Abstract
Tumour recurrence and drug resistance in hepatocellular carcinoma remain challenging. Cancer stem cells (CSCs) are responsible for tumour initiation because of their stemness characteristics. CSCs accounting for drug resistance and tumour relapse are promising therapeutic targets. We report that Abelson interactor 2 (ABI2) is a novel therapeutic target of HCC CSCs. First, ABI2 was upregulated in HCC tissues compared with liver tissues and was associated with tumour size, pathological grade, liver cirrhosis, worse prognosis and a high recurrence rate. Functional studies illustrate that ABI2 knockdown suppresses cell growth, migration, invasion and sorafenib resistance in vitro. Furthermore, ABI2 knockdown inhibited HCC sphere formation and decreased the CD24+, CD133+ and CD326+ CSCs populations, suggesting the suppression of HCC stemness characteristics. A tumour xenograft model and limiting dilution assay demonstrated the inhibition of tumorigenicity and tumour initiation. Moreover, molecular mechanism studies showed that ABI2 recruits and directly interacts with the transcription factor MEOX2, which binds to the KLF4 and NANOG promoter regions to activate their transcription. Furthermore, overexpression of MEOX2 restored HCC malignant behaviour and the CSC population. The ABI2‐mediated transcriptional axis MEOX2/KLF4‐NANOG promotes HCC growth, metastasis and sorafenib resistance by maintaining the CSC population, suggesting that ABI2 is a promising CSC target in HCC treatment.
Keywords: ABI2, CSCs, HCC, recurrence, sorafenib
Abbreviations
- HCC
hepatocellular carcinoma
- CSCs
cancer stem cells
- ABI2
The Abelson interactors 2
- MEOX2
Mesenchyme Homeo Box 2
- ABC
ATP‐binding cassette
- ChIP assay
chromatin immunoprecipitation assay
- NOD‐SCID
non‐obese diabetic severe combined immunodeficiency
- ELDA
extreme limiting dilution analysis
- OS
overall survival
- DFS
disease‐free survival
Lay summary.
Cancer stem cells (CSCs) are responsible for tumour initiation and became promising therapeutic targets. Here, we report that ABI2‐mediated MEOX2/KLF4‐NANOG axis promotes HCC growth, metastasis and sorafenib resistance by maintaining the CSCs population, suggesting that ABI2 is a promising CSCs target in HCC treatment.
1. INTRODUCTION
The burden of liver cancer is increasing rapidly worldwide. In 2020, the incidence rate of liver cancer was ranked sixth and became the third leading cause of cancer death. 1 Hepatocellular carcinoma (HCC) accounts for the largest proportion of primary liver cancer (approximately 75%–85%). The main risk factors relating to HCC include hepatitis virus infection, alcohol‐related hepatic injury, ingestion of aflatoxin‐contaminated food, obesity, smoking and diabetes. 2 The poor prognosis of HCC contributes to its high mortality. However, satisfactory treatment options for HCC are lacking. Surgical treatment is reported to improve survival, but only for early‐stage patients (approximately 5%–15%). For patients with advanced stages, trans‐arterial chemoembolization (TACE) or chemotherapeutic drugs were considered. However, few patients can achieve cancer‐free survival after these treatments due to the high rate of recurrence. Drug (such as sorafenib) resistance and recrudescence are common challenges in advanced HCC. 3 Further research to identify effective treatments and improve survival is necessary.
Cancer stem cells (CSCs) are defined as a subpopulation of tumour cells that maintain intratumor heterogeneity and drive tumour initiation or relapse. 4 CSCs have the characteristics of self‐renewal and tumorigenicity. Evidence shows that CSCs are related to tumour initiation, recurrence and drug resistance. 5 A quiescent state causes CSCs to acquire resistance under clinical treatment. 6 Moreover, CSCs are located far away from tumour blood vessels, so they are not easily targeted by drug agents delivered through nanoparticles. 7 CSCs also express ATP‐binding cassette (ABC) transporters by unidirectional intracellular pumps, causing tumours to resist high concentrations of chemotherapeutic drugs by increasing drug efflux. Therefore, the cellular drug concentration is not sufficient to eliminate cancer cells. 8 , 9 Moreover, overexpression of antiapoptotic genes in CSCs results in the failure of cells to enter the apoptotic process. 10 The high expression level of free radical scavengers in CSCs leads to a decrease in reactive oxygen species in cells after radiotherapy, which results in radiation resistance of tumour cells. 11 , 12 Unregulated DNA repair is another possible reason for treatment resistance in CSCs. 9 Therefore, exploring the regulatory mechanism of CSCs can help to find effective methods for tumour treatment.
Abelson interactors (ABIs) are a group of proteins that act as the substrate of the eponymous Abelson non‐receptor tyrosine kinase (c‐ABL). The ABI proteins are activated through the phosphorylation of the Abl kinase. 13 ABI exist in multiple forms including ABI1 and ABI2, which are predominant in mammalian cells. 14 Previous study proved that the expression of ABI1 mainly distributed in the central nervous system. 15 Steinestel and his colleagues reported that ABI1 plays an important role in synaptic maturation and cytoskeletal dynamic. 16 ABI2 is almost ubiquitously expressed. CRISPR screening uncovered that ABI2 is a key determinant of multiple drug resistance in pancreatic cancer cells. 17 Kyoung Lim's research demonstrated that the Doxo‐induced HCC cell senescence can be attenuated by TIS21 through inhibiting linear actin nucleation, which is regulated by Nox4‐ROS‐ABI2‐DRF signal cascade. 18 These studies indicated that ABI2 might play a crucial role in drug resistance and relapse of tumour. However, the potential functions and underlying mechanisms of ABI2 in malignant tumours, especially HCC, are largely unclear.
In our current study, we reported that ABI2 can promote the phenotype of HCC stem cells and induce the recurrence of HCC, which is mediated by interacting with the transcription factor MEOX2.
2. MATERIALS AND METHODS
2.1. HCC specimens
The paraffin‐embedded HCC tissue microarray was obtained from Shanghai Outdo Biotech Company (Shanghai, China). Written informed consent and clinical characteristic statistics of all patients were obtained. All the protocols have been complied with the Helsinki Declaration and approved by the Medical Ethics Committee of Shanghai Outdo Biotech Company and the Medical Ethics Committee of the Fourth Affiliated Hospital, Nanchang University.
2.2. Cell culture and treatment
The human HCC cell lines PLC/PRF/5 and Huh7 were purchased from the Chinese Academy of Sciences, Shanghai Branch. Cells were cultured in Dulbecco's modified Eagle medium (Thermo Fisher Scientific) supplemented with 10% foetal bovine serum (FBS; Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma–Aldrich) and maintained in an incubator at 37°C with 5% CO2. HCC cells in the ABI2 knockdown and control groups were treated with 5, 7 and 10 μm sorafenib. Then, live real‐time IncuCyte ZOOM was used to evaluate cell proliferation as described. 19 For foci formation assay, 1000 HCC cells were seeded on 6‐well plates and cultured for 10 days. Colonies were further stained and counted with 1% crystal violet.
2.3. Establishment of overexpression and knockdown HCC cells
To establish MEOX2‐overexpressing cells, full‐length human MEOX2 cDNA was cloned into pCDH‐CMV‐MCS‐EF1‐Puro. To obtain ABI2 knockdown cells, short hairpin RNA specifically targeting ABI2 was cloned into the pLKO vector (Addgene). Puromycin (Sigma–Aldrich) was used to select stably transduced cells.
2.4. Flow cytometry
APC‐conjugated CD133 antibody was purchased from Miltenyi (cat. no. 130‐113‐184). PE‐conjugated CD24 antibody was obtained from BD Biosciences (cat. no. 555428). FITC‐conjugated CD326 antibody was purchased from STEMCELL (cat. no. 60136FI). Flow cytometry was performed according to the manufacturer's instructions.
2.5. ChIP assay
The ChIP assay was conducted as previously described. 20 The primers were purchased from Sangon Biotech (Shanghai), and the primer sequences of KLF4 and NANOG are shown in Table S1.
2.6. Immunoprecipitation
An immunoprecipitation kit (ab206996) was used to confirm the interaction between ABI2 and MEOX2 according to the instructions. There are four steps in the process. The first step is antibody binding, followed by bead preparation. After bead capture, the target proteins are eluted. Western blot was used to examine the target proteins.
2.7. Cell invasion and sphere formation assays
Cell invasion was evaluated by Transwell assay. Firstly, cells were pretreated with 0.5 μg/ml mitomycin C for 24 h. 200 μl of DMEM containing 1 × 105 cells was instilled into the upper chamber, and then 500 ml of DMEM containing 20% FBS was instilled into the lower chamber and maintained at 37°C for 48 h. Cells were fixed with paraformaldehyde for 10 min, and then PBS was used to wipe and rinse the stroma and cells that failed to cross the membrane. Finally, 0.1% crystal violet stain solution was used to stain the cells for 10 min, and the cells were observed with a microscope. For the foci formation assay, firstly, the cell suspension was prepared with complete DMEM medium. Then one thousand cells from different groups were seeded into a six‐well plate and cultured for 2–3 weeks. Finally, the cells were fixed with 4% paraformaldehyde for 20 min followed by staining with 0.5% crystal violet at room temperature for 10 min. The colonies containing a cell count of 50 or more per dish were counted. For the sphere formation assays, the collected cells were dispersed into a single cell suspension with DMEM supplemented with bFGF (20 ng/ml), EGF (20 ng/ml), 1x B27, and 1x N2. Subsequently, ultralow attachment 12‐well plates were used to culture these cells at a density of 1000 cells per well. 21
2.8. RNA extraction and quantitative RT–PCR
TRIzol (Life Technologies) was used to extract the total RNA from cultured cells according to the manufacturer's instructions. To investigate the relative gene expression levels, SYBR Green (TaKaRa, Japan) was utilized on a BIO‐RAD Real‐Time PCR system. All primer sequences shown in Table 1 were supplied by Sangon Biotech. All experiments were conducted in triplicate.
TABLE 1.
Correlation between the clinicopathologic variables and ABI2 in HCC
| Variables | Total | ABI2 expression | p value | |
|---|---|---|---|---|
| Low expression | High expression | |||
| Age (years) | ||||
| ≤50 | 40 (44.4%) | 20 (50.0%) | 20 (50.0%) | .254 |
| >50 | 50 (55.6%) | 19 (38.0%) | 31 (62.0%) | |
| Gender | ||||
| Male | 10 (11.1%) | 5 (50.0%) | 5 (50.0%) | .652 |
| Female | 80 (88.9%) | 34 (42.5%) | 46 (57.5%) | |
| Pathological grades | ||||
| I | 3 (3.3%) | 3 (100.0%) | 0 (0%) | .005* |
| II | 54 (60%) | 28 (51.9%) | 26 (48.1%) | |
| III | 33 (36.7%) | 8 (24.2%) | 25(75.8%) | |
| AJCC grades | ||||
| I | 63 (70%) | 30 (47.6%) | 33 (52.4%) | .404 |
| II | 25 (27.8%) | 8 (32.0%) | 17 (68.0%) | |
| III | 2 (2.2%) | 1 (50.0%) | 1 (50.0%) | |
| Tumour size, cm | ||||
| ≤5 cm | 55 (61.1%) | 30 (54.5%) | 25 (45.5%) | .007* |
| >5 cm | 35 (38.9%) | 9 (25.7%) | 26 (74.3%) | |
| Cirrhosis | ||||
| No | 9 (10%) | 8 (88.9%) | 1 (11.1%) | .004* |
| Yes | 81 (90%) | 31 (38.3%) | 50 (61.7%) | |
| Cirrhosis nodule | ||||
| Solitary | 9 (10%) | 4 (44.4%) | 5 (55.6%) | .943 |
| Multiple | 81 (90%) | 35 (43.2%) | 46 (56.8%) | |
| Tumour number | ||||
| Solitary | 79 (87.8%) | 36 (45.6%) | 43 (54.4%) | .251 |
| Multiple | 11 (12.2%) | 3 (27.3%) | 8 (72.7%) | |
| Encapsulation | ||||
| Complete | 42 (41%) | 23 (54.8%) | 19 (45.2%) | .041* |
| None | 48 (59%) | 16 (33.3%) | 32 (66.7%) | |
| Tumour recurrence | ||||
| No | 41 (45.6%) | 22 (53.7%) | 19 (46.3%) | .071 |
| Yes | 49 (54.4%) | 17 (34.7%) | 32 (65.3%) | |
| ALT | ||||
| Normal | 53 (58.9%) | 21 (39.6%) | 32 (60.4%) | .395 |
| Increase | 37 (41.1%) | 18 (48.6%) | 19 (51.4%) | |
| TB | ||||
| Normal | 76 (84.4%) | 32 (42.1%) | 44 (57.9%) | .584 |
| Increase | 14 (15.6%) | 7 (50.0%) | 7 (50.0%) | |
| AFP | ||||
| <20 | 36 (40%) | 17 (47.2%) | 19 (52.8%) | .543 |
| ≥20 | 54 (60%) | 22 (40.7%) | 32 (59.3%) | |
| HBsAg | ||||
| Negative | 19 (21.1%) | 7 (36.8%) | 12 (63.2%) | .520 |
| Positive | 71 (78.9%) | 32 (45.1%) | 39 (54.9%) | |
| HBcAb | ||||
| Negative | 7 (7.8%) | 3 (42.9%) | 4 (57.1%) | .979 |
| Positive | 83 (92.2%) | 36 (43.4%) | 47 (56.6%) | |
| GGT | ||||
| Normal | 44 (48.9%) | 19 (43.2%) | 25 (56.8%) | .977 |
| Decrease | 46 (51.1%) | 20 (43.5%) | 26(56.5%) | |
Abbreviations: ALT, Alanine aminotransferase; GGT, γ‐glutamyl transpeptidase; HBcAb, hepatitis B core antigen AFP, alpha‐fetoprotein; HBsAg, hepatitis B surface antigen; TB, total bilirubin. Bold values and * represent p < .05.
2.9. Western blotting
RIPA buffer was used to extract proteins. Then, western blotting was performed as previously described. 22 The protein bands were evaluated using ChemiDoc MP (Bio–Rad). The primary antibodies were purchased from Abcam.
2.10. Animal experiments
Male nonobese diabetic severe combined immunodeficiency (NOD‐SCID) mice aged 4–6 weeks were obtained from Shanghai Model Organisms Center. All experiments were conducted in line with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Ethics Committee of the Fourth Affiliated Hospital of Nanchang University. For the tumour formation assay, NOD‐SCID mice were randomly divided into three groups (control, ABI2 knockdown and ABI2 knockdown MEOX2 overexpression groups). Each group was treated with two different concentrations of cells (50 000 and 20 000). HCC cells were injected subcutaneously into the mouse flanks. The monitoring of tumour formation was performed as previously described. 23 , 24 The terminal of monitoring was 12 weeks after injection. A negative result was considered if no tumour had formed at the injection site.
Five‐week‐old male BALB/c nude mice were purchased from the Shanghai Model Organisms Center. HCC cells (PLC/PRF/5) with ABI2 knockdown and MEOX2 overexpression in 100 μl of phosphate‐buffered saline (PBS) were injected into nude mice. Tumour volumes were calculated as volume (mm3) = L × W 2 × 0.5.
2.11. Immunohistochemical staining
The tissue sections were incubated with ABI2 antibody (Abcam) at 4°C for 8 h. Normal goat serum was used to replace the primary antibody in the negative control group. The immunohistochemically stained tissue sections were scored by two independent and experienced pathologists, and the biochemical and clinical patient information was not provided. The scoring method was conducted as described. 25 HCC specimens were assigned an H‐score method, which multiplies the score corresponding to the percentage of tumour cells stained by the score corresponding to the strength of staining (from 0 for negative staining, 1 for weak, 2 for moderate and 3 for strong staining). The median value in 90 patients served as a cut‐off to categorize all cases as high or low ABI2 expression.
2.12. Statistical analysis
Statistical data were analysed using SPSS version 13.0 (Chicago, IL, USA). Overall survival (OS) and disease‐free survival (DFS) curves were plotted by Kaplan–Meier survival analysis and log‐rank test. Student's t test and ANOVA were used to evaluate the differences amongst groups. Extreme limiting dilution analysis (ELDA) was performed in software (http://bioinf.wehi.edu.au/software/elda/). Pearson chi‐square test, univariate and multivariate analyses were used for the categorical variables. We considered differences to be statistically significant when the p values were <.05.
3. RESULTS
3.1. ABI2 overexpression is associated with poor prognosis in HCC patients
An analysis in genome‐wide CRISPR activation library screening for Temozolomide resistant shown that ABI2 ranked 15th of 308 hits, implying that expression of ABI2 may play a central role in drug resistance of tumour (Figure 1A). Moreover, by analysing TCGA data using UALCAN, we identified that the mRNA expression level of ABI2 was higher and that the promoter methylation level of ABI2 was significantly lower in HCC tissues than in normal tissues (Figure 1B,C). Survival analysis suggested high expression of ABI2 was correlated with poor overall survival and disease‐free survival in HCC patients (Figure 1D–F). Furthermore, the ABI2 expression level was upregulated in stages II and III compared with stage I, respectively (Figure 1G). These results suggest that ABI2 may play a role in HCC progression. Meanwhile, we further utilized the Oncomine database to explore the expression of ABI2 in HCC tissues and normal tissues. As expected, the ABI2 expression level was significantly increased in HCC tissues compared with normal tissues (Figure 1H–K). Subsequently, we analysed the association between the ABI2 protein expression level and clinicopathological features in an HCC tissue microarray. Figure 1L displays representative graph of weak, moderate and strong staining in HCC tissues. We evaluated ABI2 expression levels by H‐score for further analyses. Kaplan–Meier survival curves demonstrated that high expression of ABI2 was correlated with worse overall survival and higher recurrence rate in HCC patients (Figure 1M,N). Moreover, ABI2 overexpression was positively correlated with tumour size, pathological grade, liver cirrhosis and tumour encapsulation in chi‐square analysis (Table 1). In univariate and multivariate analyses, ABI2 overexpression significantly correlated with tumour size, pathological grade and liver cirrhosis (Table 2). Taken together, these findings indicated that ABI2 was highly expressed in HCC and that aberrant expression of ABI2 was associated with worse prognosis in HCC patients.
FIGURE 1.
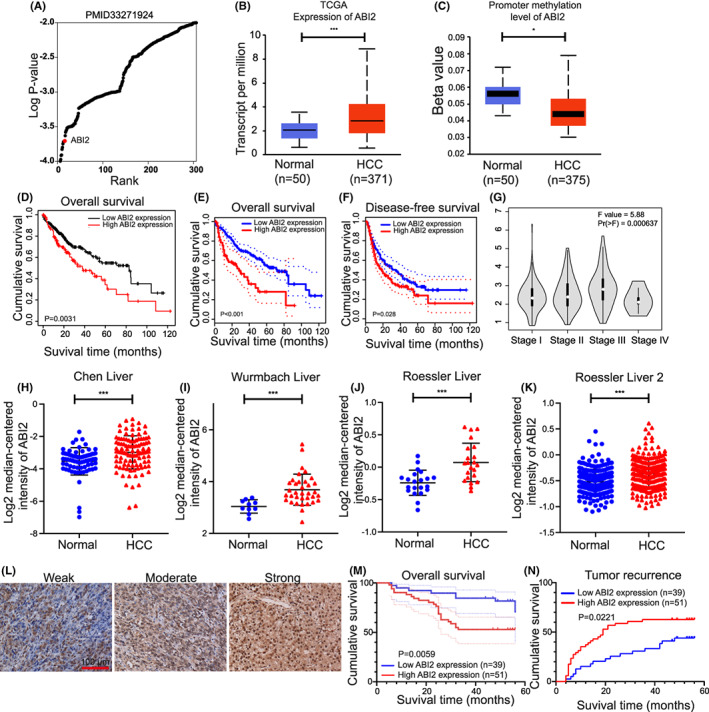
ABI2 overexpression predicts the poor survival of HCC patients. (A) ABI2 was the top‐ranking gene from genome‐wide CRISPR activation library screening for Temozolomide resistant. (B, C) The expressions level and promoter methylation level of ABI2 in HCC tumour and normal liver tissues were analysed based on TCGA database (GEPIA). Data are presented as the mean ± SD (*p < .05, **p < .01, ***p < .001). (D–F) Survival curves of patients with HCC showed that ABI2 expression level was negatively correlated with prognosis prediction analysed by Kaplan–Meier Plotter and GEPIA. (G) ABI2 expression in different stages of HCC patients. (H–K) The expressions of ABI2 in HCC tumour and normal liver tissues were analysed by oncomine. Data are presented as the mean ± SD (*p < .05, **p < .01, ***p < .001). (L) The expressions of ABI2 in HCC liver tissues were tested by IHC staining. Scale bars, 100 μm. (M) Kaplan–Meier survival curves showed that ABI2 expression level was negatively correlated with prognosis prediction of HCCs analysed by IHC in HCC tissue. (N) ABI2 overexpression was positively correlated with tumour recurrence.
TABLE 2.
Univariate and multivariate analyses of clinicopathologic variables and ABI2 in HCC
| Variables |
ABI2 univariate p value |
ABI2 multivariate | |
|---|---|---|---|
| 95%CI | p value | ||
| Age (≤50/>50 years) | .255 | ||
| Gender (male/female) | .653 | ||
| Pathological grades | .003* | 1.646–11.701 | .003* |
| ajcc grades | .290 | ||
| Tumour size (≤5/>5 cm) | .008* | 1.245–9.852 | .018* |
| Cirrhosis (no/yes) | .018* | 1.007–84.184 | .049* |
| Cirrhosis nodule (solitary/multiple) | .943 | ||
| Tumour number (solitary/multiple) | .260 | ||
| Encapsulation (complete/none) | .042* | ||
| Tumour recurrence (no/yes) | .072 | ||
| ALT (normal/increase) | .386 | ||
| TB (normal/increase) | .585 | ||
| AFP (<20/≥20) | .544 | ||
| HBsAg (negative/positive) | .521 | ||
| HBcAb (negative/positive) | .979 | ||
| GGT (normal/decrease) | .977 | ||
Abbreviations: ALT, Alanine aminotransferase; CI, confidence interval; GGT, γ‐glutamyl transpeptidase; HBcAb, hepatitis B core antigen AFP, alpha‐fetoprotein; HBsAg, hepatitis B surface antigen; TB, total bilirubin. Bold values and * represent p < 0.05.
3.2. Knockdown of ABI2 inhibits HCC cell growth, sorafenib resistance, migration and invasion
Considering that ABI2 was overexpressed in HCC and may act as a potential oncogene in HCC, we tried to clarify the effect of ABI2 on HCC cell malignant biological behaviours through ABI2 knockdown. Two cell lines (PLC/PRF/5 and Huh 7) were stably transfected with lentiviral sh‐ABI2 and scramble. qRT–PCR and western blot verified that shRNA against ABI2 significantly knocked down the mRNA and protein expression levels of ABI2 in PLC/PRF/5 and Huh7 cells (Figure 2A,B). Additionally, 2D cell growth assays showed that knockdown of ABI2 significantly suppressed HCC cell proliferation (Figure 2C,E). Consistently, ABI2 knocking down supressed HCC cell growth in foci formation assays (Figure 2G,H). Furthermore, transwell assays demonstrated that downregulated ABI2 expression significantly inhibit tumour migration and invasion in PLC/PRF/5 and Huh7 cells (Figure 2I,J). Studies suggested drug resistance play a part in tumour recurrence and our results suggested that high ABI2 expression was correlated with HCC recurrence. We further cultured HCC cells treated with sorafenib at different concentrations as previous studies suggested (10, 7 and 5 μM). 26 The scramble groups exhibited relatively higher tolerance to sorafenib than the sh‐ABI2 groups, which strongly suppressed the proliferation rate (Figure 2D,F). Therefore, these data indicated that inhibition of ABI2 impeded HCC cell growth, sorafenib resistance, migration and invasion.
FIGURE 2.
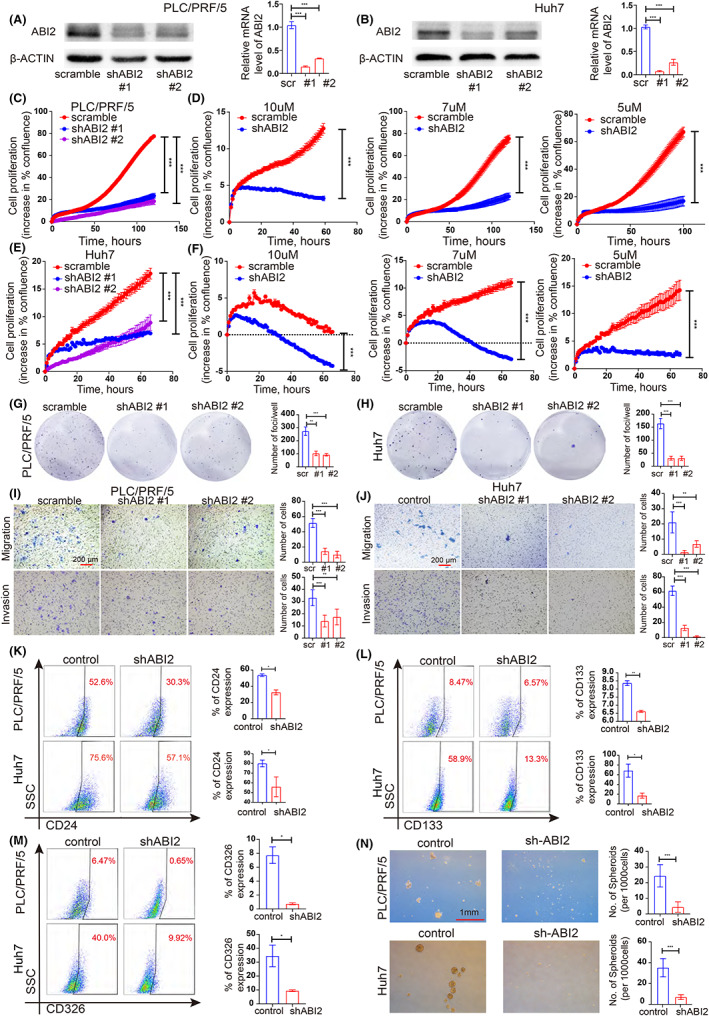
Knockdown of ABI2 inhibits HCC cell growth, sorafenib resistance, migration, invasion and cancer stem cells expansion. (A, B) shRNA against ABI2 significantly down‐regulated the expression of ABI2 in PLC/PRF/5 and Huh7 cells detected with western blotting and Quantitative RT‐PCR. Scrambled shRNA was used as negative control. (C, E) Summary graph showing the HCC cell 2D growth rate. (D, F) Summary graph showing the PLC/PRF/5 and Huh7 cells 2D growth rate in 10, 7 and 5μM sorafenib. (G, H) Foci formation assays were performed in HCC cell lines to evaluate cell proliferation ability. (I, J) Down‐regulated ABI2 expression could inhibit cell migration and invasion in PLC/PRF/5 and Huh7 cells. (K–M) Flow cytometric analysis of CD24+, CD133+ and CD326+ cell properties in PLC/PRF/5 and Huh7 cells. (N) Sphere formation assays were performed to investigate HCC cancer stem cell properties in PLC/PRF/5 and Huh7 cells. In all panels, data are presented as the mean ± SD. (*p < .05, **p < .01, ***p < .001, ns: no statistical significance).
3.3. ABI2 promotes the stemness characteristics of HCC
Accumulating evidence indicates that HCC CSCs are responsible for HCC initiation, recurrence and drug resistance. We considered ABI2 influence HCC CSC properties to regulate cell growth, sorafenib resistance, migration and invasion, leading to tumour recurrence and drug resistance. Flow cytometry proved that knockdown of ABI2 significantly suppressed the populations of CD24+, CD133+ and CD326+ CSCs in PLC/PRF/5 and Huh7 cells (Figure 2K–M). Sphere formation assays indicated fewer and smaller spheroids in the sh‐ABI2 group than in the control group (Figure 2N). Collectively, these results demonstrated that ABI2 played a key role in maintaining the CSCs population.
3.4. ABI2 recruits the transcription factor MEOX2 to promote HCC progression
To further study the potential mechanism by which ABI2 triggers the progression of HCC, we investigated the proteins which were directly regulated by ABI2 using the IntAct database. Several ABI2‐interacting proteins were obtained from the IntAct database (Figure 3A). Interestingly, a previous study showed that the transcription factor MEOX2 was associated with poor survival and cellular chemoresistance to oncologic drugs in non‐small‐cell lung cancer patients, 27 suggesting that MEOX2 may play a role in drug resistance. Through analysing correlation from TCGA data using the GEPIA web server, we identified that MEOX2 was positively correlated with ABI2 in HCC patients (Figure 3B). Knockdown of ABI2 significantly down‐regulated the expression of MEOX2 in PLC/PRF/5 and Huh7 cells (Figure 3C,D). Moreover, the GEPIA online tool revealed that high expression of MEOX2 was associated with poor overall survival in HCC patients (Figure 3E). In addition, the Oncomine database identified that MEOX2 was significantly overexpressed in HCC tissues compared with normal liver tissues (Figure 3F–I). Furthermore, we tried to verify whether MEOX2 interacted with ABI2 in HCC cells. Coimmunoprecipitation showed that ABI2 can directly bind to MEOX2 in PLC/PRF/5 and Huh7 cells (Figure 3J,K). Collectively, these results demonstrated that ABI2 recruits the transcription factor MEOX2 to promote HCC progression.
FIGURE 3.
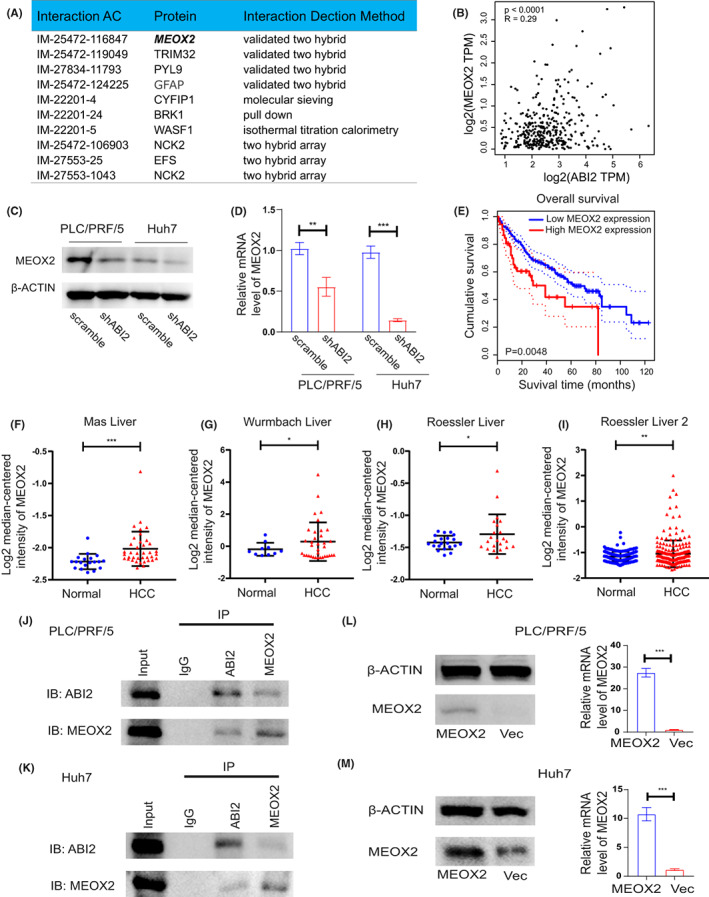
ABI2 interacts with MEOX2 to promote HCC progression. (A) The datasheet showed ABI2 interacting proteins obtained from IntAct database. (B) Correlation analysis between the expressions of ABI2 and MEOX2 in HCC using GEPIA web server. (C, D) Western blot and quantitative RT‐PCR analysis showed that ABI2 knocking down suppressed MEOX2 expression level in HCC cell lines. (E) Survival curves of patients with HCC showed that MEOX2 expression level was negatively correlated with prognosis prediction analysed by Kaplan–Meier Plotter. (F–I) The expressions of MEOX2 in HCC tumour and normal liver tissues were analysed by oncomine. (J, K) Co‐immunoprecipitation showed that ABI2 directly bound to transcription factor MEOX2 in PLC/PRF/5 and Huh7 cells. (L, M) Western blot and quantitative RT‐PCR analysis showing expression of MEOX2 in MEOX2 transfected PLC/PRF/5 and Huh7 cells. In all panels, data are presented as the mean ± SD. (*p < .05, **p < .01, ***p < .001, ns: no statistical significance).
3.5. ABI2 enhances CSC properties through the MEOX2/KLF4‐Nanog axis
Given the tumour‐promoting effects of ABI2 and MEOX2, we tried to characterize the underlying molecular mechanism of ABI2‐MEOX2. First, we transfected the MEOX2 plasmid and control plasmid (vector) into HCC cells and further confirmed MEOX2 expression (Figure 3L,M). Previous studies revealed that KLF4 and Nanog played important roles in regulating CSC properties. Through Danalysing correlation analyses from TCGA data using the GEPIA web server, we identified that KLF4 and Nanog were positively correlated with ABI2 and MEOX2 in HCC patients (Figure 4A–D). Additionally, MEOX2 ChIP‐seq analysis using Cistrome Data Browser indicated that MEOX2 was enriched in the promoters of KLF4 and Nanog in human colon cells (Figure 4E–F). ChIP‐qPCR showed that MEOX2 bound to the promoters of KLF4 and Nanog in PLC/PRF/5 and Huh7 cells (Figure 4G–J). Furtherly, luciferase assay verified that the MEOX2 recruited by ABI2 promoted KLF4 and Nanog expression in PLC/PRF/5 and Huh7 cells (Figure 4K–N). Subsequently, we studied whether ABI2 regulated HCC CSC properties through the MEOX2/KLF4‐Nanog axis. We found that knockdown of ABI2 significantly inhibited the mRNA levels of KLF4 and Nanog, which could be partly reversed by MEOX2 overexpression in PLC/PRF/5 and Huh7 cells (Figure 4O–R). Cell growth assays and Transwell assays showed that silencing ABI2 significantly inhibited cell proliferation migration and invasion in PLC/PRF/5 and Huh7 cells, which could be partly mitigated by MEOX2 overexpression (Figure 5A–F). Similarly, flow cytometry and sphere formation assays revealed that inhibition of ABI2 suppressed the CD24+, CD133+ and CD326+ CSC populations and the formation volume in PLC/PRF/5 and Huh7 cells, which could be partly relieved by MEOX2 overexpression (Figure 5G–K). Collectively, these findings indicated that ABI2 promotes HCC CSC properties through the MEOX2/KLF4‐Nanog axis.
FIGURE 4.
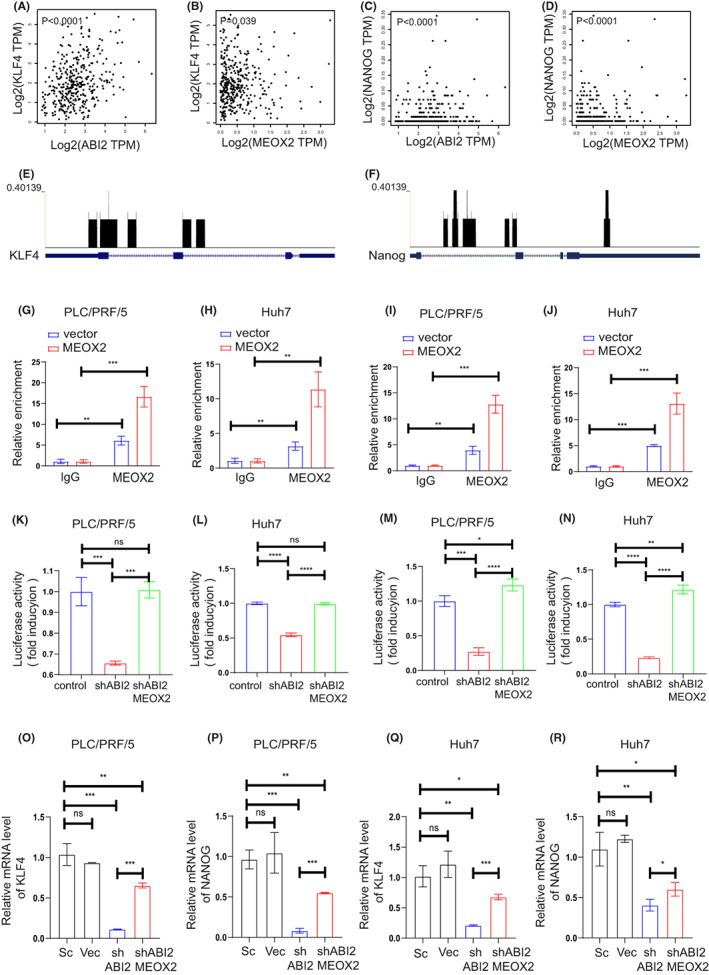
KLF4 And Nanog are key performers in ABI2 promoting HCC cancer stem cell by transcription factor MEOX2. (A–D) Correlation analysis between the expressions of ABI2, MEOX2, KLF4 and Nanog in HCC using GEPIA web server. (E, F) MEOX2 CHIP‐seq analysis in human colon cells using Cistrome Data Browser. (G, H) MEOX2 binding to the promoters of KLF4 detected by ChIP‐qPCR in PLC/PRF/5 and Huh7 cells. (I, J) MEOX2 binding to the promoters of Nanog detected by ChIP‐qPCR in PLC/PRF/5 and Huh7 cells. (K, L) PLC/PRF/5 and Huh7 cells were transfected with KLF4 promoter luciferase reporter or co‐transfected with KLF4 promoter luciferase reporter and MEOX2 plasmids followed by analysis of luciferase activity. (M, N) PLC/PRF/5 and Huh7 cells were transfected with Nanog promoter luciferase reporter or co‐transfected with Nanog promoter luciferase reporter and MEOX2 plasmids followed by analysis of luciferase activity. (O–R) Quantitative RT‐PCR assessment of KLF4 and Nanog expression in PLC/PRF/5 and Huh7 cells. Transcription levels were normalized to those of β‐Actin. In all panels, data are presented as the mean ± SD. (*p < .05, **p < .01, ***p < .001, ns: no statistical significance).
FIGURE 5.
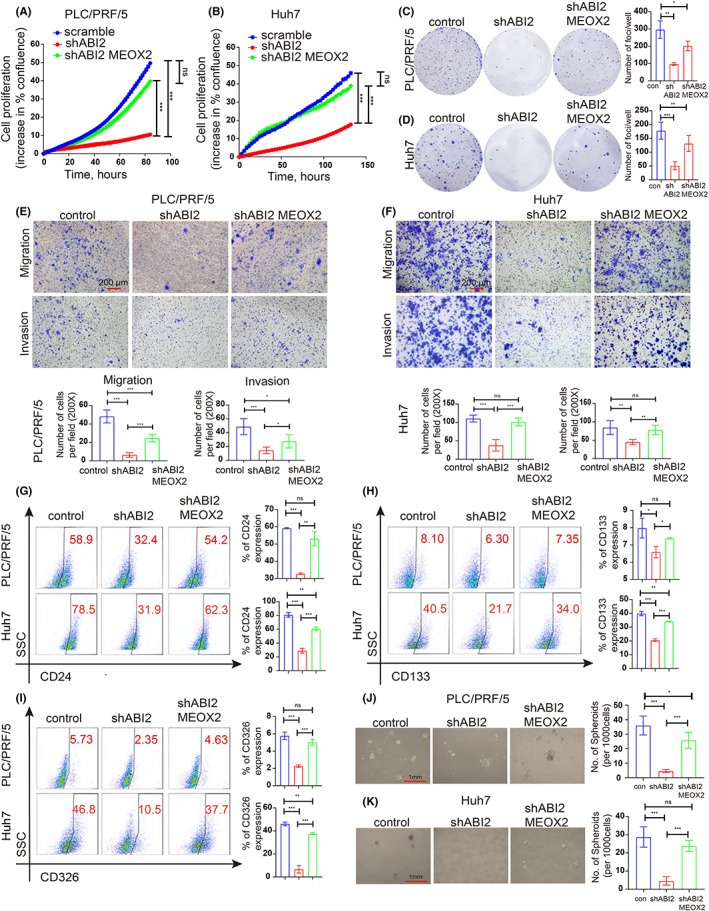
Knockdown of ABI2 inhibits HCC malignant behaviour and cancer stem cell properties, which can be rescued by MEOX2 overexpression. (A, B) Summary graph showing the HCC cell 2D growth rate. (C, D) Representative images of foci formation. (E, F) Transwell assay of HCC cells in PLC/PRF/5 and Huh7 cells. (G, I). Flow cytometric analysis of CD24+, CD133+ and CD326+ cancer stem cell properties in PLC/PRF/5 and Huh7 cells. (J, K). Sphere formation assays were performed to investigate HCC cancer stem cell properties in PLC/PRF/5 and Huh7 cells. In all panels, data are presented as the mean ± SD. (*p < .05, **p < .01, ***p < .001, ns: no statistical significance).
3.6. ABI2 contributes to tumour growth, initiation and self‐renewal potential in vivo
The above results suggested that ABI2 regulated HCC proliferation, invasion, migration and CSC properties in vitro. We further investigated whether ABI2 contributed to tumour growth, initiation and self‐renewal potential through MEOX2 in vivo. Xenografted tumour results showed that treatment with sh‐ABI2 resulted in significant reductions in tumour weight and tumour volume compared with the control group, which could be partly rescued by MEOX2 overexpression (Figure 6A–C). An in vivo LDA (limiting dilution assay) was performed using HCC cells to examine whether ABI2 mediates MEOX2 to promote the CSC population and enhance HCC tumorigenicity. As expected, ABI2 knockdown suppressed tumorigenicity and decreased the CSC frequency of HCC cells, improving mouse disease‐free survival. Moreover, MEOX2 overexpression restored the CSC population, proving that ABI2 regulates MEOX2 to maintain CSC tumorigenicity (Figure 6D–H). Taken together, ABI2 promotes the CSC population, improving HCC proliferation and tumorigenicity through MEOX2.
FIGURE 6.
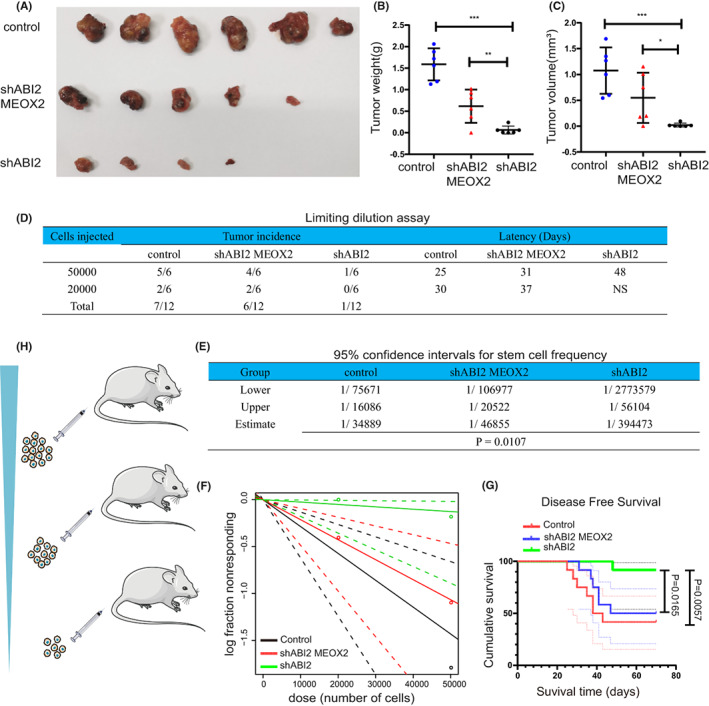
ABI2 contribute to tumour growth, initiation and self‐renewal potential in vivo. (A) Representative xenograft tumours derived from PLC/PRF/5 control, PLC/PRF/5‐shABI2 and shABI2 MEOX2. (B, C) Tumour weight and tumour volume of xenograft tumour. (D–F) Cells were inoculated subcutaneously in NOD‐SCID mice. The cancer stem cell frequency was calculated by Extreme Limiting Dilution Analysis (ELDA). (G) The survival curves of mice. In all panels, data are presented as the mean ± SD. (*p < .05, **p < .01, ***p < .001, ns: no statistical significance).
4. DISCUSSION
In clinical practice, HCC recurrence and drug resistance remain challenging. The CSC theory illustrates that a small population of tumour cells called CSCs is responsible for tumour initiation because of their stemness characteristics. These populations (CSCs) account for worse clinical phenomena, such as drug resistance, tumour relapse, radiation therapy resistance and tumour dormancy and metastasis. 28 , 29 However, we know little about how CSCs maintain stemness characteristics to initiate tumours. Therefore, uncovering a promising CSC target to suppress tumour stemness characteristics may be an effective therapeutic strategy.
Regardless of the important roles of ABI2 in cell pathophysiology, the potential functions of ABI2 in malignant tumours remain to be identified. In this study, the analysis in genome‐wide CRISPR activation library screening for Temozolomide resistant shown that the expression of ABI2 may play a key role in drug resistance of tumour. Furthermore, we found that ABI2 was highly expressed in HCC and that ABI2 overexpression was positively correlated with HCC recurrence, tumour size, pathological grade and poor prognosis in HCC patients. Furtherly, knockdown of ABI2 inhibited HCC cell growth, sorafenib resistance, migration and invasion by repressing CSC self‐renewal and tumorigenicity. Therefore, our findings revealed that ABI2 exerted its pro‐proliferation and pro‐tumorigenicity effects by inducing stemness characteristics in HCC.
Through searching the IntAct database and analysing the correlation from TCGA data, we found that Mesenchyme Homeo Box 2 (MEOX2) was an important mediator of ABI2 in HCC patients. As one of the homeobox gene families, MEOX2 is regarded as a growth arrest‐specific homeobox that activates the cyclin‐dependent kinase inhibitors p21 and p16. An increasing number of studies have indicated that MEOX2 participates in the development and progression of several malignant tumours. Wang et al. found that MEOX2 could promote macrophage infiltration through CSF‐1/CSF‐1R signalling in oesophageal squamous cell carcinoma and other kinds of digestive system carcinomas. 30 Additionally, Tachon and colleagues showed that MEOX2 was upregulated in glioblastoma and that high expression of MEOX2 was associated with poor survival in patients with gliomas, which suggested that MEOX2 may serve as an interesting prognostic biomarker, especially for lower grade glioma. 31 However, Tian and coworkers found that the expression of MEOX2 was lower in laryngeal carcinoma tissues than in control tissues. Furthermore, they revealed that overexpression of MEOX2 facilitated laryngeal cancer cell apoptosis by suppressing the PI3K/Akt pathway, which suggested that MEOX2 may be a significant suppressor gene in laryngeal carcinoma. 32 Importantly, our analysis from the TCGA and Oncomine database revealed that MEOX2 was significantly overexpressed in HCC tissues and high expression of MEOX2 was associated with low overall survival in HCC patients. Irrespective of the dual role of MEOX2 in the pathogenesis of malignant tumours, the potential functions and mechanisms of MEOX2 in HCC remain largely unclear. In this study, we identified that ABI2 was directly bound to MEOX2 by coimmunoprecipitation. ABI2 promoted MEOX2 expression and MEOX2 overexpression effectively restore sh‐ABI2 suppression effects in tumour proliferation, migration, invasion and CSCs expansion. These results revealed that ABI2 mediated CSCs promoting effects by recruiting MEOX2. However, sh‐ABI2 suppression effects in tumour cells were partially mitigated by MEOX2 overexpression, which suggested there are some other factors interacted with MEOX2 and promoted tumour malignant effects, which remained to be investigated.
KLF4, a group of Spl/Kruppel‐like zinc finger‐containing transcription factors, participates in regulating several important cellular pathophysiological processes. Additionally, accumulating studies have shown that KLF4 serves as an oncogene in several cancers, including breast cancer, 33 osteosarcoma 34 and colorectal cancer. 35 These studies also indicated that KLF4 promoted cancer cell proliferation, migration and invasion by maintaining cancer stem cell‐like features. Similarly, Nanog was also reported to be an important transcription factor associated with the development of tumour cells, drug resistance, migration and stemness. 36 Further studies indicated that Nanog promoted CSC development via multiple pathways, including angiogenesis and reduced E‐cadherin expression levels. 37 For instance, Yuan and coworkers found that overexpression of Nanog significantly facilitated cancer stem cell‐like features and promoted the migration of glioblastoma multiforme cells. 38 Serej and colleagues showed that suppression of Nanog inhibited malignant phenotypes and cancer stem cell‐like features of BC cells. In this study, we found that KLF4 and Nanog were positively correlated with ABI2 and MEOX2 in HCC patients through Danalysing correlation analyses from TCGA data. The transcriptional level of KLF4 and Nanog were down‐regulated by the knockdown of ABI2, which suppressed the tumour proliferation, migration, invasion and CSCs expansion. However, these suppression effects were partially restored by MEOX2 overexpression. Furtherly, we verified that KLF4 and Nanog were key performers in ABI2‐promoting HCC cancer stem cells by binding of the transcription factor MEOX2 to the promoter regions and mediating their transcription levels.
Furthermore, we verified that knockdown of ABI2 significantly inhibited the tumour weight and tumour volume of xenograft tumours. As expected, HCC stem cell fluency was dramatically decreased in the sh‐ABI2 group, which was restored by MEOX2 overexpression in a limiting dilution assay. These results demonstrate that HCC CSCs were maintained by ABI2 via recruiting its performer MEOX2, suggesting that ABI2 is a promising CSC target in HCC treatment.
In summary, our findings indicated that ABI2 was significantly upregulated in HCC tissues and that high expression of ABI2 was associated with poorer survival. Moreover, we demonstrated that knockdown of ABI2 inhibited HCC cell growth, sorafenib resistance, migration and invasion by repressing HCC CSC self‐renewal and tumorigenicity. Mechanistically, we identified that ABI2 promoted HCC malignant behaviour and CSC properties via the MEOX2/KLF4‐Nanog axis. Additionally, inhibition of ABI2 suppressed the progression of HCC in vivo. These results revealed the important roles of the ABI2/MEOX2/KLF4‐Nanog axis in the pathogenesis of HCC progression, which may provide novel biomarkers and promising therapeutic targets for HCC treatment (Figure 7).
FIGURE 7.
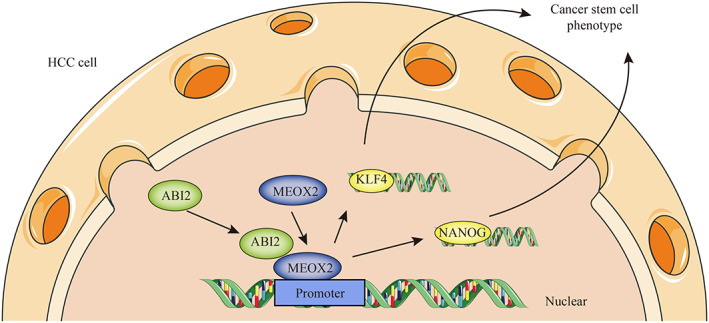
Summary of pathological and underlying regulatory mechanisms of ABI2 in hepatocellular carcinoma.
AUTHOR CONTRIBUTIONS
Shuixing Zhang and Xiangnan Zhu conceived and designed the experiments. Jiandi Chen, Huizi Li performed the experiment, assisted with data interpretation and wrote of the paper. Bin Zhang, Zhiyuan Xiong, Zhe Jin, Jiaxi Chen, Yang Zheng helped in data interpretation and article preparation. All authors have read and approved the final manuscript.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China (81871323, 81801665, 81901709) and the Natural Science Foundation of Guangdong Province (2018B030311024, 2019A1515011918).
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
Supplementary Figure 1
Supplementary Table 1
Supplemental Table 2
ACKNOWLEDGEMENTS
Dr. Jing Zhang in Jinan University helped in manuscript revision.
Chen J, Li H, Zhang B, et al. ABI2‐mediated MEOX2/KLF4‐NANOG axis promotes liver cancer stem cell and drives tumour recurrence. Liver Int. 2022;42:2562‐2576. doi: 10.1111/liv.15412
Jiandi Chen and Huizi Li contributed equally to this work.
Handling Editor: Alejandro Forner
Contributor Information
Xiangnan Zhu, Email: zxndoctor@162.com.
Shuixing Zhang, Email: zsx7515@jnu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Petrick JL, Florio AA, Znaor A, et al. International trends in hepatocellular carcinoma incidence, 1978–2012. Int J Cancer. 2020;147:317‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47‐76. [DOI] [PubMed] [Google Scholar]
- 5. Desai A, Yan Y, Gerson SL. Concise reviews: cancer stem cell targeted therapies: toward clinical success. Stem Cells Transl Med. 2019;8:75‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bai X, Ni J, Beretov J, Graham P, Li Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152‐163. [DOI] [PubMed] [Google Scholar]
- 7. Zuo ZQ, Chen KG, Yu XY, et al. Promoting tumor penetration of nanoparticles for cancer stem cell therapy by TGF‐β signaling pathway inhibition. Biomaterials. 2016;82:48‐59. [DOI] [PubMed] [Google Scholar]
- 8. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275‐284. [DOI] [PubMed] [Google Scholar]
- 9. Dianat‐Moghadam H, Heidarifard M, Jahanban‐Esfahlan R, et al. Cancer stem cells‐emanated therapy resistance: implications for liposomal drug delivery systems. J Control Release. 2018;288:62‐83. [DOI] [PubMed] [Google Scholar]
- 10. Zakaria N, Mohd Yusoff N, Zakaria Z, Widera D, Yahaya BH. Inhibition of NF‐κB signaling reduces the stemness characteristics of lung cancer stem cells. Front Oncol. 2018;8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells ‐ a clinical update. Nat Rev Clin Oncol. 2020;17:204‐232. [DOI] [PubMed] [Google Scholar]
- 12. Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, notch, and hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97‐106. [DOI] [PubMed] [Google Scholar]
- 13. Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan PD, Cong F, Goff SP. Homo‐ and hetero‐oligomerization of the c‐Abl kinase and Abelson‐interactor‐1. Cancer Res. 2003;63:873‐877. [PubMed] [Google Scholar]
- 15. Courtney KD, Grove M, Vandongen H, Vandongen A, LaMantia AS, Pendergast AM. Localization and phosphorylation of Abl‐interactor proteins, Abi‐1 and Abi‐2, in the developing nervous system. Mol Cell Neurosci. 2000;16:244‐257. [DOI] [PubMed] [Google Scholar]
- 16. Steinestel K, Brüderlein S, Lennerz JK, et al. Expression and Y435‐phosphorylation of Abelson interactor 1 (Abi1) promotes tumour cell adhesion, extracellular matrix degradation and invasion by colorectal carcinoma cells. Mol Cancer. 2014;13:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramaker RC, Hardigan AA, Gordon ER, Wright CA, Myers RM, Cooper SJ. Pooled CRISPR screening in pancreatic cancer cells implicates co‐repressor complexes as a cause of multiple drug resistance via regulation of epithelial‐to‐mesenchymal transition. BMC Cancer. 2021;21:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lim IK, Choi JA, Kim EY, et al. TIS21(/BTG2) inhibits doxorubicin‐induced stress fiber‐vimentin networks via Nox4‐ROS‐ABI2‐DRF‐linked signal cascade. Cell Signal. 2017;30:179‐190. [DOI] [PubMed] [Google Scholar]
- 19. Martin JL, Julovi SM, Lin MZ, de Silva HC, Boyle FM, Baxter RC. Inhibition of basal‐like breast cancer growth by FTY720 in combination with epidermal growth factor receptor kinase blockade. Breast Cancer Res. 2017;19:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma B, Yuan Z, Zhang L, et al. Long non‐coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta Mol Cell Res. 2017;1864:1393‐1404. [DOI] [PubMed] [Google Scholar]
- 21. Shen W, Xie J, Zhao S, et al. ICAM3 mediates inflammatory signaling to promote cancer cell stemness. Cancer Lett. 2018;422:29‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xue Y, Petrovic N, Cao R, et al. Hypoxia‐independent angiogenesis in adipose tissues during cold acclimation. Cell Metab. 2009;9:99‐109. [DOI] [PubMed] [Google Scholar]
- 23. Zhou SL, Yin D, Hu ZQ, et al. A positive feedback loop between cancer stem‐like cells and tumor‐associated neutrophils controls hepatocellular carcinoma progression. Hepatology. 2019;70:1214‐1230. [DOI] [PubMed] [Google Scholar]
- 24. Ma XL, Sun YF, Wang BL, et al. Sphere‐forming culture enriches liver cancer stem cells and reveals stearoyl‐CoA desaturase 1 as a potential therapeutic target. BMC Cancer. 2019;19:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Specht E, Kaemmerer D, Sänger J, Wirtz RM, Schulz S, Lupp A. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology. 2015;67:368‐377. [DOI] [PubMed] [Google Scholar]
- 26. Wei L, Lee D, Law CT, et al. Genome‐wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for sorafenib resistance in HCC. Nat Commun. 2019;10:4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ávila‐Moreno F, Armas‐López L, Álvarez‐Moran AM, et al. Overexpression of MEOX2 and TWIST1 is associated with H3K27me3 levels and determines lung cancer chemoresistance and prognosis. PLoS ONE. 2014;9:e114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takebe N, Miele L, Harris PJ, et al. Targeting notch, hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670‐1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Z, Yang H, Zhang R, et al. MEOX2 serves as a novel biomarker associated with macrophage infiltration in oesophageal squamous cell carcinoma and other digestive system carcinomas. Autoimmunity. 2021;54:373‐383. [DOI] [PubMed] [Google Scholar]
- 31. Tachon G, Masliantsev K, Rivet P, et al. Prognostic significance of MEOX2 in gliomas. Mod Pathol. 2019;32:774‐786. [DOI] [PubMed] [Google Scholar]
- 32. Tian L, Tao ZZ, Ye HP, Li GY, Zhan ZF, Tuo HW. Over‐expression of MEOX2 promotes apoptosis through inhibiting the PI3K/Akt pathway in laryngeal cancer cells. Neoplasma. 2018;65:745‐752. [DOI] [PubMed] [Google Scholar]
- 33. Yu F, Li J, Chen H, et al. Kruppel‐like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161‐2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi XT, Li YL, Zhang YQ, et al. KLF4 functions as an oncogene in promoting cancer stem cell‐like characteristics in osteosarcoma cells. Acta Pharmacol Sin. 2019;40:546‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leng Z, Li Y, Zhou G, et al. Krüppel‐like factor 4 regulates stemness and mesenchymal properties of colorectal cancer stem cells through the TGF‐β1/Smad/snail pathway. J Cell Mol Med. 2020;24:1866‐1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khosravi N, Shahgoli VK, Amini M, et al. Suppression of Nanog inhibited cell migration and increased the sensitivity of colorectal cancer cells to 5‐fluorouracil. Eur J Pharmacol. 2021;894:173871. [DOI] [PubMed] [Google Scholar]
- 37. Najafzadeh B, Asadzadeh Z, Motafakker Azad R, et al. The oncogenic potential of NANOG: an important cancer induction mediator. J Cell Physiol. 2021;236:2443‐2458. [DOI] [PubMed] [Google Scholar]
- 38. Yuan Y, Zhang M, Yan G, et al. Nanog promotes stem‐like traits of glioblastoma cells. Front Biosci (Landmark ed). 2021;26:552‐565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Table 1
Supplemental Table 2
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


