student T‐test if values followed a Gaussian distribution, Mann–Whitney test otherwise. Significance is assigned by * for , ** for , *** for .
3. RESULTS
3.1. Geometrical representation of typical astrocyte processes
In order to investigate the role of the nano‐morphology of astrocytic processes on the spatio‐temporal properties of microdomains, we have designed geometries of typical astrocyte processes, derived from our recent characterization of their ultrastructure at a high spatial resolution (50 nm in x–y) in organotypic hippocampal culture as well as in acute slices and in vivo (Arizono et al. 2020) (Figure 1a). Geometries consist of alternations of bulbous structures, nodes, connected to each other via cylindrical structures, referred to as shafts. At this spatial scale, cell swelling resulting from hypo‐osmotic conditions is not homogeneous: node width is constant while shaft width increases (Figure 3 in Arizono, Inavalli, et al. 2021). To test the effect of such morphological changes of node‐shaft structures on astrocytic activity, we designed idealized node‐shaft geometries in three spatial dimensions in which we could perform reaction–diffusion simulations of astrocytic calcium signaling. In those geometries, node width was d 0 = 380 nm, consistent with median values of node width reported previously (Arizono et al. 2020). The range of shaft width tested was based on node‐shaft width ratios that we measured in organotypic hippocampal culture using STED microscopy (Figure 1a2). The resulting 3D geometries are presented in Figure 1b. To model astrocytic activity with a high spatial resolution while taking into account the randomness of reactions in small volumes, we used the stochastic voxel‐based model from Denizot et al. (2019). The reactions included in the model are presented in Figure 1d and in the Methods section. As the majority of signals in astrocytes result from the opening of Inositol 3‐Phosphate receptors (IP3Rs), located at the membrane of the endoplasmic reticulum (ER) (Srinivasan et al. 2015), signals in the model result from the opening of IP3 3Rs, while IP3 3R‐independent fluxes into the cytosol are lumped into a single rate (see Denizot et al. (2019) for more details on the model's assumptions). Neuronal stimulation was simulated as an injection of IP3 molecules in the cytosol and a influx at channels at the plasma membrane at a rate = 0–1000 s−1. Astrocytic activity was quantified by measuring peak probability, frequency, amplitude, duration and time to 1st peak (see Figure 1 and Methods).
3.2. Validation of the geometrical model of astrocyte processes
In order to test whether the geometries designed in this study are a good approximation of the ultrastructure of the gliapil, we have compared molecular diffusion flux in those geometries with those reported experimentally. To do so, we simulated photobleaching experiments and compared our results to experimental results from Arizono et al. (2020). The principle of bleaching simulations is presented in the Methods section and in Movie S1. Here, we refer to an increased node compartmentalization when the time to recovery after bleaching, , increases (see Figure 2b).
Bleaching traces in simulations are both qualitatively (Figure 2b) and quantitatively (Figure 2c) similar to experimental bleaching traces, for shaft width and . Indeed, no significant difference of (Figure 2c1), (Figure 2c2) and (Figure 2c3) was observed between simulations and experimental traces. Simulations were also performed with . Our simulations successfully reproduce experimental bleaching experiments and suggest that , and thus node compartmentalization, increases when shaft width decreases (Figure 2c3). This result is not surprising as a decreased shaft width results in a smaller size of the exit point for diffusing molecules from the node. This is similar to, for example, dendritic spines, which compartmentalization is increased for thinner spine necks (Santamaria et al. 2011; Tonnesen et al. 2014). The geometries that we have designed can thus be considered as a reasonable approximation of the ultrastructure of the gliapil observed experimentally in live tissue.
3.3. Thin shafts enhance activity in nodes
About 80% of astrocyte activity occurs in the gliapil (Bindocci et al. 2017), which suggests that most neuron‐astrocyte communication occurs at fine astrocytic processes. As we observed that a decreased shaft width is associated with a decreased diffusion flux, that is, an increased compartmentalization of nodes, we have tested whether this effect influences activity upon neuronal stimulation. To do so, we have first analyzed signals resulting from a single neuronal stimulation, which was simulated as an infusion of IP3 and in the stimulated node, node 1 (see Methods). Those parameters encompass the IP3 production by phospholipase C following the activation of G q‐G‐protein‐coupled receptors (GPCRs) resulting from the binding of neuronal glutamate, ATP and noradrenaline to proteins, and entry at the plasma membrane through channels, ionotropic receptors or the sodium/calcium exchanger (NCX) functioning in reverse mode (Ahmadpour et al. 2021; Semyanov et al. 2020). Signals were recorded both in the stimulated node, node 1, and in the neighboring node, node 2 (Movie S2). Representative traces in node 2 for , and are displayed in Figure 3a. Our first result is that peak probability increases when decreases (Figure 3b1). The time to 1st peak increases with (Figure 3b2). By contrast, peak amplitude (Figure 3b3) and duration (Figure 3b4) increase when decreases. Interestingly, time to 1st peak was similar in the stimulated and neighboring node, which probably results from the homogenization of IP3 concentration throughout the branchlet by diffusion prior to peak detection. To better understand the mechanisms responsible for the increased peak probability, amplitude and duration when decreases, we measured the frequency of opening in nodes 1 and 2. The frequency of opening increases when decreases (Figure 3b5). Note that the duration of opening and the number of s open per peak did not vary with . The increased opening frequency associated with small values of probably results from the increased residency time of molecules in nodes connected to thin shafts (Figure 3c). Indeed, a thin shaft can “trap” and longer in the node, thus locally increasing the probability of IP 3 Rs to open, resulting in larger peaks. For more details, the reader can refer to the theoretical work investigating the narrow escape problem for diffusion in microdomains (Schuss et al. 2007). Nodes connected to thinner shafts, despite being characterized by a lower diffusion flux (Figure 2), could thus consist of signal amplification units, favoring the generation of larger signals, therefore increasing peak probability, amplitude and duration both in the stimulated and in neighboring nodes.
To identify the cause of the increased activity when shaft width decreases (Figure 3), we performed simulations in which we altered the stimulated node (Figure 3S2D‐E), influx at the plasma membrane (Figure S3) and boundary conditions (Figure S4). Those parameters did not affect the observed effects of on dynamics. Furthermore, we repeated simulations of Figure 3 with constant cytosolic volume and constant number of IP3Rs, irrespective of the value of (Figure S5). Our results highlight that the relevant parameter responsible for the observed effects of on signal characteristics is the node/shaft width ratio or the cytosolic volume rather than itself. Spontaneous signals were affected by shaft width in the same way as neuronal‐induced signals (Figure S6) and, in particular, reproduced the increase of the amplitude ratio of spontaneous signals between node 2 and node 1 with shaft width observed in hippocampal organotypic cultures (Arizono et al. 2020). Note that ER morphology, in particular ER surface area (Figures S7, S8 and S9), and buffering by indicators (Figure S10), consistent with previous reports (Denizot et al. 2019; Bartol et al. 2015; Majewska et al. 2000), also altered local activity. Overall, our results suggest that a decreased shaft width, resulting in a decreased diffusion efflux from nodes, increases peak probability, amplitude and duration. Conversely, the swelling of fine processes, resulting in an increase of shaft width, attenuates local peak probability, amplitude, and duration.
3.4. imaging confirms that swelling attenuates local spontaneous activity
To validate the role of thin shafts suggested by our model's predictions, we tried to recreate the widening of shaft width in experimental conditions. Coincidentally, our recent super‐resolution study revealed that the nano‐architecture of fine processes is remodeled in hypo‐osmotic conditions, where shaft width increased while node width remained unaltered (see Figure 3 in Arizono, Inavalli, et al. (2021)). While hypo‐osmotic conditions undoubtedly cause many physiological changes to astrocytes, it is the closest experimentally available model to test our model predictions. We thus performed experimental measurements of activity in fine astrocytic processes under basal and hypo‐osmotic conditions. To record signals in fine branchlets, we used confocal microscopy in organotypic cultures, which provide a high level of optical access and sample stability in live tissue (resolution 200 nm in x–y versus 500 nm for two‐photon microscopy). In accordance with our model's predictions, peak amplitude and duration were lower in hypo‐osmotic compared to basal conditions (Figure 4a–d). Such differences were not observed in the absence of HOC (Figure 4e, f). Overall, our experimental results confirm that the complex nano‐architecture of fine astrocytic processes and its alteration, such as cell swelling, shapes local activity.
FIGURE 4.
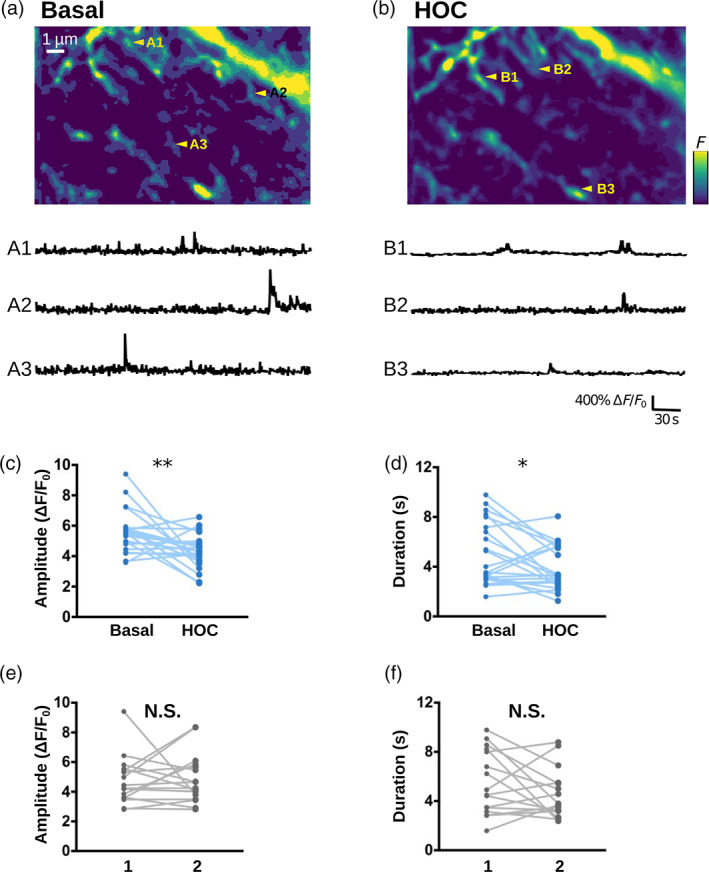
imaging confirms that swelling attenuates local spontaneous activity. (a, b) (top) Confocal images of the astrocytic spongiform domain expressing GCaMP6s at baseline (basal, a) and in hypo‐osmotic condition (HOC, b), measured in organotypic hippocampal cultures (resolution: 200 nm in x–y, 600 nm in z). (bottom) representative traces of spontaneous events from ROIs indicated in (a) (a1–a3) and (b) (b1–b3). (c, d) peak amplitude (c) and duration (d) of spontaneous events are significantly smaller in hypo‐osmotic conditions (HOC) compared to basal conditions (Basal). (e, f) Amplitude (e) and duration (f) of spontaneous events do not significantly vary when measured twice in a row (1, 2) in the absence of HOC. Lines represent measurements in the same cell, before and after applying hypo‐osmotic stress.
3.5. Thin shafts favor more robust signal propagation
As a single branchlet communicates with multiple dendritic spines, which can function independently or belong to a cluster of co‐active synapses (Arizono et al. 2020; Reichenbach et al. 2010; Witcher et al. 2007; Cali et al. 2019; Semyanov et al. 2020), the frequency of node stimulation within the branchlet can vary drastically. Thus, we have tested how node stimulation frequency affects activity in the branchlet. To do so, we have performed simulations in which neighboring nodes were repeatedly stimulated after a time period , that varied from 50 ms to 5 s, while signals were recorded in a remote node, node 5 (Figure 5a). In this stimulation protocol, node 1 is stimulated at t = t 0 = 5 s, node 2 at , node 3 at and node 4 at . Neuronal stimulation is simulated as an infusion of 50 IP3 molecules in the stimulated node. Representative traces in node 5 in branchlets with various shaft widths are presented in Figure 5a for = 250 and 3000 ms. Our first notable result is that the time to 1st peak in node 5 decreases with , whatever the value of (Figure 5b1). More specifically, time to 1st peak is higher for = compared to both = and , while differences between = and are not as striking. Moreover, the difference between = , and increases with . This suggests that geometries with = better discriminate slow from fast frequency of node stimulation compared to geometries with thinner shafts. Geometries with = are further characterized by a lower peak probability in node 5 compared to geometries with = and (Figure 5b2). More precisely, peak probability decreases as increases for = , which was observed independently of our boundary conditions (Figure S11, see Methods). Interestingly, the same effect was observed with various node stimulation patterns, where nodes were stimulated in a different order (Figure S12a) or where a single node was repeatedly stimulated (Figure S12b, c). This suggests that geometries with larger shafts could be associated with decreased signal propagation to remote nodes in case of repeated node stimulation at low frequency ( > 2 s).
For = 4 s and = , signals were detected in node 5 11.55 ± 3.89 s after the stimulation of node 1, which means that they occurred before the stimulation of node 4 (t = + 12 s for = 4 s). This phenomenon was not observed for = , for which time to 1st peak when = 4 s was 23.67 ± 0.47 s. This suggests that for = , contrary to = , one node stimulation could be omitted without having any consequence on peak probability in node 5. In order to test this hypothesis, we have performed simulations in which the stimulation of nodes 2, 3 and 4 occurred with a given probability of failure . Simulations were performed for = 0, 0.05, 0.1, 0.15, 0.25 and 0.75. peak probability in node 5, depending on and on is presented in Figure 5c, for = (Figure 5c1), = (Figure 5c2) and = (Figure 5c3). As expected, peak probability, despite high values of , increases when decreases. Thus, thin shafts can favor signal propagation by allowing the omission of a node stimulation. In that sense, geometries displaying thin shafts are characterized by a more robust signal propagation (Figure 5d).
Together, our results suggest that, in the context of repeated node stimulation, thin shafts are associated with an increase of peak probability in more remote nodes, with an earlier signal onset, suggesting increased signal propagation. Astrocytic processes with thicker shafts (here = ), such as observed in hypo‐osmotic conditions (Arizono, Inavalli, et al. 2021), are associated with lower signal propagation in case of low stimulation frequency (time period >2 s), potentially favoring the formation of local hotspots. Our results suggest that geometries with thick shafts could impair signal propagation when a branchlet is stimulated at a low frequency. In that sense, astrocyte branchlets with thicker shafts would be better detectors of the surrounding level of neuronal activity. By contrast, branchlets with thin shafts would be less discriminating and provide more robust signal propagation.
4. DISCUSSION
Fine astrocytic processes are responsible for most astrocytic signals (Bindocci et al. 2017) and are preferential sites of neuron‐astrocyte communication (Arizono et al. 2020). A better understanding of the mechanistic link between their morphology and the spatio‐temporal properties of local signals is crucial, yet hard to test experimentally. Here, we perform reaction–diffusion simulations in idealized morphologies of astrocytic processes derived from 3D super‐resolution microscopy to investigate the effect of astrocyte nanoscale morphology on activity in the gliapil. Our simulation results indicate that the nanoscale morphological features of astrocytic processes effectively increase the peak probability, duration, amplitude and propagation of signals. Conversely, the alteration of the node‐shaft arrangement of the spongiform domain associated with astrocyte swelling attenuates local activity and signal propagation. Our simulation results, in accordance with experimental data, suggest that thin shafts effectively decrease diffusion flux, resulting in an increased compartmentalization of biochemical signals in nodes. Thus, nodes, similarly to dendritic spines (Santamaria et al. 2011), act as diffusion traps when shaft width is low. Note that, more than the value of shaft width itself, our results emphasize the effect of the ratio between node and shaft diameter on activity. The simple geometries that we have evaluated in this study could be used to build a more comprehensive model of the spongiform structure to simulate activity in the entire astrocyte. By recording the molecular interactions resulting in signals upon neuronal stimulation in small cellular compartments of the gliapil, which cannot be performed experimentally, our simulation results shed light on the mechanisms by which the nano‐architecture of astrocytic processes influences the frequency, amplitude and propagation of local signals at tripartite synapses in health and disease.
Experimental recordings of astrocyte activity have established that astrocyte processes display both highly localized microdomain signals and propagating waves (Srinivasan et al. 2015; Bindocci et al. 2017). Our simulations suggest that the morphology of the cell and of its organelles can strongly influence the formation of these patterns of astrocytic signaling. Notably, thinner shafts allow less discriminating and more robust signal propagation upon repeated stimuli compared to larger shafts. On the contrary, geometries with thick shafts seem to be more discriminating, potentially favoring the propagation of signals resulting from repeated stimuli from co‐active synapses. Cellular morphology thus emerges as a key parameter that regulates the active propagation of signals. The ultrastructure of the spongiform domain of astrocytes is very complex, characterized by abundant branching points, conferring a reticular morphology (Arizono et al. 2020). Those branching points are reportedly sometimes arranged into ring‐like structures, although their occurrence and shape are still debated and could differ depending on the brain region under study (Arizono et al. 2020; Panatier et al. 2014; Kiyoshi et al. 2020; Salmon et al. 2021; Arizono and Nägerl 2021). The effect of this reticular ultrastructure on the propagation of signals remains to be uncovered. Further characterization of the shape of fine astrocytic processes of the spongiform domain, their variability as well as their connectivity to the neighboring synapses are thus required. Pairing those observations with biophysically‐detailed models such as the one presented in this study stands to deepen our understanding of the roles of astrocytic and neuronal morphology at tripartite synapses on neuron‐astrocyte communication.
In neurons, both experimental (Yuste et al. 2000; Noguchi et al. 2005; Tonnesen et al. 2014) and modeling (Schmidt and Eilers 2009; Biess et al. 2007; Simon et al. 2014; Bell et al. 2019; Holcman and Schuss 2005, 2011; Santamaria et al. 2011; Cugno et al. 2019) studies have suggested that thin spine necks favor the compartmentalization of signals within the spine head. This compartmentalization of synapses allows neurons to discriminate various inputs and to process information locally (Wybo et al. 2019, Poirazi and Papoutsi 2020), increasing the computational power of the neuronal circuits. According to our simulation results, nodes connected to thin shafts could favor the emergence of large signals at the site of neuron‐astrocyte communication. Interestingly, we further propose that those amplified signals in nodes, instead of resulting in hotspots, favor active signal propagation. Fine astrocytic processes encounter morphological rearrangements that are activity‐dependent, which notably influence synaptic maturation, efficacy and spine stability (Theodosis et al. 2008; Zhou et al. 2019; Henneberger et al. 2020). Our study sheds light on the influence of rearrangements of the reticular morphology of fine processes on signal computation by astrocytes. Further investigation manipulating astrocyte morphology in situ as well as in vivo is required to better characterize the variability of astrocyte ultrastructure and the associated integration of signals.
The morphology of the complex spongiform domain of astrocytes is highly dynamic, subject to activity‐dependent as well as pathological remodeling. Our study, providing mechanisms by which an altered astrocyte morphology influences neuron‐astrocyte communication at the nanoscale, gives new insights into the involvement of astrocytes in brain function in health and disease.
AUTHOR CONTRIBUTION
AD, HB and EDS designed the study. UVN, EDS and HB supervised the work. AD implemented the code, conducted the simulations, and analyzed the computational data. Calcium imaging and analysis was performed by MA. AD wrote the first draft of the manuscript, which was revised and approved by all authors.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
Supporting information
Appendix S1 Supporting Information
Movie S1
Movie S2
ACKNOWLEDGMENTS
This work was funded by the Okinawa Institute of Science and Technology Graduate University and by JSPS (Japan Society for the Promotion of Science) Postdoctoral Fellowship for Research in Japan (Standard, P21733). We thank Iain Hepburn and Weiliang Chen of the Computational Neuroscience Unit, OIST, Japan for discussion on 3D meshes and STEPS.
Denizot, A. , Arizono, M. , Nägerl, U. V. , Berry, H. , & De Schutter, E. (2022). Control of Ca2+ signals by astrocyte nanoscale morphology at tripartite synapses. Glia, 70(12), 2378–2391. 10.1002/glia.24258
Funding information Japan Society for the Promotion of Science, Grant/Award Number: P21733; Okinawa Institute of Science and Technology Graduate University; European Research Council Synergy grant (ENSEMBLE), Grant/Award Number: 951294
DATA AVAILABILITY STATEMENT
The simulation code and meshes used in this study are available at http://modeldb.yale.edu/266928.
REFERENCES
- Agarwal, A. , Wu, P.‐H. , Hughes, E. G. , Fukaya, M. , Tischfield, M. A. , Langseth, A. J. , Wirtz, D. , & Bergles, D. E. (2017, February). Transient opening of the mitochondrial permeability transition pore induces microdomain calcium transients in astrocyte processes. Neuron, 93(3), 587–605.e7. 10.1016/j.neuron.2016.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadpour, N. , Kantroo, M. , & Stobart, J. L. (2021). Extracellular calcium influx pathways in astrocyte calcium microdomain physiology. Biomolecules, 11(10), 1467. 10.3390/biom11101467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizono, M. , Bancelin, S. , Bethge, P. , Chéreau, R. , Idziak, A. , Inavalli, V. V. G. K. , Pfeiffer, T. , Tønnesen, J. , & Nägerl, U. V. (2021). Nanoscale imaging of the functional anatomy of the brain. Neuroforum, 27(2), 67–77. 10.1515/nf-2021-0004 [DOI] [Google Scholar]
- Arizono, M. , Inavalli, V. V. G. K. , Bancelin, S. , Fernández‐Monreal, M. , & Nägerl, U. V. (2021). Super‐resolution shadow imaging reveals local remodeling of astrocytic microstructures and brain extracellular space after osmotic challenge. Glia, 69(6), 1605–1613. 10.1002/glia.23995 [DOI] [PubMed] [Google Scholar]
- Arizono, M. , Inavalli, V. V. G. K. , Panatier, A. , Pfeiffer, T. , Angibaud, J. , Levet, F. , Ter Veer, M. J. T. , Stobart, J. , Bellocchio, L. , Mikoshiba, K. , Marsicano, G. , Weber, B. , Oliet, S. H. R. , & Nägerl, U. V. (2020). Structural basis of astrocytic Ca2+ signals at tripartite synapses. Nature Communications, 11(1), 1–15. 10.1038/s41467-020-15648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizono, M. , & Nägerl, U. V. (2021). Deciphering the functional nano‐anatomy of the tripartite synapse using stimulated emission depletion microscopy. Glia, 70, 607–618. 10.1002/glia.24103 [DOI] [PubMed] [Google Scholar]
- Bartol, T. M. , Keller, D. X. , Kinney, J. P. , Bajaj, C. L. , Harris, K. M. , Sejnowski, T. J. , & Kennedy, M. B. (2015). Computational reconstitution of spine calcium transients from individual proteins. Frontiers in Synaptic Neuroscience, 7, 17. 10.3389/fnsyn.2015.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, M. , Bartol, T. , Sejnowski, T. , & Rangamani, P. (2019). Dendritic spine geometry and spine apparatus organization govern the spatiotemporal dynamics of calcium. The Journal of General Physiology, 151(8), 1017–1034. 10.1085/jgp.201812261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny, I. , Watras, J. , & Ehrlich, B. E. (1991). Bell‐shaped calcium‐response curves of ins(1,4,5)P3‐ and calcium‐gated channels from endoplasmic reticulum of cerebellum. Nature, 351(6329), 751–754. 10.1038/351751a0 [DOI] [PubMed] [Google Scholar]
- Biess, A. , Korkotian, E. , & Holcman, D. (2007). Diffusion in a dendritic spine: The role of geometry. Physical Review E, 76(2), 21922. 10.1103/PhysRevE.76.021922 [DOI] [PubMed] [Google Scholar]
- Bindocci, E. , Savtchouk, I. , Liaudet, N. , Becker, D. , Carriero, G. , & Volterra, A. (2017). Threedimensional imaging advances understanding of astrocyte biology. Science, 356(6339), eaai8185. 10.1126/science.aai8185 [DOI] [PubMed] [Google Scholar]
- Brazhe, A. R. , Postnov, D. E. , & Sosnovtseva, O. (2018). Astrocyte calcium signaling: Interplay between structural and dynamical patterns. Chaos: An Interdisciplinary Journal of Nonlinear Science, 28(10), 106320. 10.1063/1.5037153 [DOI] [PubMed] [Google Scholar]
- Bushong, E. A. , Martone, M. E. , Jones, Y. Z. , & Ellisman, M. H. (2002). Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 22(1), 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali, C. , Agus, M. , Kare, K. , Boges, D. J. , Lehvaslaiho, H. , Hadwiger, M. , & Magistretti, P. J. (2019). 3D cellular reconstruction of cortical glia and parenchymal morphometric analysis from serial block‐face electron microscopy of juvenile rat. Progress in Neurobiology, 183, 101696. 10.1016/j.pneurobio.2019.101696 [DOI] [PubMed] [Google Scholar]
- Cugno, A. , Bartol, T. M. , Sejnowski, T. J. , Iyengar, R. , & Rangamani, P. (2019, August). Geometric principles of second messenger dynamics in dendritic spines. Scientific Reports, 9(1), 1–18. 10.1038/s41598-019-48028-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Young, G. W. , & Keizer, J. (1992). A single‐pool inositol 1,4,5‐trisphosphate‐receptorbased model for agonist‐stimulated oscillations in Ca2+ concentration. Proceedings of the National Academy of Sciences, 89(20), 9895–9899. 10.1073/pnas.89.20.9895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot, A. , Arizono, M. , Nägerl, U. V. , Soula, H. , & Berry, H. (2019). Simulation of calcium signaling in fine astrocytic processes: Effect of spatial properties on spontaneous activity. PLoS Computational Biology, 15(8), e1006795. 10.1371/journal.pcbi.1006795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Castro, M. A. , Chuquet, J. , Liaudet, N. , Bhaukaurally, K. , Santello, M. , Bouvier, D. , Tiret, P. , & Volterra, A. (2011). Local Ca2+ detection and modulation of synaptic release by astrocytes. Nature Neuroscience, 14(10), 1276–1284. 10.1038/nn.2929 [DOI] [PubMed] [Google Scholar]
- Francis, M. , Qian, X. , Charbel, C. , Ledoux, J. , Parker, J. C. , & Taylor, M. S. (2012). Automated region of interest analysis of dynamic Ca2+ signals in image sequences. American Journal of Physiology‐Cell Physiology, 303(3), C236–C243. 10.1152/ajpcell.00016.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gähwiler, B. H. (1981). Organotypic monolayer cultures of nervous tissue. Journal of Neuroscience Methods, 4(4), 329–342. 10.1016/0165-0270(81)90003-0 [DOI] [PubMed] [Google Scholar]
- Giaume, C. , & Venance, L. (1998). Intercellular calcium signaling and gap junctional communication in astrocytes. Glia, 24(1), 50–64. [DOI] [PubMed] [Google Scholar]
- Gillespie, D. T. (1977). Exact stochastic simulation of coupled chemical reactions. The Journal of Physical Chemistry, 81(25), 2340–2361. 10.1021/j100540a008 [DOI] [Google Scholar]
- Grosche, J. , Matyash, V. , Möller, T. , Verkhratsky, A. , Reichenbach, A. , & Kettenmann, H. (1999). Microdomains for neuron–glia interaction: Parallel fiber signaling to Bergmann glial cells. Nature Neuroscience, 2(2), 139–143. 10.1038/5692 [DOI] [PubMed] [Google Scholar]
- Haustein, M. D. , Kracun, S. , Lu, X.‐H. , Shih, T. , Jackson‐Weaver, O. , Tong, X. , Xu, J. , Yang, X. W. , O'Dell, T. J. , Marvin, J. S. , Ellisman, M. H. , Bushong, E. A. , Looger, L. L. , & Khakh, B. S. (2014). Conditions and constraints for astrocyte calcium signaling in the hippocampal mossy fiber pathway. Neuron, 82(2), 413–429. 10.1016/j.neuron.2014.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger, C. , Bard, L. , Panatier, A. , Reynolds, J. P. , Kopach, O. , Medvedev, N. I. , Minge, D. , Herde, M. K. , Anders, S. , Kraev, I. , Heller, J. P. , Rama, S. , Zheng, K. , Jensen, T. P. , Sanchez‐Romero, I. , Jackson, C. J. , Janovjak, H. , Ottersen, O. P. , Nagelhus, E. A. , … Rusakov, D. A. (2020). LTP induction boosts glutamate spillover by driving withdrawal of Perisynaptic Astroglia. Neuron, 108(5), 919–936.e11. 10.1016/j.neuron.2020.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn, I. , Chen, W. , Wils, S. , & De Schutter, E. (2012). STEPS: Efficient simulation of stochastic reaction–diffusion models in realistic morphologies. BMC Systems Biology, 6(1), 36. 10.1186/1752-0509-6-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcman, D. , & Schuss, Z. (2005). Modeling calcium dynamics in dendritic spines. SIAM Journal on Applied Mathematics, 65(3), 1006–1026. 10.1137/S003613990342894X [DOI] [Google Scholar]
- Holcman, D. , & Schuss, Z. (2011). Diffusion laws in dendritic spines. The Journal of Mathematical Neuroscience, 1(1), 10. 10.1186/2190-8567-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyoshi, C. M. , Aten, S. , Arzola, E. P. , Patterson, J. A. , Taylor, A. T. , Du, Y. , Guiher, A. M. , Philip, M. , Camacho, E. G. , Mediratta, D. , Collins, K. , Benson, E. , Kidd, G. , Terman, D. , & Zhou, M. (2020). Ultrastructural view of astrocyte‐astrocyte and astrocyte‐synapse contacts within the hippocampus. bioRxiv, 2020.10.28.358200. doi: 10.1101/2020.10.28.358200 [DOI] [PMC free article] [PubMed]
- Lafrenaye, A. D. , & Simard, J. M. (2019). Bursting at the seams: Molecular mechanisms mediating astrocyte swelling. International Journal of Molecular Sciences, 20(2), 330. 10.3390/ijms20020330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.‐G. , Wheeler, M. A. , & Quintana, F. J. (2022). Function and therapeutic value of astrocytes in neurological diseases. Nature Reviews Drug Discovery, 21, 1–20. 10.1038/s41573-022-00390-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind, B. L. , Brazhe, A. R. , Jessen, S. B. , Tan, F. C. C. , & Lauritzen, M. J. (2013). Rapid stimulus‐evoked astrocyte Ca2+ elevations and hemodynamic responses in mouse somatosensory cortex in vivo. Proceedings of the National Academy of Sciences, 110, E4687. 10.1073/pnas.1310065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska, A. , Brown, E. , Ross, J. , & Yuste, R. (2000). Mechanisms of calcium decay kinetics in hippocampal spines: Role of spine calcium pumps and calcium diffusion through the spine neck in biochemical compartmentalization. Journal of Neuroscience, 20(5), 1722–1734. 10.1523/JNEUROSCI.20-05-01722.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal, R. A. , Morse, T. M. , Carnevale, T. , Marenco, L. , Wang, R. , Migliore, M. , Miller, P. L. , Shepherd, G. M. , & Hines, M. L. (2017). Twenty years of ModelDB and beyond: Building essential modeling tools for the future of neuroscience. Journal of Computational Neuroscience, 42(1), 1–10. 10.1007/s10827-016-0623-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard, M. , Rodríguez, J. J. , & Verkhratsky, A. (2010). Glial calcium and diseases of the nervous system. Cell Calcium, 47(2), 140–149. 10.1016/j.ceca.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Noguchi, J. , Matsuzaki, M. , Ellis‐Davies, G. C. R. , & Kasai, H. (2005). Spine‐neck geometry determines NMDA receptor‐dependent Ca2+ signaling in dendrites. Neuron, 46(4), 609–622. 10.1016/j.neuron.2005.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu, Y. , Couchman, K. , Lyons, D. G. , Collot, M. , Agarwal, A. , Mallet, J.‐M. , Pfrieger, F. W. , Bergles, D. E. , & Charpak, S. (2015, February). Calcium dynamics in astrocyte processes during neurovascular coupling. Nature Neuroscience, 18(2), 210–218. 10.1038/nn.3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panatier, A. , Arizono, M. , & Nägerl, U. V. (2014). Dissecting tripartite synapses with STED microscopy. Philosophical Transactions of the Royal Society B, 369 (1654), 20130597. doi: 10.1098/rstb.2013.0597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panatier, A. , Vallée, J. , Haber, M. , Murai, K. K. , Lacaille, J.‐C. , & Robitaille, R. (2011). Astrocytes are endogenous regulators of basal transmission at central synapses. Cell, 146(5), 785–798. 10.1016/j.cell.2011.07.022 [DOI] [PubMed] [Google Scholar]
- Poirazi, P. , & Papoutsi, A. (2020). Illuminating dendritic function with computational models. Nature Reviews Neuroscience, 21(6), 303–321. 10.1038/s41583-020-0301-7 [DOI] [PubMed] [Google Scholar]
- Reichenbach, A. , Derouiche, A. , & Kirchhoff, F. (2010). Morphology and dynamics of perisynaptic glia. Brain Research Reviews, 63(1–2), 11–25. 10.1016/j.brainresrev.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Rusakov, D. A. (2015). Disentangling calcium‐driven astrocyte physiology. Nature Reviews Neuroscience, 16(4), 226–233. 10.1038/nrn3878 [DOI] [PubMed] [Google Scholar]
- Salmon, C. K. , Syed, T. A. , Kacerovsky, J. B. , Alivodej, N. , Schober, A. L. , Pratte, M. T. , Rosen, M. P. , Green, M. , DasGupta, A. , Vali, H. , Mandato, C. A. , Siddiqi, K. , & Murai, K. K. (2021). Organizing principles of astrocytic nanoarchitecture in the mouse cerebral cortex (Tech. Rep.). doi: 10.1101/2021.11.05.467391 [DOI] [PubMed]
- Santamaria, F. , Wils, S. , De Schutter, E. , & Augustine, G. J. (2011). The diffusional properties of dendrites depend on the density of dendritic spines. The European Journal of Neuroscience, 34(4), 561–568. 10.1111/j.1460-9568.2011.07785.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H. , & Eilers, J. (2009). Spine neck geometry determines spino‐dendritic crosstalk in the presence of mobile endogenous calcium binding proteins. Journal of Computational Neuroscience, 27(2), 229–243. 10.1007/s10827-009-0139-5 [DOI] [PubMed] [Google Scholar]
- Schuss, Z. , Singer, A. , & Holcman, D. (2007). The narrow escape problem for diffusion in cellular microdomains. Proceedings of the National Academy of Sciences, 104(41), 16098–16103. 10.1073/pnas.0706599104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semyanov, A. , Henneberger, C. , & Agarwal, A. (2020). Making sense of astrocytic calcium signals —From acquisition to interpretation. Nature Reviews Neuroscience, 21(10), 1–14. 10.1038/s41583-020-0361-8 [DOI] [PubMed] [Google Scholar]
- Sherwood, M. W. , Arizono, M. , Hisatsune, C. , Bannai, H. , Ebisui, E. , Sherwood, J. L. , Panatier, A. , Oliet, S. H. R. , & Mikoshiba, K. (2017). Astrocytic IP3Rs: Contribution to Ca2+ signalling and hippocampal LTP. Glia, 65(3), 502–513. 10.1002/glia.23107 [DOI] [PubMed] [Google Scholar]
- Shigetomi, E. , Bushong, E. A. , Haustein, M. D. , Tong, X. , Jackson‐Weaver, O. , Kracun, S. , Xu, J. , Sofroniew, M. V. , Ellisman, M. H. , & Khakh, B. S. (2013). Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno‐associated viruses. The Journal of General Physiology, 141(5), 633–647. 10.1085/jgp.201210949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi, E. , Patel, S. , & Khakh, B. S. (2016). Probing the complexities of astrocyte calcium signaling. Trends in Cell Biology, 26(4), 300–312. 10.1016/j.tcb.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi, E. , Saito, K. , Sano, F. , & Koizumi, S. (2019). Aberrant calcium signals in reactive astrocytes: A key process in neurological disorders. International Journal of Molecular Sciences, 20(4), 996. 10.3390/ijms20040996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, C. M. , Hepburn, I. , Chen, W. , & Schutter, E. D. (2014). The role of dendritic spine morphology in the compartmentalization and delivery of surface receptors. Journal of Computational Neuroscience, 36(3), 483–497. 10.1007/s10827-013-0482-4 [DOI] [PubMed] [Google Scholar]
- Spacek, J. , & Harris, K. M. (1997). Three‐dimensional Organization of Smooth Endoplasmic Reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. Journal of Neuroscience, 17(1), 190–203. 10.1523/JNEUROSCI.17-01-00190.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, R. , Huang, B. S. , Venugopal, S. , Johnston, A. D. , Chai, H. , Zeng, H. , Golshani, P. , & Khakh, B. S. (2015). Ca(2+) signaling in astrocytes from Ip3r2(−/−) mice in brain slices and during startle responses in vivo. Nature Neuroscience, 18(5), 708–717. 10.1038/nn.4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart, J. L. , Ferrari, K. D. , Barrett, M. J. P. , Glück, C. , Stobart, M. J. , Zuend, M. , & Weber, B. (2018). Cortical circuit activity evokes rapid astrocyte calcium signals on a similar timescale to neurons. Neuron, 98(4), 726–735.e4. 10.1016/j.neuron.2018.03.050 [DOI] [PubMed] [Google Scholar]
- Stobart, J. L. , Ferrari, K. D. , Barrett, M. J. P. , Stobart, M. J. , Looser, Z. J. , Saab, A. S. , & Weber, B. (2018). Long‐term in vivo calcium imaging of astrocytes reveals distinct cellular compartment responses to sensory stimulation. Cerebral Cortex (New York, N.Y.: 1991), 28(1), 184–198. 10.1093/cercor/bhw366 [DOI] [PubMed] [Google Scholar]
- Theodosis, D. T. , Poulain, D. A. , & Oliet, S. H. R. (2008). Activity‐dependent structural and functional plasticity of astrocyte‐neuron interactions. Physiological Reviews, 88(3), 983–1008. 10.1152/physrev.00036.2007 [DOI] [PubMed] [Google Scholar]
- Tonnesen, J. , Katona, G. , Rózsa, B. , & Nägerl, U. V. (2014). Spine neck plasticity regulates compartmentalization of synapses. Nature Neuroscience, 17(5), 678–685. 10.1038/nn.3682 [DOI] [PubMed] [Google Scholar]
- Tonnesen, J. , Nadrigny, F. , Willig, K. I. , Wedlich‐Söldner, R. , & Nägerl, U. V. (2011). Two‐color STED microscopy of living synapses using a single laser‐beam pair. Biophysical Journal, 101(10), 2545–2552. 10.1016/j.bpj.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, N. T. , Willig, K. I. , Hell, S. W. , & Nägerl, U. V. (2011). STED Nanoscopy of Actin dynamics in synapses deep inside living brain slices. Biophysical Journal, 101(5), 1277–1284. 10.1016/j.bpj.2011.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky, A. , & Nedergaard, M. (2018). Physiology of astroglia. Physiological Reviews, 98(1), 239–389. 10.1152/physrev.00042.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witcher, M. R. , Kirov, S. A. , & Harris, K. M. (2007). Plasticity of perisynaptic astroglia during synaptogenesis in the mature rat hippocampus. Glia, 55(1), 13–23. 10.1002/glia.20415 [DOI] [PubMed] [Google Scholar]
- Wu, Y.‐W. , Gordleeva, S. , Tang, X. , Shih, P.‐Y. , Dembitskaya, Y. , & Semyanov, A. (2018,). Morphological profile determines the frequency of spontaneous calcium events in astrocytic processes. bioRxiv, 410076. doi: 10.1101/410076 [DOI] [PubMed]
- Wybo, W. A. M. , Torben‐Nielsen, B. , Nevian, T. , & Gewaltig, M.‐O. (2019). Electrical compartmentalization in neurons. Cell Reports, 26(7), 1759–1773.e7. 10.1016/j.celrep.2019.01.074 [DOI] [PubMed] [Google Scholar]
- Yasuda, R. (2017). Biophysics of biochemical signaling in dendritic spines: Implications in synaptic plasticity. Biophysical Journal, 113(10), 2152–2159. 10.1016/j.bpj.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste, R. , Majewska, A. , & Holthoff, K. (2000). From form to function: Calcium compartmentalization in dendritic spines. Nature Neuroscience, 3(7), 653–659. 10.1038/76609 [DOI] [PubMed] [Google Scholar]
- Zhou, B. , Zuo, Y.‐X. , & Jiang, R.‐T. (2019). Astrocyte morphology: Diversity, plasticity, and role in neurological diseases. CNS neuroscience & therapeutics, 25(6), 665–673. 10.1111/cns.13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information
Movie S1
Movie S2
Data Availability Statement
The simulation code and meshes used in this study are available at http://modeldb.yale.edu/266928.


