Abstract
Bordetella pertussis produces a 73-kDa protein, BrkA (Bordetella resistance to killing), which inhibits the bactericidal activity of complement. In this study we characterized the step in the complement cascade where BrkA acts, using three strains: a wild-type strain, a strain containing an insertional disruption of brkA, and a strain containing two copies of the brkA locus. Following incubation with 10% human serum, killing was greatest for the BrkA mutant, followed by that for the wild-type strain, while the strain with two copies of brkA was the most resistant. Complement activation was monitored by enzyme-linked immunosorbent assay (ELISA) or Western blotting. ELISAs for SC5b-9, the soluble membrane attack complex, showed that production of SC5b-9 was greatest with the brkA mutant, less with the wild type, and least with the strain containing two copies of brkA. Deposition of complement proteins on the bacteria was monitored by Western blotting. A decrease in deposition on the bacteria of C4, C3, and C9 corresponded with decreased complement sensitivity. Deposition of C1, however, was not affected by the presence of BrkA. These studies show that BrkA inhibits the classical pathway of complement activation and prevents accumulation of deposited C4.
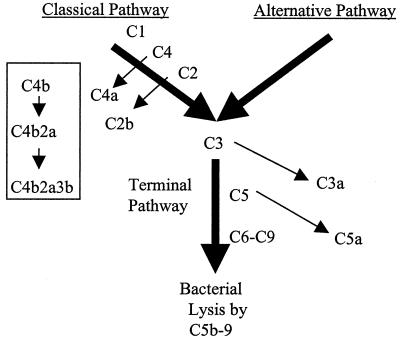
The complement system is a series of proteins that act in a defined sequence (Fig. 1) to promote immune clearance by opsonizing or killing microorganisms and augmenting the inflammatory response. Antigen-antibody complexes on the surface of a microorganism can activate the classical pathway of complement, a part of the acquired immune system (Fig. 1), by providing a binding site for C1. A number of proteolytic activation steps follow which lead to deposition of C4b, C2a, C3b, and C5b on the bacterium. Finally, a series of binding steps occur, leading to the formation of the membrane attack complex (C5b-9), which promotes lysis of the bacterium. Complement is also part of the innate immune defenses and provides a defense against pathogens that have not previously infected the host by recognizing repeating structures such as lipopolysaccharide (LPS) found on the surface of bacteria. In addition, it has become increasingly apparent that the acquired and innate immune systems are linked. Host-expressed pattern recognition molecules recognize structures commonly expressed by microorganisms. For example, the mannose binding protein is a pattern recognition molecule with structural similarity to C1 that bridges the classical and alternative pathways (18). Mannose binding protein activates the classical pathway by binding to mannose residues on microbial surfaces and activating C4 in a manner similar to that for C1.
FIG. 1.
The complement cascade. Both the classical and alternative pathways of complement activation are represented, although details are given only for the classical pathway. The nonactivated complement proteins are indicated above and to the right of the wide arrows, and the arrows pointing from them show the product that is released during activation. The smaller flowchart in the box indicates the membrane-bound products and complexes formed by the classical pathway on the bacterial surface.
Bordetella pertussis is an obligate human pathogen and must therefore survive within the hostile environment of the human host. B. pertussis produces a number of virulence factors to counter most host immune defenses (28). Although often overlooked as a defense of the respiratory tract, complement levels in this location are normally 10 to 20% of that found in serum and increase during inflammation (21). Complement resistance is common among respiratory pathogens. For example, Streptococcus pyogenes (1, 14) and Haemophilus influenzae (20) possess complement resistance mechanisms. B. pertussis must also overcome this defense in order to survive within the host. Interestingly, B. pertussis does not activate the alternative pathway of complement (7). However, an acquired immune response can lead to killing of the bacteria by activating the antibody-dependent classical pathway. The Bordetella resistance to killing (BrkA) protein has been shown to inhibit killing by this pathway (7).
To elucidate the molecular basis for complement resistance by BrkA, in this study we have attempted to determine which step in the complement cascade is affected by BrkA by monitoring the deposition of complement proteins on the surface of strains either expressing or not expressing BrkA. The presence of BrkA inhibits deposition of C4, C3, and C9 and production of the soluble membrane attack complex. In contrast, BrkA does not affect the deposition of C1. These studies suggest that BrkA acts before C4 deposition.
MATERIALS AND METHODS
Bacterial strains.
The four strains of B. pertussis used in this study were previously described. Strain BP338 is a nalidixic acid-resistant derivative of the Tohama I strain (29). RFBP2171 is a derivative of BP338 that contains two chromosomal copies of the brkAB locus and is more resistant to complement than the wild-type strain (8). Two complement-sensitive brkA mutants were used. BPM2041 is a derivative of BP338 that has a Tn5 lac insertion (30) in the brkA locus (7), while RFBP2152 is a derivative of BP338 that has a gentamicin insertion and small deletion in the brkA locus (8). Bacteria were grown on Bordet-Gengou agar (BGA) containing 15% defibrinated sheep's blood (Colorado Serum, Denver, Colo.) and 1% glycerol. The following antibiotics, at the following concentrations, were added to the BGA: 30 μg of nalidixic acid/ml for all four strains, 50 μg of kanamycin/ml for BPM2041, and 30 μg of gentamicin/ml for RFBP2152 and RFBP2171.
Serum.
Human sera were collected from adult volunteers. Heat inactivation, when appropriate, was carried out at 56°C for 30 min. All samples were stored at −80°C.
Serum killing assay.
BGA cultures of bacteria were harvested after 20 to 24 h to a concentration of about 109 bacteria per ml (optical density at 600 nm of 0.5) in warm (37°C) Stainer Scholte (SS) salts (63.4 mM l-glutamic acid monosodium salt, 2.1 mM proline, 43 mM NaCl, 3.7 mM KH2PO4, 2.7 mM KCl, 20.1 mM Tris-Cl, 48.5 mM Tris-base, 0.5 mM MgCl2, 0.135 mM CaCl2). A volume of 500 μl of bacteria was put into a microcentrifuge tube to which was added 400 μl of SS salts. A volume of 100 μl of normal or heat-inactivated human serum was added to the tube, and the tube was placed in a 37°C water bath for 15 or 120 min. The tube was then placed on ice for 5 min to stop the complement reaction. A 20-μl aliquot of the mixture was added to 180 μl of phosphate-buffered saline (PBS) (128 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 5 mM K2HPO4 [pH 7.4]) with 10 mM EDTA. Tenfold serial dilutions were performed into SS salts. The dilutions were plated on BGA and incubated at 37°C, and colonies were counted. Survival was calculated as a percentage of serum-treated colonies compared to the number of colonies from heat-inactivated serum control (nonkilling control). Killing was calculated as 100% − percent survival. Statistical analysis was performed using Student's t test.
To assay for complement deposition, the remaining 0.98 ml of the killing assay mixture was centrifuged at 4°C at 17,000 × g in an SS-34 rotor for 10 min to pellet the cells. The supernatant was filter sterilized and analyzed for SC5b-9, a soluble complement activation product. The cell pellets were washed with 500 μl of ice-cold PBS (pH 7.4) three times to remove any nonspecifically bound complement, suspended in 100 μl of SS salts, and used for analysis of C1, C4, C3, and C9 deposition. All samples were stored at −80°C.
Purified C1 deposition.
BGA cultures of bacteria were harvested after 20 h to a concentration of about 109 bacteria per ml (optical density at 600 nm of 0.5) in warm (37°C) SS salts. A volume of 100 μl of bacteria was put into a microcentrifuge tube to which was added 20 μl (1.28 mg/ml) of purified C1 (lot no. 4; Advanced Research Technologies, San Diego, Calif.) with or without 20 μl (50 mg/ml) of purified human immunoglobulin G (IgG) (lot no. 104H8872; Sigma, St. Louis, Mo.). The sample was brought to 200 μl with SS salts. The tube was placed in a 37°C water bath for 15 min and then on ice for 5 min. The bacteria were washed three times with 100 μl of ice-cold PBS (pH 7.4) to remove any unbound complement proteins. All samples were stored at −80°C.
SDS-polyacrylamide gel electrophoresis and Western blotting.
The cell suspensions were mixed with 4 volumes of sample buffer (62.5 mM Tris-HCl [pH 6.8], 20% glycerol, 2% sodium dodecyl sulfate [SDS], and 0.5% [wt/vol] bromophenol blue). Samples for determination of C1, C4, C3, and BrkA included 5% β-mercaptoethanol, while samples for C9 determination did not. Samples were boiled and subjected to electrophoresis using the Mini Protean II gel system (Bio-Rad, Hercules, Calif.) on 10% polyacrylamide gels with 5% stacking gels. For examination of C1, C4, and C3, proteins were transferred onto nitrocellulose membrane by wet transfer in a submarine apparatus (Trans-Blot Tank; Bio-Rad) using a modified Towbin buffer (26) (15.6 mM Tris-base, 120 mM glycine, 0.02% SDS, 20% methanol). For examination of C9 or BrkA, proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore; Bedford, Mass.) using a semidry apparatus (model FB-SDB-2020; Fisher, Hanover Park, Ill.). The membranes were blocked with 5% nonfat dry milk in blocking buffer (81 mM Na2HPO4, 25 mM NaH2PO4, 100 mM NaCl). Commercial antibodies to complement proteins—goat anti-human C1q (lot no. 1), goat anti-human C4 (lot no. 1H), goat anti-human C3 (lot no. 2Q), goat anti-human C9 (lot no. 1) (all from Advanced Research Technologies)—were used at a 1:500 dilution in wash buffer (blocking buffer with 0.25% nonfat dry milk and 0.50% Tween 20). Rabbit anti-BrkA antibodies (obtained from Rachel Fernandez and David Ogden) were used at a 1:50,000 dilution in wash buffer. Secondary antibodies—rabbit anti-goat horseradish peroxidase (lot no. 40786), goat anti-rabbit horseradish peroxidase (lot no. 39678) (all from ICN, Costa Mesa, Calif.)—were used at a 1:62,500 dilution in wash buffer. Proteins were detected by chemiluminescence using the Dupont Western blot Renaissance kit (NEN Life Science Products, Boston, Mass.). Protein identification was based on comparison with human serum and the Benchmark protein ladder (Gibco Life Sciences, Grand Island, N.Y.).
Densitometry.
Relative amounts of protein were determined by densitometry of the Western blots (4), using Image Quant 5.0 (Molecular Dynamics, http: //www.mdyn.com) for quantitation. In a control experiment, a dilution series of serum was detected with the antibody to C1q and found to have a coefficient of correlation (R2) of 0.99 (data not shown), suggesting a strong linear relationship between concentration and signal using this technique.
To compare deposition on different strains, results were set relative to the amount of deposition on the brkA mutant (RFBP2152), using the killing results as the expected maximum percent deposition. For example, if 91% of the RFBP2152 cells were killed in the assay, then deposition on RFBP2152 was set at 91% and deposition on the other strains was calculated relative to this value. Statistical analysis was performed using the Student t test.
ELISAs.
Quantitative sandwich enzyme-linked immunosorbent assays (ELISAs) (Quidel, San Diego, Calif.) were performed to quantify production of SC5b-9 as a measure of membrane attack complex formation, according to the manufacturer's directions. Plates were read on an enzyme immunoassay reader (model 2550; Bio-Rad) at 405 nm. Results were set relative to the brkA mutant RFBP2152 as in densitometry of Western blots. Statistical analysis was performed using Student's t test.
RESULTS
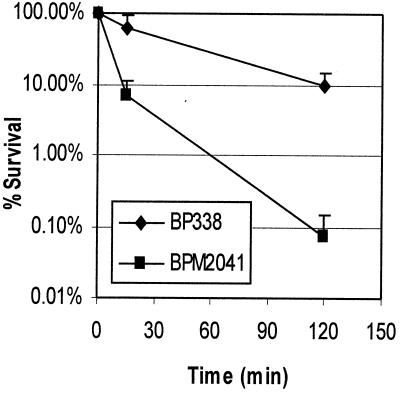
It has previously been shown that the brkA locus mediates resistance to killing by complement in B. pertussis (7). Preliminary studies to investigate this resistance phenomenon showed that after 2 h in the presence of 10% serum, viability of the wild-type strain BP338 was reduced 10-fold (Fig. 2). In contrast, under the same conditions, the viability of the BrkA mutant BPM2041 was reduced more than 1,000-fold. To determine at which step in the complement cascade BrkA mediates resistance to complement killing, we compared the amount of specific complement proteins deposited on the bacteria and the production of SC5b-9, the soluble membrane attack complex. While differences in bacterial survival can be detected over several orders of magnitude, Western blotting is sensitive only to differences in the linear range. Little difference in complement deposition could be observed when there was significant killing of the wild-type strain (data not shown). To improve the sensitivity of our assays, we wanted to establish conditions with the greatest survival of B. pertussis. From the killing curve shown in Fig. 2, we chose to examine complement deposition at 15 min, at which time BP338 survives well in complement. We also examined the influence of two variables known to affect the ability of serum to kill the wild-type strain: BrkA expression and the presence of bactericidal antibodies in human serum.
FIG. 2.
Survival of B. pertussis in complement. BP338 and BPM2041 were incubated with 10% human serum for 15 (P < 0.0038) or 120 (P < 0.0012) min. Percent survival is calculated relative to a non-killing heat-inactivated serum control. The results are the average and standard error of four separate trials. BPM2041 is the BrkA mutant; BP338 is the wild type.
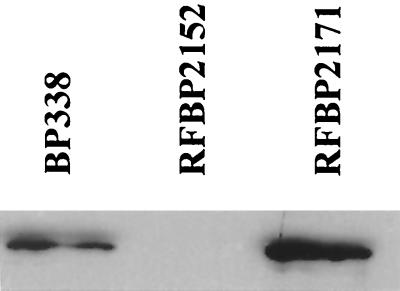
In order to establish conditions with greater distinction in killing, two new strains were tested. A strain possessing two copies of brkA, RFBP2171, was chosen because it is more resistant to killing by complement than is wild-type BP338 (8). The brkA mutant RFBP2152 was chosen because, like RFBP2171, it is gentamicin resistant. Western blotting with an antibody to BrkA confirmed that expression of BrkA is increased about 80% in strain RFBP2171 from that in BP338 (Fig. 3,) while no BrkA was detected in the brkA mutant RFBP2152. Resistance to killing correlated with BrkA expression (Fig. 4, empty bars).
FIG. 3.
BrkA expression. A representative Western blot showing the variation in expression of BrkA protein from the three strains used for the killing assays and deposition assays. BP338, wild type; RFBP2152, BrkA mutant. RFBP2171 contains two copies of brkA.
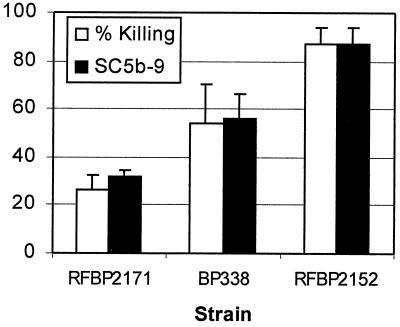
FIG. 4.
Production of the soluble membrane attack complex, SC5b-9, relative to bacterial killing. Empty bars represent percent killing; solid bars represent SC5b-9 production as determined by ELISA relative to RFBP2152 with standard deviation. Three separate experiments were performed. Values for RFBP2152 are significantly different from those for BP338 (P = 0.05) and RFBP2171 (P < 0.0005).
We also examined the effect of different serum samples on killing of B. pertussis. We have shown that some individuals mount an immune response capable of overcoming the BrkA resistance mechanism (31). To achieve the greatest discrimination in deposition by Western blotting, we chose a serum with weak bactericidal activity. When this serum was used in a 15-min killing assay, survival of RFBP2171 was 71% ± 12% (n = 8), survival of BP338 was 49% ± 8% (n = 9), and survival of RFBP2152 was 12% ± 7% (n = 9). The values were all statistically different. Complement deposition was examined under these conditions.
BrkA inhibits formation of the membrane attack complex.
The membrane attack complex is composed of activated C5 (C5b) bound to C6, C7, C8, and C9 (Fig. 1). Upon binding of C5b and C6 to C7, the complex becomes lipophilic and can insert into the membrane. After membrane insertion, C8 and C9 bind the complex, followed by binding of additional C9 molecules to form poly-C9. The poly-C9 forms pores in the membrane of gram-negative bacteria and leads to death of the bacteria. Alternatively, prior to membrane insertion, C5b-7 can bind a soluble complement regulatory protein, protein S, and form a complex, SC5b-7. This complex cannot associate with the membrane but can bind C8 and C9, leading to the formation of the soluble, inactive SC5b-9 complex (17). The formation of SC5b-9 is often measured to determine activation of the terminal complement cascade (9, 24, 25).
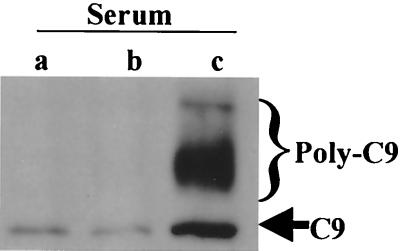
An ELISA for SC5b-9 was used to measure activation of the terminal pathway of complement (Fig. 4). Less of SC5b-9 was detected in supernatants from BP338 than in supernatants from BPM2152 (P = 0.05; n = 3), and even less was detected in supernatants from RFBP2171 than in those from RFBP2152 (P < 0.0005; n = 3). Reduced activation of the terminal pathway of complement by strains possessing the BrkA protein was also observed by Western blot analysis for deposition of either mono-C9 or poly-C9 on the bacterial membrane (Fig. 5). These results explain why strains expressing BrkA resist killing by complement and suggest that BrkA acts prior to activation of the terminal pathway.
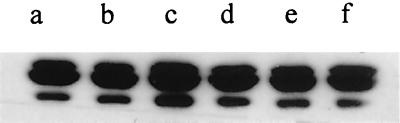
FIG. 5.
C9 deposition. Bacteria were incubated in 10% serum for 15 min. Equal numbers of cells were subjected to Western blotting with a goat anti-human C9 antibody. Strains were RFBP2171 (a), BP338 (b), and RFBP2152 (c). The bracket indicates the region of poly-C9 deposition; the arrow indicates mono-C9 deposition.
BrkA inhibits the deposition of C3 onto the bacterial surface.
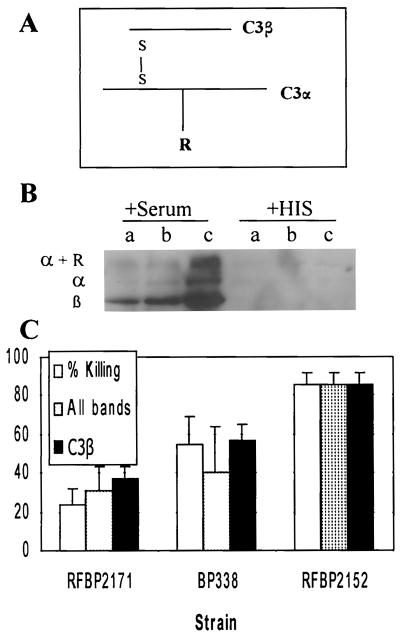
C3 plays a central role in the complement cascade. It is present at the convergence of the classical and alternative pathways and leads to formation of the membrane attack complex (Fig. 1). The molecule has two disulfide-bonded chains, an α chain (119 kDa) and a β chain (75 kDa) (Fig. 6A). Activation by the C3 convertase leads to the release of a small fragment, C3a (9 kDa). Cleavage also exposes an unstable thioester bond on the C3α chain that can form a covalent bond to the bacterial surface.
FIG. 6.
C3 deposition. (A) C3 is composed of an α chain and β chain connected by disulfide bonds (S—S). The α chain can bind covalently to the bacterial surface (represented by R). (B) Bacteria were incubated in 10% normal human serum or heat-inactivated serum (HIS) for 15 min and were analyzed by Western blotting under reducing conditions using goat anti-human C3 antibodies. RFBP2171 (a), BP338 (b), and RFBP2152 (c) were used. The α and β chains were determined by a comparison to bacterium-free serum run as a size control. α + R indicates larger bands than those seen in a serum control. (C) Densitometry was performed on the Western blots, and results are graphed in arbitrary units relative to RFBP2152 with standard error from four separate experiments. “All bands” indicates densitometry of all bands observed in the Western blot, while “C3β” indicates densitometry of only the lower, C3β band. Densitometry of “All bands” and “C3β ” for RFBP2152 was significantly higher than for BP338 (P < 0.04 and 0.03, respectively) or for RFBP2171 (P < 0.0005 and 0.004, respectively).
C3 deposition was examined by Western blotting using reducing conditions (Fig. 6B). The C3β chain was detected as a 75-kDa fragment which comigrated with the β chain seen in normal human sera without bacteria (data not shown). This peptide is not cleaved during activation of C3 and reflects the total amount of C3 deposited on the bacteria. Deposition of C3α and C3α covalently bound to bacteria was indicated by the higher-molecular-weight bands. None of these bands were present on bacteria incubated with heat-inactivated serum, indicating that they were generated by complement activation.
Densitometry revealed a gradient of deposition of C3 on the three strains, with less deposition associated with increased BrkA expression (Fig. 6C). Deposition of C3 (α and β) was significantly greater on RFBP2152 than on BP338 (P < 0.04; n = 4) or on RFBP2171 (P < 0.0005; n = 4) (Fig. 6C). In addition, deposition of C3β was significantly greater on RFBP2152 than on BP338 (P < 0.03; n = 4) or RFBP2171 (P < 0.004; n = 4). These results show that the presence of BrkA inhibits deposition of all C3 fragments, suggesting that BrkA inhibits the complement cascade prior to the deposition of C3.
BrkA inhibits the deposition of C4.
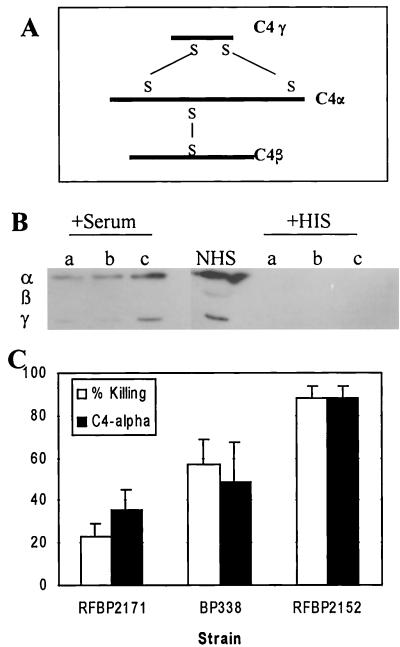
C4 is the second component of the classical pathway to be activated. This protein is made up of three polypeptides that are held together by disulfide bonds (Fig. 7A). The three chains are the α (90 kDa), β (78 kDa), and γ (33 kDa) chains. Upon activation, the α chain is cleaved, releasing the 9-kDa C4a fragment. C4 deposition was examined by Western blotting using reducing conditions (Fig. 7B). The antibody used recognized C4α much better than it recognized C4β or C4γ. Deposition of the C4α chain was greater on RFBP2152 than on BP338 (P < 0.02; n = 3) or on RFBP2171 (P < 0.03; n = 3) (Fig. 7C). This deposition was not observed on bacteria incubated with heat-inactivated serum. These data demonstrate that BrkA prevents deposition of C4 on the bacteria or promotes the degradation of C4 after binding, thereby blocking further steps in the complement cascade.
FIG. 7.
C4 deposition. (A) C4 is composed of three chains, α, β and γ, connected by disulfide bonds (S—S). (B) Bacteria were incubated in 10% serum or heat-inactivated serum (HIS) for 15 min and were analyzed by Western blotting under reducing conditions with goat anti-human C4 antibodies. RFBP2171 (a), BP338 (b), and RFBP2152 (c) were tested. NHS, normal human sera. The three chains were determined by a comparison with bacterium-free serum that was run as a size control. (C) Densitometry was performed, and results are graphed as arbitrary units relative to RFBP2152 with standard error of three separate experiments. Densitometry results for RFBP2152 were significantly higher than for BP338 (P < 0.02) or for RFBP2171 (P < 0.03).
C1q deposition is not antibody dependent or altered by heat inactivation.
The initiation of the classical pathway of complement activation requires the binding of C1q to antibody. This binding facilitates a change in structure of C1q, leading to activation of C1r and C1s, which activates C4 and C2. C1q is made up of three chains, A (24 kDa), B (23 kDa), and C (22 kDa), connected by disulfide bonds.
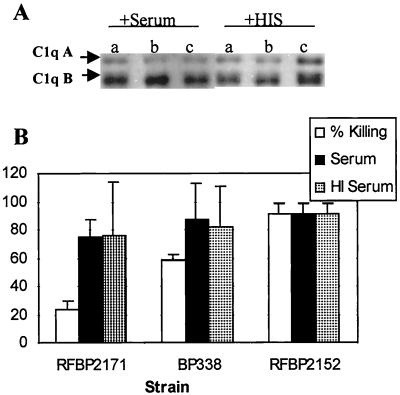
Western blotting was performed using reducing conditions to monitor C1q deposition during the serum killing assay (Fig. 8). Deposition of C1q was equivalent on all three strains. However, unlike the other complement proteins characterized in this study, deposition occurred even when heat-inactivated serum was used. Previous studies have shown that C1 activity is destroyed by heat inactivation (13). C1 possesses an enzymatic activity in addition to its binding activity, and inactivation could be achieved without affecting the binding activity.
FIG. 8.
C1 deposition. (A) Bacteria were incubated in 10% serum or heat-inactivated serum (HIS) for 15 min and were analyzed under reducing conditions using goat anti-human C1q antibodies. RFBP2171 (a), BP338 (b), and RFBP2152 (c) were used. (B) Densitometry was performed, and results are graphed as arbitrary units relative to RFBP2152 with standard error determined from five independent experiments. Differences were not significant. HI, heat inactivated.
C1 binding was investigated further. Incubation of B. pertussis with purified C1 alone, purified C1 with purified IgG, or heat-inactivated purified C1 all showed equivalent deposition on RFBP2171 or BP338 (Fig. 9) or RFBP2152 (data not shown). These results suggest that binding occurs in the absence of antibody; however, we cannot determine if C1 has an affinity for B. pertussis or whether B. pertussis has an affinity for C1. The equivalent binding to the wild type and the BrkA mutant suggests that BrkA is not involved in binding to C1.
FIG. 9.
Deposition of purified C1. Bacteria were incubated with purified C1 (a and d), purified C1 with purified IgG (b and e), or heat-inactivated purified C1 (c and f) for 15 min and were analyzed under reducing conditions using goat anti-human C1q antibodies. a to c, BP338; d to f, RFBP2171.
DISCUSSION
Pathogenic organisms have evolved mechanisms to interfere with the complement system at many steps in the cascade (6, 10, 16, 19). One strategy of resistance depends on prevention of complement function. For example, Pseudomonas aeruginosa produces proteases specific for C1q and C3 (12). Enteric pathogens, such as Escherichia coli, avoid killing by complement by making highly polymerized LPS to prevent complement deposition close enough to the membrane to promote lysis (5). Another avoidance mechanism is to recruit host regulatory proteins. For example, S. pyogenes (11, 14) and Yersinia enterocolitica (22) bind the host regulatory protein, factor H, which regulates the C3 and C5 convertases. Host cells resist activating complement by presenting a high density of sialic acid on their surface, and Neisseria gonorrhoeae mimics this by incorporating sialic acid into its LPS (15).
BrkA provides complement resistance to B. pertussis (7). In this study, we wanted to explore the mechanism by which BrkA provides this protection. Deposition of C1, C4, C3, and C9 on the bacteria and production of SC5b-9 were monitored. Deposition of C4, C3, and C9 and production of SC5b-9 all increased with increased killing. Deposition of C1 was not affected by the presence of BrkA or antibody or by heat treatment of the serum. In total, these data suggest that BrkA inhibits activation or promotes degradation of C4 after deposition.
The mechanism by which BrkA inhibits complement has not been determined, but in theory it could work by either of the two paradigms discussed above: preventing complement function or recruiting a complement-inhibitory protein. BrkA could prevent complement function by acting as a protease against C1r, C1s, or C4. Inactivation of C1r or C1s would inhibit deposition of C4, as observed. Alternatively, proteolysis of activated C4 would also result in decreased C4 deposition.
Alternatively, BrkA may recruit a complement inhibitor. While there are many inhibitors of the complement cascade, only four act at the C1/C4 interface: soluble C4 binding protein and C1 inhibitor or membrane-bound CD46 (membrane cofactor protein) and CR1. The soluble complement regulators present the likeliest binding partners for BrkA. However, E. coli bas been shown to recruit protectin (CD59) (23), another complement inhibitor which is usually considered to be bound by the host membrane.
Purified C4 binding protein has been shown to bind filamentous hemagglutinin (FHA) (2); however, FHA is not required for resistance to complement (8), suggesting that C4 binding protein bound to FHA is not protective. While deposition of C4 binding protein was greatly reduced on an FHA mutant, even less deposition was observed on a bvg mutant lacking expression of all virulence factors, including BrkA. This further decrease in binding leaves open the possibility that BrkA can also bind C4 binding protein and that C4 binding protein bound to BrkA may be protective.
Binding of C1 inhibitor is another attractive mechanism of action since BrkA has two SGXG proteoglycan binding motifs (7) that could allow efficient binding of the highly glycosylated C1 inhibitor (3). Recruitment of either C4 binding protein or C1 inhibitor by BrkA would result in a decrease in the accumulation of C4 on the surface of the bacteria as observed in this study.
By preventing activation of the complement cascade beyond C4, BrkA protects the bacterium against all the effects of the complement activation products, such as upregulation of the immune response by C4a, C3a, and C5a; opsonization of pathogens by C3b and iC3b; and direct killing of gram-negative bacteria by the membrane attack complex. Inhibiting these immune functions has obvious benefits for the bacteria. It should be noted that antibody alone is usually effective at opsonizing bacteria and promoting phagocytosis; however, in the case of B. pertussis, opsonization with antibodies does not promote phagocytosis by human neutrophils since adenylate cyclase toxin poisons this process (27).
BrkA is not a component of the present acellular pertussis vaccines. However, given its role in mediating protection from complement, it should be considered for inclusion in future generations of acellular vaccines.
ACKNOWLEDGMENTS
This work was supported in part by grant RO1 AI38415 to A.A.W. M.G.B. was the recipient of a Graduate Student Summer Research Fellowship from the University of Cincinnati.
We thank Rachel Fernandez and David Ogden for the antibodies to BrkA.
REFERENCES
- 1.Akesson P, Sjoholm A G, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Berggard K, Johnsson E, Mooi F R, Lindahl G. Bordetella pertussis binds the human complement regulator C4BP: role of filamentous hemagglutinin. Infect Immun. 1997;65:3638–3643. doi: 10.1128/iai.65.9.3638-3643.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock S C, Skriver K, Nielsen E, Thogersen H C, Wiman B, Donaldson V H, Eddy R L, Marrinan J, Radziejewska E, Huber R, et al. Human C1 inhibitor: primary structure, cDNA cloning, and chromosomal localization. Biochemistry. 1986;25:4292–4301. doi: 10.1021/bi00363a018. [DOI] [PubMed] [Google Scholar]
- 4.Brzezinski R M, Boutelet-Bochan H, Person R E, Fantel A G, Juchau M R. Catalytic activity and quantitation of cytochrome P-450 2E1 in prenatal human brain. J Pharmacol Exp Ther. 1999;289:1648–1653. [PubMed] [Google Scholar]
- 5.Burns S M, Hull S I. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun. 1998;66:4244–4253. doi: 10.1128/iai.66.9.4244-4253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper N R. Complement evasion strategies of microorganisms. Immunol Today. 1991;12:327–331. doi: 10.1016/0167-5699(91)90010-Q. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez R C, Weiss A A. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun. 1994;62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez R C, Weiss A A. Serum resistance in bvg-regulated mutants of Bordetella pertussis. FEMS Microbiol Lett. 1998;163:57–63. doi: 10.1111/j.1574-6968.1998.tb13026.x. [DOI] [PubMed] [Google Scholar]
- 9.Fitch J C, Rollins S, Matis L, Alford B, Aranki S, Collard C D, Dewar M, Elefteriades J, Hines R, Kopf G, Kraker P, Li L, O'Hara R, Rinder C, Rinder H, Shaw R, Smith B, Stahl G, Shernan S K. Pharmacology and biological efficacy of a recombinant, humanized, single-chain antibody C5 complement inhibitor in patients undergoing coronary artery bypass graft surgery with cardiopulmonary bypass. Circulation. 1999;100:2499–2506. doi: 10.1161/01.cir.100.25.2499. [DOI] [PubMed] [Google Scholar]
- 10.Frank M M. The mechanism by which microorganisms avoid complement attack. Curr Opin Immunol. 1992;4:14–19. doi: 10.1016/s0952-7915(92)90117-w. [DOI] [PubMed] [Google Scholar]
- 11.Hong K, Kinoshita T, Takeda J, Kozono H, Pramoonjago P, Kim Y U, Inoue K. Inhibition of the alternative C3 convertase and classical C5 convertase of complement by group A streptococcal M protein. Infect Immun. 1990;58:2535–2541. doi: 10.1128/iai.58.8.2535-2541.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong Y Q, Ghebrehiwet B. Effect of Pseudomonas aeruginosa elastase and alkaline protease on serum complement and isolated components C1q and C3. Clin Immunol Immunopathol. 1992;62:133–138. doi: 10.1016/0090-1229(92)90065-v. [DOI] [PubMed] [Google Scholar]
- 13.Horowitz P M, Wasylewski Z, Kolb W P. Fluorometric detection of low temperature thermal transitions in the C1Q component of human complement. Biochem Biophys Res Commun. 1980;96:382–387. doi: 10.1016/0006-291x(80)91226-7. [DOI] [PubMed] [Google Scholar]
- 14.Horstmann R D, Sievertsen H J, Knobloch J, Fischetti V A. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci USA. 1988;85:1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis G A, Vedros N A. Sialic acid of group B Neisseria meningitidis regulates alternative complement pathway activation. Infect Immun. 1987;55:174–180. doi: 10.1128/iai.55.1.174-180.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joiner K A. Complement evasion by bacteria and parasites. Annu Rev Microbiol. 1988;42:201–230. doi: 10.1146/annurev.mi.42.100188.001221. [DOI] [PubMed] [Google Scholar]
- 17.Kolb W P, Muller-Eberhard H J. The membrane attack mechanism of complement. Isolation and subunit composition of the C5b-9 complex. J Exp Med. 1975;141:724–735. [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita M, Fujita T. Activation of the classical complement pathway by mannose-binding protein in association with a novel C1s-like serine protease. J Exp Med. 1992;176:1497–1502. doi: 10.1084/jem.176.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffitt M C, Frank M M. Complement resistance in microbes. Springer Semin Immunopathol. 1994;15:327–344. doi: 10.1007/BF01837364. [DOI] [PubMed] [Google Scholar]
- 20.Noel G J, Brittingham A, Granato A A, Mosser D M. Effect of amplification of the Cap b locus on complement-mediated bacteriolysis and opsonization of type b Haemophilus influenzae. Infect Immun. 1996;64:4769–4775. doi: 10.1128/iai.64.11.4769-4775.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson C G. Plasma exudation in the airways: mechanisms and function. Eur Respir J. 1991;4:1268–1274. [PubMed] [Google Scholar]
- 22.Pilz D, Vocke T, Heesemann J, Brade V. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect Immun. 1992;60:189–195. doi: 10.1128/iai.60.1.189-195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rautemaa R, Jarvis G A, Marnila P, Meri S. Acquired resistance of Escherichia coli to complement lysis by binding of glycophosphoinositol-anchored protectin (CD59) Infect Immun. 1998;66:1928–1933. doi: 10.1128/iai.66.5.1928-1933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinder C S, Rinder H M, Smith M J, Tracey J B, Fitch J, Li L, Rollins S A, Smith B R. Selective blockade of membrane attack complex formation during simulated extracorporeal circulation inhibits platelet but not leukocyte activation. J Thorac Cardiovasc Surg. 1999;118:460–466. doi: 10.1016/S0022-5223(99)70183-2. [DOI] [PubMed] [Google Scholar]
- 25.Sorace J M, Rollins S, Aniagolu J U, Mergner W J, Cole K, Swartz G M, Jr, Green S J. Role of atheroma liposomes and malondialdehyde-modified low-density lipoproteins in complement activation. Pathobiology. 1996;64:73–78. doi: 10.1159/000164012. [DOI] [PubMed] [Google Scholar]
- 26.Stibitz S. Mutations in the bvgA gene of Bordetella pertussis that differentially affect regulation of virulence determinants. J Bacteriol. 1994;176:5615–5621. doi: 10.1128/jb.176.18.5615-5621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhl A M, Miller J F. BvgAS is sufficient for activation of the Bordetella pertussis ptx locus in Escherichia coli. J Bacteriol. 1995;177:6477–6485. doi: 10.1128/jb.177.22.6477-6485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weingart C L, Weiss A A. Bordetella pertussis virulence factors affect phagocytosis by human neutrophils. Infect Immun. 2000;68:1735–1739. doi: 10.1128/iai.68.3.1735-1739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss A. Mucosal immune defenses and the response of Bordetella pertussis. ASM News. 1997;63:22–28. [Google Scholar]
- 31.Weiss A A, Hewlett E L, Myers G A, Falkow S. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect Immun. 1983;42:33–41. doi: 10.1128/iai.42.1.33-41.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss A A, Melton A R, Walker K E, Andraos-Selim C, Meidl J J. Use of the promoter fusion transposon Tn5 lac to identify mutations in Bordetella pertussis vir-regulated genes. Infect Immun. 1989;57:2674–2682. doi: 10.1128/iai.57.9.2674-2682.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss A A, Mobberley P S, Fernandez R C, Mink C M. Characterization of human bactericidal antibodies to Bordetella pertussis. Infect Immun. 1999;67:1424–1431. doi: 10.1128/iai.67.3.1424-1431.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]