Abstract
Objective
The importance of interleukin‐17A (IL‐17A) in the pathogenesis of axial spondyloarthritis (SpA) has been demonstrated by the success of IL‐17A blockade. However, the nature of the cell populations that produce this important proinflammatory cytokine remains poorly defined. We undertook this study to characterize the major IL‐17A–producing blood cell populations in the peripheral blood of patients with axial SpA, with a focus on mucosal‐associated invariant T (MAIT) cells, a population known to be capable of producing IL‐17.
Methods
We evaluated IL‐17A production from 5 sorted peripheral blood cell populations, namely, MAIT cells, γδ T cells, CD4+ T cells, CD8+ T cells, and neutrophils, before and after stimulation with phorbol myristate acetate, the calcium ionophore A23187, and β‐1,3‐glucan. Expression of IL‐17A transcripts and protein were determined using nCounter and ultra‐sensitive Simoa technology, respectively. MAIT cells from the axial entheses of non‐axial SpA control patients (n = 5) were further characterized using flow cytometric immunophenotyping and quantitative polymerase chain reaction, and the production of IL‐17 was assessed following stimulation.
Results
On a per‐cell basis, MAIT cells from peripheral blood produced the most IL‐17A compared to CD4+ T cells (P < 0.01), CD8+ T cells (P < 0.0001), and γδ T cells (P < 0.0001). IL‐17A was not produced by neutrophils. Gene expression analysis also revealed significantly higher expression of IL17A and IL23R in MAIT cells. Stimulation of peripheral blood MAIT cells with anti‐CD3/CD28 and IL‐7 and/or IL‐18 induced strong expression of IL17F. MAIT cells were present in the normal, unaffected entheses of control patients who did not have axial SpA and showed elevated AHR, JAK1, STAT4, and TGFB1 transcript expression with inducible IL‐17A protein. IL‐18 protein expression was evident in spinal enthesis digests.
Conclusion
Both peripheral blood MAIT cells and resident MAIT cells in normal axial entheses contribute to the production of IL‐17 and may play important roles in the pathogenesis of axial SpA.
INTRODUCTION
Within the last 15 years, a clear role of the interleukin‐23 (IL‐23)/IL‐17 axis underpinning the pathophysiology of axial spondyloarthritis (SpA) has emerged (1) as many of the genetic variants associated with ankylosing spondylitis (AS) susceptibility have been identified through genome‐wide association studies. Among the genes found to be associated with AS are those in the IL‐23/IL‐17 pathway, including IL23R, IL12B, IL6R, JAK2, and TYK2 (2). As IL‐17 is the terminal cytokine of this pathophysiologic pathway, the development of new treatments initially focused on blocking this cytokine (3, 4). In support of this hypothesis and beyond the findings of genetic association studies, an increased prevalence of IL‐17–producing T helper (Th17) cells have been reported in the peripheral blood of axial SpA patients (5) and in the normal entheses of individuals who do not have axial SpA (6). Mucosal‐associated invariant T (MAIT) cells and γδ T cells have been further described as alternative sources of IL‐17A in the blood of patients with axial SpA (7, 8). Appel et al also suggested that neutrophils might be the main producers of IL‐17A in the facet joints of patients with AS (9).
IL‐23 plays a crucial role in maintaining the differentiation state of Th17 cells. Nevertheless, recent studies have identified alternative pathways, independent of IL‐23, that can stimulate IL‐17 production, including T cell receptor (TCR) signaling through the engagement of major histocompatibility complex class I–related proteins by MAIT cells (10), IL‐7 signaling in group 3 innate lymphoid cells (ILCs) (11), transforming growth factor β (TGFβ) and IL‐1β signaling in invariant natural killer T (iNKT) cells (12, 13) and γδ T cells (specifically the Vδ1 T cell subset) (14), and, recently, the combination of IL‐12 and IL‐18 together with anti‐CD3/CD28 triggering in MAIT cells (10). Such observations have substantial translational relevance, given that antagonism of IL‐23 has thus far shown no efficacy in the treatment of AS (15, 16).
Considering that the growing body of evidence suggests a role for IL‐17 in the pathogenesis of SpA, we wished to study the capability of MAIT cells, CD4+ T cells, CD8+ T cells, and γδ T cells to express IL‐17 in patients with axial SpA. We first analyzed the cell type–specific expression patterns of AS‐associated genes in those cells, with a particular focus on genes belonging to the IL‐23/IL‐17 pathway. We further compared the respective IL‐17 production capacity of these different cell subsets from the adaptive and innate immune systems and identified MAIT cells as potent IL‐17–secreting cells in patients with axial SpA. MAIT cells from healthy individuals have been reported to express high levels of IL‐7 receptor (IL‐7R) and IL‐18R (17). In this study, we found that MAIT cells can produce IL‐17 with combined TCR triggering and stimulation by IL‐7 and IL‐18, and independently of IL‐23 stimulation. Additionally, we identified resident MAIT cells in the axial entheses of normal, healthy individuals, a site at which the unaffected tissue is targeted in the inflammatory process that leads to severe axial inflammation and later spinal fusion in patients with axial SpA. Taken together, our data highlight the crucial role of MAIT cells in the pathophysiology of axial SpA.
PATIENTS AND METHODS
Patients and samples
Axial SpA
Blood samples from 18 patients naive to treatment with synthetic medications and biologics and with a clinical diagnosis of axial SpA who fulfilled the Assessment of SpondyloArthritis international Society criteria (18) were included in the study.
Enthesis tissue and peripheral blood samples from controls
As controls, human interspinous processes and matched peripheral blood samples were obtained from 5 non‐axial SpA patients who underwent elective spinal surgery for either decompression or scoliosis correction using methods previously reported (19).
Before enrollment in the study, all patients provided written informed consent as approved by the French Ethics Committee and the North West–Greater Manchester West Research Ethics Committee. The clinical characteristics of the axial SpA patients and of the non‐axial SpA control patients who underwent spinal surgery are summarized in Supplementary Table 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.42090.
Cell sorting and stimulation
The isolation of cell populations from peripheral blood samples and entheseal samples was undertaken as previously described (8, 20) and is further described in the Supplementary Materials (http://onlinelibrary.wiley.com/doi/10.1002/art.42090).
Gene expression analysis
Gene expression profiles from the sorted cells of axial SpA patients were assessed using the nCounter Autoimmune Discovery panel (NanoString Technologies), and samples from the normal entheses of non‐axial SpA patients were assessed using a focused gene card. Profiling of the transcription factors of entheseal and blood MAIT cells (CD3+, CD45+, CD161+, and TCRVα7.2+) was performed using the entheseal and peripheral blood samples from non‐axial SpA patients. The basal transcript expression of cytokines, chemokines, growth factors, signaling molecules, and tissue residency markers were assessed using a focused gene card. A description of the RNA preparation and gene expression analysis is available in the Supplementary Materials (http://onlinelibrary.wiley.com/doi/10.1002/art.42090).
Protein expression analyses
The concentrations of IL‐17A (expressed in fg/ml) in cell culture supernatants from axial SpA patients were determined using the Quanterix Simoa IL‐17A Advantage Kit and HD‐1 platform. Stimulated cells from perientheseal bone entheseal mononuclear cells (EMCs) were intracellularly stained for tumor necrosis factor (TNF) and IL‐17 and analyzed using a flow cytometry gating system, as described previously (8, 20). Following 24‐hour stimulation with lipopolysaccharides (100 ng/ml), IL‐18 protein was analyzed in the supernatant using BioLegend LEGENDplex Human Inflammation Panel 1 and the Beckman Coulter CytoFLEX LX Flow Cytometer, according to the manufacturer's instructions (Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.42090). Serum levels of IL‐17A, IL‐17F, IL‐7, and IL‐18 were quantified using the Olink Proximity Extension Assay (Supplementary Materials, http://onlinelibrary.wiley.com/doi/10.1002/art.42090).
Statistical analyses
GraphPad Prism software was used for statistical analyses. Detailed information regarding methods and statistical analyses is provided in the Supplementary Materials (http://onlinelibrary.wiley.com/doi/10.1002/art.42090).
RESULTS
Differential expression of genes associated with AS susceptibility in innate and adaptive T cell populations isolated from peripheral blood of axial SpA patients
Following stimulation with phorbol myristate acetate, the calcium ionophore A23187, and β‐1,3‐glucan, the expression of 36 genes from a panel of 45 genes associated with AS was analyzed in 4 T cell populations (MAIT cells, CD8+ T cells, CD4+ T cells, and γδ T cells) isolated from 9 axial SpA patients. The expression pattern observed after hierarchical clustering showed a clear distinction between the innate and adaptive T cell groups, as shown in the Figure 1A heatmap. Gene clusters consisting of members of the pathway network for major histocompatibility complex class I–mediated antigen processing and presentation (NPEPPS and UBE2L3) were up‐regulated in CD4+ and CD8+ T cells. We observed that genes were expressed at relatively high levels in specific cell types, such as PTGER4 in CD4+ T cells and TYK2 in CD8+ T cells.
Figure 1.
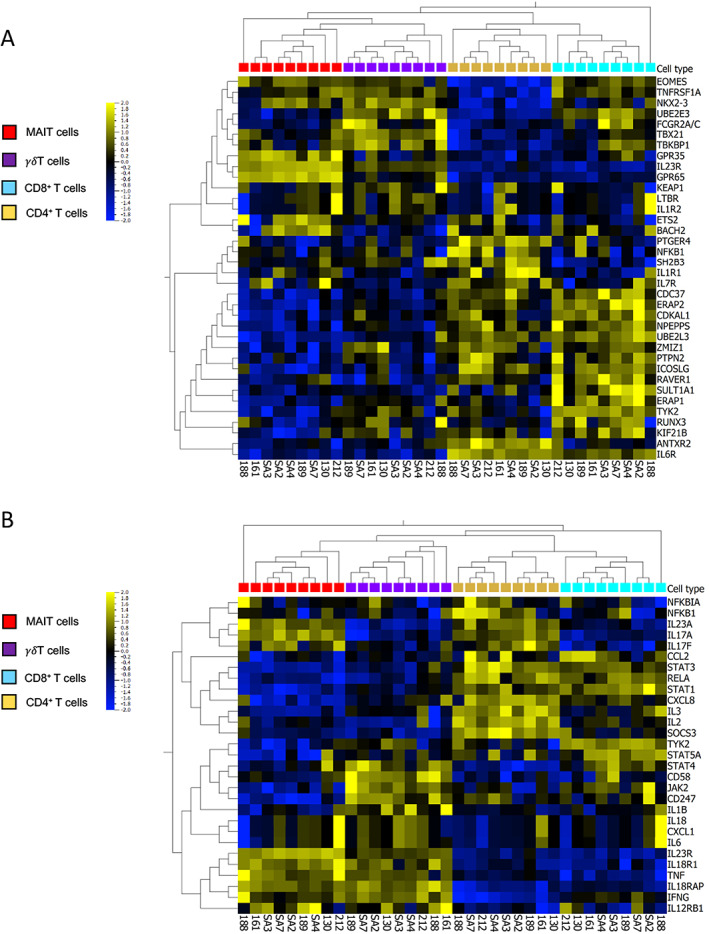
Heatmaps showing the expression patterns of genes associated with ankylosing spondylitis (AS) and of genes of the interleukin‐23 (IL‐23)/IL‐17 pathway in T cell subpopulations isolated from the peripheral blood of patients with axial spondyloarthritis (SpA). A, Expression levels of 36 genes associated with AS susceptibility in T cells from axial SpA patients. B, Messenger RNA expression levels of 29 genes associated with the IL‐23/IL‐17 pathway, selected from the Molecular Signatures Database. T cells were stimulated for 2 hours with phorbol myristate acetate (50 ng/ml), calcium ionophore A23187 (5 μM), and β‐1,3‐glucan (50 μg/ml). Heatmaps show hierarchical clustering of genes among T cell populations from individual patient samples (n = 9). Gene expression data are log2 transformed, centered to a mean value of 0, and scaled to unit variance. The color key on the left denotes the scale of gene expression, ranging from lower levels (blue) to higher levels (yellow).
MAIT cells expressed high levels of IL23R and the G protein–coupled receptors GPR35 and GPR65. We also noted cell type–specific expression of several IL‐23/IL‐17 pathway genes (Figure 1B). IL17F was expressed at higher levels (~1 log‐fold difference in expression) in CD4+ T cells and MAIT cells compared to the other T cell populations, while IL23R expression was higher (2 log‐fold difference) in MAIT cells and γδ T cell expression was higher (1 log‐fold difference) than that in CD4+ and CD8+ T cells. NFKB1, RELA, and NFKBIA were preferentially expressed in CD4+ T cells, while several genes encoding cytokines and their receptors (IL23A, IL23R, IL12RB1, IL18R1, IL18RAP, TNF, and IFNG) were expressed at higher levels in innate‐like MAIT cells and γδ T cells when compared to adaptive CD4+ and CD8+ T cells. IL1R1, TYK2, and RUNX3 were expressed at high levels in CD8+ T cells. Nevertheless, many other genes not belonging to the IL‐23/IL‐17 pathway participated in cell clustering, suggesting that those different cell types were involved in AS susceptibility beyond their relative role in the IL‐23/IL‐17 pathway.
High potential for IL‐17A and IL‐17F secretion in peripheral blood–derived MAIT cells
To better define the relative role of MAIT cells, CD4+ T cells, CD8+ T cells, and γδ T cells in IL‐17 expression, we further sorted these cell types together with neutrophils, whose secretion of IL‐17A is still controversial (21). Cell sorting was performed on samples from 18 axial SpA patients (see Supplementary Figure 1 for the gating strategy, available at http://onlinelibrary.wiley.com/doi/10.1002/art.42090). The production of IL‐17A by MAIT cells was significantly higher than that of CD4+ T cells (P < 0.05), γδ T cells (P < 0.01), CD8+ T cells (P < 0.01), and neutrophils (P < 0.01). Although lower than in MAIT cells (mean 478.60 fg/1,000 cells), IL‐17A production by CD4+ T cells was significant (mean 128.65 fg/1000 cells), while γδ T cells produced a smaller amount of IL‐17A (mean 13.71 fg/1,000 cells), albeit in the same range as IL‐17A production by CD8+ T cells (mean 4.66 fg/1,000 cells). The main component of Aspergillus fumigatus hyphae, β‐glucan, has been demonstrated to have a potential effect on the production of IL‐17A by human neutrophils (22). Despite strong β‐glucan–associated stimulation, the expression of IL‐17A by most neutrophils in the axial SpA patients did not exceed the detection limit (Figure 2A).
Figure 2.
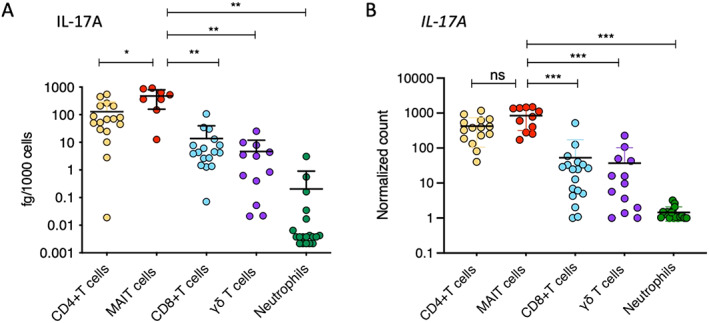
IL‐17A protein production after 18 hours of stimulation (A) and IL17A transcript levels (normalized expression) after 2 hours of stimulation (B) were assessed in sorted CD4+ T cells, mucosal‐associated invariant T (MAIT) cells, CD8+ T cells, γδ T cells, and neutrophils from the peripheral blood of axial SpA patients. Cells were stimulated with phorbol myristate acetate (50 ng/ml), calcium ionophore A23187 (5 μM), and β‐1,3‐glucan (50 μg/ml). Symbols represent individual samples. Bars show the mean ± SEM. * = P < 0.05; ** = P < 0.01, by Mann‐Whitney test. *** = P < 0.001, by Wilcoxon‐Mann‐Whitney test. ns = not significant (see Figure 1 for other definitions).
Gene expression analysis confirmed the findings of the IL‐17A protein analysis, with MAIT cells and CD4+ T cells displaying the highest levels of IL17A expression (no significant difference between IL17A expression in MAIT cells and CD4+ T cells). A low level of IL17A expression was observed in γδ T cells and CD8+ T cells. In neutrophils, expression of IL17A was undetectable, which was consistent with the findings of the IL‐17A protein analysis (Figure 2B).
Next, we expanded our analysis to include the expression of IL17F, IL23R, and IFNG. We observed that the 5 cell populations were similarly ranked in their expression levels of IL17F as they were in their expression levels of IL17A (Figure 3A), but there was no significant difference in the expression levels of IL17F between CD4+ T cells and MAIT cells. We also observed a 10‐fold lower level of expression of IL17F compared to that of IL17A in all cell subsets. Regarding IL23R (Figure 3B), MAIT cells had the highest level of expression, followed by γδ T cells and CD4+ T cells, while CD8+ T cells and neutrophils did not express significant levels of IL23R. In contrast to IL17A and IL17F expression, IFNG was highly expressed in all T cell populations (Figure 3C), suggesting cell‐specific expression profiles for IL17A and IL17F. Collectively, these data indicate that MAIT cells are the major producers of IL‐17A on a per‐cell level and express high levels of both IL17F and IL23R compared to other IL‐17A–producing cells in patients with axial SpA.
Figure 3.
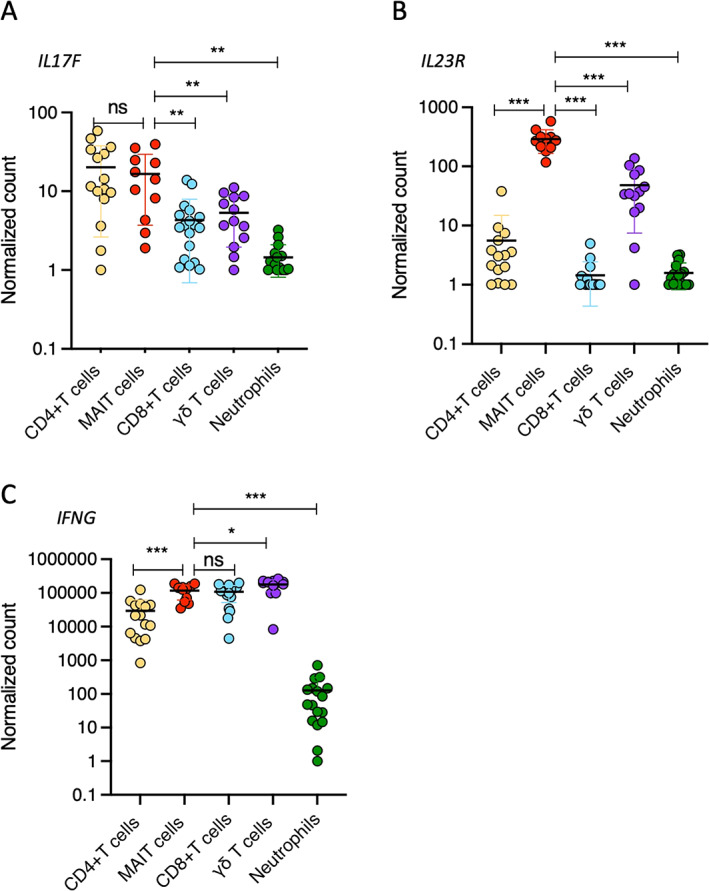
Sorted CD4+ T cells, MAIT cells, CD8+ T cells, γδ T cells, and neutrophils from the peripheral blood of axial SpA patients were analyzed for gene expression of IL17F (A), IL23R (B), and IFNG (C). Results are presented as normalized transcript levels. Cells were stimulated with phorbol myristate acetate (50 ng/ml), calcium ionophore A23187 (5 μM), and β‐1,3‐glucan (50 μg/ml). Symbols represent individual samples. Bars show the mean ± SEM. * = P < 0.05; ** = P < 0.01; *** = P < 0.001, by Wilxocon‐Mann‐Whitney test. See Figure 2 for definitions.
Enhancement of IL17F expression by IL‐7 and IL‐18 in peripheral blood–derived MAIT cells
We further assessed which stimulation conditions induced IL‐17A and IL‐17F protein expression by MAIT cells. Sorted MAIT cells (see Supplementary Figure 2 for the gating strategy, available at http://onlinelibrary.wiley.com/doi/10.1002/art.42090) were stimulated for 36 hours with anti‐CD3/anti‐CD28, anti‐CD3/anti‐CD28 in combination with either IL‐7 or IL‐18, and anti‐CD3/anti‐CD28 combined with both IL‐7 and IL‐18. We used CD4+CCR6+ T cells (“Th17‐like” T cells) as controls (Figure 4). We observed that stimulation with anti‐CD3/anti‐CD28 combined with IL‐7 or IL‐18 induced high expression levels of IL17F but not IL17A in MAIT cells (Figures 4A and B). Furthermore, we identified increased IL18R1 expression (Figure 1B) by MAIT cells, which supports the finding that IL‐18 induces IL17F production. The combination of both IL‐7 and IL‐18 with anti‐CD3/anti‐CD28 further increased IL17F expression. Expression levels of IFNG were also remarkably high after MAIT cells were stimulated with anti‐CD3/anti‐CD28 and either IL‐7 or IL‐18 and when they were stimulated with anti‐CD3/anti‐CD28 and both IL‐7 and IL‐18 combined (Figure 4C).
Figure 4.
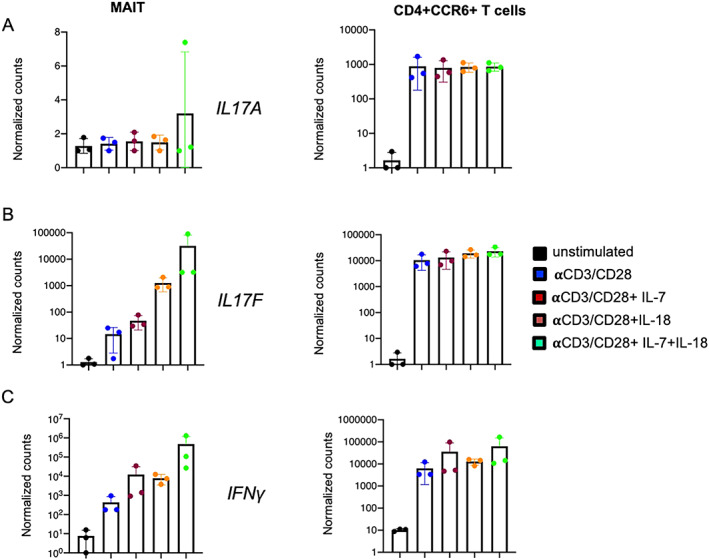
Gene expression of IL17A (A), IL17F (B), and IFNG (C) in sorted mucosal‐associated invariant T (MAIT) cells and CD4+CCR6+ T cells from non‐axial SpA patients (n = 3) after 36 hours of stimulation in 6 different conditions, including unstimulated, stimulation with anti‐CD3/anti‐CD28, stimulation with anti‐CD3/anti‐CD28 and interleukin‐7 (IL‐7) (20 ng/ml), stimulation with anti‐CD3/anti‐CD28 and IL‐18 (50 ng/ml), and stimulation with anti‐CD3/anti‐CD28 and both IL‐7 (20 ng/ml) and IL‐18 (50 ng/ml). Results are presented as the mean ± SEM normalized transcript levels. Symbols represent individual samples.
Basal levels of IL‐18 expression were demonstrated in MAIT cells isolated from the normal entheses of patients who did not have axial SpA (Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.42090), which arguably supports the importance of IL‐18 in MAIT cell modulation. In peripheral blood samples, the comparison between axial SpA patients and non‐axial SpA control patients did not reveal a significant difference in the serum production of IL‐17A and IL‐18, but a trend toward increased expression of IL17F and IL7 in axial SpA patients was observed (P = 0.0508 and P = 0.06, respectively) (Supplementary Figure 6, http://onlinelibrary.wiley.com/doi/10.1002/art.42090). In MAIT cells, no significant difference in IL‐17A secretion or IL17A gene expression was observed (Supplementary Figures 7 and 8, http://onlinelibrary.wiley.com/doi/10.1002/art.42090).
Presence of MAIT cells in entheses unaffected by axial SpA
Considering that the hallmark of axial SpA pathogenesis is entheseal inflammation, we investigated and confirmed the presence of MAIT cells in the axial entheses of individuals who did not have axial SpA (Figure 5A). Within both entheseal soft tissue and perientheseal bone, MAIT cells mainly expressed the resident memory marker CD69, while MAIT cells from the peripheral blood mainly expressed CD45RA, which corresponds to a naive/circulating phenotype (Figure 5A).
Figure 5.
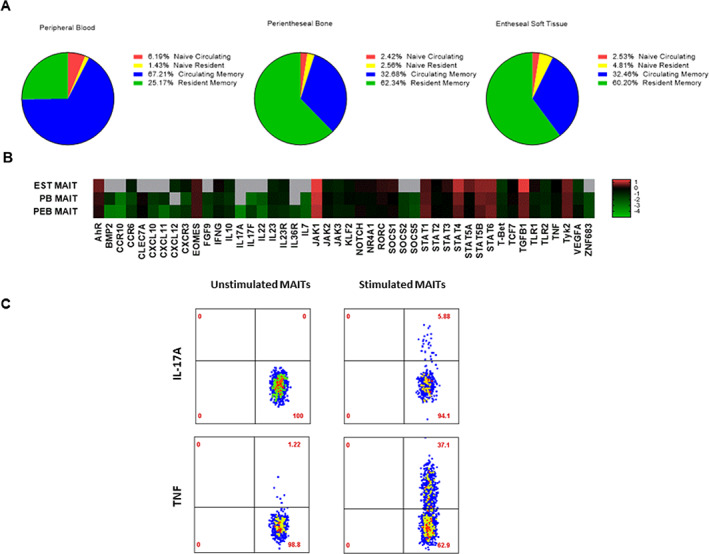
Transcriptional profiling of entheseal mucosal‐associated invariant T (MAIT) cells and proinflammatory cytokine induction. A, Stratification of MAIT cell subsets based on their expression of T cell receptor Vα7.2 and CD161 in entheseal soft tissue (EST), perientheseal bone (PEB), and peripheral blood (PB) from non‐axial spondyloarthritis patients. MAIT cells expressing tissue resident/memory markers were identified by CD69 expression, and naive/circulating MAIT cells were identified by CD45RA expression. Results are shown as the mean percentage (n = 5). B, Basal expression of cytokines, chemokines, growth factors, signaling molecules, and tissue residency markers. Expression values are the log10 change in threshold cycle relative to the values for hypoxanthine guanine phosphoribosyltransferase (n = 7). The color key denotes differential gene expression, in which values <–1 indicate lower relative expression and values >1 indicate higher relative expression. Gray indicates absence of expression. P = 0.038 by 2‐tailed t‐test for independent samples for the difference in expression of CCR6 by MAIT cells from peripheral blood and MAIT cells from perientheseal bone. C, Intracellular tumor necrosis factor (TNF) and interleukin‐17 (IL‐17) cytokine expression in conditions with or without stimulation with phorbol myristate acetate (50 ng/ml) and ionomycin (1 μg/ml) for 3 hours in the presence of BD GolgiPlug protein transport inhibitor in perientheseal bone–derived MAIT cells (n = 2).
Transcriptional profiling of entheseal MAIT cells compared to peripheral blood–derived MAIT cells
The comparison of peripheral blood–derived and enthesis‐derived (from both perientheseal bone and entheseal soft tissue) MAIT cell transcription factors (TCRVα7.2+ and CD161+) showed that entheseal soft tissue MAIT cells had a phenotype characterized by higher AHR, JAK1, STAT4, and TGFβ1 transcript expression (Figure 5B). Furthermore, transcription factors indicative of circulating T cells, such as KLF2 and TBX21, showed higher expression in peripheral blood–derived MAIT cells (Figure 5B). Enthesis‐derived MAIT cells also showed higher expression of growth factors and molecules associated with tissue repair and homeostasis, such as VEGFA and IL10, when compared to matched peripheral blood–derived MAIT cells (Figure 5B).
In comparison to blood‐derived MAIT cells, enthesis‐derived MAIT cells showed higher CXCR3 and CCR6 expression. These findings suggest that enthesis‐derived MAIT cells are better equipped to mediate proinflammatory signals and tissue migration.
Induction of TNF and IL‐17 expression in entheseal MAIT cells
Following on previous findings that blood‐derived MAIT cells produced IL‐17, we next investigated whether entheseal MAIT cells also had the ability to produce this cytokine. Following stimulation with phorbol myristate acetate and ionomycin, MAIT cells showed elevated expression of TNF and IL‐17A through intracellular flow (Figure 5C). Overall, 3.2% of stimulated MAIT cells expressed IL‐17A and 17.9% of stimulated MAIT cells expressed TNF (n = 2 independent samples with MAIT cells). Given that IL‐18 was shown to enhance IL‐17 production, we also investigated the expression of IL‐18. Our results confirmed that IL‐18 was expressed at basal levels in the normal entheses (Supplementary Figure 5, http://onlinelibrary.wiley.com/doi/10.1002/art.42090).
DISCUSSION
The findings presented herein suggest a potentially important role for MAIT cells in the setting of axial SpA. Gene expression profiles based on gene polymorphisms associated with AS showed substantial clustering in MAIT cells. Furthermore, MAIT cells had at least as much IL‐17A production capacity as CD4+ T cells. In addition, MAIT cells were able to produce IL17A and IL17F under conditions of combined stimulation with IL‐7 and IL‐18 together with TCR triggering. Finally, MAIT cells were present in the spinal entheses of healthy controls, where they were mainly of the resident memory cell phenotype with inducible IL‐17A protein production. Collectively, these findings substantially add to the body of evidence incriminating innate‐like lymphocytes in the pathogenesis of axial SpA.
We first analyzed the gene expression profiles of 45 genes whose polymorphisms were significantly associated with the predisposition to AS in 4 T cell populations: CD4+ T cells, CD8+ T cells, γδ T cells, and MAIT cells. Eighty percent of these genes were expressed among these 4 cell types, with differential expression from one cell population to another leading to cell subset clustering. We observed higher expression levels of prostaglandin receptor EP4 (PTGER4) in CD4+ T cells, through which prostaglandin E2 regulates Th17 cell differentiation and functioning (23). This up‐regulation of the receptor EP4 in axial SpA patients could promote CD4 differentiation toward the Th17 pathway, as has been previously reported (23). TYK2 was expressed at higher levels in adaptive CD8+ T cells, followed by innate‐like γδ T cells. Tyk2 is a crucial type 3 immunity mediator in SpA, and its inhibition can prevent disease progression by reducing the expansion of Th17 cells in murine models of SpA (24). Inhibition of Tyk2 also showed promising results in a phase II trial in psoriasis (25).
We observed an up‐regulation of IL18R1 and IL18RAP in MAIT cells, in accordance with the sensitivity of MAIT cells to IL‐18 stimulation for the induction of IL‐17. High expression of IL‐18R and IL‐12R by MAIT cells has been shown to facilitate their activation in a TCR‐independent manner, such as during viral infections (26). The relatively high expression of the AS‐associated G protein–coupled receptor genes GPR35 and GPR65 in MAIT cells could indicate their pathogenic role in the setting of axial SpA, as increased expression of GPR65 in granulocyte–macrophage colony‐stimulating factor–positive CD4+ T cells has been associated with “pathogenic” Th17 cells in SpA patients (27). These results could help us to design functional analyses specifically for MAIT cells.
To better decipher the relative contribution of MAIT cells to IL‐17A/IL‐17F expression in comparison to CD4+ T cells, we assessed the secretion of IL‐17A in cell culture supernatants after stimulation. Generally, MAIT cell frequency is relatively low compared to CD4+ T cells and CD8+ T cells. However, on a per‐cell basis, MAIT cells were the major producers of IL‐17A in axial SpA patients when compared to CD4+ T cells, CD8+ T cells, and γδ T cells. Analysis on a per‐cell basis allowed us to precisely characterize the production capacity of each cell type, which is generally challenging for small cell subsets.
MAIT cells strongly expressed IL23R, but our work shows that cytokines other than IL‐23, such as IL‐7/IL‐18 alone or in combination, can induce strong expression of IL17F messenger RNA. We found particularly high levels of IL18R1 and IL18RAP expression in MAIT cells, suggesting that IL‐18 is an important cytokine in the modulation of their function. Furthermore, we found IL‐18 production at the basal level in unstimulated MAIT cells derived from the perientheseal bone of non‐axial SpA patients, which demonstrates the importance of IL‐18 in MAIT cell regulation. IL‐18, which is associated with IL‐12 and IL‐15, induces IFNγ secretion by Th1 cells (28) and has been shown to synergize with IL‐12 to promote the production of IL‐17A/IL‐17F by MAIT cells independently of IL‐23 (10).
Here we show that another cytokine combination (IL‐7 and IL‐18) particularly potentiates the production of IL‐17F by MAIT cells. To our knowledge, this is the first time that a combination of cytokines has been shown to induce the expression of IL17F by MAIT cells in the setting of axial SpA. IL‐7 is a key cytokine of the adaptive immune system not only for the development of T cells and dendritic cells, but also for the expansion and survival of immature B cells. While stromal and epithelial cells are known to be the main producers of IL‐7, a study by Ciccia et al demonstrated that intestinal Paneth cells could also produce IL‐7 (29). IL‐7 is constitutively produced at low levels, but elevated levels of IL‐7 have been observed in the sacroiliac joint fluid of patients with SpA (30). Recently, IL‐17F has gained interest with the approval of bimekizumab for the treatment of psoriasis (31), and bimekizumab has also shown promising results in patients with axial SpA (32). In the present study, we did not focus on the role of IL23R in the pathogenesis of axial SpA. Additional studies with specific approaches to better deciphering the complexity of the interactions in the different IL‐17–producing cells are needed.
In this work, the contribution of γδ T cells and CD8+ T cells from peripheral blood to the production of IL‐17A was minimal, but it is possible that tissue‐specific expression of IL‐17A is much higher at sites targeted during the inflammatory process in axial SpA. While neutrophils have previously been described as IL‐17–producing cells (22), our work with 2 robust techniques (nCounter and Simoa technology) did not confirm this, even when strong stimulation combining phorbol myristate acetate, the calcium ionophore A23187, and β‐1,3‐glucan was used. This supports the previous finding that neutrophils do not substantially contribute to the production of IL‐17 (21).
Several reports have suggested that in the peripheral form of axial SpA, MAIT cells are not the only cells which produce IL‐17. Both iNKT and γδ T cells are increased in the synovial fluid of patients with SpA and contribute to IL‐17 expression, but they contribute through an IL‐23–dependent mechanism (13). This may be in part due to the shared transcription factor promyelocytic leukaemia zinc‐finger (PLZF; encoded by ZBTB16), which is present in MAIT cells, iNKTs, and γδ T cells (33). Kenna et al observed high expression of IL‐23R on the surface of γδ T cells (6), but this cell population did not appear to be the main source of IL‐17 production in the axial form of SpA. Group 3 innate lymphoid cells are also capable of producing IL‐17A, but Blijdorp et al (34) recently demonstrated that these cells produced IL‐22 rather than IL‐17A in the joints of patients with peripheral SpA.
This study had some limitations which can be addressed in future research, such as the assessment of local production of IL‐7 and IL‐18 within the entheseal tissues of axial SpA patients. It would be ideal to use matched MAIT cells from the blood and entheses of patients with axial SpA, but entheseal tissue samples from axial SpA patients are difficult to access.
In this study, we used data from genome‐wide association studies to examine transcript expression in cells relevant to the pathogenesis of axial SpA, and given the recent and unexpected developments in translational therapeutics for the treatment of SpA where IL‐17 inhibition rather than IL‐23 inhibition was effective, we focused our research on the IL‐23/IL‐17 pathway. Our findings support the hypothesis of major involvement of conventional T cell subsets and innate‐like lymphocytes in the pathogenesis of SpA. However, MAIT cells from peripheral blood were shown to be the main producers of IL‐17A in axial SpA patients when compared to CD4+ T cells, CD8+ T cells, and γδ T cells. Moreover, the stimulation of MAIT cells with combined IL‐7 and IL‐18 was able to strongly induce IL‐17F expression. A trend toward increased expression of IL‐17F and IL‐7 was also observed in the serum of axial SpA patients compared to control patients. The importance of MAIT cells was highlighted by their identification within entheseal tissues where basal IL‐18 protein expression was also found. Further studies are needed to assess whether similar findings hold true in the entheseal tissues from axial SpA patients and to what extent the in situ production of IL‐7 and IL‐18 induces IL‐17F expression.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Miceli‐Richard had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Rosine, Rowe, Koturan, Yahia‐Cherbal, Leloup, Watad, Berenbaum, Sellam, Dougados, Aimanianda, Cuthbert, Bridgewood, Newton, Bianchi, Rogge, McGonagle, Miceli‐Richard.
Acquisition of data
Rosine, Rowe, Koturan, Yahia‐Cherbal, Leloup.
Analysis and interpretation of data
Rosine, Rowe, Koturan, Cuthbert, Bridgewood, Newton, Bianchi, Rogge, McGonagle, Miceli‐Richard.
Supporting information
Disclosure Form:
Appendix S1 Supporting Information
Figure S1 Flow cytometry gating strategy 1. Flow cytometry gating strategy for the isolation of MAIT, γδ T‐cells, CD4+ and CD8+ T‐cells. For T cells, PBMCs were isolated from blood using lymphocyte separation medium (Eurobio®). After isolation, PBMCs were labeled with CD3 BUV395 (BD Biosciences®), CD4 VioBright FITC (Milteny Biotec®), CD8 PerCP vio700 (Milteny Biotec®), TCR Vδ PE (Milteny Biotec®), TCR Vδ2 PE (Milteny Biotec®), TCR Vα7.2 APC (BioLegend®), CD161 BV421 (Sony Biotecnology®).
Figure S2: Flow cytometry gating strategy 2. Flow cytometry gating strategy, after CD3 positive magnetic sorting, of MAIT and CD4+ CCR6 + T‐cells. The CD3 positive fraction was labeled with CD3 APC Vio770 (Milteny Biotec®) CD4 VioBright FITC (Milteny Biotec®), Vα7.2 APC (BioLegend®), CD161 BV421 (Sony Biotechnology®) and CCR6 BV786 (BD Biosciences®).
Figure S3: Transcripts levels of CD28 and CLEC7A presented as normalized counts in sorted CD4+ T‐cells (in yellow), MAIT cells (in red), CD8 + T‐cells (in blue), 𝛾𝛿 T‐cells (in purple) and neutrophils (in green) after 2h of stimulation by PMA (50ng/ml) + A23187(μM) + βglucan (50μg/ml).
Figure S4 Flow cytometry gating strategy for phenotypic identification of MAIT cells in entheseal tissues and peripheral blood cells. Doublet excluded EMCs (PEB and EST) or PBMCs were stained with zombie aqua (live/dead discrimination, Biolegend), anti‐CD45 (to exclude non‐leucocytes) and anti‐CD3 (T‐cell inclusion). MAIT cells were identified by CD161+ and Vα7.2 TCR+ staining. PEB EMCs were plated out at a minimum of 4x106/mL in RPMI (containing 10% FCS, 1% penicillin/streptomycin) and stimulated with PMA (50 ng/ml) and Ionomycin (1 μg/ml) for 3 hrs in the presence of Golgi Plug (BD).
Figure S5: Basal production of IL‐18 in MAIT cells from entheseal tissue. Protein expression of IL‐18 (pg/mL) in unstimulated and LPS stimulated (100ng/mL) EMCs from PEB Mean ± SEM (n = 6). PEB EMCs were plated out at 5x106/mL in RPMI (containing 10% FCS, 1% penicillin/streptomycin) and stimulated with LPS (100ng/mL) for 24 hrs
Figure S6 Comparison of IL‐17A, IL‐17F, IL‐7, IL‐18 serum levels between 9 patients (in light blue) and 31 controls (in red). The p values were calculated using a Mann‐Whitney test. The differences are considered significant for P values <0.05.
Figure S7: Comparison of IL‐17A secretion by CD4 + T, CD8 + T, 𝛾𝛿 T, MAIT and neutrophils between patients and controls depending on cell type. (A) IL‐17A protein expression levels are presented in femtograms per 1000 cells in sorted CD4 + T cells (in orange), MAIT cells (in light blue), CD8 + T cells (in dark blue), 𝛾𝛿 T cells (in purple) and neutrophils (in green) after 18h of stimulation by PMA (50ng / ml) + A23187 (μM) + βglucan (50 μg / ml). The p values were calculated using a Mann‐Whitney test. Horizontal bars indicate the median and standard deviation. The differences are considered significant for P values <0.05. ****, P < 0.0001, ***, P < 0.001, **, P < 0.01, *, P < 0.05, ns; not significant. (B) Diagram showing the distribution of IL17A secretion in patients and controls according to cell type.
Figure S8: Gene expression analysis in MAIT cells from 10 patients and 10 controls. (A) The principal component analysis was performed on the gene expression data of MAIT cells stimulated for 2 hours with PMA (50 ng / ml) + A23187 (5 μM) + βglucan (50 μg / ml). Each dot represents a sample colored according to group of patients (dark blue) or controls (yellow). (B) Heatmap showing the expression of genes differentially expressed by MAITs between patients and controls after 2 h of stimulation with PMA (50 ng / ml) + A23187 (μM) + βglucan (50 μg / ml). In the heatmap, the columns represent the samples belonging either to the patient group in blue (n = 10) or to the control group in yellow (n = 10) and the lines represent the genes differentially expressed between patients and controls, the samples are ordered by hierarchical clustering. T‐test with false discovery rate correction q = 0.29. Yellow indicates high levels of expression and blue indicates low levels of expression. The values for each gene were logarithmically transformed, centered on a mean value of zero, and adjusted to a variance of 1.
ACKNOWLEDGMENT
We thank the members of Cytometry and Biomarkers UTechS from the Institut Pasteur for their technical assistance.
Supported by grants from MSDAVENIR (Project iCARE‐SpA), by a Sanofi Innovation award Europe, by the FOREUM Foundation for Research in Rheumatology, and by the Société Française de Rhumatologie. Dr. Rosine's work was supported by a Poste d'Accueil de l'AP‐HP and by a grant from the Société Française de Rhumatologie. Ms. Rowe's work was supported by investigator‐initiated nonclinical research funding from Novartis UK. Dr. Watad's work was supported by the PARTNER fellowship program. Dr. Cuthbert's work was supported by an investigator‐initiated research grant from Pfizer. Dr. Bridgewood's work was supported by investigator‐initiated nonclinical research funding from Novartis UK. Dr. McGonagle's work was supported by the Leeds NIHR Biomedical Research Centre.
Dr. Rosine, Ms. Rowe, and Dr. Koturan contributed equally to this work. Drs. Rogge, McGonagle, and Miceli‐Richard contributed equally to this work.
Author disclosures are available at https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fart.42090&file=art42090‐sup‐0001‐Disclosureform.pdf.
Contributor Information
Dennis McGonagle, Email: d.g.mcgonagle@leeds.ac.uk.
Corinne Miceli‐Richard, Email: corinne.miceli@aphp.fr.
REFERENCES
- 1. Smith JA, Colbert RA. The interleukin‐23/interleukin‐17 axis in spondyloarthritis pathogenesis: Th17 and beyond. Arthritis Rheumatol 2014;66:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ellinghaus D, Jostins L, Spain SL, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease‐specific patterns at shared loci. Nat Genet 2016;48:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baeten D, Baraliakos X, Braun J, et al. Anti‐interleukin‐17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: a randomised, double‐blind, placebo‐controlled trial. Lancet 2013;382:1705–13. [DOI] [PubMed] [Google Scholar]
- 4. Van der Heijde D, Wei JC, Dougados M, et al. Ixekizumab, an interleukin‐17A antagonist in the treatment of ankylosing spondylitis or radiographic axial spondyloarthritis in patients previously untreated with biological disease‐modifying anti‐rheumatic drugs (COAST‐V): 16‐week results of a phase 3 randomised, double‐blind, active‐controlled and placebo‐controlled trial. Lancet 2018;398:2441–51. [DOI] [PubMed] [Google Scholar]
- 5. Jandus C, Bioley G, Rivals JP, et al. Increased numbers of circulating polyfunctional Th17 memory cells in patients with seronegative spondylarthritides. Arthritis Rheum 2008;58:2307–17. [DOI] [PubMed] [Google Scholar]
- 6. Kenna TJ, Davidson SI, Duan R, et al. Enrichment of circulating interleukin‐17–secreting interleukin‐23 receptor–positive γ/δ T cells in patients with active ankylosing spondylitis. Arthritis Rheum 2012;64:1420–9. [DOI] [PubMed] [Google Scholar]
- 7. Gracey E, Qaiyum Z, Almaghlouth I, et al. IL‐7 primes IL‐17 in mucosal‐associated invariant T (MAIT) cells, which contribute to the Th17‐axis in ankylosing spondylitis. Ann Rheum Dis 2016;75:2124–32. [DOI] [PubMed] [Google Scholar]
- 8. Watad A, Rowe H, Russell T, et al. Normal human enthesis harbours conventional CD4+ and CD8+ T cells with regulatory features and inducible IL‐17A and TNF expression. Ann Rheum Dis 2020;79:1044–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Appel H, Maier R, Wu P, et al. Analysis of IL‐17(+) cells in facet joints of patients with spondyloarthritis suggests that the innate immune pathway might be of greater relevance than the Th17‐mediated adaptive immune response. Arthritis Res Ther 2011;13:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cole S, Murray J, Simpson C, et al. Interleukin (IL)‐12 and IL‐18 synergize to promote MAIT Cell IL‐17A and IL‐17F production independently of IL‐23 signaling. Front Immunol 2020;11:585134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spits H, Artis D, Colonna M, et al. Innate lymphoid cells—a proposal for uniform nomenclature [review]. Nat Rev Immunol 2013;13:145–9. [DOI] [PubMed] [Google Scholar]
- 12. Monteiro M, Almeida CF, Agua‐Doce A, et al. Induced IL‐17‐producing invariant NKT cells require activation in presence of TGF‐β and IL‐1β. J Immunol 2013;190:805–11. [DOI] [PubMed] [Google Scholar]
- 13. Venken K, Jacques P, Mortier C, et al. RORγt inhibition selectively targets IL‐17 producing iNKT and γδ‐T cells enriched in Spondyloarthritis patients. Nat Commun 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cuthbert RJ, Bridgewood C, Watad A, et al. Evidence that tissue resident human enthesis γδT‐cells can produce IL‐17A independently of IL‐23R transcript expression. Ann Rheum Dis 2019;78:1559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baeten D, Østergaard M, Wei JC, et al. Risankizumab, an IL‐23 inhibitor, for ankylosing spondylitis: results of a randomised, double‐blind, placebo‐controlled, proof‐of‐concept, dose‐finding phase 2 study. Ann Rheum Dis 2018;77:1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wendling D, Prati C, Chouk M, et al. Effects of anti‐IL‐23 and anti‐IL‐17: the hidden side of spondyloarthritis polymorphism? [editorial]. Joint Bone Spine 2020;87:5–7. [DOI] [PubMed] [Google Scholar]
- 17. Tang XZ, Jo J, Tan AT, et al. IL‐7 licenses activation of human liver intrasinusoidal mucosal‐associated invariant T cells. J Immunol 2013;190:3142–52. [DOI] [PubMed] [Google Scholar]
- 18. Rudwaleit M, van der Heijde D, Landewé R, et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis 2009;68:777–83. [DOI] [PubMed] [Google Scholar]
- 19. Bridgewood C, Russell T, Weedon H, et al. The novel cytokine Metrnl/IL‐41 is elevated in psoriatic arthritis synovium and inducible from both entheseal and synovial fibroblasts. Clin Immunol 2019;208:108253. [DOI] [PubMed] [Google Scholar]
- 20. Menegatti S, Guillemot V, Latis E, et al. Immune response profiling of patients with spondyloarthritis reveals signalling networks mediating TNF‐blocker function in vivo. Ann Rheum Dis 2020;80:475–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamassia N, Arruda‐Silva F, Calzetti F, et al. A reappraisal on the potential ability of human neutrophils to express and produce IL‐17 family members in vitro: failure to reproducibly detect it. Fron Immunol 2018;9:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor PR, Roy S, Leal SM, et al. Activation of neutrophils by autocrine IL‐17A‐IL‐17RC interactions during fungal infection is regulated by IL‐6, IL‐23, RORγt and dectin‐2. Nat Immunol 2014;15:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klasen C, Meyer A, Wittekind PS, et al. Prostaglandin receptor EP4 expression by Th17 cells is associated with high disease activity in ankylosing spondylitis. Arthritis Res Ther 2019;21:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gracey E, Hromadová D, Lim M, et al. TYK2 inhibition reduces type 3 immunity and modifies disease progression in murine spondyloarthritis. J Clin Invest 2020;130:1863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Papp K, Gordon K, Thaçi D, et al. Phase 2 trial of selective tyrosine kinase 2 inhibition in psoriasis. N Engl J Med 2018;379:1313–21. [DOI] [PubMed] [Google Scholar]
- 26. Ussher JE, Willberg CB, Klenerman P. MAIT cells and viruses. Immunol Cell Biol 2018;96:630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Al‐Mossawi MH, Chen L, Fang H, et al. Unique transcriptome signatures and GM‐CSF expression in lymphocytes from patients with spondyloarthritis. Nat Commun 2017;8:1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leng T, Akther HD, Hackstein CP, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Reports 2019;28:3077–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ciccia F, Guggino G, Rizzo A, et al. Type 3 innate lymphoid cells producing IL‐17 and IL‐22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis 2015;74:1739–47. [DOI] [PubMed] [Google Scholar]
- 30. Rihl M, Kellner H, Kellner W, et al. Identification of interleukin‐7 as a candidate disease mediator in spondylarthritis. Arthritis Rheum 2008;58:3430–5. [DOI] [PubMed] [Google Scholar]
- 31. Papp KA, Merola JF, Gottlieb AB, et al. Dual neutralization of both interleukin 17A and interleukin 17F with bimekizumab in patients with psoriasis: results from BE ABLE 1, a 12‐week randomized, double‐blinded, placebo‐controlled phase 2b trial. J Am Acad Dermatol 2018;79:277–86. [DOI] [PubMed] [Google Scholar]
- 32. Van der Heijde D, Gensler LS, Deodhar A, et al. Dual neutralisation of interleukin‐17A and interleukin‐17F with bimekizumab in patients with active ankylosing spondylitis: results from a 48‐week phase IIb, randomised, double‐blind, placebo‐controlled, dose‐ranging study. Ann Rheum Dis 2020;79:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang S, Laouar A, Denzin LK, et al. Zbtb16 (PLZF) is stably suppressed and not inducible in non‐innate T cells via T cell receptor‐mediated signaling. Sci Rep 2015;5:12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blijdorp IC, Menegatti S, Mens LJ, et al. Expansion of interleukin‐22– and granulocyte–macrophage colony‐stimulating factor–expressing, but not interleukin‐17A–expressing, group 3 innate lymphoid cells in the inflamed joints of patients with spondyloarthritis. Arthritis Rheumatol 2019;71:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclosure Form:
Appendix S1 Supporting Information
Figure S1 Flow cytometry gating strategy 1. Flow cytometry gating strategy for the isolation of MAIT, γδ T‐cells, CD4+ and CD8+ T‐cells. For T cells, PBMCs were isolated from blood using lymphocyte separation medium (Eurobio®). After isolation, PBMCs were labeled with CD3 BUV395 (BD Biosciences®), CD4 VioBright FITC (Milteny Biotec®), CD8 PerCP vio700 (Milteny Biotec®), TCR Vδ PE (Milteny Biotec®), TCR Vδ2 PE (Milteny Biotec®), TCR Vα7.2 APC (BioLegend®), CD161 BV421 (Sony Biotecnology®).
Figure S2: Flow cytometry gating strategy 2. Flow cytometry gating strategy, after CD3 positive magnetic sorting, of MAIT and CD4+ CCR6 + T‐cells. The CD3 positive fraction was labeled with CD3 APC Vio770 (Milteny Biotec®) CD4 VioBright FITC (Milteny Biotec®), Vα7.2 APC (BioLegend®), CD161 BV421 (Sony Biotechnology®) and CCR6 BV786 (BD Biosciences®).
Figure S3: Transcripts levels of CD28 and CLEC7A presented as normalized counts in sorted CD4+ T‐cells (in yellow), MAIT cells (in red), CD8 + T‐cells (in blue), 𝛾𝛿 T‐cells (in purple) and neutrophils (in green) after 2h of stimulation by PMA (50ng/ml) + A23187(μM) + βglucan (50μg/ml).
Figure S4 Flow cytometry gating strategy for phenotypic identification of MAIT cells in entheseal tissues and peripheral blood cells. Doublet excluded EMCs (PEB and EST) or PBMCs were stained with zombie aqua (live/dead discrimination, Biolegend), anti‐CD45 (to exclude non‐leucocytes) and anti‐CD3 (T‐cell inclusion). MAIT cells were identified by CD161+ and Vα7.2 TCR+ staining. PEB EMCs were plated out at a minimum of 4x106/mL in RPMI (containing 10% FCS, 1% penicillin/streptomycin) and stimulated with PMA (50 ng/ml) and Ionomycin (1 μg/ml) for 3 hrs in the presence of Golgi Plug (BD).
Figure S5: Basal production of IL‐18 in MAIT cells from entheseal tissue. Protein expression of IL‐18 (pg/mL) in unstimulated and LPS stimulated (100ng/mL) EMCs from PEB Mean ± SEM (n = 6). PEB EMCs were plated out at 5x106/mL in RPMI (containing 10% FCS, 1% penicillin/streptomycin) and stimulated with LPS (100ng/mL) for 24 hrs
Figure S6 Comparison of IL‐17A, IL‐17F, IL‐7, IL‐18 serum levels between 9 patients (in light blue) and 31 controls (in red). The p values were calculated using a Mann‐Whitney test. The differences are considered significant for P values <0.05.
Figure S7: Comparison of IL‐17A secretion by CD4 + T, CD8 + T, 𝛾𝛿 T, MAIT and neutrophils between patients and controls depending on cell type. (A) IL‐17A protein expression levels are presented in femtograms per 1000 cells in sorted CD4 + T cells (in orange), MAIT cells (in light blue), CD8 + T cells (in dark blue), 𝛾𝛿 T cells (in purple) and neutrophils (in green) after 18h of stimulation by PMA (50ng / ml) + A23187 (μM) + βglucan (50 μg / ml). The p values were calculated using a Mann‐Whitney test. Horizontal bars indicate the median and standard deviation. The differences are considered significant for P values <0.05. ****, P < 0.0001, ***, P < 0.001, **, P < 0.01, *, P < 0.05, ns; not significant. (B) Diagram showing the distribution of IL17A secretion in patients and controls according to cell type.
Figure S8: Gene expression analysis in MAIT cells from 10 patients and 10 controls. (A) The principal component analysis was performed on the gene expression data of MAIT cells stimulated for 2 hours with PMA (50 ng / ml) + A23187 (5 μM) + βglucan (50 μg / ml). Each dot represents a sample colored according to group of patients (dark blue) or controls (yellow). (B) Heatmap showing the expression of genes differentially expressed by MAITs between patients and controls after 2 h of stimulation with PMA (50 ng / ml) + A23187 (μM) + βglucan (50 μg / ml). In the heatmap, the columns represent the samples belonging either to the patient group in blue (n = 10) or to the control group in yellow (n = 10) and the lines represent the genes differentially expressed between patients and controls, the samples are ordered by hierarchical clustering. T‐test with false discovery rate correction q = 0.29. Yellow indicates high levels of expression and blue indicates low levels of expression. The values for each gene were logarithmically transformed, centered on a mean value of zero, and adjusted to a variance of 1.


